Abstract
We have analyzed the functional activity of the p53 tumor suppressor in human T-cell lymphotropic virus type 2 (HTLV-2)-transformed cells. Abundant levels of the p53 protein were detected in both HTLV-2A and -2B virus-infected cell lines. The p53 was functionally inactive, however, both in transient-transfection assays using a p53 reporter plasmid and in induction of p53-responsive genes in response to gamma irradiation. We further investigated HTLV-2A Tax and HTLV-2B Tax effects on p53 activity. Interestingly, although Tax-2A and -2B inactivate p53, the Tax-2A protein appears to inhibit p53 function less efficiently than either Tax-1 or Tax-2B. In transient-cotransfection assays, Tax-1 and Tax-2B inactivated p53 by 80%, while Tax2A reduced p53 activity by 20%. In addition, Tax-2A does not increase the steady-state level of cellular p53 as well as Tax-1 or -2B does in the same assays. Cotransfection assays demonstrated that Tax-2A could efficiently transactivate CREB-responsive promoters to the same level as Tax-1 and Tax-2B, indicating that the protein was functional. This report provides evidence of the first functional difference between the HTLV-2A and -2B subtypes. This comparison of the action of HTLV-1 and HTLV-2 Tax proteins on p53 function will provide important insights into the mechanism of HTLV transformation.
Human T-cell lymphotropic virus type 1 (HTLV-1) and type 2 (HTLV-2) are closely related viruses which infect T cells and share an important number of biological properties (22, 47). HTLV-1 is the etiological agent of two different diseases, an aggressive T-cell leukemia (ATL) (34) and a myelopathy tropical spastic paraparesis/HTLV-1-associated myelopathy [TSP/HAM]) (15). HTLV-2 has not been clearly demonstrated as the agent of any T-cell malignancy (12). At the amino acid level, the viral transactivator Tax from HTLV-1 and HTLV-2 have approximately 77 to 85% homology depending on the part of the protein studied, the N-terminal part being the most conserved between the two proteins. Tax-1 and Tax-2 can transactivate their respective long terminal repeats (LTR) (2, 3, 7, 37). The existence of different HTLV-2 subtypes has been uncovered only recently (11, 19, 20, 30, 48). Strikingly, the Tax-2 proteins of the four different HTLV-2 subtypes (named A, B, C, and D) have different lengths. Tax-2A is 331 amino acids (aa) long, Tax-2B and Tax-2C are 356 aa, and Tax-2D is 344 aa long. Moreover, Tax-2B and -2C, whose lengths are very similar to that of Tax-1 (353 aa), have totally different C-terminal sequences (11, 18, 48). Conflicting studies have reported as to the ability of Tax-1 and Tax-2 to activate heterologous promoters (2, 3, 38). The functional regions or domains important for transactivation through the CREB/ATF and NF-κB signaling pathways are similar but not identical between the two proteins (38).
HTLV-2A has been shown to be predominant among intravenous drug users in North America and Europe and is widespread worldwide (29). In contrast, HTLV-2B predominates in Paleo-Indian groups, while HTLV-2C and -2D are detected in remote populations of Brazil and Central African Pygmies, respectively (11, 18, 25, 29, 43, 44, 48). The geographical locations of the HTLV-2B, -2C, and -2D-infected populations and the relatively small number of infected individuals studied do not allow for easy follow-up to determine whether they are at higher risk for developing a disease.
Several studies have compared the transactivation and transformation properties of the Tax-1 and Tax-2 proteins. Semmes et al. (41) have reported that, in comparison to Tax-1, the Tax-2A protein lacks the ability to induce micronuclei in infected cells. In another study, Tanaka et al. reported that Tax-2A was able to activate the ICAM-1 promoter in HeLa cells but not in T-cell lines, whereas Tax-1 could activate the ICAM-1 promoter in all cell lines tested (46). Finally, it has been reported that Tax-2B is a more potent transactivator than -2A (11). In transformation studies, the Tax-2A protein has been shown to be essential for HTLV-mediated transformation of human T lymphocytes in culture (39), but there are no published data concerning HTLV-2B, -2C, and -2D. Immortalized interleukin 2 (IL-2)-dependent cell lines obtained from cell cultures of human T lymphocytes obtained from healthy carriers infected with HTLV-2A and -2B, but not -2C or -2D have been reported (17, 24).
Several laboratories have shown that wild-type p53 was transcriptionally inactive in HTLV-1-infected cells and that Tax protein alone could inactivate p53 function (1, 6, 14, 26, 31, 32, 51). Several models have been proposed to account for the p53 inhibition. Pise-Masison et al. (32) have recently demonstrated that this inactivation was associated with hyperphosphorylation of serine 15, a residue located in the N-terminal activation domain of the protein. This modification was sufficient for inhibiting the binding of p53 to TFIID. In contrast, the laboratories of Nyborg and Yoshida have reported that inhibition of p53 function is due to squelching of CREB-binding protein (CBP) by the HTLV-1 Tax protein (42, 49). In either case p53 inactivation, which results in both impairment of the DNA repair pathway and inhibition of apoptosis, could be one of the primary events which leads to the clonal expansion of the HTLV-1-infected cells (4, 5, 50).
We have investigated the ability of Tax-2 proteins to regulate p53 function. We demonstrate here that HTLV-2A- and -2B-immortalized and -transformed cells contained an increased level of nuclear, wild-type, but transcriptionally inactive p53 protein. We further demonstrate that in HTLV-2-immortalized and -transformed cells, p53 activity is inhibited. Interestingly, the Tax-2A protein inhibits p53 tumor suppressor function less efficiently than either Tax-1 or Tax-2B. These studies may provide a molecular correlation for a lower transformation frequency of the HTLV-2 subtype A in vivo.
MATERIALS AND METHODS
Cell lines.
The human myeloid cell line ML-1, the T-lymphocyte cell line Jurkat, and the HTLV-1-transformed cell lines MT2 and C8166 were grown in RPMI medium supplemented with 10% fetal bovine serum. HTLV-2-immortalized or -transformed cell lines C19 (13), MO (24), and Pyg220 (17) were grown in the same medium except with 20% fetal bovine serum and with 10% IL-2 for Pyg220. Peripheral blood lymphocytes (PBLs) were grown in RPMI medium supplemented with 20% fetal bovine serum and 10% IL-2. H1299 lung carcinoma cells and MCF-7 breast cancer cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum.
p53 direct sequencing.
Mutation detection analysis was performed by determining the nucleotide sequence of the three cDNA PCR products from the three HTLV-2 cell lines and from mammary gland polyadenylated RNA used as a wild-type sequence control. The first-strand cDNA synthesis was made by incubating 50 ng of oligo(dT)12–18 with 5 μg of total RNA from the three cell lines (CO, MO, and Pyg19) and with 5 μg of mammary gland polyadenylated RNA (Clontech). After heating at 70°C for 10 min, the reverse transcription was carried out at 42°C using Superscript RT (Gibco-BRL) according to the manufacturer's instruction. Following DNA extension, the RNA template was digested with RNase H for 20 min at 37°C. Ten percent of the cDNA product was used for PCR amplification. The PCR mixture contained 50 mM KCl, 10 mM Tris (pH 8.4), 1.5 mM MgCl2, 10 mM each of the primers, 50 mM each of the four deoxynucleoside triphosphates, and 5 U of Taq polymerase (Boehringer) in a final volume of 100 μl. After an initial denaturation cycle at 94°C for 3 min, amplification was carried out for 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min.
