Abstract
The first Citrus tristeza virus (CTV) genomes completely sequenced (19.3-kb positive-sense RNA), from four biologically distinct isolates, are unexpectedly divergent in nucleotide sequence (up to 60% divergence). Understanding of whether these large sequence differences resulted from recent evolution is important for the design of disease management strategies, particularly the use of genetically engineered mild (essentially symptomless)-strain cross protection and RNA-mediated transgenic resistance. The complete sequence of a mild isolate (T30) which has been endemic in Florida for about a century was found to be nearly identical to the genomic sequence of a mild isolate (T385) from Spain. Moreover, samples of sequences of other isolates from distinct geographic locations, maintained in different citrus hosts and also separated in time (B252 from Taiwan, B272 from Colombia, and B354 from California), were nearly identical to the T30 sequence. The sequence differences between these isolates were within or near the range of variability of the T30 population. A possible explanation for these results is that the parents of isolates T30, T385, B252, B272, and B354 have a common origin, probably Asia, and have changed little since they were dispersed throughout the world by the movement of citrus. Considering that the nucleotide divergence among the other known CTV genomes is much greater than those expected for strains of the same virus, the remarkable similarity of these five isolates indicates a high degree of evolutionary stasis in some CTV populations.
Citrus tristeza virus (CTV), a member of the Closteroviridae, is one of the more economically important plant viruses, mainly because of the severity of the damage that it causes and the high value of individual citrus trees, which have a productive life that spans up to 100 years (4). CTV phenotypes have been characterized based on symptoms produced in a range of hosts. Generally, CTV induces two types of severe phenotypes in the field: decline, which consists of the rapid death of sweet orange [Citrus sinensis (L.) Osb.] trees on sour orange [C. aurantium (L.)] rootstock, and stem pitting, which reduces vigor and yield of sweet orange and grapefruit (C. paradisi Macf.) trees on any rootstock (4). During the last century, CTV has destroyed entire citrus industries in several countries, thus, the name tristeza (Portuguese for sadness). However, some isolates are mild, essentially symptomless, in citrus hosts (4). Historically, when the efficient aphid vector (Toxoptera citricidus Kirkaldy) is present, more damaging phenotypes of CTV tend to become prevalent (37). It is not known whether this finding results from an accumulation of mutations in the viral genomes or from shifts in population structure. A better understanding of this phenomenon would be valuable in the design of disease management strategies. Genetically engineered mild-strain cross protection and RNA-mediated transgenic resistance, two strategies being considered, require targeting of specific CTV sequences. If the virus changes rapidly, these approaches could lose effectiveness during the life expectancy of a citrus tree. Little is known about the rates of evolution of closteroviruses.
It is well known that RNA viruses are capable of rapid evolution (9). RNA replication is error prone (12). Thus, a strain consists of a swarm of sequence variants around a consensus sequence (i.e., quasispecies) (11). Additionally, recombination can occur, producing defective RNAs (dRNAs) and possibly chimeric genotypes (24, 41). While these two processes increase population diversity, different selection pressures and bottlenecks may reduce it (10). RNA viruses undergo substantial changes through these processes. A major question for any given virus is, what is the time scale? Some viral populations, such as human immunodeficiency virus or influenza virus, are notorious for sequence changes within short time periods (14, 18). However, the potential for variation does not necessarily ensure rapid changes. Tobamoviruses and tymoviruses are thought to have evolved much more slowly (5, 15, 17, 35, 36, 42). It has been argued that the time period of evolution of these viruses has paralleled the evolution of the host plants (17).
CTV has a single-stranded, positive-sense RNA genome of 19.3 kb, encapsidated in filamentous flexuous particles that are assembled with two coat proteins, of 25 and 27 kDa (4, 13). The CTV genome consists of 12 open reading frames (ORFs), in addition to a nontranslated region (NTR) at each terminus (Fig. 1A). The 10 3′-proximal ORFs are translated from 3′ coterminal subgenomic mRNAs. ORF 1 (replicase) is translated from the genomic RNA into a 349-kDa polyprotein (ORF 1a) that is thought to occasionally continue through a +1 ribosomal frameshift into the RNA-dependent RNA polymerase-like (RdRp) domain (ORF 1b) (22).
FIG. 1.
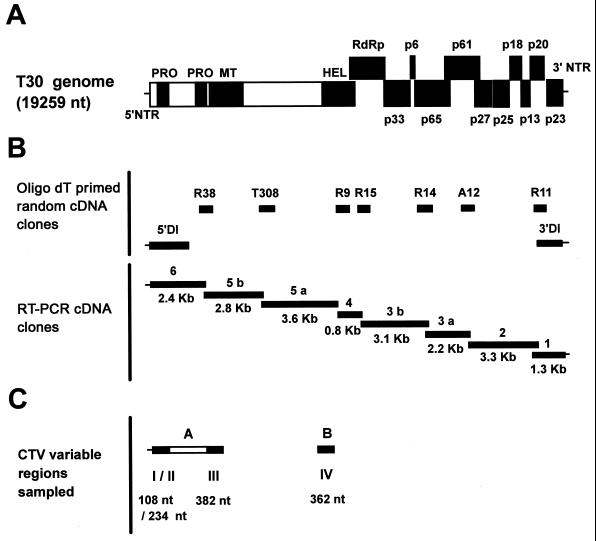
cDNA cloning strategy for mild CTV isolate sequences. (A) Diagram of the organization of the CTV T30 genome (19,259 nt). PRO, papain protease-like domain; MT, methyltransferase-like domain; HEL, helicase-like domain. (B, top panel) Schematic representation of the localization of the oligo(dT)-primed random T30 cDNA clones, the naturally occurring dRNA in isolate T30 (DI) (M. Mawassi et al., unpublished data), and the cDNA clone T308 (20) used to design T30-specific primers. (B, bottom panel) Schematic representation of the localization and size of the eight overlapping RT-PCR T30 cDNA clones used to determine the T30 genomic sequence. (C) Schematic representation of the localization of RT-PCR CTV clones A and B and locations (solid boxes) of the four hypervariable regions sequenced for comparing CTV isolates B252, B272, and B354 with T30 and T385. These CTV cDNA clones were obtained as indicated in Material and Methods.
