Abstract
Human adenovirus (Ad) is extensively used for a variety of gene therapy applications. However, the utility of Ad vectors is limited due to the low efficiency of Ad-mediated gene transfer to target cells expressing marginal levels of the Ad fiber receptor. Therefore, the present generation of Ad vectors could potentially be improved by modification of Ad tropism to target the virus to specific organs and tissues. The fact that coxsackievirus and adenovirus receptor (CAR) does not play any role in virus internalization, but functions merely as the virus attachment site, suggests that the extracellular part of CAR might be utilized to block the receptor recognition site on the Ad fiber knob domain. We proposed to design bispecific fusion proteins formed by a recombinant soluble form of truncated CAR (sCAR) and a targeting ligand. In this study, we derived sCAR genetically fused with human epidermal growth factor (EGF) and investigated its ability to target Ad infection to the EGF receptor (EGFR) overexpressed on cancer cell lines. We have demonstrated that sCAR-EGF protein is capable of binding to Ad virions and directing them to EGFR, thereby achieving targeted delivery of reporter gene. These results show that sCAR-EGF protein possesses the ability to effectively retarget Ad via a non-CAR pathway, with enhancement of gene transfer efficiency.
Adenovirus (Ad) represents a large family of nonenveloped viruses containing a double-stranded DNA genome of approximately 36 kb (19, 32). Human Ad includes 49 known viral serotypes grouped into six distinct subgroups, A to F. Ad has been widely used as a vector for both in vitro and in vivo gene delivery, largely because of its relatively high infection efficiency in a variety of cell types and tissues (37, 54). However, the broad tropism of the virus represents a drawback when gene delivery to a specific tissue is needed. Most of the studies on the mechanism of Ad infection have concluded that the host range of Ad seems to be dependent to a large extent on the interaction with primary binding receptor. In this regard, the initial steps of Ad infection involve at least two sequential virus-cell interactions, each mediated by a specific capsid protein of the viral particle. Ad infection is initiated by the formation of the complexes between globular knob domain of the fiber protein and a host cell primary receptor (12, 25, 39). Three putative Ad fiber receptors have been described to date. A fiber receptor for Ad of groups A, C, D, E, and F has been identified as the coxsackievirus group B and Ad receptor (CAR) (3, 34, 42). In addition to CAR, the major histocompatibility complex class I (MHC-I) α2 subunit was also proposed as a cell receptor for subgroup C (18). However, CAR has been suggested to mediate high-affinity binding to Ad fiber, while the MHC-I α2 subunit has been hypothesized to facilitate Ad attachment and permissivity to cells with little or no CAR expression (7). It was shown recently that Ad serotype 37 (Ad37) of subgroup D uses α(2→3)-linked sialic acid saccharides on glycoproteins as the cellular receptor moiety instead of CAR or MHC-I α2 (1). After binding to the fiber receptor, penton base interaction with αvβ integrins facilitates internalization via receptor-mediated endocytosis (14, 27, 50). These data suggest that, being expressed in a wide range of human and murine cell types (42), CAR may serve as a primary cellular receptor for the majority of representatives of known Ad serotypes. CAR is an integral membrane protein of unknown cellular function consisting of two extracellular immunoglobulin (Ig) superfamily domains, a single membrane-spanning region, and one carboxy-terminal cytoplasmic domain (3, 4, 42). According to the recent crystallography study of Ad12 fiber knob domain complexed with CAR amino-terminal Ig1 domain, three CAR monomers bind per knob trimer, indicating the location of CAR-binding sites on the knob (5). Furthermore, it was demonstrated that the extracellular domain of CAR is sufficient to allow virus attachment and infection (11, 34), while the transmembrane and intracellular regions appear to be dispensable for these functions (44).
Although Ad vectors can infect most cells, a few cell types including endothelial (23), lung epithelial (38), smooth muscle (33), neural (22), and T (2) cells are poorly infected by Ad apparently due to the scarcity of an appropriate cell surface receptor. The limitations associated with broad native tropism of Ad and low-efficiency gene delivery to Ad receptor-deficient cells could be solved by redirecting the binding of virus to a specific cellular receptor present at sufficient magnitude on target cells. Several strategies are currently being considered to redirect the Ad in order to confer targeting capability or to enhance vector infectivity. In this regard, the incorporation of a targeting peptide ligand by genetic virion modifications offers a rational approach (8, 35, 51, 53) but has several limitations; the capacity for peptide substitution or addition to capsid proteins is size limited, and such modifications can often interfere with correct protein folding and consequent virus assembly (53). On the other hand, the technical achievement of Ad retargeting via bispecific molecules has been approached by a variety of methods. Chemically conjugated bispecific antibodies have been used, with viral linkage accomplished via a peptide epitope incorporated in the penton base (49, 52) or via specific recognition for the knob domain of the fiber protein (9, 13, 29, 41). Further refinement of the strategy of the retargeting complexes has been achieved by the engineering of recombinant proteins consisting of an antiknob single chain fragment variable (scFv) of antibody fused with human epidermal growth factor (EGF) (45) or anti-EGF receptor (EGFR) scFv (16). Recombinant molecules such as these may offer advantages for Ad retargeting, since use of the chemical conjugation method, as well as antibody-containing molecules, increases the difficulties of producing such retargeting complexes, making this approach relatively complex and expensive to develop. Consequently, a simple and efficient method of targeting Ad infection to specific cells would be of great utility for significantly improving present Ad vectors for gene therapy.
In this study, we have developed a targeting approach based on achieving a linkage to the vector particle through a soluble form of its own cellular receptor. Specifically, we have derived a bispecific targeting protein consisting of the ectodomain of CAR in fusion with human EGF. This recombinant fusion protein has the ability to effectively retarget the vector via non-CAR pathways, with enhancement of gene transfer efficiency. In addition, this approach may allow the derivation of different fusion proteins that are capable of Ad retargeting to other cellular receptors by a simple substitution of targeting ligand.
MATERIALS AND METHODS
Cells.
The 293 human kidney cell line transformed with Ad5 DNA was purchased from Microbix (Toronto, Ontario, Canada). The human ovarian carcinoma cell line SKOV3.ip1 was obtained from Janet Price (M. D. Anderson Cancer Center, Houston, Tex.). The human epidermoid carcinoma cell line (A-431), human squamous carcinoma (SCC-4) cells, and human mammary gland (MDA-MB-453) cells were from the American Type Culture Collection (Manassas, Va.). All cell lines were grown at 37°C in media recommended by the suppliers in a humidified atmosphere of 5% CO2.
Enzymes.
Restriction endonucleases, Klenow enzyme, T4 DNA ligase, and proteinase K were from either New England Biolabs (Beverly, Mass.) or Boehringer Mannheim (Indianapolis, Ind.).
Antibodies.
Murine monoclonal antibody (MAb) 4D2 to the tail domain of Ad5 fiber protein (20) and murine polyclonal serum to baculovirus-produced human soluble CAR (sCAR) protein were generated at the University of Alabama at Birmingham Hybridoma Core Facility. Murine MAb 425 to human EGFR was a generous gift from Zenon Steplewski (Thomas Jefferson University, Philadelphia, Pa.).
Viruses.