The oligonucleotide primer sequences were 5′-TGCCAGAGGCTGCTCCCCGCG-3′ (forward) and 5′-AACATCTCGAAGCGCTCACG-3′ (reverse). The PCR products (800 bp spanning exons 4 through 9) were purified using a PCR product presequencing kit (Amersham Life Science). The PCR sequencing reaction was performed using two primers, spanning exons 4 and 8, respectively: 5′-TGCCAGAGGCTGCTCCCCGCG-3′ and 5′-ACCATCTCGAAGCGCTCACG-3′. The reaction was performed using the Thermo-sequenase radiolabeled terminator cycle sequencing kit (Amersham). The sequence cycling was carried out as follows: 95°C for 30 s, 55°C for 30 s, and 72°C for 60 s, for a total of 35 cycles. After heating at 70°C for 2 to 10 min, samples were loaded in a 6% denaturing gel (HR-1000, Genomix) and run on the GenomixLR sequencer (Genomix-Beckman) for 2 h at 2,750 V, 125 W, and 50°C. After drying, the gel was exposed to X-ray film (Bio-Max; Kodak) for 12 to 18 h at room temperature, developed, and analyzed.
Irradiation, RNA isolation and RPA.
Exponentially growing cells were γ-irradiated with 6 Gy and incubated for 3 h. Cells were lysed in RNAzol B solution (Tel-Test, Inc.), and total RNA was isolated according to the manufacturer's instructions. Fifteen micrograms of total RNA obtained from the different cells was used for the RNase protection assay (RPA) experiment. The RiboQuant kit was used according to the manufacturer's instructions (Pharmingen) using the human cytokine/chemokine multiprobe template hSTRESS-1. Samples were loaded then on a 5% acrylamide–urea gel and run at a constant 65 W for 1 h. Gels were subsequently dried and placed on a PhosphorImager cassette for an overnight exposure. Signals were quantitated using the ImageQuant program (Molecular Dynamics).
Transfections and luciferase assays.
Transient-transfection experiments with 5 × 106 cells of Jurkat or HTLV-2-infected cells per sample were performed using the Superfect procedure according to the manufacturer's instructions (Qiagen). Adherent cells (H1299) were transfected using the Effectene procedure as recommended by the manufacturer with an enhancer-DNA ratio of 8:1 and an Effectene-DNA ratio of 5:1. The amount of DNA transfected was equalized by addition of a control vector. All the transfections were carried out in the presence of a pRL-TK vector in order to normalize the results for the transfection efficiencies. Reporter activities were assayed 24 h posttransfection using the Dual-Luciferase reporter assay system (Promega). Luciferase assays were performed with a Berthold LB9500C luminometer as described elsewhere (31). PG13pyLuc contains 13 p53 consensus binding sites. pCMV-53, pcTAX, pG104-Tax2A, pCG-Tax2B6wt, and pCG-Tax2B5 (cytomegalovirus [CMV]-driven constructs) as well as HTLV-1-luc were described previously (11, 31, 38).
Western blots.
Cells were washed in phosphate-buffered saline and then lysed in radio immunoprecipitation assay buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml, 1 μg of leupeptin per ml, and 5 mM sodium fluoride at 0°C for 30 min. Lysates were cleared by centrifugation in a microcentrifuge at 14,000 rpm for 15 min. For each cell lysate, a total of 30 μg of protein, as determined by the Bio-Rad protein assay, was separated on an SDS-acrylamide denaturing gel, transferred to an Immobilon membrane (Millipore, Bedford, Mass.), and probed with antibodies against p53 (DO-1; Oncogene) or β-tubulin (Boehringer Mannheim). Detection was performed with an enhanced chemiluminescence system (Amersham Corp., Arlington Heights, Ill.).
P-Ser15 and P-Ser392 antibody characterization.
Whole-cell extracts were made as described previously (32). Using agarose-linked DO-1 antibody, p53 complexes were immunoprecipitated from 500 μg of cell extract, separated on a 4 to 20% Tris–glycine gel, and analyzed by Western blot using anti-Ser15P, anti-Ser392P, and anti-DO-1. The specificity of these antibodies has been described previously (32, 48).
Tax expression in HTLV-2-infected cells.
Total RNA was isolated from HTLV-2-infected (MO, C19, and Pyg220) and HTLV-1-infected (MT2) growing cell cultures. Reverse transcription was carried out using the Superscript preamplification system (Life Technologies). PCR was carried out as previously described using degenerated primers which allow the amplification of all Tax subtypes: primer RM1 (5′ ATCCCGTGGMGAYTCCTSAA 3′) and primer RM2 (5′ AACACGTAGACKGGGTATCC) (16), where Y stands for C or T, M stands for A or C, S stands for G or C, and K stands for G or T. The 146-bp PCR product was then resolved on a 2.5% agarose gel. We also determined Tax-1 and -2B expression by Western blot using either a Tax-2B monoclonal or a Tax-1 monoclonal antibody.
RESULTS
Nuclear localization and protein and sequence analysis of p53 in HTLV-2-infected, immortalized or transformed cell lines.
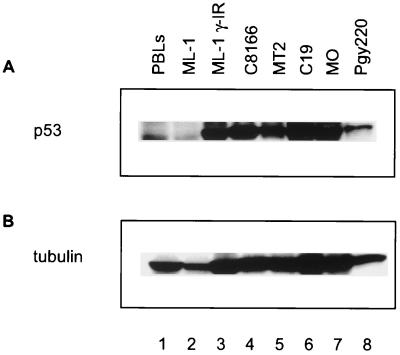
To analyze the level of p53 expression in HTLV-infected cell lines, cellular extracts were prepared from two HTLV-1-transformed lymphocytic lines (MT-2 and C8166), three HTLV-2-infected lines (MO, C19, and Pyg220), and from control ML-1 and PBL cells, which contain wild-type p53. Cell extracts were separated on an SDS-polyacrylamide gel, transferred to an Immobilon membrane, and probed with the p53 monoclonal antibody DO-1. PBLs as well as untreated and γ-irradiated ML-1 cell extracts were used as controls to show low and induced (high) levels of p53 (Fig. 1A, lanes 1 to 3). As previously reported, in comparison to control cells, the steady-state level of p53 protein is elevated in HTLV-1-infected cells (Fig. 1, compare lanes 4 and 5 with lane 1) (31, 36). Similarly, we observed that the level of p53 protein in all three HTLV-2 cell lines was elevated (Fig. 1A, lanes 6 to 8). The same Western blot membrane was then reprobed for β-tubulin (Fig. 1B). The results of this experiment demonstrate that similar amounts of extract were analyzed for each sample.
FIG. 1.
Western blot analysis of (A) p53 and (B) β-tubulin in HTLV-infected and control (ML-1) cells. Lanes 1, PBLs; lane 2, ML-1, no γ-irradiation; lane 3, ML-1, γ-irradiation; lane 4, C8166; lane 5, MT2; lane 6, C19; lane 7, MO; lane 8, Pyg220.