Most CTV isolates are mixed populations of numerous different genotypes (3) and dRNAs (1, 26, 28). However, some CTV isolates consist principally of one genotype, having minor concentrations of other genotypes (3). Complete genomic sequences of four CTV isolates from different geographic areas and inducing different phenotypes in Citrus have been determined. These include one from a heterogeneous CTV isolate (a sweet orange stem pitting and decline isolate, SY568, imported into California [47]) and three from relatively homogeneous CTV isolates (a severe decline isolate, T36, from Florida [22, 34, 40], a grapefruit stem pitting and decline isolate, VT, from Israel [27], and an essentially symptomless isolate, T385, from Spain [46]) (Table 1). These CTV genomic sequences differ markedly, with as little as 50 to 80% nucleotide identity in much of the genome (27, 46). In addition, while the identity between some sequences is nearly uniform throughout the genome, the identity of other sequences is asymmetrical and progressively decreases toward the 5′ terminus to as little as 42% within the 5′ NTR (25, 27, 46). Additionally, recombination also may be frequent, based on the many dRNAs present in most populations (28). Likewise, it has been suggested that the asymmetrical diversity of the T36 genotype resulted from recombination between CTV and a different closterovirus (27). Thus, the products of evolution of CTV are among the most diverse of the RNA viruses.
TABLE 1.
CTV isolates used for sequence comparisons
| Isolate | Origin | Phenotype |
|---|---|---|
| T36 | Florida | Declinea |
| VT | Israel | Declinea and grapefruit stem pitting |
| SY568 | California | Declinea and sweet orange stem pitting |
| T385 | Spain | Symptomless |
| T30 | Florida | Symptomless |
| B252 | Taiwan | Symptomless |
| B272 | Colombia | Symptomless |
| B354 | California | Symptomless |
Decline in sweet orange trees grafted on sour orange rootstock.
In this paper, we report that the complete genomic sequences of the major component of CTV isolates T30 from Florida and T385 (46) from Spain, which have been maintained in isolation in different environments for at least 24 years and probably for more than 100 years, are essentially identical. Additionally, sequence samples of hypervariable regions of the CTV genome from three other isolates biologically similar to T30 and T385 and also separated by time and space (B252 [Taiwan], B272 [Colombia], and B354 [California]) are nearly identical to the T30 sequence. The sequence variability observed between these CTV isolates was near or within the range of T30 population variability. The possibility that the T30, T385, B252, B272, and B354 CTV isolates have a common origin is discussed.
MATERIALS AND METHODS
Virus isolates.
CTV T30 was isolated from a Valencia orange [C. sinensis (L.) Osb.] tree in Polk County, Fla., in 1976. This tree was free of CTV when it was planted; subsequently, it was naturally infected by aphids. T30 was sequentially propagated in confinement in a greenhouse. First, it was aphid transmitted to Mexican lime [C. aurantifolia (Christm) Swingle]; second, it was graft inoculated to Madam Vinous sweet orange; and third, it was graft inoculated to Mexican lime, Madam Vinous sweet orange, and Etrog citron (C. medica L.) plants, which were used as a source of viral double-stranded RNA (dsRNA) for CTV cDNA synthesis.
CTV isolates B252, B272, and B354 were obtained from a collection of exotic citrus pathogens maintained at the quarantine facilities of the U.S. Department of Agriculture in Beltsville, Md. Isolate B252 was collected by H. J. Su in 1990 from an originally virus-free Ponkan mandarin tree (C. reticulata Blanco) that had been planted 2 years earlier in a commercial citrus region near Hsinchu (Taiwan). Isolate B272 was collected from a 20-year-old sweet orange tree on a sour orange rootstock from northern Colombia in 1991. Isolate B354 was collected from a naturally infected tree from the central valley of California in 1993. A pool of Mexican lime or sweet orange tissue infected with B252, B272, or B354 was used for CTV cDNA cloning purposes. The virus isolates used for sequence comparisons are shown in Table 1.
CTV T30 genome cloning and sequencing.
Random cDNA clones (Fig. 1B) from CTV isolate T30 were obtained from viral dsRNA purified from young bark tissue (31). T30 dsRNA was separated by electrophoresis in a 5% polyacrylamide gel, and the band containing the replicative form of the genomic RNA was excised under UV light, extracted overnight with (1:1) 0.5× TBE (4.45 mM Tris, 4.45 mM boric acid, 1 mM EDTA [pH 8.0])–buffer-saturated phenol, ethanol precipitated, and denatured by treatment with methylmercuric hydroxide (38). Genomic T30 dsRNA was polyadenylated using yeast poly(A)-polymerase (U.S. Biochemicals) and primed with an oligo(dT) primer containing an XhoI site (M-111) (20). Single-stranded cDNAs of both polarities were synthesized using avian myeloblastosis virus reverse transcriptase (Life Technologies) according to the supplier's instructions. For PCR amplification of T30 cDNA, Taq DNA polymerase (Promega Corporation) and the M-111 primer, which served as the forward and reverse primer, were used under standard conditions. PCR-amplified cDNA was digested with XhoI and ligated into XhoI-digested pGEM-7fZ (Promega). Sequencing of oligo(dT)-primed random cDNA clones was carried out on both strands using universal forward and reverse primers and a T7 Sequenase sequencing kit (U.S. Biochemicals) according to the manufacturer's recommendations. The sequences obtained were mapped in the CTV genome by comparison with T36 (22) and VT (27) sequences.
In order to obtain a set of overlapping reverse transcription (RT)-PCR cDNA clones covering the entire genome of T30 (Fig. 1B), T30-specific forward and reverse primers were designed based on sequences obtained from the oligo(dT)-primed random cDNA clones (Table 2). The forward primer and the reverse primer containing the exact 5′ (C198) and 3′ (C118) terminal sequences (Table 2), respectively, were designed based on the sequence of a dRNA naturally occurring in isolate T30 (M. Mawassi, unpublished data). Primers C257 and C258 (Table 2) were designed based on the sequence of the T30 clone T308 (20). Genomic T30 dsRNA was denatured as indicated above and reverse transcribed using Superscript II reverse transcriptase (Life Technologies) and T30-specific reverse primers (Table 2), following supplier recommendations. Eight overlapping T30 cDNA fragments (Fig. 1B) were amplified using Vent DNA polymerase (New England Biolabs) and T30-specific forward and reverse primers (Table 2). Annealing and elongation times were standardized for each pair of primers and specific DNA product. PCR products were cloned into pGEM-7fZ or pUC119 using the restriction sites indicated in Table 2 and standard cloning techniques (38). Sequencing of cloned RT-PCR products was performed with an automatic sequencer (Applied Biosystems model 373) at the Interdisciplinary Center for Biotechnology Research DNA sequencing core facility of the University of Florida, Gainesville.
TABLE 2.