A recombinant Ad5 vector, AdCMVLuc, containing a firefly luciferase-expressing cassette in place of the E1 region of the Ad genome, was obtained from R. D. Gerard (University of Texas Southwestern Medical Center, Dallas). Ad was propagated on 293 cells and purified by centrifugation in CsCl gradients by a standard protocol. Virus particle titer was determined spectrophotometrically by the method of Maizel et al. (26), using a conversion factor of 1.1 × 1012 viral particles per absorbance unit at 260 nm. To determine the titer of infectious viral particles, the plaque assay on 293 cells was performed by the method of Mittereder et al. (30). Radiolabeled Ad was made by adding 50 μCi of [methyl-3H]thymidine (Amersham Pharmacia Biotech, Piscataway, N.J.) per ml to the medium of infected cells at 20 h postinfection at a multiplicity of infection (MOI) of 2.5 PFU/cell. The infected cells were then harvested at 50 h postinfection, and the virus was purified as described above. The activity of the labeled virus was approximately 10−4 cpm/virus particle.
Indirect immunofluorescence.
Confluent cells were released with EDTA and resuspended in HEPES-buffered saline (20 mM HEPES [pH 7.4], 1% bovine serum albumin (BSA)] at 2 × 106 cells/ml. Cells (2 × 105) were incubated with either MAb 425 (5 μg/ml) or murine anti-CAR serum (1:250) for 1 h at 4°C. Either an isotype-matched IgG (4D2) or normal murine serum was used as a negative control. Cells were then washed with buffer and incubated with secondary goat anti-mouse IgG labeled with fluorescein isothiocyanate (Jackson Laboratories, West Grove, Pa.) at a concentration 5 μg/ml for 1 h at 4°C. After washing, 104 cells per sample were analyzed by flow cytometry performed at the University of Alabama at Birmingham FACS Core Facility. Data were expressed as the geometric mean fluorescence intensity of the entire gated population. The positive population cells was determined by gating the right-hand tail of the distribution of the negative control sample for each cell line at 1%. This gate setting was then used to determine the percentage of CAR- or EGFR-positive cells in each cell line.
Construction of recombinant plasmids.
To introduce the six-His purification tag into the carboxy terminus of sCAR, oligonucleotides 5′GAT CCC CCC GAT ATC ACC ATC ACC ATC ACT AAT AAA 3′ and 5′ GAT CTT TAT TAG TGA TGG TGA TGG TGA TAT CGG GGG 3′ were designed to form DNA duplex coding for histidines followed by two in-frame stop codons. In addition, the generated DNA duplex contained BamHI-compatible cohesive ends and an EcoRV restriction site designed to fuse the CAR open reading frame with six-His coding sequence. The oligonucleotide duplex was cloned into BamHI-digested pQBI-AdCMV5 (Quantum Biotechnologies Inc., Montreal, Quebec, Canada). Plasmid clones were then sequenced in the region of the insert, and the plasmid containing the duplex in the correct orientation was designated pQBI-AdCMV5.6h. To generate a gene encoding the extracellular domain of human CAR, PCR was used. Sense primer (5′ AAA CCG CCT ACC TGC AGC CG 3′) complementary to the position 20 of the 5′ untranslated region of human CAR cDNA (3) and antisense primer (5′ GAG CTT TAT TTG AAG GAG GGA CAA CG 3′) complementary to position 767 were designed to fuse the CAR open reading frame with DNA sequence coding for six histidines incorporated in pQBI-AdCMV5.6h. To construct the plasmid containing the sCAR-His6 gene, a 751-bp PCR fragment was cloned into PmeI- and EcoRV-digested pQBI-AdCMV5.6h, resulting in plasmid pQBIshCAR.6h. This plasmid encodes 236 amino-terminal amino acids (aa) of an extracellular domain of human CAR, including signal sequence, fused with a carboxy-terminal six-His purification tag. To express human sCAR, the Bac-to-Bac baculovirus expression system (Life Technologies, Grand Island, N.Y.) was used. The recombinant donor plasmid for the generation of baculovirus expressing human sCAR was made as follows. The base donor plasmid pFastBac1 was cleaved with Acc65I, and 3′ recessed ends were filled in with the Klenow fragment of Escherichia coli DNA polymerase I. The Klenow fragment was heat inactivated, and plasmid was then cleaved with PstI. Plasmid pQBI-shCAR was cleaved with BsrDI and treated with Klenow fragment to remove the 3′ overhang. After inactivation of DNA polymerase, the plasmid was cleaved with PstI and a PstI-BsrDI fragment (808 bp) and gel purified for cloning into Acc65I- and PstI-digested pFastBac1. After transformation of E. coli strain DH5α (Life Technologies), the resultant plasmid pFBshCAR6h was isolated and used for generation of the recombinant baculovirus genome.
To create the gene for the sCAR-EGF fusion protein, the DNA sequence coding for a short flexible linker and human EGF was amplified from plasmid pBsF5slEGF (unpublished data) using the primers 5′ CCC ATT GGC CAT CAG CCT CCG CAT C 3′ and 5′ GCC CCC GCT CGA GGT CGA CGG TAT C 3′. The PCR-derived DNA fragment contained a unique 5′ MscI site and 3′ SalI site introduced into the molecule to facilitate subsequent cloning. The PCR product was cleaved with MscI and SalI, and a 282-bp DNA fragment was gel purified for further cloning. To construct the plasmid containing the gene coding for sCAR-EGF, primers 5′ CCC ACG GTC CGG CAG CCA CCA TG 3′ and 5′ TCG GGG GAT CTT TAC ACG TGA TGG TGA TGG 3′ were used to reamplify DNA sequence coding for sCAR-His6 using pFBshCAR as the template. The PCR product cleaved at the RsrII restriction site introduced into the 5′ end of the DNA molecule was then cloned into RsrII- and StuI-digested pFastBac1, resulting in plasmid pFBshCARfuse. To derive the plasmid containing the recombinant gene encoding sCAR-EGF, the MscI-SalI PCR fragment was ligated with PmlI- and SalI-digested pFBshCARfuse. After transformation of E. coli DH5α, plasmid clones were sequenced in the region of the insert and the resultant plasmid pFBshCAR-EGF was selected. The constructed plasmid encoding recombinant sCAR fused with EGF and tagged with internal His6 was then used for generation of the recombinant baculovirus genome.
Expression and purification of six-His-tagged recombinant proteins.
Recombinant sCAR-His6 and sCAR-EGF proteins were expressed in High Five cells (Invitrogen, Carlsbad, Calif.) infected with recombinant baculovirus by the method recommended by the manufacturer. Briefly, High Five cells were maintained in suspension culture and infected with recombinant baculovirus at an MOI of 10 PFU/cell. The cell suspension was harvested 72 to 96 h postinfection, and cells were pelleted by centrifugation. Cleared supernatant medium was concentrated 10-fold and dialyzed against phosphate-buffered saline (PBS; 0.01 M PBS [pH 7.4], 138 mM NaCl, 2.7 mM KCl) using a Hemoflow capillary dialyzer (Fresenius Medical Care AG, Bad Homburg vor der Höhe, Germany). Recombinant proteins were then purified by immobilized metal ion affinity chromatography on Ni-nitrilotriacetic acid (NTA)-Sepharose (Qiagen, Valencia, Calif.) as recommended by the manufacturer. Protein concentrations were determined by the Bradford protein assay (Bio-Rad, Hercules, Calif.) with bovine gamma globulin as the standard.
ELISA.