The cellular localization of p53 in the HTLV-2-infected cell lines was determined by immunofluorescent staining and confocal microscopy. HTLV-2 MO cells were fixed and stained with DAPI (4′,6′-diamidino-2-phenylindole) (nuclear staining) and with the p53 DO-1 antibody. Confocal imaging of the anti-p53-stained MO cells clearly demonstrated that the p53 was localized in the nucleus of the cell (data not shown).
To determine if the increase in the level of p53 was due to p53 mutation, we next looked at the p53 sequence in the three HTLV-2 cell lines. Total RNA was extracted from the exponentially growing HTLV-2 cell lines, and cDNA from exons 4 through 9 was directly sequenced as described in Materials and Methods. This method offers the advantage of detecting heterozygous sequences in which one of the alleles may be mutated. No mutations were found in the six exons sequenced in any of the three HTLV-2-infected cell lines (data not shown). This result suggests that the increased level of p53 was not due to mutation in the protein coding sequences.
p53 is inactive in HTLV-2-immortalized and -transformed cell lines.
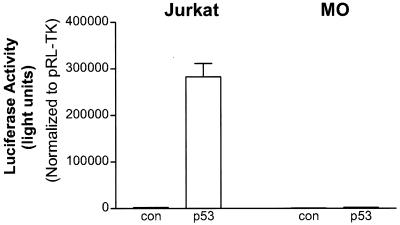
We next looked at the p53 transcriptional activity in these cell lines as well as in Jurkat control cells. When the p53 reporter construct was transfected into the p53-negative Jurkat cell line, we did not detect any activity (Fig. 2). When the p53 expression plasmid was cotransfected, we observed a 185-fold increase in activity. In the MO HTLV-2 line, the levels of endogenous p53 activity were very low (Fig. 2), similar to the p53-negative Jurkat cell line. Moreover, when the cells were cotransfected with a p53 expression vector, p53 transactivation was not observed in the MO HTLV-2 cell line. The results of these studies were normalized for transfection efficiency by inclusion of the pRL-TK plasmid in the transfection mix. The activity of the pRL-TK expression plasmid further demonstrated that the lack of p53 activity was not due to cell death. These results suggest that (i) the wild-type p53 present in the HTLV-2 cell lines is transcriptionally inactive and (ii) similar to HTLV-1-transformed cells, the cells are capable of rapidly inhibiting exogenous p53 produced from the expression plasmid (31). Although transfection efficiency was low, similar results were observed in the C19 HTLV-2 cell line (data not shown).
FIG. 2.
Transcriptional stimulation of p53 activity and Tax activity in different cell lines. The p53 reporter plasmid PG13pyLuc (4 μg) was transfected into HTLV-2 (MO) as well as Jurkat control cell lines in the presence (2 μg) or absence (control) of pCMV-p53. In all experiments, the amount of total DNA was adjusted to 6 μg by adding carrier DNA. pRL-TK (0.15 μg) was cotransfected in all experiments. Transfection results are the mean ± standard deviation (SD) of three independent experiments, normalized to control (con) activity.
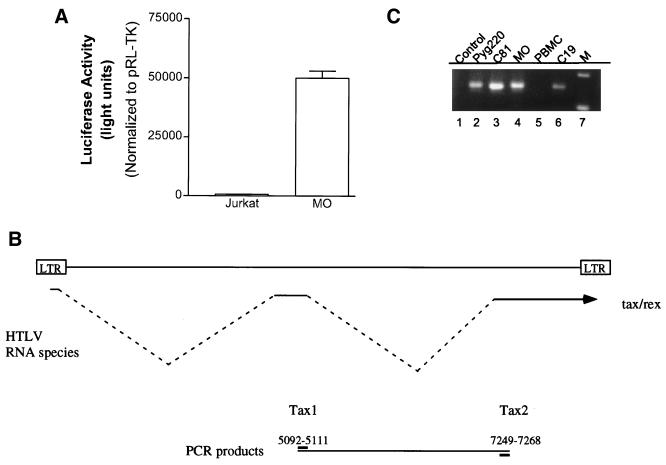
To investigate Tax transcription activity in the HTLV-2-infected cells, we transfected the MO HTLV-2 cell line with the HTLV-1-luc reporter plasmid. Upon transfection of HTLV-1-luc, we detected a significant activity in MO and C19 cells, but not in the control Jurkat cell line (Fig. 3A, and data not shown). These results demonstrate the presence of a transcriptionally active Tax-2 protein in the cells.
FIG. 3.
(A) HTLV-1-luc plasmid (4 μg) was transfected into HTLV-2 MO or Jurkat cells. The amount of total DNA was adjusted to 6 μg by adding carrier DNA. pRL-TK (0.15 μg) was cotransfected in all experiments. Transfection results are the mean ± SD of three independent experiments, normalized to control activity. (B) Oligonucleotide primers and strategy for PCR following reverse transcription of RNA from HTLV-1 and HTLV-2 cell lines. (C) RT-PCR analysis. Lane 1, no RT added, lane 2, Pyg220; lane 3, C8166; lane 4, MO; lane 5, peripheral blood mononuclear cells (PBMC); lane 6, C19; lane 7, size markers.
To analyze Tax expression in the HTLV-2 cell lines, we performed a series of reverse transcription (RT)-PCR experiments. The design of the primers allows amplification of the cDNA of all Tax subtypes (Fig. 3B). Under these conditions, we detected the expression of the Tax RNA for all HTLV-1 or HTLV-2 cell lines tested (Fig. 3C, lanes 2 to 4 and 6), but not in the control PBLs (Fig. 3C, lane 5). Importantly, no PCR product was observed in the absence of reverse transcription (Fig. 3C, lane 1). The results of this experiment demonstrate that Tax RNA is expressed in each of the HTLV-2-transformed cell lines.
We next attempted to analyze the level of Tax protein expression using a panel of Tax antibodies including four Tax monoclonals (Tab 169, 170, 171, and 172) from our laboratory, three antibodies (IF7, 4C5, and 9F11) from C. Z. Giam against different relatively conserved domains in Tax-1 and Tax-2, and Tax antibodies from J. Semmes and W. Hall. Although each of the antibodies worked well against the respective Tax protein, none of the antibodies was able to detect Tax-1, Tax-2A, and Tax-2B at the same time (data not shown). In light of these experiments, we cloned each of the Tax coding sequences into a common expression vector (pCMV-Tag) containing a Flag tag. In transient-transfection assays, we were able to demonstrate that similar amounts of Tax fusion protein were expressed (data not shown). Unfortunately, the presence of the Flag tag at either the amino- or carboxy-terminal end of the Tax protein abolished the activity of the Tax protein on the HTLV-1 LTR in transient assays (data not shown). Similar results have been obtained in the laboratory of Patrick Green (personal communication). At this point, we are not aware of any antibody that will allow one to detect Tax-1, -2A, and -2B.
p53 function in HTLV-2 cell lines is not responsive to γ-irradiation.