RT-PCR primers used to generate specific CTV cDNA clones
| Forward primer | Location (nt) | Cloning site | Reverse primer | Location (nt) | Cloning site | cDNA clone generated |
|---|---|---|---|---|---|---|
| C198 | 1–19 | HindIII | C227 | 2427–2449 | NcoI | 6 |
| C226 | 2402–2430 | NdeI | C258 | 5169–5193 | Blunt | 5b |
| C257 | 5077–5097 | NdeI | C225 | 8654–8677 | NcoI | 5a |
| C229 | 8631–8654 | HindIII | C228 | 9460–9492 | BamHI | 4 |
| C213 | 9439–9457 | BamHI | C239 | 12625–12648 | XhoI | 3b |
| C240 | 12562–12584 | BamHI | C214 | 14766–14787 | XhoI | 3a |
| C215 | 14746–14766 | HindIII | C216 | 18079–18100 | XhoI | 2 |
| C217 | 17903–17922 | XhoI | C118 | 19237–19259 | NcoI | 1 |
| C326 | 1–28 | Blunt | C323 | 3686–3706 | NotI | A |
| C295 | 8291–8310 | Blunt | C225 | 8630–8654 | Blunt | B |
Cloning and sequencing of B252, B272, and B354 cDNA fragments.
dsRNA was extracted (31) from citrus bark tissue infected with isolate B252 from Taiwan, B272 from Colombia, or B354 from California. Two independent cDNA clones (Fig. 1C) were synthesized by RT-PCR with Superscript II reverse transcriptase and a proofreading thermostable polymerase (Pfu Turbo; Stratagene) by using the primers listed in Table 2. The sequences for variable regions I, II, and III were obtained from CTV cDNA clone A, and those for variable region IV were obtained from CTV cDNA clone B (Fig. 1C). A and B cDNA fragments were cloned in NotI/SmaI-digested pUC119 and SmaI-digested pGEM-7fZ, respectively. Sequencing of these cDNA clones was carried out using an automatic sequencer as indicated above.
Nucleotide and amino acid sequence comparisons.
The Genetics Computer Group package (8) programs SEQED to edit sequences, GAP to compare nucleotide and amino acid sequences, and TRANSLATE to obtain amino acid sequences were used for sequence analysis. Multiple sequence alignments were performed using the ClustalW program (45), and sequences were assembled using the DNA STRIDER program.
Nucleotide sequence accession numbers.
The complete nucleotide sequence of CTV isolate T30 was deposited in the GenBank database under accession no. AF260651. CTV nucleotide sequences for regions I, II, III, and IV of isolates B354, B272, and B252 were deposited in the GenBank database under accession numbers AF260652 to AF260663.
RESULTS
Comparison of the complete genomic sequences of CTV isolates T30 (Florida) and T385 (Spain).
It is not known whether the symptoms induced by a CTV isolate in citrus plants are induced by the predominant genomic sequence, the viral population, a combination of genomic RNAs and dRNAs, or other factors. The genomic sequences available at the moment correspond to isolates inducing different phenotypes. To examine whether different isolates inducing similar phenotypes might also have similar sequences, the complete sequence of the mild CTV isolate T30 (from Florida) was determined and compared to the sequence of the Spanish mild CTV isolate T385 (46). Neither isolate induces noticeable symptoms in field trees, and they cause only inconspicuous symptoms in sensitive indicator plants (Mexican lime). T30 and T385 have been separated from each other for a long time, as the T30 parental strain was imported into Florida via infected budwood more than 100 years ago and, to our knowledge, there was no citrus trading between Spain and Florida. Presently, T30 is representative of one of the two predominant Florida CTV strains. T30-like genotypes have been spread in the field by graft and aphid transmission, thereby becoming endemic in most commercial citrus groves. In addition, T30 and T385 were placed in isolation in greenhouses in different countries in 1976 and 1982, respectively, and were exposed to different sequences of host, graft, and aphid transmission passages (see Materials and Methods and references 30 and 46).
The entire sequence of CTV T30 was determined from the cDNA clones shown in Fig. 1B. Random cDNA clones were generated by polyadenylation of genomic dsRNA and RT with oligo(dT) as a primer (Fig. 1B). Sequences obtained from these random cDNA clones were mapped in relation to the T36 and VT genomes and were used to generate a set of T30-specific primers (Table 2). These primers were used to amplify eight overlapping RT-PCR cDNA fragments that covered the entire T30 genome (Fig. 1B).
Analysis of the resulting complete T30 sequence revealed a genome organization (Fig. 1A) identical to that reported for the other CTV sequences (T36 [22], VT [27], T385 [46], and SY568 [47]). Nucleotide sequence alignment (Table 3) of the genome of T30 with those of T36, VT, SY568, and T385 revealed that in contrast to the high sequence variability found between the T30 and the T36, VT, and SY568 genomes, the T30 genome was nearly identical to that of T385 (Table 3, roman boldface values). The genomic RNAs of both isolates were the same size (19,259 nucleotides [nt]), and their mean nucleotide identity was 99.3%. The identities between the different ORFs ranged from 98.7% to 100%. Their 3′ NTRs were 99.6% identical, and their 5′ NTRs were 96.3% identical (Table 3, roman boldface values). Compared to T36, VT, and SY568, T30 had the same nucleotide insertions and deletions and similar patterns of nucleotide variability throughout the genome as T385 (data not presented). Nucleotide differences between T30 and T385 were constant throughout the genomes (71 and 72 differences in the 3′ and 5′ halves of the genomes, respectively), whereas sequence differences were greater in the 5′ halves of the genomes of other CTV genotypes (Table 3). The similarity between the predicted amino acid sequences of T30 and T385 ranged from 98.6% (p23) to 100% (p20, p6, and RdRp domain in ORF 1) (Table 3, roman boldface values). Analysis of the amino acid sequences indicated that 52.9% of the nucleotide changes were silent, 41.2% encoded similar amino acids, and only 5.9% encoded dissimilar amino acids (in p23, p33, and ORF 1a proteins).
TABLE 3.