Solid-phase binding enzyme-linked immunosorbent assay (ELISA) was performed by a method previously described (8). Either purified sCAR-His6 or sCAR-EGF was diluted in 50 mM carbonate-bicarbonate buffer (pH 9.6) to a concentration of 8.0 pmol/ml, and 100-μl aliquots were added to wells of a 96-well Nunc-Maxisorp ELISA plate. Plates were incubated overnight at 4°C and then blocked for 2 h at room temperature by the addition of 200 μl of blocking buffer (0.01 M PBS [pH 7.4], 138 mM NaCl, 2.7 mM KCl, 0.05% Tween 20, 2% BSA) to each well. Wells were then washed three times with washing buffer (0.01 M PBS [pH 7.4], 138 mM NaCl, 2.7 mM KCl, 0.05% Tween 20). Purified Ad5 fiber protein diluted in binding buffer (0.01 M PBS [pH 7.4], 138 mM NaCl, 2.7 mM KCl, 0.05% Tween 20, 0.5% BSA) to concentrations ranging from 0.46 to 11 ng/ml was added to the wells in 100-μl aliquots. After 1 h of incubation at room temperature, the wells were washed three times and bound fiber was detected by incubation with 1:1,000 dilution of MAb 4D2. Following incubation at room temperature for 1 h, the wells were washed again and incubated with a 1:10,000 dilution of goat anti-mouse IgG conjugated to alkaline phosphatase (Sigma, St. Louis, Mo.) for 45 min. The wells were then washed four times, and the plate was developed with p-nitrophenyl phosphate (Sigma) as recommended by the manufacturer. Plates were read in a microtiter plate reader set at 405 nm; results are presented as mean ± standard deviation (SD).
Competitive inhibition analysis.
The ability of sCAR-EGF to bind to EGFR was evaluated by competition analysis of radiolabeled EGF binding to EGFR-positive cells in the presence of increasing concentrations of sCAR-EGF. Briefly, A-431 cells were harvested and resuspended in binding buffer (PBS [pH 7.2], 0.1% BSA) at 5 × 106 cells/ml. The cells were aliquoted (100 μl per sample) in triplicate into polystyrene tubes followed by addition of 10-fold dilutions (∼1 pM to 20 μM) of unlabeled human EGF (Pepro Tech, Inc., Rocky Hill, N.J.), sCAR-EGF, or sCAR-His6 used as a negative control. [125I]EGF (100 μl; ∼0.1 nM; Amersham Pharmacia Biotech) was then added, and the cells were incubated at 4°C for 90 min. The cells were then rinsed once with ice-cold buffer and centrifuged at 1,700 × g for 10 min, and the supernatant was removed. The cells were then counted in a gamma counter to determine the amount of bound radioactivity.
Radiolabeled Ad binding assay.
Binding of 3H-labeled Ad to 293, MDA-MB-453, A-431, SCC-4, or SKOV3.ip1 cells was assayed as follows. Three microliters of 3H-labeled AdCMVLuc (∼5.6 × 105 cpm) was preincubated with either sCAR-His6 or sCAR-EGF for 1 h at room temperature. Confluent cells were released with EDTA, washed once with PBS, pelleted, and resuspended to a final concentration of 107 cells/ml in binding medium (Dulbecco modified Eagle medium-Ham's F12, [DMEM-F12], 20 mM HEPES, 0.5% BSA). Then 100-μl aliquots of the cells were transferred to 5-ml test tubes and kept at 4°C. Virus mixtures were then diluted with binding medium to 100 μl, and 25-μl aliquots were added to cell samples and incubated at 4°C with shaking to allow binding. After a 1-h incubation, the cells were washed with 4 ml of binding buffer and centrifuged. Supernatant containing unbound virus was aspirated and cell pellets were solubilized in EcoLume scintillation cocktail (ICN Biomedicals, Costa Mesa, Calif.); then cell-associated radioactivity was measured in a liquid scintillation analyzer (Packard, Downers Grove, Ill.).
Purification of Ad/sCAR-EGF complexes by gel filtration.
A column of Sephadex G-100 (Sigma) was prepared with a bed volume of 10 ml (size exclusion volume, 3.6 ml) and washed with equilibration buffer (PBS containing 0.5% BSA). AdCMVLuc preincubated in the presence or absence 0.4 μg of sCAR-EGF for 30 min was diluted to 2 ml with equilibration buffer, and a 1-ml sample was loaded onto the column at gravity flow. Once the sample was loaded onto the column, an additional 1.5 ml of equilibration buffer was applied. The fraction containing the high-molecular-weight Ad or Ad/sCAR-EGF complexes was eluted with 3.3 ml of equilibration buffer solution. The eluted fraction was used immediately for the gene transfer assay.
Ad-mediated gene transfer assay.
Ad-mediated infection experiments using cell lines were performed as follows. Cell monolayers grown in a 24-well plate (5 × 105 cells/well) were washed with PBS. AdCMVLuc (5 × 106 PFU) was preincubated with either sCAR-His6 or sCAR-EGF for 1 h at room temperature. Virus mixtures were then diluted with DMEM-F12 (Mediatech, Herndon, Va.) containing 2% fetal bovine serum to a final concentration of 5 × 106 PFU/ml, and 200-μl aliquots were added to wells (at an MOI of 2 PFU/cell) to allow internalization of AdCMVLuc for 45 min at room temperature. Virus complexes were then aspirated, the cells were washed with PBS, and 1 ml of complete growth medium containing 10% fetal bovine serum and 2 mM glutamine was added to each well. The cells were incubated at 37°C to allow expression of the luciferase gene. Forty hours after the addition of virus, cells were lysed with 250 μl of lysis buffer and analyzed for luciferase expression. Luciferase activity in the cell lysates was analyzed by using the Promega (Madison, Wis.) luciferase assay system and a Berthold (Gaithersburg, Md.) luminometer.
RESULTS
Design and generation of sCAR-ligand protein.
As was recently demonstrated, Ad of subgroups A, C, D, E, and F use CAR as a cellular fiber receptor. It was also shown that sCAR bound to representatives of all Ad subgroups except subgroup B (34). To apply this finding to Ad retargeting, we proposed to design protein molecules consisting of the extracellular domain of CAR in fusion with a targeting ligand (Fig. 1A). Our goal was to generate a bispecific CAR-ligand molecule that could block the cell-binding domain of the fiber knob as well as target the Ad vector to a novel cellular receptor present at a sufficient level on target cells. Therefore, the infection of cells by this virus complex would not be dependent on the presence of CAR on a target cell membrane. The targeting ligand that we chose to use was EGF because it is well established that the EGFR is overexpressed on a variety of cancer cells (46).
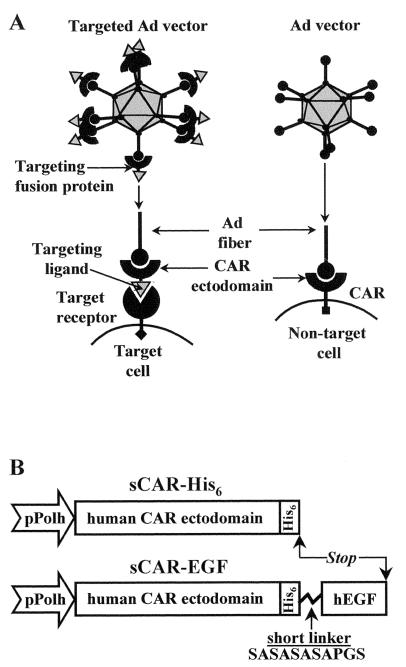
FIG. 1.
(A) Utilization of sCAR-ligand fusion proteins for receptor-specific targeting of Ad vectors. Ad vectors normally achieve cell binding via interaction between the knob domain of viral fiber protein with CAR. To redirect an Ad vector to an alternative cell surface receptor, a genetically engineered targeting fusion protein consisting of the CAR ectodomain fused to a receptor-specific targeting ligand was used. By virtue of its dual binding capacity, this complex serves as a bridge between an Ad virion and a cell-specific receptor molecule, thereby providing novel cell-binding capacity to the virion. (B) Construction of sCAR fusion proteins. The gene coding for either human sCAR-His6 or sCAR-EGF was constructed in a baculovirus expression vector. Expression is driven from the polyhedrin promoter (pPolh). A His6 tag was introduced for purification purposes into the carboxy terminus of the extracellular CAR domain (236 aa). To construct sCAR-EGF protein, human EGF (53 aa) was fused with the CAR ectodomain by a flexible linker (SASASASAPGS) and tagged with His6. See Materials and Methods for details of construction.