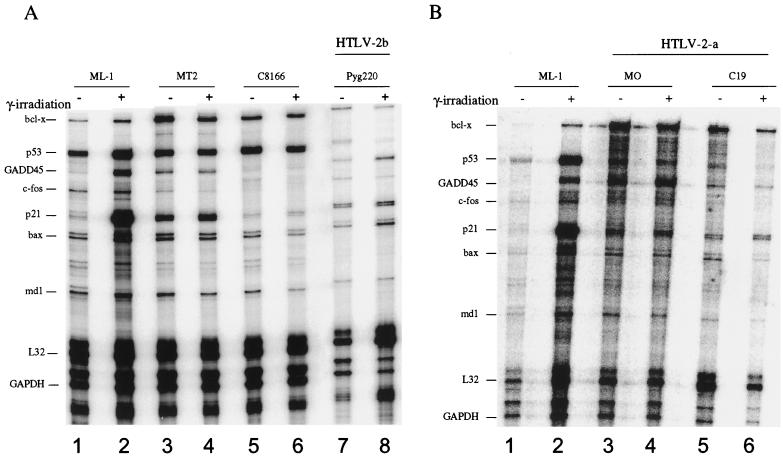
To further test the transcriptional activity of p53 in the HTLV-2 cells, we conducted a series of RPAs (Fig. 4A and B). Exponentially growing cells were γ-irradiated and harvested 3 h later. Total RNA was isolated from HTLV-1-infected cells (MT-2 and C8166), ML-1 control cells, which contain a wild-type p53, and three HTLV-2 lines (MO, C19, and Pyg220) before and after γ-irradiation. The RNA was then subjected to hybridization with the human hSTRESS-1 probe set to look at the induction of the p53-responsive genes bcl-x, GADD45, p21, bax, and bcl-2, as well as p53 expression. This probe set also contains two housekeeping genes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and L32, which are used as internal controls for each sample. Quantitative analysis of the RPA profile is presented in Table 1. Upon γ-irradiation, GADD45 and p21 expression was induced 4.5- and 19-fold, respectively, in p53-positive control ML-1 cells. In contrast, there was no induction of the p53-responsive genes in the three HTLV-2 cell lines (Table 1 and Fig. 4A, lanes 7 and 8; 4B, lanes 3 to 6). The lack of induction was not due to a problem in preparation or analysis of the RNA sample, since the control GAPDH and L32 levels were fairly constant between the samples. Consistent with previous results, we did not detect a significant change in the levels of GADD45 and p21 expression in HTLV-1 C8166 and MT2 cells (Table 1; Fig. 4A, lanes 3 to 6) (31).
FIG. 4.
Effect of γ-irradiation on the expression of various p53-inducible genes. Fifteen micrograms of total RNA obtained from either (A) p53-inactive HTLV-2 (Pyg220) or HTLV-1 (MT2 and C8166) and (B) p53-inactive HTLV-2 (MO and C19) or (A and B) p53-active control (ML-1) cells were used before (–) and after (+) γ-irradiation. The RiboQuant kit was used according to the manufacturer's instructions (Pharmingen) using the human cell cycle regulator multiprobe template hSTRESS-1. Samples were then loaded on a 5% acrylamide–urea gel and run at a constant 65 W for 1 h. Gels were subsequently dried and placed on a PhosphorImager cassette for overnight exposure.
TABLE 1.
Effect of γ-irradiation on the expression of various DNA damage-inducible genes
| Infecting virus | Cell line | Fold increase in relative mRNAa
|
|||
|---|---|---|---|---|---|
| p21 | GADD45 | Bax | GAPDH | ||
| HTLV-1 | ML-1 | 19 | 4.5 | 2 | 1 |
| MT-2 | 1.5 | 1 | 1 | 1 | |
| C-8166 | 1.5 | 1 | 1 | 1 | |
| HTLV-2a | MO | 0.5 | 0.5 | 0.5 | 1 |
| C19 | 1 | 1 | 1 | 1 | |
| HTLV-2b | Pyg220 | 1 | 1 | 1 | 1 |
Fold increase was determined by comparing the intensity of the signal before and after γ-irradiation. Each signal was normalized to GAPDH and L32 signals.
Hyperphosphorylation of p53 at Ser15 and Ser392 in HTLV-2 cell lines.
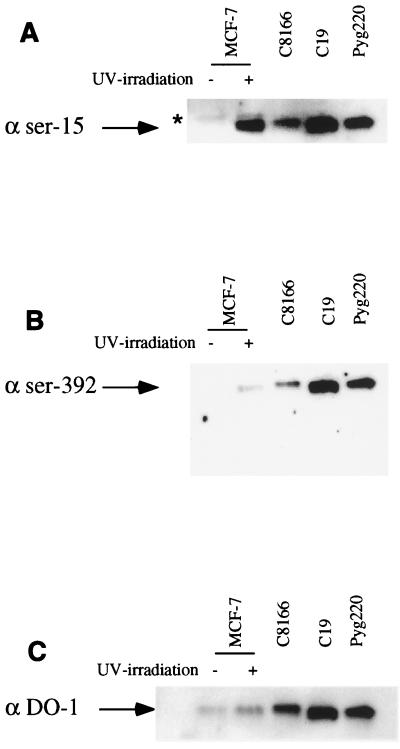
Hyperphosphorylation of serines 15 and 392 has been previously shown to correlate with the transcriptional inactivation of p53 in HTLV-1-infected cells (32). Since the p53 sequence is wild type in the HTLV-2-immortalized or -transformed cell lines tested (MO, C19, and Pyg220), but there is no response to ionizing radiation, we checked whether this phenomenon also correlated with the hyperphosphorylation of these specific residues. For that purpose, p53 was immunoprecipitated from HTLV-1 (C8166), HTLV-2 (C19 and Pyg220), and control (MCF-7) cell lines and separated by gel electrophoresis. We then used affinity-purified antibodies specific for P-ser15, P-ser392, or WTp53 (32) (Fig. 5A, B, and C, respectively). The specificity of the antibodies for phospho-p53 protein has been demonstrated previously (32, 40). Using the DO-1 antibody as a control, we detected the presence of p53 in the UV-treated wild-type p53-containing MCF-7 cells, as well as in C8166 and two HTLV-2 cell lines (Fig. 5C). We also detected a hyperphosphorylated residue 15 in all HTLV-infected cells, but not in the noninduced MCF-7 control cells (Fig. 5A). Finally, and consistent with what has been reported for HTLV-1 cells, we detected hyperphosphorylated serine 392 in both HTLV-2 cell lines tested (Fig. 5B). Similar results were also obtained for the MO cell line (data not shown). These results are therefore similar to those reported for HTLV-1-transformed cell lines in which hyperphosphorylation is correlated with an inactive p53.
FIG. 5.
p53 phosphorylation is increased on Ser15 and Ser392 in HTLV-1- and HTLV-2-immortalized or -transformed cells. Antibodies specific for P-ser15 (A) and P-ser392 (B) and antibody DO-1, which reacts with phosphorylated and nonphosphorylated p53 (C), were used in Western blot analyses of lysates from C19, Pyg220, C8166, and MCF-7 cells (without [−] and with [+] UV treatment) after immunoprecipitation with the DO-1 antibody as described in Materials and Methods. ★, immunoglobulin G heavy chain.
Comparison of the ability of HTLV-1 and HTLV-2 Tax to inactivate p53.