Nucleotide and deduced amino acid sequence differences between the CTV genomic sequences from T30 and isolate T385, VT, T36, or SY568a
| T30 genome
|
T385 (Spain)
|
VT (Israel)
|
T36 (Florida)
|
SY568 (California)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Size (nt) | Nucleotide I | Amino acid
|
Nucleotide I | Amino acid
|
Nucleotide I | Amino acid
|
Nucleotide I | Amino acid
|
||||
| S | I | S | I | S | I | S | I | ||||||
| 5′ NTR | 108 | 96.3 | 70.2 | 41.1 | 64.2 | ||||||||
| ORF 1a | 9,348 | 99.2 | 99.6 | 99.1 | 89.6 | 93.8 | 89.0 | 72.4 | 88.7 | 70.1 | 90.7 | 96.0 | 92.0 |
| RdRp | 1,434 | 99.5 | 100 | 99.8 | 90.0 | 96.6 | 94.8 | 79.4 | 95.2 | 91.0 | 91.4 | 96.9 | 96.4 |
| p33 | 912 | 99.2 | 99.3 | 99.3 | 84.6 | 90.5 | 85.2 | 84.1 | 92.4 | 86.1 | 99.4 | 99.7 | 99.3 |
| p6 | 156 | 100 | 100 | 100 | 90.9 | 98.1 | 92.3 | 88.4 | 94.2 | 92.3 | 100 | 100 | 100 |
| p65 | 1,785 | 99.2 | 99.3 | 98.8 | 87.3 | 93.9 | 90.9 | 93.7 | 97.1 | 95.3 | 99.6 | 99.5 | 99.2 |
| p61 | 1,608 | 99.4 | 99.1 | 98.7 | 87.9 | 94.6 | 90.7 | 94.8 | 97.4 | 95.3 | 99.4 | 99.3 | 99.0 |
| p27 (CPm) | 730 | 99.3 | 99.6 | 99.2 | 88.7 | 95.4 | 93.3 | 93.6 | 97.5 | 96.3 | 99.1 | 97.9 | 97.1 |
| p25 (CP) | 672 | 99.6 | 99.6 | 99.1 | 92.4 | 97.8 | 94.6 | 93.0 | 98.7 | 96.0 | 93.1 | 98.2 | 96.0 |
| p18 | 504 | 99.4 | 99.4 | 98.8 | 93.0 | 94.6 | 92.2 | 95.2 | 97.0 | 95.2 | 93.1 | 96.4 | 95.2 |
| p13 | 360 | 99.4 | 99.1 | 99.1 | 91.1 | 96.6 | 94.1 | 91.7 | 95.8 | 90.6 | 95.8 | 90.8 | |
| p20 | 549 | 99.6 | 100 | 100 | 90.3 | 96.2 | 93.4 | 88.8 | 96.7 | 93.4 | 93.1 | 98.9 | 97.7 |
| p23 | 630 | 98.7 | 98.6 | 98.1 | 89.6 | 93.3 | 89.0 | 90.9 | 95.2 | 91.4 | 89.7 | 95.2 | 91.4 |
| 3′ NTR | 274 | 99.6 | 99.0 | 97.0 | 97.8 | ||||||||
I, percent identity; S, percent similarity. CP, major coat protein; CPm, minor coat protein. See text for explanation of boldface, italic, and underlined values.
Comparison of additional mild CTV sequences to the CTV T30 sequence.
The homology between the complete sequences of CTV isolates T30 and T385 was greater than expected. To examine whether there were other CTV isolates highly similar to T30, we examined isolates B252, B272, and B354, which induce the same phenotype as T30. These isolates originated from different environments and were thought to have been separated for a long time. B252 has its origins in the first importation of Citrus and CTV isolates to Taiwan from mainland China 100 to 500 years earlier. There has been no known movement of citrus into or out of this area since that time. B272 was collected in an area of Colombia where the citrus bud wood is thought to have been imported from Spain more than 20 years earlier. B354 was found in a Californian citrus field into or out of which there has been no known movement of citrus during the past 60 years.
Four hypervariable CTV genome regions, based on the T36, T385, and VT sequences, were chosen for sequence comparison (Fig. 1C). These regions were cloned and sequenced from dsRNA of isolates B252, B272, and B354 and compared with those of T30, T385, VT, and T36 (Table 1 and Fig. 2).
FIG. 2.
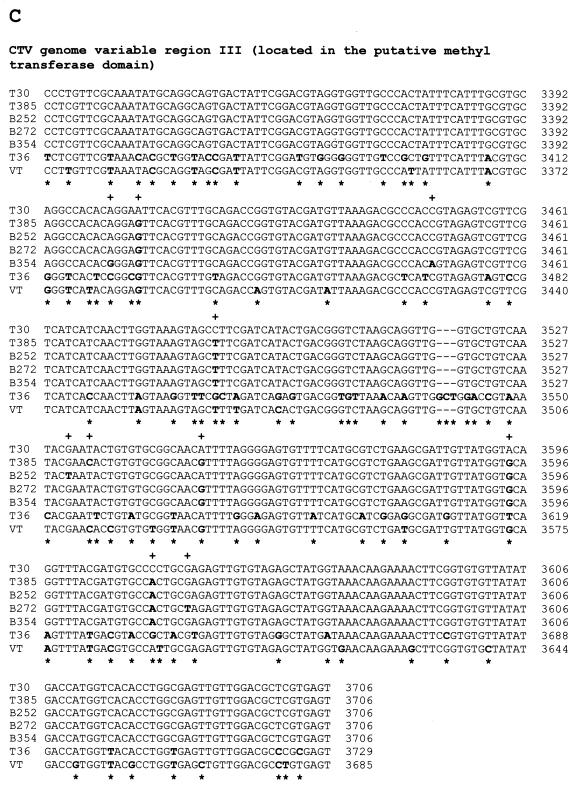
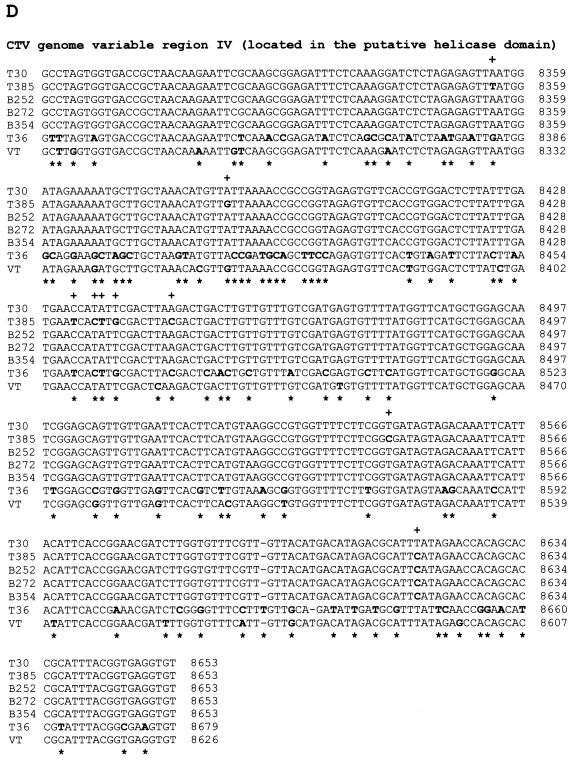
Nucleotide sequence alignments of CTV hypervariable genomic regions (ClustalW program alignments; see Materials and Methods). (A) Alignment of variable region I (5′ NTR, 108 nt) sequences. (B) Alignment of variable region II (located from nt 109 to nt 324 of the T30 genome in the 5′-proximal region of the replicase gene) sequences. (C) Alignment of variable region III (located from nt 3324 to nt 3706 of the T30 genome in the putative methyltransferase domain) sequences. (D) Alignment of variable region IV (located from nt 8265 to nt 8627 of the T30 genome in the putative helicase domain of the replicase ORF) sequences. In all panels, nucleotide differences in aligned sequences are showed in boldface. +, nucleotide differences between T30 and the mild CTV isolates T385, B252, B272, and B354; ∗, nucleotide differences between T30 and severe isolates T36 and VT.