To express the sCAR-EGF fusion protein, we designed a gene sequence coding for the ectodomain of human CAR, six histidines, a short flexible linker, and human EGF (Fig. 1B). The sequence encoding the internal six-His tag was incorporated into the recombinant CAR fusion gene in order to facilitate downstream purification of the product. The gene coding for sCAR-His6 was created to express the relevant control protein (Fig. 1B). To produce sCAR-EGF and sCAR-His6, recombinant baculoviruses containing the genes of interest were created and used to infect insect cells. Infection of High Five insect cells with recombinant baculoviruses resulted in a high level of sCAR-His6 as well as sCAR-EGF protein expression in a secreted soluble form. Baculovirus-expressed sCAR-His6 and sCAR-EGF proteins were recovered from the infected cell culture media by means of Ni-NTA affinity chromatography and were then analyzed for purity by gel electrophoresis.
Analysis of sCAR-EGF protein.
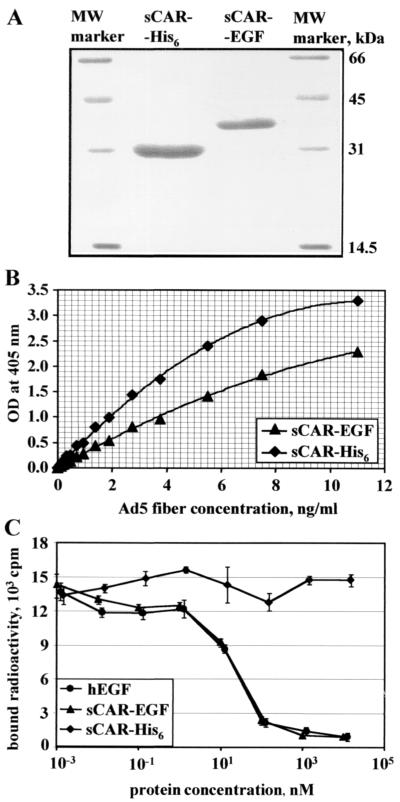
Analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis of both sCAR-His6 and sCAR-EGF (Fig. 2A) shows that purified proteins have molecular masses close to expected of 24.9 and 31.9 kDa, respectively. Thus, by using the baculovirus expression system and Ni-NTA affinity purification, we were able to obtain preparative amounts of homogeneously purified sCAR-EGF fusion protein for subsequent analysis.
FIG. 2.
Characterization of sCAR fusion proteins. (A) Analysis of recombinant sCAR-EGF protein by polyacrylamide gel electrophoresis. Soluble CAR-His6 and sCAR-EGF six-histidine-tagged proteins expressed in insect cells were purified on a Ni-NTA-Sepharose column and analyzed by electrophoresis on a 12% polyacrylamide gel at denaturing conditions. The bands were visualized by GELCODE blue stain reagent. Numbers on the right indicate molecular masses of marker proteins in kilodaltons. (B) Analysis of interaction between recombinant sCAR-EGF protein and Ad fiber protein by ELISA. Baculovirus-expressed sCAR-His6 and sCAR-EGF proteins absorbed on an ELISA plate were incubated with various concentrations of purified recombinant Ad5 fiber protein. Bound fiber protein was then detected with antifiber monoclonal antibody 4D2. Each point represents the cumulative mean ± SD of triplicate determinations. Error bars depicting SDs are smaller than the symbols. OD, optical density. (C) Competitive inhibition analysis. The binding of sCAR-EGF to EGFR was quantified by competition analysis of radiolabeled EGF binding to A-431 EGFR-overexpressing cells in the presence of various concentrations of sCAR-EGF. The cells were mixed with 10-fold dilutions (1 pM to 20 μM) of unlabeled human EGF, sCAR-EGF, or sCAR-His6 (negative control). [125I]EGF (∼0.1 nM) was then added, and incubation was continued 4°C. The cells were then rinsed and counted in a gamma counter to determine the amount of bound radioactivity. Each point represents the cumulative mean ± SD of triplicate determinations. Some error bars depicting SDs are smaller than the symbols.
We first chose to characterize the sCAR-EGF fusion protein with respect to its ability to bind Ad fiber knob. Therefore, we used it in an ELISA (Fig. 2B) with purified Ad5 fiber expressed in insect cells (8). This assay showed that fiber protein efficiently bound to immobilized sCAR-EGF in a wide range of concentrations. Compared to the sCAR-His6 protein used as a control, the fiber-binding affinity of sCAR-EGF fusion protein was slightly lower, most probably due to changes in the molecular conformation of the CAR ectodomain. Based on the obtained result, whereby generated sCAR-EGF fusion protein is able to efficiently interact with Ad fiber knob, we hypothesized that the affinity of this interaction is sufficient to block the cell-binding site on the knob domain in order to block viral infection.
To evaluate the ability of sCAR-EGF to bind to EGFR, we performed competition analysis of radiolabeled EGF binding to EGFR-positive A-431 cells in the presence of increasing concentrations of sCAR-EGF. Equimolar concentrations of unlabeled human EGF or sCAR-His6 protein (negative control) were tested in a parallel. Figure 2C shows that EGF and sCAR-EGF inhibited the binding of [125I]EGF to A-431 EGFR-positive cells (47), while sCAR-His6 did not. The level of inhibition was similar for each, with EGF having a 50% inhibitory concentration of 24.1 nM, compared to 19.5 nM for sCAR-EGF.
sCAR-EGF inhibits Ad binding and gene transfer to 293 cells.
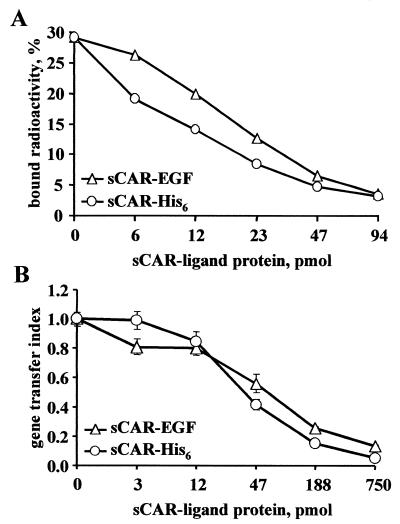
Having established that the sCAR-EGF fusion protein could bind to both Ad fiber and cellular EGFR, we examined whether the formation of bonds between Ad and sCAR protein results in any changes in the capacity of such virus complexes to infect cells. To this end, we first compared the cell-binding efficiencies of Ad/sCAR fusion complexes. To do so, we preincubated 3H-radiolabeled Ad with either sCAR-His6 or sCAR-EGF and then used the formed complexes in a cell- binding assay. 293 human kidney cells were selected for this analysis because they express high levels of CAR and readily support Ad infection (8). The binding assay was performed under conditions (4°C) allowing the viruses to bind the cells but preventing virus internalization. As shown in Fig. 3A, the cell-binding capacity of both Ad/sCAR-His6 and Ad/sCAR-EGF complexes was less than that of Ad alone and dependent on the sCAR-fusion protein dose; 94 pmol of both sCAR fusion proteins could block 90% of Ad binding to CAR-positive 293 cells. According to flow cytometry analysis, 293 cells express high levels of CAR and EGFR (data not shown). Because of relatively equivalent expression of CAR and EGFR on the cell membrane, binding of Ad/CAR-EGF complexes to 293 cells may occur via any combination of CAR- and EGFR-dependent routes. Therefore, the capacity of sCAR-EGF molecules to block Ad cell binding through a CAR-dependent pathway might be diminished by the alternative ability to mediate binding to EGFR and result in cumulative inhibition of Ad binding and subsequent gene delivery. As can be seen in Fig. 3A, sCAR-EGF protein indeed displayed less Ad-blocking ability than the control sCAR-His6, particularly in the low concentration range.