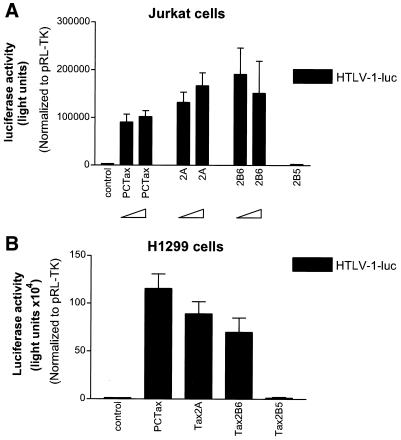
We next wanted to determine whether the Tax protein was directly responsible for the p53 inactivation detected in the three HTLV-2 cell lines. It was important in these comparative studies to demonstrate the relative activity of HTLV-1 and HTLV-2 Tax in the transfection assay. To this end, we took advantage of the observation that Tax-1 and Tax-2A proteins transactivate the HTLV-1 promoter with roughly similar efficiency (38, 41, 46). Wild-type Tax, Tax-2A, and Tax-2B CMV-driven expression plasmids were cotransfected with the HTLV-1-luc reporter, which contains the HTLV-1 LTR upstream of the luciferase gene. So that the activity of the proteins could be analyzed in different cell types, both Jurkat T lymphocytes and H1299 lung carcinoma cells were used in the studies. All results were normalized for transfection efficiency by cotransfection of the pRL-TK plasmid. The results of these assays demonstrate that all three proteins efficiently transactivated the HTLV-1 LTR in both the Jurkat (Fig. 6A) and H1299 (Fig. 6B) cells. It is worth noting that in both cell types, Tax-2A was as active as Tax-1 and -2B for transactivating the HTLV-1 promoter. We also used as a control the Tax-2B5 mutant, which encodes a deleted (aa 1 to 301), inactive Tax protein. Indeed, upon transfection of this mutant, we did not detect any transactivation of the HTLV-1-luc promoter (Fig. 6A and B). These results are of particular importance to the conclusions reached in this study. Since there is no available antibody that will detect Tax-1, Tax-2A, and Tax-2B, we must rely on the biological activity of the proteins as an indication of expression level. Along these lines, it is important to point out that three earlier studies (38, 41, 46) all report that Tax-1 and Tax-2 proteins have similar transactivation activity on the HTLV-1 LTR, which is exactly the result we report.
FIG. 6.
HTLV-1 LTR activation by the different HTLV-1, HTLV-2A, and HTLV-2B Tax cDNA constructs. Tax-2B5 is a Tax-2B deletion mutant that retains no activity, Tax-2B6 represents the Tax-2B wild-type sequence. (A and B) Titration of the different constructs on the HTLV-1-luc reporter plasmid. Jurkat (A) or H1299 (B) cells were transfected with increasing amounts of Tax (2 and 4 μg) together with HTLV-1-luc (2 μg) and pRL-TK (0.15 μg) as described in Materials and Methods with Superfect and Effectene reagents, respectively. DNA concentrations were adjusted with vector control to that equivalent amounts of DNA were used in all experiments. Results were normalized to Renilla activity. The results presented in panels A and B are the mean ± SD of three experiments.
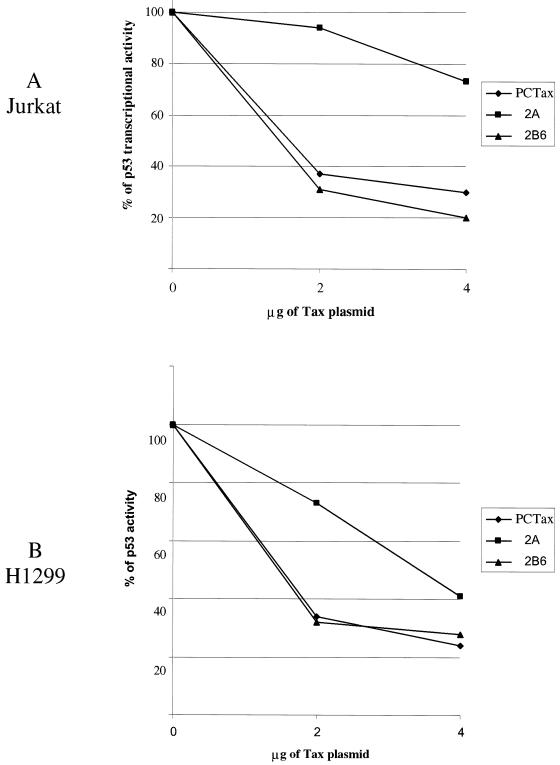
We next wanted to determine the relative activities of the different Tax proteins on p53 inactivation. Plasmids encoding Tax-1, Tax-2A, and Tax-2B were titrated with the p53- and the PG13pyLuc p53-responsive plasmids in Jurkat and H1299 cells in the presence of an pRL-TK plasmid to normalize for transfection efficiency. The results of these experiments demonstrate that the Tax-1 and Tax-2B proteins were able to repress the p53 activity in both cell types (Fig. 7A and B). At the highest concentration of Tax-1 plasmid transfected, p53 activity was reduced by approximately 80%. Similarly, Tax-2B was also able to inhibit p53 activity in both Jurkat and H1299 cells (Fig. 7B). Strikingly, we reproducibly found that the ability of Tax-2A to inhibit p53 was cell type dependent. In H1299 cells, Tax-2A inhibited p53 activity by 60%, whereas Tax-1 and Tax-2B inhibited p53 by 70 to 75% at 4 μg of Tax plasmid. Remarkably, Tax-2A protein was significantly less efficient in its ability to suppress p53 activity in Jurkat T cells, even at high concentrations of plasmid (4 μg) (Fig. 7A).
FIG. 7.
Dose-dependent repression of p53 transactivation by Tax. Jurkat (A) or H1299 (B) cells were transfected with increasing amounts of Tax-1, -2A, or -2B6 (2 and 4 μg) together with PG13pyLuc (2 μg [A] or 0.5 μg [B]), pCMV-p53 (2 μg [A] or 0.3 μg [B]), and pRL-TK (0.15 μg) as described in Materials and Methods with Superfect and Effectene reagents, respectively. DNA concentrations were adjusted with vector control so that an equivalent amount of DNA was used in all experiments. Transfection results were normalized to Renilla activity. The results presented in panels A and B are representative of three independent experiments.
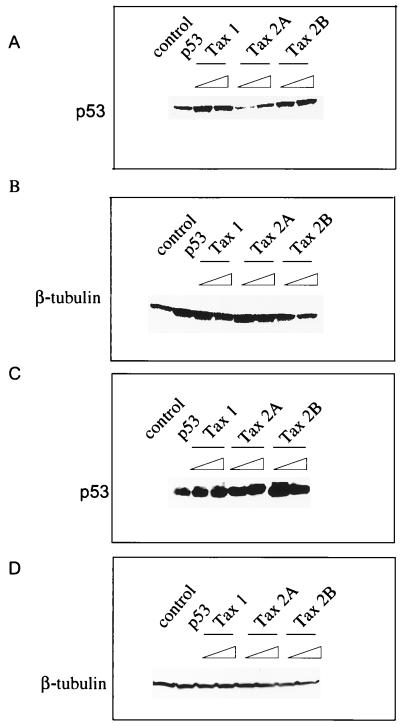
We also performed Western blots of the transfected cell extracts to examine the ability of Tax to increase the cellular level of p53. Tax-1 and Tax-2B were found to increase the steady-state level of p53 protein in Jurkat and H1299 cells (Fig. 8A and 8C), respectively. In agreement with the results obtained in the p53 transactivation assays, we found that Tax-2A was significantly weaker in its ability to increase the steady-state level of p53 in Jurkat T lymphocytes (Fig. 8A). Again, consistent with the ability of Tax-2A to inhibit p53 activity slightly better in the H1299 cell, the Tax-2A protein was more efficient at increasing the level of p53 in these cells (Fig. 8C). A control Western blot with an antibody specific for β-tubulin demonstrated that equivalent amounts of protein were analyzed in each of the samples (Fig. 8B and D).