Variable region I (108 nt), which comprised the 5′ NTR (Fig. 1C), is the most variable region among reported CTV sequences (25, 46) (Table 3, underlined values). T30 had 41.1% identity (43 nucleotide differences, six gaps, and four insertions) and 70.2% identity (23 nucleotide differences, four gaps, and three insertions) with T36 and VT, respectively, in variable region I (Fig. 2A). However, B252, B272, and B354 had two to four nucleotide differences relative to the sequences of T30 and T385 (with identities ranging from 97.2% to 99.1%) (Fig. 2A). Moreover, the 5′ NTR sequences of B354 (California) and B252 (Taiwan) were identical (Fig. 2A).
Variable region II included the first 234 nt of ORF 1a (Fig. 1C). In this region, T30 had 72.24% identity (59 nucleotide differences and one insertion) and 92.2% identity (18 nucleotide differences and one insertion) with T36 and VT, respectively (Fig. 2B). In contrast, the variable region II sequence of B252 was identical to that of T30 and the variable region II sequence of B272 was identical to that of B354. The single nucleotide change observed between the T30 (or B252) and B272 (or B354) sequences was silent. Maximum differences (2 nt) occurred between T385 (Spain) and T30 (Florida) or B252 (Taiwan) (Fig. 2B). One of these nucleotide changes was silent, whereas the other resulted in a change to a codon for a dissimilar amino acid.
Variable region III is located between nt 3324 and 3706 of the T30 genome, consisting of part of the putative methyltransferase domain (Fig. 1C). Nucleotide identities of T30 compared to T36 and VT were 81.2% (74 nucleotide differences and one deletion) and 90.3% (39 nucleotide differences), respectively (Fig. 2C). Alignment of this region between paired B252, B272, B354, T30, and T385 isolates showed identities of 98.3 to 99.7%. Maximum differences occurred between T30 (Florida) and the other four isolates, differing by 5 or 6 nt (Fig. 2C). Minimum differences were found between B354 (California), B252 (Taiwan), B272 (Colombia), and T385 (Spain), differing by 2 or 3 nt (Fig. 2C). Ten nucleotide differences were found in total. Six of them were silent, two resulted in changes to codons for similar amino acids, and two resulted in a change to a codon for a dissimilar amino acid.
Variable region IV is located from nt 8265 to nt 8627 of the T30 genome, in the helicase-like domain (Fig. 1D). In this region, nucleotide identities between T30 and T36 or VT were 75.5% (85 nucleotide differences, one deletion, and one insertion) and 93.9% (22 nucleotide differences), respectively (Fig. 2D). Region IV also was the area of lowest nucleotide identity between T30 and T385 (97.5% identity). Yet, only two of the nine nucleotide changes resulted in dissimilar amino acid changes, and the other seven were silent. Variable region IV sequences from B252, B272, and B235 cDNA clones were identical to the T30 sequence (Fig. 2D).
Total nucleotide identities of variable CTV genome regions I to IV among B354 (California), B252 (Taiwan), B272 (Colombia), and T30 (Florida) ranged from 98.3% to 100%, demonstrating genomic variability in the same range as that for T30 and T385 (97.5 to 98.4% identity).
T30 viral quasispecies variation and comparison with T30, T385, B252, B272, and B354 sequence variability.
T30 and T385 complete sequences and B354, B252, and B272 sequence samples were nearly identical. To assess whether the level of divergence between these mild isolates was within the range of variation present in a CTV quasispecies, we examined the population variability of isolate T30.
The sequences of 22 oligo(dT)-primed random cDNA clones which mapped to nine different regions in the CTV genome and the T30 cDNA clone (T308) previously obtained (20) (Table 4) were compared with the homologous sequence in the eight RT-PCR cDNA clones that comprised the T30 genomic sequence (Fig. 1B) (Table 4). The random cDNA clones plus the T308 cDNA clone covered 3,704 nt, corresponding to 19% of the CTV genome. These comparisons revealed nucleotide identities of 98.6 to 100% (Table 4). Most of the nucleotide changes were silent (30%) or encoded similar amino acids (60%). Additionally, overlapping sequences of oligo(dT)-primed cDNA clones (B10 and B18F; R3, R10, and R11; and R20 and R6) (Table 4) were 98 to 100% identical (data not shown). Likewise, overlapping regions between neighboring RT-PCR cDNA clones (between 30 and 100 nt) were also identical (data not shown). The small amount of sequence divergence between paired T30 cDNA clones indicated that the T30 population is principally composed of one genotype.
TABLE 4.
Nucleotide differences between the T30 oligo(dT)-primed random cDNA clone sequences and the T30 genomic sequence
| T30 cDNA clone | Size (nt) | Locationa | % Nucleotide identity | No. of nucleotide differences |
|---|---|---|---|---|
| R36 | 139 | 658, Replicase | 98.6 | 2 |
| R8 | 100 | 1861, Replicase | 100 | |
| R38 | 198 | 2378, Replicase | 99.5 | 1 |
| T308b | 339 | 4885, Replicase | 99.4 | 2 |
| R9 | 163 | 8598, Replicase | 99.4 | 1 |
| R27 | 134 | 8951, Replicase | 100 | |
| R15 | 120 | 9576, RdRp | 100 | |
| R19 | 225 | 9783, RdRp | 99.1 | 1 |
| B18R | 133 | 13911, p61 | 99.5 | 1 |
| B30 | 105 | 14271, p61 | 100 | |
| A12 | 181 | 14635, p61 | 100 | |
| B14 | 134 | 15497, p27 | 100 | |
| B24 | 149 | 15705, p27 | 100 | |
| B10 | 155 | 15976, p27-p25 | 99.4 | 1 |
| B18F | 154 | 15977, p27-p25 | 100 | |
| R31 | 187 | 17148, p18 | 100 | |
| R3 | 143 | 17850, p20 | 100 | |
| R10 | 178 | 17877, p20 | 98.9 | 2 |
| R11 | 187 | 17940, p20 | 99.5 | 1 |
| R2 | 113 | 18050, p20 | 100 | |
| A9 | 173 | 18364, p23 | 99.4 | 1 |
| R20 | 152 | 19107, 3′ NTR | 99.3 | 2 |
| R6 | 142 | 19120, 3′ NTR | 99.3 | 2 |
Nucleotide position of the 5′ end of the T30 oligo(dT)-primed random cDNA clone sequence in the T30 genome.