FIG. 3.
Functional characterization of sCAR fusion proteins. (A) Inhibition of Ad binding. 3H-labeled Ad was preincubated with different amounts of either sCAR-His6 or sCAR-EGF, and then 3H-Ad/sCAR-ligand complex samples (105 cpm) were mixed with 293 cells (106 cells per aliquot) and allowed to bind at 4°C. Cell-bound radioactivities were determined as described in Materials and Methods. Data are presented as the percentage of input 3H-Ad bound after washing and calculated as the cumulative mean ± SD of triplicate determinations. Error bars depicting SDs are smaller than the symbols. (B) Inhibition of Ad-mediated gene transfer. Recombinant Ad vector AdCMVLuc, expressing the firefly luciferase reporter gene, was preincubated with various amounts of either sCAR-His6 or sCAR-EGF. Monolayers of 293 cells were then exposed to AdCMVLuc/sCAR-ligand complexes and assayed for luciferase activity as described in Materials and Methods. Gene transfer indices were calculated from the ratio of the mean luciferase activity documented in cells infected with either AdCMVLuc/sCAR-EGF or AdCMVLuc/sCAR-His6 to those treated with AdCMVLuc alone. Each point represents the cumulative mean ± SD of triplicate determinations. Some error bars depicting SDs are smaller than the symbols.
To evaluate the ability of the derived fusion protein to block Ad infection, we performed an infection inhibition assay. The results showed that sCAR-EGF protein is able to block AdCMVLuc-mediated luciferase gene transfer to 293 cells, demonstrating an inhibition profile similar to that for control sCAR-His6 protein (Fig. 3B). Therefore, these experiments confirmed the utility of sCAR-EGF to efficiently inhibit Ad binding as well as gene transfer to CAR-positive 293 cells and provided a rationale for further studies.
Analysis of different human cell lines for the expression of CAR and EGFR.
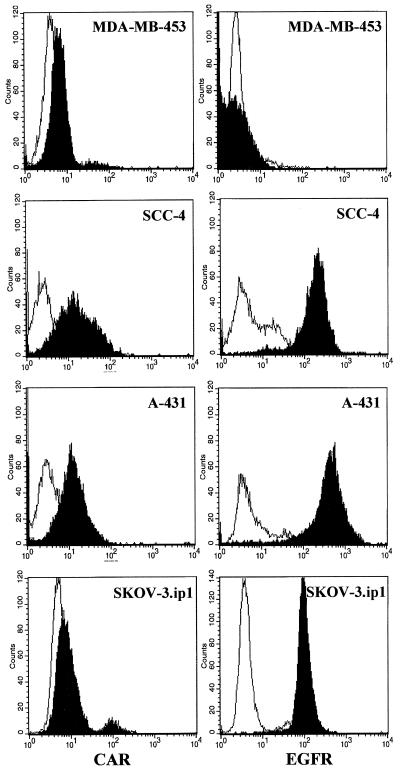
Several human cell lines of different origins, including SKOV3.ip1 ovarian carcinoma cells, A-431 epidermoid carcinoma cells, SCC-4 squamous carcinoma cells, and MDA-MB-453 mammary gland cells, were analyzed for cell surface expression of CAR and EGFR by indirect immunofluorescence assay (Fig. 4). The indicated cell lines were chosen based on previously published data on levels of CAR and EGFR expression and varying susceptibility to Ad infection (6, 8). Flow cytometry showed that SCC-4 cells express moderate levels of CAR and rather large amounts of EGFR (Fig. 4). SKOV3.ip1 cells were CAR negative but high EGFR expressors. A431 cells display a low level of CAR expression while being highest in levels of EGFR. This agrees with previous reports showing that A-431 cells express as many as 3 × 106 EGFR molecules/cell (47). MDA-MB-453 cells, known to be EGFR negative (28), also demonstrated a low level of CAR and were thus selected as a negative control. Flow cytometry data indicate the following order for the tested cell lines with respect to relative expression of EGFR: MDA-MB-453 < SKOV3.ip1 < SCC-4 < A-431. Therefore, for our subsequent experiments, we established a set of cell lines covering a wide range of EGFR expression.
FIG. 4.
Relative expression of CAR and EGFR on different human cell lines. Flow cytometric analysis of MDA-MB-453, SCC-4, SKOV3.ip1, or A-431 cells using either murine polyclonal serum to CAR or anti-EGFR MAb was performed as described in Materials and Methods. Positive staining with anti-CAR or anti-EGFR antibody (black) is seen relative to an isotype control (white). A representative of three separate experiments is shown. Flow cytometry assay revealed that SCC-4, SKOV3.ip1, and A-431 cells express high levels of cell surface EGFR but moderate to low levels of CAR. MDA-MB-453 cells demonstrated dramatically lower levels of both CAR and EGFR and were selected as a negative control.
sCAR-EGF can mediate EGFR-specific cell binding.
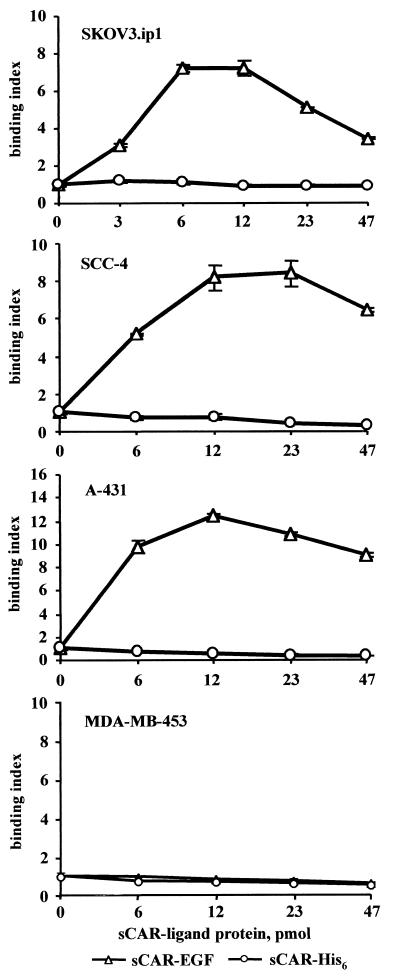
Having established that sCAR-EGF demonstrates sufficient ability to block Ad infection, we investigated the ability of the Ad/sCAR-EGF complex to infect cells through a CAR-independent pathway. To address this issue, we studied the ability of sCAR-EGF to target Ad binding to EGFR. Retargeting Ad infection through EGFR on cells normally refractory to Ad due to lack of CAR expression on their cell membranes may enhance the level of gene delivery by facilitating Ad binding to EGFR. To investigate this hypothesis, we estimated the capacity of sCAR-EGF fusion protein to mediate binding of 3H-radiolabeled Ad to EGFR-positive cells. To determine the optimum concentration of sCAR-EGF providing maximal Ad binding, 3H-Ad was preincubated with increasing amounts of either sCAR-EGF or sCAR-His6 to allow complex formation. Suspensions of MDA-MB-453, SCC-4, SKOV3.ip1, or A-431 cells were then exposed to either 3H-Ad/sCAR-EGF or 3H-Ad/sCAR-His6 complexes at 4°C to prevent virus internalization. As shown in Fig. 5, preincubation with sCAR-EGF resulted in significant increase of 3H-Ad binding to EGFR-positive SCC-4, SKOV3.ip1, and A-431 cells compared to EGFR-negative MDA-MB-453 cells. Preincubation of Ad in the presence of sCAR-His6 had no effect on the level of binding to EGFR-positive cells compared to Ad alone. Complexing 3H-Ad with increasing amounts of targeting sCAR-EGF fusion protein increased the level of cell-bound radioactivity in a dose-dependent manner. Maximal EGFR-targeted Ad binding occurred with an sCAR-EGF/virus ratio of 12 pmol of sCAR-EGF protein per 6 × 109 viral particles. Increasing the sCAR-EGF/virus ratio further proved inhibitory to binding, presumably because of competition for EGFR binding by excess sCAR-EGF protein. As shown in Fig. 5, calculated binding indices for Ad targeted to EGFR on SKOV3.ip1, SCC-4, and A-431 cells were increased 7-, 8-, and 12-fold, respectively. Bound radioactivities registered in cell samples incubated with EGF-targeted Ad/sCAR-EGF complexes were dependent on sCAR-EGF protein dose and significantly higher than in the case of Ad/sCAR-His6 complexes or Ad alone. These experiments clearly demonstrated that sCAR-EGF protein is capable of inducing changes in the initial steps of virus-cell interaction and suggest that formation of the Ad/sCAR-EGF complexes allows for Ad binding to EGFR.