FIG. 8.
(A and C) p53 Western blot and (B and D) β-tubulin Western blot. Protein extracts (50 μg) from the lysates used for the luciferase assay after Jurkat (A) or H1299 (B) cell transfection were ran on a 4 to 20% SDS gel and probed for p53 with the DO-1 antibody or for β-tubulin. Lanes: 1, no p53 transfected, 2, p53 transfected; 3 to 8, p53 expression in the presence of increasing (2 and 4 μg) doses of Tax-1, -2A, and -2B expression plasmids.
DISCUSSION
HTLV-1 and HTLV-2 are highly related at both the nucleotide and protein sequence level (3). For example, the Tax protein sequence is highly conserved between the two viruses, with more than 77% identity at the amino acid level (41). Strikingly, however, HTLV-2 is not clearly associated with any malignancy. In fact, there have been only sporadic associations of HTLV-2 with diseases such as a rare hairy T-cell variant hairy cell leukemia (23) and some neurological diseases (27, 28). It is probable that differences in pathogenicity between HTLV-1 and HTLV-2 are accounted for, in part, by the differential activity of the tax gene on cellular promoters. Along these lines, there have been a few studies reporting phenotypic differences between the HTLV-1 and HTLV-2 Tax proteins. Semmes et al. (41) reported that the Tax-2A protein lacks the ability to induce micronuclei in infected cells. Particularly relevant to the Tax-2A cell type specificity reported here, Tanaka et al. (46) also reported that Tax-2A was able to activate the ICAM-1 promoter in HeLa cells but not in T-cell lines, whereas Tax-1 could activate the ICAM-1 promoter in all cell lines tested. Finally, recent data suggest that HTLV-1 Tax and HTLV-2A Tax might activate transcription from their homologous LTRs by utilizing different transcription factors (H. Fan, personal communication).
It is important to address the apparent paradox between inactivation of p53 in HTLV-2-transformed cells versus the relatively weak inactivation of p53 by Tax-2A in transfected Jurkat lymphocytes. If Tax-2A protein is not a strong repressor of p53, how can one obtain HTLV-2A-transformed T-cell lines containing an inactivated p53? One hypothesis could be that the cells adapt to the presence of the HTLV-2 virus and undergo metabolic changes. Changes in the cell metabolism may allow the Tax-dependent regulation of transcription factors and/or kinases required to inactivate p53. Alternatively, it is possible that other viral proteins contribute to the inactivation of p53. While we have shown that Tax is able to inactivate p53, we cannot rule out the possibility that the expression of other viral proteins contributes to the efficiency of p53 inactivation. For example, adenovirus encodes several proteins, including E1B and E4orf6, which inactivate p53 function (10, 35).
HTLV-1 causes clonal expansion of the infected cells in the carrier of the virus. This result can be shown by PCR amplification of the HTLV integration sites and explains the remarkable genetic stability and the high proviral loads (50). One explanation for that phenomenon could be inactivation of p53 functions. Indeed, if p53 functions are inactivated, the infected cell might become more resistant to apoptosis. If this hypothesis is valid, and if HTLV-2A is a weaker p53 inactivator, one might then expect a lower viral load in HTLV-2A-infected individuals than in HTLV-2B or HTLV-1 carriers. Although there are very few studies aimed at looking at viral load as well as at the clonal expansion level in HTLV-2-infected cells in the context of the molecular subtype (8, 9), the available data suggest that HTLV-2A-infected people have a lower viral load than HTLV-2B-infected people (8, 9). The levels of Tax-2 expression in vivo in these patients were not determined.
As mentioned above, the discovery of HTLV-2 subtypes took place only 7 years ago, not allowing prospective studies on the possible higher incidence of hematological diseases in HTLV-2B- versus -2A-infected individuals (19, 21, 30). ATL occurs in 3 to 5 out of 100 HTLV-1-infected individuals, and therefore, the association between ATL and HTLV-1 infection was uncovered only because of the large number of infected individuals living in Japan (45). There is not really such a population infected with HTLV-2B, perhaps giving the false impression that 2B viruses are nonpathogenic. Future analysis of HTLV-2-infected populations will show a better assessment of virus-associated diseases.
In conclusion, we have presented evidence for an important and novel phenotypic difference between HTLV-1 and HTLV-2 Tax proteins. We show that the HTLV-2A Tax protein is a weaker inactivator of p53 than HTLV-1 or HTLV-2B Tax in T cells. It will be of interest to follow dynamic epidemiological studies on HTLV-2A versus -2B-infected populations to see if the latter are at higher risk of developing hematological diseases. These studies will provide valuable information on the pathogenicity of the HTLV viruses. It will also be of interest to analyze the molecular mechanism by which HTLV-2B inactivates p53 function. Two recent reports by Van Orden et al. (49) and Suzuki et al. (42) have suggested that Tax-1 inhibits p53 function by binding and sequestering the coactivator CBP from p53. In contrast, results from Pise-Masison et al. (33) suggest that Tax inhibits p53 function through induction of the NF-κB transcription pathway, not through squelching of the coactivator CBP. It will be of interest to determine the mechanism of p53 inhibition by Tax-2B.
ACKNOWLEDGMENTS
We thank Tom Misteli and Christopher Baumann for assistance in confocal microscopy of p53 and the generous gift of reagents. We also thank Y. Yao, C.-Z. Giam, and John Semmes for providing antibody reagents.