T30 cDNA clone previously obtained by Hilf et al. (20).
The number of samples used to examine the sequence variability within the viral variants of the T30 population was small but sufficient to provide an estimate of 0.5% nucleotide variability. A comparison of the T30 and T385 genome sequences yielded 0.7% nucleotide variability. Comparisons between T30, B252, B272, and B354 sequences in the most divergent regions of the CTV genome (Fig. 2) yielded 0.4 to 0.8% nucleotide variability. These data show that the variation between the complete sequences of T30 and T385 and the sequence samples of B252, B272, and B354 (less than 1% nucleotide variability) was similar to the sequence variation for the genomic variants of isolate T30.
DISCUSSION
The high degree of variability found between CTV genotypes, up to 20 to 60% nucleotide differences throughout the 5′ half of the genome plus a large number of deletions and insertions (27, 46), demonstrates that sequence divergence of the CTV genome has been extensive. Furthermore, CTV population structures are complex, and the apparent prevalence of recombination based on the omnipresence of multiple dRNAs (1, 26, 28) provides more opportunity for change. Surprisingly, we found that the consensus sequence of CTV T30 from Florida was nearly identical to that of CTV T385 from Spain. This result was not expected, since T30 and T385 were geographically isolated in different environments for at least 24 years and likely for more than 100 years, with aphids mixing different local populations of CTV variants. This striking similarity between distant CTV genotypes was extended by sampling hypervariable regions within the genomes of CTV isolates from Taiwan, Colombia, and California, which also were almost identical to the T30 genome. Another example of remarkable similarity between CTV isolates was recently reported (46) for a portion of the sequence of isolate SY568 (47). This virus was introduced into the citrus variety collection at the University of California, Riverside. A 6-kb central region of the SY568 genome, between the RdRp domain in ORF 1 and the p27 ORF, was 99.1 to 100% identical to the T30 genome (Table 3, italic values), although the rest of the sequence was more divergent from the T30 sequence; these findings support the hypothesis of Vives et al. (46) that SY568 probably resulted from recombination between two different genotypes. Furthermore, these data indicate the presence of a genotype almost identical to that of T30 and T385 in the original citrus host.
It should be noted that all of these isolates were chosen because of their similar mild phenotypes. However, it is likely that there are other strains of CTV with similar mild phenotypes but with divergent sequences, and there is evidence that there are isolates with similar predominant genotypes but with severe phenotypes (M. E. Hilf, unpublished results).
The absence of proofreading in the viral RNA polymerases (12) is thought to generate mutant genomes making up the viral quasispecies distribution (11). Pairwise comparisons between independent cDNA sequences from the T30 population provided an estimate of 0.5% nucleotide variability. This degree of nucleotide variability was supported by similar measurements within other CTV populations (3) and between CTV dRNAs and their helper (26). It also falls within the range of nucleotide variability of the quasispecies distribution in other RNA viruses (2, 6, 14, 19, 39, 44). Comparisons of the genomic sequence of T30 with those of four other mild isolates revealed that in spite of these isolates being separated in time and space, the nucleotide sequence variability was less than 1% and within or near the range of variability in the T30 population. Thus, the exceptional similarity of these five mild isolates suggests that they could be considered variants of the same CTV genotype.
Although a great capacity for rapid evolution is a common feature of RNA viruses (9), there are examples of genetic stability in viral RNA populations (19, 21, 23). Some of the more stable examples are the putative “rapidly evolving” viruses, such as vesicular stomatitis rhabdovirus (33, 44) and a strain of H1N1 influenza A orthomyxovirus (32) that were nearly identical after 500 passages and 27 years, respectively. Other examples of high genetic stability are found within the plant viruses tobacco mild green mosaic tobamovirus (16) and turnip yellow mosaic tymovirus (5, 42), which were reported to be nearly identical after 100 years and 13,000 to 14,000 years, respectively. This genetic stability has been explained as a consequence of strong selection and competition between the mutants that arise in each replication cycle, creating an equilibrium in the viral quasispecies distribution (11). However, viral populations are dynamic. Founder effects or bottlenecks can allow newly arising mutants to shift the quasispecies distribution, promoting rapid evolution (7, 43). CTV genotypes were dispersed to different environments around the world by vegetative propagation of citrus. Additionally, different CTV genotypes were further spread and mixed by graft and aphid transmission, which may create bottlenecks (1, 29) allowing minority viral variants (sometimes, the most virulent ones [30]) to become prevalent in CTV populations (37). These continuous changes in selective pressures probably have accounted for much of the variation between and within other CTV isolates.
The remarkable sequence similarity of these five CTV isolates could have resulted from any of a number of possibilities. One is that all five isolates converged to a similar sequence as a result of host selection. However, several studies of CTV populations have indicated that unrelated CTV sequences can coexist in the same area, same host, and same environment. For example, T30- and T36-like genotypes are endemic in the same citrus areas in Florida and differ greatly in symptoms induced and genome sequences. In fact, most CTV isolates contain minor components of disparate sequences (3, 30). Thus, it appears unlikely that selection was sufficiently strong to cause convergence of these five CTV isolates within relatively short periods of time. Since the only natural host for CTV is citrus, a more likely possibility is that these isolates evolved in one of the native citrus parents at its point of origin in Asia and were dispersed around the world with citrus budwood within the last 200 years. The fact that all five isolates are essentially invisible (nonsymptomatic) in infected trees would have aided this spread. This hypothesis is supported by the fact that most of the nucleotide changes in the five CTV isolates were silent or resulted in changes to codons for similar amino acids, suggesting that this CTV genotype is well adapted to the citrus environment and perhaps has changed little since exportation from its origin in the last several hundred years. Since there are at least four progenitor species that are thought to be the origin of all of the current varieties in agriculture, it is possible that the divergent CTV strains evolved in different progenitors prior to citrus agriculture. If other CTV genotypes have remained stable over time, it should be possible to trace isolates with similar sequences to their original sources. From a standpoint of management, if CTV sequences tend to remain relatively stable over periods of years, sequence-based control strategies, such as transgenic plants with posttranscriptional gene silencing directed against specific viral sequences or cross protection based on mild strains excluding superinfection by severe strains with similar sequences, have a higher probability of success.
ACKNOWLEDGMENTS
We thank Adrian Gibbs, Fernando García Arenal, Isabel Novella, and Chris Kearney for critically reviewing the manuscript.