FIG. 5.
Comparison of 3H-labeled Ad binding to MDA-MB-453, SCC-4, SKOV3.ip1, and A-431 cells. 3H-labeled Ad was preincubated for 30 min at room temperature with different amounts of sCAR-His6 or sCAR-EGF. 3H-Ad/sCAR-ligand mixtures (105 cpm per sample) were then added to cells aliquots (106) and allowed to bind for 1 h at 4°C. Bound radioactivity was determined after pelleting the cells by centrifugation. Binding indices were calculated from the ratio of the mean bound radioactivity of 3H-Ad preincubated in presence of sCAR-ligand versus 3H-Ad preincubated in absence of sCAR-ligand protein. Each point represents the cumulative mean ± SD of triplicate determinations. Some error bars depicting SDs are smaller than the symbols.
sCAR-EGF mediates specific Ad binding to cellular EGFR.
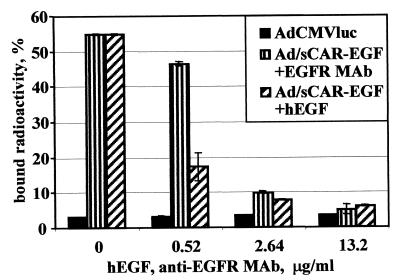
Since our ultimate goal was the targeting of Ad vectors to EGFR, we conducted a competition assay to prove that sCAR-EGF-mediated virus-cell interactions occurred specifically via EGFR as the alternative cellular receptor. By blocking Ad/sCAR-EGF interaction with a specific competitor, the level of EGFR-dependent binding could be determined. In this regard, analysis of binding of 3H-Ad/sCAR-EGF complexes to the cells was accomplished at 4°C in the presence of either human EGF or anti-EGFR neutralizing MAb capable of blocking the binding to EGFR. To perform this analysis, the optimum sCAR-EGF/virus ratio determined in the binding assay was used to form 3H-Ad/sCAR-EGF complexes prior to the binding to A-431 cells in the presence of increasing concentrations of either human EGF or anti-EGFR MAb. As shown in Fig. 6, binding of 3H-Ad in the absence of competitors was not significantly different from binding in the presence of any tested concentrations of EGF or anti-EGFR MAb. When binding of the 3H-Ad/sCAR-EGF complex was assayed (Fig. 6), the presence of EGF as well as anti-EGFR MAb decreased the level of binding in a dose-dependent manner. Due to significant differences in molar concentrations of the blocking agents used, EGF protein displayed higher blocking efficiency than MAb at low concentrations but similar efficiency at high concentrations tested, blocking 90% of the binding. These results demonstrate that derived sCAR-EGF protein can be effectively used to direct Ad binding via a non-CAR pathway to a novel cellular receptor. Of note, the increased Ad binding efficiency was shown to occur through specific interaction of the sCAR-EGF targeting protein with EGFR.
FIG. 6.
Specific inhibition of sCAR-EGF-mediated Ad binding. 3H-Ad was preincubated for 30 min at room temperature with 0.4 μg of sCAR-EGF. Human epidermoid carcinoma A-431 cells, overexpressing EGFR, were preincubated for 30 min at 4°C in the presence or absence of either human EGF or anti-EGFR MAb at different concentrations (0.52 to 13.2 μg/ml). 3H-Ad/sCAR-EGF samples (105 cpm) were then added and allowed to bind for 1 h at 4°C. Cells were washed by centrifugation, and radioactivities of cell pellets were determined in a beta counter. Data are presented as the percentage of input 3H-Ad bound after washing and calculated as the cumulative mean ± SD of triplicate determinations.
sCAR-EGF mediates enhanced gene transfer to human cancer cells.
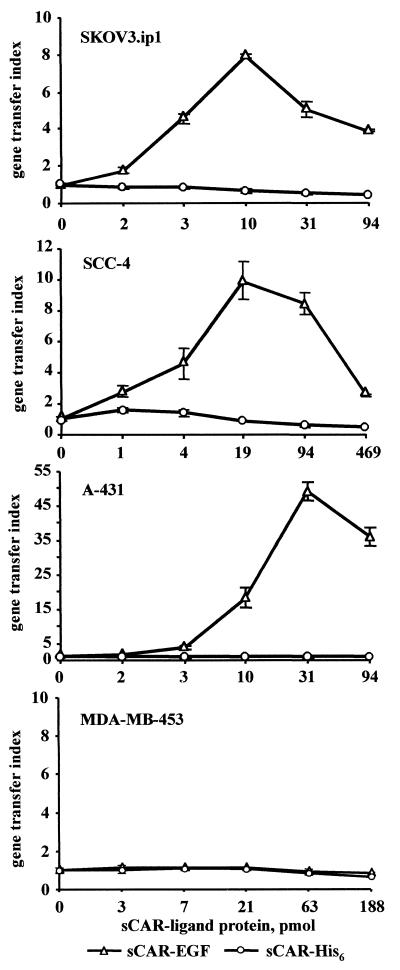
We used the same strategy to evaluate the ability of sCAR-EGF targeting protein to mediate Ad gene delivery to cultured human cancer cell lines SCC-4, SKOV3.ip1, A-431, and MDA-MB-453. Our previous study showed that these cells are relatively difficult to infect with Ad vectors (6, 8). These findings were corroborated by our flow cytometry data, which showed either modest or low levels of CAR expression. Importantly, rather high levels of EGFR detected in three of these cell lines suggested that low Ad susceptibility due to relative lack of CAR may be overcome by targeting to EGFR present at sufficient magnitude. The sCAR-EGF protein was titered against Ad to ascertain the optimal ratio of targeting protein to virus as measured by improvements in gene transfer. The magnitude of gene expression mediated by the sCAR-EGF-complexed AdCMVLuc was demonstrated on EGFR-positive SCC-4, SKOV3.ip1, and A-431 cells versus EGFR-negative MDA-MB-453 cells. As shown in Fig. 7, compared with AdCMVLuc alone, AdCMVLuc complexed with sCAR-EGF targeting protein (EGFR-targeted Ad) mediated 8-, 10-, and 50-fold enhancements of luciferase expression in SKOV3.ip1, SCC-4, and A-431 cells, respectively. As evidence that the sCAR-EGF promoted gene transfer by an EGFR-specific mechanism, no enhancement was observed in cells exposed to AdCMVLuc complexed with sCAR-His6 (untargeted Ad). Further, the specificity of sCAR-EGF-mediated Ad targeting was illustrated by the failure of sCAR-EGF to enhance Ad-based gene transfer to EGFR-negative MDA-MB-453 cells. Thus, this set of assays demonstrated that the sCAR-EGF targeting protein enables retargeting of an Ad vector via a CAR-independent pathway, with severalfold enhancement of gene transfer efficiency specifically to EGFR-positive cells.