REFERENCES
- 1.Akagi T, Ono H, Tsuchida N, Shimotohno K. Aberrant expression and function of p53 in T-cells immortalized by HTLV-1 Tax-1. FEBS Lett. 1997;406:263–266. doi: 10.1016/s0014-5793(97)00280-9. [DOI] [PubMed] [Google Scholar]
- 2.Cann A J, Rosenblatt J D, Wachsman W, Chen I S. In vitro mutagenesis of the human T-cell leukemia virus types I and II tax genes. J Virol. 1989;63:1474–1479. doi: 10.1128/jvi.63.3.1474-1479.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cann A J, Rosenblatt J D, Wachsman W, Shah N P, Chen I S. Identification of the gene responsible for human T-cell leukaemia virus transcriptional regulation. Nature. 1985;318:571–574. doi: 10.1038/318571a0. [DOI] [PubMed] [Google Scholar]
- 4.Cavrois M, Gessain A, Wain-Hobson S, Wattel E. Proliferation of HTLV-1 infected circulating cells in vivo in all asymptomatic carriers and patients with TSP/HAM. Oncogene. 1996;12:2419–2423. [PubMed] [Google Scholar]
- 5.Cavrois M, Leclercq I, Gout O, Gessain A, Wain-Hobson S, Wattel E. Persistent oligoclonal expansion of human T-cell leukemia virus type 1-infected circulating cells in patients with tropical spastic paraparesis/HTLV-1 associated myelopathy. Oncogene. 1998;17:77–82. doi: 10.1038/sj.onc.1201906. [DOI] [PubMed] [Google Scholar]
- 6.Cereseto A, Diella F, Mulloy J C, Cara A, Michieli P, Grassmann R, Franchini G, Klotman M E. p53 functional impairment and high p21waf1/cip1 expression in human T-cell lymphotropic/leukemia virus type I-transformed T cells. Blood. 1996;88:1551–1560. [PubMed] [Google Scholar]
- 7.Chen I S, Slamon D J, Rosenblatt J D, Shah N P, Quan S G, Wachsman W. The x gene is essential for HTLV replication. Science. 1985;229:54–58. doi: 10.1126/science.2990037. [DOI] [PubMed] [Google Scholar]
- 8.Cimarelli A, Duclos C A, Gessain A, Casoli C, Bertazzoni U. Clonal expansion of human T-cell leukemia virus type II in patients with high proviral load. Virology. 1996;223:362–364. doi: 10.1006/viro.1996.0487. [DOI] [PubMed] [Google Scholar]
- 9.Cimarelli A, Duclos C A, Gessain A, Cattaneo E, Casoli C, Biglione M, Mauclere P, Bertazzoni U. Quantification of HTLV-II proviral copies by competitive polymerase chain reaction in peripheral blood mononuclear cells of Italian injecting drug users, central Africans, and Ameridians. J Acquir Immune Defic Syndr Hum Retroviril. 1995;10:198–204. doi: 10.1097/00042560-199510020-00014. [DOI] [PubMed] [Google Scholar]
- 10.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 11.Eiraku N, Novoa P, da Costa F, Monken C, Ishak R, Zhu S W, Lorenco R, Ishak M, Azvedo V, Guerreiro J, de Oliveira M P, Loureiro P, Hammerschlak N, Ijichi S, Hall W M. Identification and characterization of a new and distinct molecular subtype of human T-cell lymphotropic virus type 2. J Virol. 1996;70:1481–1492. doi: 10.1128/jvi.70.3.1481-1492.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouchard N, Flageul B, Bagot M, Avril M F, Hermine O, Sigaux F, Merle-Beral H, Troussard X, Delfraissy J F, de The G. Lack of evidence of HTLV-I/II infection in T CD8 malignant or reactive lymphoproliferative disorders in France: a serological and/or molecular study of 169 cases. Leukemia. 1995;9:2087–2092. [PubMed] [Google Scholar]
- 13.Gallo D, Penning L M, Hanson C V. Detection and differentiation of antibodies to human T-cell lymphotropic virus types I and II by the immunofluorescence method. J Clin Microbiol. 1991;29:2345–2347. doi: 10.1128/jcm.29.10.2345-2347.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gartenhaus R B, Wang P. Functional inactivation of wild-type p53 protein correlates with loss of IL-2 dependence in HTLV-I transformed human T lymphocytes. Leukemia. 1995;9:2082–2086. [PubMed] [Google Scholar]
- 15.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calender A, de The G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 16.Gessain A, Louie A, Gout O, Gallo R C, Franchini G. Human T-cell leukemia-lymphoma virus type I (HTLV-I) expression in fresh peripheral blood mononuclear cells from patients with tropical spastic paraparesis/HTLV-I-associated myelopathy. J Virol. 1991;65:1628–1633. doi: 10.1128/jvi.65.3.1628-1633.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gessain A, Mauclere P, Froment A, Biglione M, Le Hesran J Y, Tekaia F, Millan J, de The G. Isolation and molecular characterization of a human T-cell lymphotropic virus type II (HTLV-II), subtype B, from a healthy Pygmy living in a remote area of Cameroon: an ancient origin for HTLV-II in Africa. Proc Natl Acad Sci USA. 1995;92:4041–4045. doi: 10.1073/pnas.92.9.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall W W, Ishak R, Zhu S W, Novoa P, Eiraku N, Takahashi H, da C. Ferreira M, Azevedo V, Ishak M O, da C. Ferreira O, Monken C, Kurata T. Human T lymphotropic virus type II (HTLV-II): epidemiology, molecular properties, and clinical features of infection. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13(Suppl. 1):S204–S214. doi: 10.1097/00042560-199600001-00031. [DOI] [PubMed] [Google Scholar]
- 19.Hall W W, Takahashi H, Liu C, Kaplan M H, Scheewind O, Ijichi S, Nagashima K, Gallo R C. Multiple isolates and characteristics of human T-cell leukemia virus type II. J Virol. 1992;66:2456–2463. doi: 10.1128/jvi.66.4.2456-2463.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heneine W, Switzer W M, Busch M, Khabbaz R F, Kaplan J E. Molecular subtyping of human T-cell lymphotropic virus type 2 by single-strand conformation polymorphism analysis. Retrovirus Epidemiology Donor Study Group. J Clin Microbiol. 1995;33:3260–3263. doi: 10.1128/jcm.33.12.3260-3263.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hjelle B, Mills R, Swenson S, Mertz G, Key C, Allen S. Incidence of hairy cell leukemia, mycosis fungoides, and chronic lymphocytic leukemia in first known HTLV-II-endemic population. J Infect Dis. 1991;163:435–440. doi: 10.1093/infdis/163.3.435. [DOI] [PubMed] [Google Scholar]
- 22.Ijichi S, Ramundo M B, Takahashi H, Hall W W. In vivo cellular tropism of human T cell leukemia virus type II (HTLV-II) J Exp Med. 1992;176:293–296. doi: 10.1084/jem.176.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalyanaraman V S, Sarngadharan M G, Robert-Guroff M, Miyoshi I, Golde D, Gallo R C. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science. 1982;218:571–573. doi: 10.1126/science.6981847. [DOI] [PubMed] [Google Scholar]
- 24.Kalyanaraman V S, Sarngadharan M G, Robert-Guroff M, Miyoshi I, Golde D, Gallo R C. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science. 1982;218:571–573. doi: 10.1126/science.6981847. [DOI] [PubMed] [Google Scholar]