This work was supported in part by an endowment from the J. R. and Addie S. Graves family and grants from the Florida Citrus Production Research Advisory Council, USDA/ARS cooperative agreement 58-6617-4-018, the U.S.-Israel BARD, and the National Citrus Research Council. M. R. Albiach-Martí was the recipient of a postdoctoral fellowship from the Ministerio de Educación y Ciencia (Spain).
Footnotes
University of Florida Agricultural Experiment Station journal series no. R-G7529.
REFERENCES
- 1.Albiach-Martí R M, Guerri J, Hermoso de Mendoza A, Laigret F, Ballester Olmos J F, Moreno P. Aphid transmission alters the genomic and defective RNA populations of citrus tristeza virus isolates. Phytopathology. 1999;90:134–138. doi: 10.1094/PHYTO.2000.90.2.134. [DOI] [PubMed] [Google Scholar]
- 2.Arias C F, López S, Espejo R T. Heterogeneity in base sequence among different DNA clones containing equivalent sequence of rotavirus double-stranded RNA. J Virol. 1986;57:1207–1209. doi: 10.1128/jvi.57.3.1207-1209.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayllón M A, Rubio L, Moya A, Guerri J, Moreno P. The haplotype distribution of two genes of citrus tristeza virus is altered after host change or aphid transmission. Virology. 1999;255:32–39. doi: 10.1006/viro.1998.9566. [DOI] [PubMed] [Google Scholar]
- 4.Bar-Joseph M, Marcus R, Lee R F. The continuous challenge of citrus tristeza virus control. Annu Rev Phytopathol. 1989;27:291–316. [Google Scholar]
- 5.Blok J, Mackenzie A, Guy P, Gibbs A. Nucleotide sequence comparisons of turnip yellow mosaic virus from Australia and Europe. Arch Virol. 1987;97:283–295. doi: 10.1007/BF01314427. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo R, Schmid A, Rebmann G, Baczko K, ter Meulen V. Accumulated measles virus mutations in a case of subacute sclerosing panencephalitis: interrupted matrix protein reading frame and transcription alteration. Virology. 1986;154:97–107. doi: 10.1016/0042-6822(86)90433-2. [DOI] [PubMed] [Google Scholar]
- 7.Clarke D K, Duarte E A, Clarke D K, Moya A, Elena S F, Domingo E, Holland J J. Genetic bottlenecks and population passages cause profound fitness differences in RNA viruses. J Virol. 1993;67:222–228. doi: 10.1128/jvi.67.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domingo E, Holland J J. Mutation rates and rapid evolution of RNA viruses. In: Morse S S, editor. The evolutionary biology of viruses. New York, N.Y: Raven Press, Ltd.; 1994. pp. 161–184. [Google Scholar]
- 10.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 11.Domingo E, Holland J J, Biebricher C, Eigen M. Quasi-species: the concept and the word. In: Gibbs A J, Calisher C H, García-Arenal F, editors. Molecular basis of virus evolution. Cambridge, England: Cambridge University Press; 1995. pp. 181–191. [Google Scholar]
- 12.Drake J W, Holland J J. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Febres V J, Ashoulin L, Mawassi M, Frank A, Bar-Joseph M, Manjunath K L, Lee R F, Niblett C L. The p27 protein is present at one end of citrus tristeza virus particles. Phytopathology. 1996;86:1331–1335. [Google Scholar]
- 14.Fields S, Winter G. Nucleotide sequence heterogeneity and sequence rearrangements in influenza virus cDNA. Gene. 1981;15:207–214. doi: 10.1016/0378-1119(81)90130-x. [DOI] [PubMed] [Google Scholar]
- 15.Fraile A, Malpica J M, Aranda M A, Rodríguez-Cerezo E, García-Arenal F. Genetic diversity in tobacco mild green mosaic tobamovirus infecting the wild plant Nicotiana glauca. Virology. 1996;223:148–155. doi: 10.1006/viro.1996.0463. [DOI] [PubMed] [Google Scholar]
- 16.Fraile A, Escriu F, Aranda M A, Malpica J M, Gibbs A J, García-Arenal F. A century of tobamovirus evolution in an Australian population of Nicotiana glauca. J Virol. 1997;71:8316–8320. doi: 10.1128/jvi.71.11.8316-8320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibbs A. Evolution and origins of tobamoviruses. Philos Trans R Soc London B. 1999;354:517–685. doi: 10.1098/rstb.1999.0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn B H, Shaw G M, Taylor M E, Redfield R R, Markham P D. Genetic variation in HTLV-III/LAV over time in patients with AIDS or at risk for AIDS. Science. 1986;232:1548–1553. doi: 10.1126/science.3012778. [DOI] [PubMed] [Google Scholar]
- 19.Hedges J F, Balasuriya U B R, Timoney P J, McCollum W H, MacLachlan N J. Genetic divergence with emergence of novel phenotypic variants of equine arteritis virus during persistent infection of stallions. J Virol. 1999;73:3672–3681. doi: 10.1128/jvi.73.5.3672-3681.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hilf M E, Karasev A V, Albiach-Martí M R, Dawson W O, Garnsey S M. Two paths of sequence divergence in the citrus tristeza virus complex. Phytopathology. 1999;89:336–342. doi: 10.1094/PHYTO.1999.89.4.336. [DOI] [PubMed] [Google Scholar]
- 21.Hillman B J, Anzola J V, Halpern B T, Cavileer T D, Nuss D L. First field isolation of wound tumor virus from a plant host: minimal sequence divergence from the type strain isolated from an insect vector. Virology. 1991;185:896–900. doi: 10.1016/0042-6822(91)90568-v. [DOI] [PubMed] [Google Scholar]
- 22.Karasev A V, Boyko V P, Gowda S, Nikolaeva O V, Hilf M E, Koonin E V, Niblett C L, Cline K, Gumpf D J, Lee R F, Garnsey S M, Lewandowski D J, Dawson W O. Complete sequence of the citrus tristeza virus RNA genome. Virology. 1995;208:511–520. doi: 10.1006/viro.1995.1182. [DOI] [PubMed] [Google Scholar]
- 23.Kruse M, Koening R, Hoffmann A, Kaufmann A, Commandeur U, Solovyev A G, Savenkov I, Burgermeister W. Restriction fragment length polymorphism analysis of reverse transcription-PCR products reveals the existence of two major strain groups of beet necrotic yellow vein virus. J Gen Virol. 1994;75:1835–1842. doi: 10.1099/0022-1317-75-8-1835. [DOI] [PubMed] [Google Scholar]