FIG. 7.
Characterization of sCAR-EGF-mediated Ad gene transfer to EGFR-positive cell lines. AdCMVLuc was preincubated with various amounts of either sCAR-EGF targeting protein, or sCAR-His6 as a control, prior to incubation with cells. Then monolayers of MDA-MB-453, SCC-4, SKOV3.ip1, or A-431 cells were exposed to Ad/sCAR-ligand complexes mixtures at 2 PFU/cell. Targeting index is defined as the ratio between mean luciferase activity for Ad preincubated in the presence of sCAR-ligand versus Ad preincubated in absence of sCAR-ligand protein. Each point represents the cumulative mean ± SD of triplicate determinations. Some error bars depicting SDs are smaller than the symbols.
Efficiency of sCAR-EGF-mediated Ad gene delivery.
To further investigate the phenomenon of sCAR-EGF-mediated Ad targeting, we attempted to evaluate the stability of Ad/sCAR-EGF complexes. As we observed previously, exceeding the optimal sCAR-EGF/virus ratio proved to be inhibitory to binding and gene transfer, presumably because of competition for EGFR binding by uncomplexed sCAR-EGF protein. Alternatively, a relative excess of targeting protein is required for optimal formation of Ad/sCAR-EGF complexes. To address this issue, we purified Ad/sCAR-EGF complexes by gel filtration in order to remove unbound sCAR-EGF protein. Using the optimum sCAR-EGF/virus ratio determined with A-431 cells, gene transfer mediated by purified versus unpurified EGFR-targeted AdCMVLuc was compared among the panel of EGFR-positive cell lines. It was shown that EGFR-targeted AdCMVLuc significantly enhanced gene transfer to tested cells (data not shown). Purified Ad/sCAR-EGF complexes were somewhat less effective than unpurified EGFR-targeted virus, demonstrating 50% less efficient gene transfer. In contrast, gene transfer mediated by AdCMVLuc was barely affected by purification, indicating that there was no significant loss of Ad during the purification procedure. Nevertheless, even after purification, Ad-mediated gene transfer efficiency was enhanced when targeted to EGFR compared to control AdCMVLuc in all three cell types examined. Specifically, 3-, 4.5-, and 9-fold increases of luciferase activity were observed in SKOV-3.ip1, SCC-4, and A-431 cells, respectively. The degree of gene transfer enhancement correlates with our flow cytometry and binding data and is likely to be dependent on the CAR/EGFR ratio on the cell surface and on EGFR affinity (29). The fact that purification of formed Ad/sCAR-EGF complexes did not ablate enhanced gene transfer capacity indicates that the sCAR-EGF targeting protein can maintain its association with Ad in the context of vector purification schemes. This relative stability provides an empiric means to derive vector/complex particles optimized with respect to gene transfer applications.
DISCUSSION
The infection spectrum of human Ad is wide with respect to different types of tissues and different age groups of patients (40). However, the present generation of Ad vectors suffer from three important limitations which have prevented the realization of their full potential. One disadvantage is related to vector-induced inflammatory and immune responses precluding readministration of the same vector. Second, Ad can infect a wide range of different cells, which makes it impossible to deliver genes to specific target cell types. The third limitation is the inability of the vector to infect the cells which do not express CAR or express it at low levels. Thus, the primary requirement for the application to gene therapy is the availability of a vector capable of accomplishing effective and selective gene delivery. In this regard, there is increasing evidence that the efficiency of Ad vectors can be limited by a deficiency of appropriate binding and entry mechanisms on the target cell (10, 48, 50, 55). While CAR is widely expressed in vivo (42), low CAR levels (17, 24, 31) or its localization on inaccessible parts of cell (43) can prevent efficient infection by Ad vectors. The requirement for expression of CAR on the target cell represents a hurdle to genetic modification of cells that lack CAR, leading to the strategy of modifying the tropism of Ad vectors. Therefore, the utility of the present generation of Ad vectors for gene therapy may be significantly improved by achieving targeted infection of specific cell types by the virus. To develop a targeted Ad vector, it is necessary both to ablate broad native Ad tropism and to introduce novel tropism, which will allow targeting of certain cell types, including cells that are inherently not sensitive to Ad infection due to a lack of CAR. In this regard, abrogation of Ad fiber binding to its natural receptor is therefore a prerequisite for Ad application particularly in vivo. This goal has been addressed by the development of retargeting complexes which simultaneously recognize the specific capsomer of the viral particle and the targeted cell surface molecule. The technical achievement of Ad retargeting via bispecific molecular complexes has been approached by a variety of methods. In this regard, chemically conjugated bispecific antibodies have been used to recognize a FLAG peptide epitope incorporated in the penton base (49, 52). Such bispecific antibodies with specific recognition for the knob domain of the fiber protein that block the knob-receptor interactions and simultaneously serve to cross-link the virus to alternate cellular receptors were designed (9). Abrogation of Ad native tropism was achieved by the use of the antiknob antibody, or its Fab fragment, conjugated with ligands specific for target cell surface receptors such as folate (9), fibroblast growth factor 2 (13, 36), CD40 (41), and EpCAM antigen (15), as well as antibodies for EGFR (6, 29). These recent advances in Ad vector targeting illustrate the potential utility of employing chemically conjugated bispecific molecular complexes to achieve both an abrogation of Ad native tropism and delivery of Ad vectors to specific cell types. However, the use of chemical conjugates increases production difficulties, making this approach relatively complex and expensive to develop. The strategy that we have developed could be of great utility to avoid at least some of these limitations. Recently, refinements of the strategy of antifiber retargeting complexes have been proposed. The ability to engineer recombinant antibodies has facilitated the production of bispecific antibodies or fusion proteins in bacteria. For example, an “adenobody” consisting of an antiknob scFv and EGF has been derived and used to target Ad to EGFR (45). An analogous approach was successfully used to produce recombinant bispecific scFv comprising both antiknob and anti-EGF scFvs in a eukaryotic expression system (16). Recombinant molecules such as these may indeed offer advantages for Ad retargeting in terms of vector production and validation. However, there is likely to be an immune response directed against virus-encoded antigens and possibly against the scFv molecules.
The novelty of the approach developed in this study is based on the utilization of native Ad-CAR interaction to provide a linkage between a targeting ligand and the viral particle. The affinity of CAR binding to the fiber knob (Kd = 4.75 nM) (21) is comparable with those determined for the highest-binding antiknob scFvs (3 to 12 nM) (45). Based on the crystal structure of Ad12 knob complexed with the Ig1 structural domain of human CAR, three Ig1 monomers bind per knob trimer (5). The predicted CAR/knob binding ratio combined with the high affinity of the interaction may contribute to the efficiency of linkage between Ad particles and sCAR-ligand and consequently to the target receptor. The sCAR-EGF protein is expected to have a very low immunogenic potential in humans because of the endogenous origin of its structural components; therefore, it might provide a high-affinity nonimmunogenic linkage to the viral particle compatible with in vivo gene delivery applications. In addition, carboxy-terminal localization of targeting ligand in the context of sCAR fusion protein mimics the native mechanism of virion binding to its high-affinity receptor.