- 25.Lairmore M D, Jacobson S, Gracia F, De B K, Castillo L, Larreategui M, Roberts B D, Levine P H, Blattner W A, Kaplan J E. Isolation of human T-cell lymphotropic virus type 2 from Guaymi Indians in Panama. Proc Natl Acad Sci USA. 1990;87:8840–8844. doi: 10.1073/pnas.87.22.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mulloy J C, Kislyakova T, Cereseto A, Casareto L, LoMonico A, Fullen J, Lorenzi M V, Cara A, Nicot C, Giam C, Franchini G. Human T-cell lymphotropic/leukemia virus type 1 Tax abrogates p53-induced cell cycle arrest and apoptosis through its CREB/ATF functional domain. J Virol. 1998;72:8852–8860. doi: 10.1128/jvi.72.11.8852-8860.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy E L. The clinical epidemiology of human T-lymphotropic virus type II (HTLV-II) J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13(Suppl. 1):S215–S219. doi: 10.1097/00042560-199600001-00032. [DOI] [PubMed] [Google Scholar]
- 28.Murphy E L, Engstrom J W, Miller K, Sacher R A, Busch M P, Hollingsworth C G. HTLV-II associated myelopathy in 43-year-old woman. REDS Investigators. Lancet. 1993;341:757–758. doi: 10.1016/0140-6736(93)90529-p. [DOI] [PubMed] [Google Scholar]
- 29.Murphy E L, Mahieux R, de The G, Tekaia F, Ameti D, Horton J, Gessain A. Molecular epidemiology of HTLV-II among United States blood donors and intravenous drug users: an age-cohort effect for HTLV-II RFLP type aO. Virology. 1998;242:425–434. doi: 10.1006/viro.1997.9009. [DOI] [PubMed] [Google Scholar]
- 30.Pardi D, Kaplan J E, Coligan J E, Folks T M, Lal R B. Identification and characterization of an extended Tax protein in human T-cell lymphotropic virus type II subtype b isolates. J Virol. 1993;67:7663–7667. doi: 10.1128/jvi.67.12.7663-7667.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pise-Masison C A, Choi K S, Radonovich M, Dittmer J, Kim S J, Brady J N. Inhibition of p53 transactivation function by the human T-cell lymphotropic virus type 1 Tax protein. J Virol. 1998;72:1165–1170. doi: 10.1128/jvi.72.2.1165-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pise-Masison C A, Radonovich M, Sakaguchi K, Appella E, Brady J N. Phosphorylation of p53: a novel pathway for p53 inactivation in human T-cell lymphotropic virus type 1-transformed cells. J Virol. 1998;72:6348–6355. doi: 10.1128/jvi.72.8.6348-6355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pise-Masison C A, Mahieux R, Jiang H, Ashcroft M, Radonovich M, Duvall J, Guillerm C, Brady J N. Inactivation of p53 by human T-cell lymphotropic virus type 1 Tax requires activation of the NF-κB pathway and is dependent upon phosphorylation. Mol Cell Biol. 2000;20:3377–3386. doi: 10.1128/mcb.20.10.3377-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prives C, Manfredi J J. The p53 tumor suppressor protein: meeting review. Genes Dev. 1993;7:529–534. doi: 10.1101/gad.7.4.529. [DOI] [PubMed] [Google Scholar]
- 36.Reid R L, Lindholm P F, Mireskandari A, Dittmer J, Brady J N. Stabilization of wild-type p53 in human T-lymphocytes transformed by HTLV-I. Oncogene. 1993;8:3029–3036. [PubMed] [Google Scholar]
- 37.Rosenblatt J D, Cann A J, Slamon D J, Smalberg I S, Shah N P, Fujii J, Wachsman W, Chen I S. HTLV-II transactivation is regulated by the overlapping tax/rex nonstructural genes. Science. 1988;240:916–919. doi: 10.1126/science.2834826. [DOI] [PubMed] [Google Scholar]
- 38.Ross T M, Minella A C, Fang Z Y, Pettiford S M, Green P L. Mutational analysis of human T-cell leukemia virus type 2 Tax. J Virol. 1997;71:8912–8917. doi: 10.1128/jvi.71.11.8912-8917.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross T M, Pettiford S M, Green P L. The tax gene of human T-cell leukemia virus type 2 is essential for transformation of human T lymphocytes. J Virol. 1996;70:5194–5202. doi: 10.1128/jvi.70.8.5194-5202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakaguchi K, Herrera J E, Saito S, Bustin M, Vassilev A, Anderson C W, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semmes O J, Majone F, Cantemir C, Turchetto L, Hjelle B, Jeang K T. HTLV-I and HTLV-II Tax: differences in induction of micronuclei in cells and transcriptional activation of viral LTRs. Virology. 1996;217:373–379. doi: 10.1006/viro.1996.0126. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki T, Uchida-Toita M, Yoshida M. Tax protein of HTLV-I inhibits CBP/p300-mediated transcription by interfering with recruitment of CBP/p300 onto DNA elements of E-box or p53 binding site. Oncogene. 1999;18:4137–4143. doi: 10.1038/sj.onc.1202766. [DOI] [PubMed] [Google Scholar]
- 43.Switzer W M, Owen S M, Pieniazek D A, Nerurkar V R, Duenas-Barajas E, Heneine W, Lal R B. Molecular analysis of human T-cell lymphotropic virus type II from Wayuu Indians of Colombia demonstrates two subtypes of HTLV-IIb. Virus Genes. 1995;10:153–162. doi: 10.1007/BF01702596. [DOI] [PubMed] [Google Scholar]
- 44.Switzer W M, Pieniazek D, Swanson P, Samdal H H, Soriano V, Khabbaz R F, Kaplan J E, Lal R B, Heneine W. Phylogenetic relationship and geographic distribution of multiple human T-cell lymphotropic virus type II subtypes. J Virol. 1995;69:621–632. doi: 10.1128/jvi.69.2.621-632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tajima K, Kuroishi T. Estimation of rate of incidence of ATL among ATLV (HTLV-I) carriers in Kyushu, Japan. Jpn J Clin Oncol. 1985;15:423–430. [PubMed] [Google Scholar]
- 46.Tanaka Y, Hayashi M, Takagi S, Yoshie O. Differential transactivation of the intercellular adhesion molecule 1 gene promoter by Tax-1 and Tax-2 of human T-cell leukemia viruses. J Virol. 1996;70:8508–8517. doi: 10.1128/jvi.70.12.8508-8517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarsis S L, Yu M T, Parks E S, Persaud D, Munoz J L, Parks W P. Human T-lymphocyte transformation with human T-cell lymphotropic virus type 2. J Virol. 1998;72:841–846. doi: 10.1128/jvi.72.1.841-846.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandamme A M, Salemi M, Van Brussel M, Liu H F, Van Laethem K, Van Ranst M, Michels L, Desmyter J, Goubau P. African origin of human T-lymphotropic virus type 2 (HTLV-2) supported by a potential new HTLV-2d subtype in Congolese Bambuti Efe Pygmies. J Virol. 1998a;72:4327–4340. doi: 10.1128/jvi.72.5.4327-4340.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Orden K, Giebler H A, Lemasson I, Gonzales M, Nyborg J K. Binding of p53 to the KIX domain of CREB binding protein. J Biol Chem. 1999;274:26321–26328. doi: 10.1074/jbc.274.37.26321. [DOI] [PubMed] [Google Scholar]
- 50.Wattel E, Cavrois M, Gessain A, Wain-Hobson S. Clonal expansion of infected cells: a way of life for HTLV-I. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13(Suppl. 1):S92–S99. doi: 10.1097/00042560-199600001-00016. [DOI] [PubMed] [Google Scholar]
- 51.Yamato K, Oka T, Hiroi M, Iwahara Y, Sugito S, Tsuchida N, Miyoshi I. Aberrant expression of the p53 tumor suppressor gene in adult T-cell leukemia and HTLV-I-infected cells. Jpn J Cancer Res. 1993;84:4–8. doi: 10.1111/j.1349-7006.1993.tb02775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]