- 24.Lai M M C. Recombination and its evolutionary effect on viruses with RNA genomes. In: Gibbs A J, Calisher C H, García-Arenal F, editors. Molecular basis of virus evolution. Cambridge, England: Cambridge University Press; 1995. pp. 119–132. [Google Scholar]
- 25.López C, Ayllón M A, Navas-Castillo J, Guerri J, Moreno P, Flores R. Molecular variability of the 5′ and 3′ terminal regions of citrus tristeza virus RNA. Phytopathology. 1998;88:685–691. doi: 10.1094/PHYTO.1998.88.7.685. [DOI] [PubMed] [Google Scholar]
- 26.Mawassi M, Karasev A V, Mietkiewska E, Gafny R, Lee R F, Dawson W O, Bar-Joseph M. Defective RNA molecules associated with citrus tristeza virus. Virology. 1995;208:383–387. doi: 10.1006/viro.1995.1165. [DOI] [PubMed] [Google Scholar]
- 27.Mawassi M, Mietkiewska E, Gofman R, Yang G, Bar-Joseph M. Unusual sequence relationships between two isolates of citrus tristeza virus. J Gen Virol. 1996;77:2359–2364. doi: 10.1099/0022-1317-77-9-2359. [DOI] [PubMed] [Google Scholar]
- 28.Mawassi M, Mietkiewska E, Hilf M E, Ashoulin L, Karasev A V, Gafny R, Lee R F, Garnsey S M, Dawson W O, Bar-Joseph M. Multiple species of defective RNAs in plants infected with citrus tristeza virus. Virology. 1995;214:264–268. doi: 10.1006/viro.1995.9930. [DOI] [PubMed] [Google Scholar]
- 29.Moreno P, Guerri J, Ballester-Olmos J F, Fuertes-Polo C, Albiach R, Martínez M E. Variations in pathogenicity and double stranded RNA (dsRNA) patterns of citrus tristeza virus isolate induced by host passage. In: Brlansky R H, Lee R F, Timmer L W, editors. Proceedings of the 12th Conference of the International Organization of Citrus Virologists. Riverside: Department of Plant Pathology, University of California; 1993. pp. 8–15. [Google Scholar]
- 30.Moreno P, Guerri J, Ballester-Olmos J F, Martínez M E. Segregation of citrus tristeza virus strains evidenced by double stranded RNA (dsRNA) analysis. In: Brlansky R H, Lee R F, Timmer L W, editors. Proceedings of the 11th Conference of the International Organization of Citrus Virologists. Riverside: Department of Plant Pathology, University of California; 1991. pp. 20–24. [Google Scholar]
- 31.Moreno P, Guerri J, Muñoz N. Identification of Spanish strains of citrus tristeza virus by analysis of double-stranded RNA. Phytopathology. 1990;80:477–482. [Google Scholar]
- 32.Nakajima K, Desselberger U, Palese P. Recent human influenza A (H1N1) viruses are closely related genetically to strains isolated in 1950. Nature. 1978;247:334–339. doi: 10.1038/274334a0. [DOI] [PubMed] [Google Scholar]
- 33.Nichol S T, Rowe J E, Fitch W M. Punctuated equilibrium and positive Darwinian evolution in vesicular stomatitis virus. Proc Natl Acad Sci USA. 1993;90:10424–10428. doi: 10.1073/pnas.90.22.10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pappu H R, Karasev A V, Anderson E J, Pappu S S, Hilf M E, Febres V J, Eckloff R M G, McCaffery M, Boyko V, Gowda S, Dolja V V, Koonin E V, Gumpf D J, Cline K, Garnsey S M, Dawson W O, Lee R F, Niblett C L. Nucleotide sequence and organization of eight 3′ open reading frames of citrus tristeza closterovirus genome. Virology. 1994;199:35–46. doi: 10.1006/viro.1994.1095. [DOI] [PubMed] [Google Scholar]
- 35.Rodríguez-Cerezo E, Moya A, García-Arenal F. Variability and evolution of the plant RNA virus pepper mild mottle virus. J Virol. 1989;63:2198–2203. doi: 10.1128/jvi.63.5.2198-2203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodríguez-Cerezo E, Elena S F, Moya A, García-Arenal F. High genetic stability in natural populations of the plant RNA virus tobacco mild green mosaic virus. J Mol Evol. 1991;32:328–332. [Google Scholar]
- 37.Roistacher C N, Moreno P. The worldwide threat from destructive isolates of citrus tristeza virus—a review. In: Brlansky R H, Lee R F, Timmer L W, editors. Proceedings of the 11th Conference of the International Organization of Citrus Virologists. Riverside: Department of Plant Pathology, University of California; 1991. pp. 7–19. [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Schubert M, Harmison G G, Meier E. Primary structure of the vesicular stomatitis virus polymerase (L) gene: evidence for a high frequency of mutations. J Virol. 1984;51:505–514. doi: 10.1128/jvi.51.2.505-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekiya M E, Lawrence S D, McCaffery M, Cline K. Molecular cloning and nucleotide sequencing of the coat protein gene of citrus tristeza virus. J Gen Virol. 1991;72:1003–1020. doi: 10.1099/0022-1317-72-5-1013. [DOI] [PubMed] [Google Scholar]
- 41.Simon A E, Bujarski J J. RNA-RNA recombination and evolution in infected plants. Annu Rev Phytopathol. 1994;32:337–362. [Google Scholar]
- 42.Skotnicki M L, Mackenzie A M, Ding S W, Mo J Q, Gibbs A J. RNA hybrid mismatch polymorphisms in Australian populations of turnip yellow mosaic tymovirus. Arch Virol. 1993;132:83–99. doi: 10.1007/BF01309845. [DOI] [PubMed] [Google Scholar]
- 43.Solé R V, Ferrer R, González-García I, Quer J, Domingo E. Red queen dynamics, competition and critical points in a model of RNA virus quasispecies. J Theor Biol. 1999;198:47–59. doi: 10.1006/jtbi.1999.0901. [DOI] [PubMed] [Google Scholar]
- 44.Steinhauer D A, de la Torre J A, Meier E, Holland J J. Extreme heterogeneity in populations of vesicular stomatitis virus. J Virol. 1989;63:2072–2080. doi: 10.1128/jvi.63.5.2072-2080.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson J D, Higgins D G, Gibson T J. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vives M C, Rubio L, López C, Navas-Castillo J, Albiach-Martí M R, Dawson W O, Guerri J, Flores R, Moreno P. The complete genome sequence of the major component of a mild citrus tristeza virus isolate. J Gen Virol. 1999;80:811–816. doi: 10.1099/0022-1317-80-3-811. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z-N, Mathews D M, Dodds J A, Mirkov T E. Molecular characterization of an isolate of citrus tristeza virus that causes severe symptoms in sweet orange. Virus Genes. 1999;19:131–142. doi: 10.1023/a:1008127224147. [DOI] [PubMed] [Google Scholar]