By complexing Ad with sCAR-EGF, we have blocked the natural cell-binding site of the fiber knob while simultaneously supplying a binding alternative in EGF. We showed that Ad modification with bifunctional sCAR-EGF molecules overcomes the barrier of inefficient gene transfer into specific cancer cell types. As expected, the enhanced binding properties of the EGFR-targeted Ad vector correlated with its ability to infect EGFR-expressing cells from a selected panel of cancer cell lines, as seen in gene transfer experiments. Cell binding of EGFR-targeted Ad could be blocked competitively by preincubation of the cells with either human EGF or anti-EGFR MAb, confirming that the redirected Ad binding was specific to EGFR. The gene transfer efficiency of the Ad when targeted through a non-CAR pathway was markedly improved in all EGFR-positive cell lines examined compared to EGFR-negative cells, suggesting that the efficiency of targeted infection is dependent on EGFR density. Purification of Ad/sCAR-EGF complexes proved our hypothesis that sCAR is capable of providing a high-affinity linkage to the viral particle, compatible with an Ad purification scheme and likely with systemic administration of Ad vectors. Use of the baculovirus system for expressing targeting molecules should overcome potential problems associated with partial or complete insolubility of some protein ligands and lack of glycosylation upon expression in E. coli. Furthermore, the observation that sCAR binds to the fiber knob domains derived from certain serotype representatives from five of the six Ad subgroups (34) suggests that sCAR may be the protein of choice to serve as a universal moiety providing a linkage to the majority of Ad serotypes. In addition, the replacement of EGF with a different ligand should enable targeting of Ad vectors to various cellular receptors. The enhancement of transgene expression that we achieved by means of sCAR-EGF-mediated infection of otherwise refractory cancer cells indicates that this approach has potential for further studies of targeting Ad gene delivery to cancer cells in vivo. In this way, it might be possible to ablate preexisting patterns of Ad infection and establish new ones that will reduce the initial dose of virus, thereby decreasing immediate toxicity and increasing the safety and efficiency of Ad vectors.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01 CA74242, R01 HL50255, R01 CA68245, and R01 CA83821, National Cancer Institute grant N01 CO-97110, and U.S. Army Department of Defense grants PC 970193 and PC 991018.
We are grateful to Robert Finberg for providing cDNA for CAR. Alex Pereboev is thanked for fruitful discussions and proofreading the manuscript.
REFERENCES
- 1.Arnberg N, Edlund K, Kidd A H, Wadell G. Adenovirus type 37 uses sialic acid as a cellular receptor. J Virol. 2000;74:42–48. [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur J F, Butterfield L H, Roth M D, Bui L A, Kiertscher S M, Lau R, Dubinett S, Glaspy J, McBride W H, Economou J S. A comparison of gene transfer methods in human dendritic cells. Cancer Gene Ther. 1997;4:17–25. [PubMed] [Google Scholar]
- 3.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 4.Bergelson J M, Krithivas A, Celi L, Droguett G, Horwitz M S, Wickham T, Crowell R L, Finberg R W. The murine CAR homolog is a receptor for coxsackie B viruses and adenoviruses. J Virol. 1998;72:415–419. doi: 10.1128/jvi.72.1.415-419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bewley M C, Springer K, Zhang Y B, Freimuth P, Flanagan J M. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286:1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 6.Blackwell J L, Miller C R, Douglas J T, Li H, Peters G E, Carroll W R, Strong T V, Curiel D T. Retargeting to EGFR enhances adenovirus infection efficiency of squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 1999;125:856–863. doi: 10.1001/archotol.125.8.856. [DOI] [PubMed] [Google Scholar]
- 7.Davison E, Kirby I, Elliott T, Santis G. The human HLA-A*0201 allele, expressed in hamster cells, is not a high-affinity receptor for adenovirus type 5 fiber. J Virol. 1999;73:4513–4517. doi: 10.1128/jvi.73.5.4513-4517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dmitriev I, Krasnykh V, Miller C R, Wang M, Kashentseva E, Mikheeva G, Belousova N, Curiel D T. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J Virol. 1998;72:9706–9713. doi: 10.1128/jvi.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douglas J T, Rogers B E, Rosenfeld M E, Michael S I, Feng M, Curiel D T. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–1578. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 10.Freimuth P. A human cell line selected for resistance to adenovirus infection has reduced levels of the virus receptor. J Virol. 1996;70:4081–4085. doi: 10.1128/jvi.70.6.4081-4085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freimuth P, Springer K, Berard C, Hainfeld J, Bewley M, Flanagan J. Coxsackievirus and adenovirus receptor amino-terminal immunoglobulin V-related domain binds adenovirus type 2 and fiber knob from adenovirus type 12. J Virol. 1999;73:1392–1398. doi: 10.1128/jvi.73.2.1392-1398.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gall J, Kass-Eisler A, Leinwand L, Falck-Pedersen E. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J Virol. 1996;70:2116–2123. doi: 10.1128/jvi.70.4.2116-2123.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman C K, Rogers B E, Douglas J T, Sosnowski B A, Ying W, Siegal G P, Baird A, Campain J A, Curiel D T. Targeted gene delivery to Kaposi's sarcoma cells via the fibroblast growth factor receptor. Cancer Res. 1997;57:1447–1451. [PubMed] [Google Scholar]
- 14.Greber U F, Willetts M, Webster P, Helenius A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell. 1993;75:477–486. doi: 10.1016/0092-8674(93)90382-z. [DOI] [PubMed] [Google Scholar]
- 15.Haisma H, Pinedo H, Rijswijk A, der Meulen-Muileman I, Sosnowski B, Ying W, Beusechem V, Tillman B, Gerritsen W, Curiel D. Tumor-specific gene transfer via an adenoviral vector targeted to the pan-carcinoma antigen EpCAM. Gene Ther. 1999;6:1469–1474. doi: 10.1038/sj.gt.3300969. [DOI] [PubMed] [Google Scholar]
- 16.Haisma, H. J., J. Grill, D. T. Curiel, S. Hoogeland, V. W. Van Beusechem, H. M. Pinedo, and W. R. Gerritsen. 2000. Targeting of adenoviral vectors through a bispecific single-chain antibody. Cancer Gene Ther. 7:in press. [DOI] [PubMed]
- 17.Hidaka C, Milano E, Leopold P L, Bergelson J M, Hackett N R, Finberg R W, Wickham T J, Kovesdi I, Roelvink P, Crystal R G. CAR-dependent and CAR-independent pathways of adenovirus vector-mediated gene transfer and expression in human fibroblasts. J Clin Investig. 1999;103:579–587. doi: 10.1172/JCI5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong S S, Karayan L, Tournier J, Curiel D T, Boulanger P A. Adenovirus type 5 fiber knob binds to MHC class I alpha2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 1997;16:2294–2306. doi: 10.1093/emboj/16.9.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horwitz M S. Adenovirus. In: Knipe D M, Fields B N, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2149–2171. [Google Scholar]
- 20.Karayan L, Gay B, Gerfaux J, Boulanger P A. Oligomerization of recombinant penton base of adenovirus type 2 and its assembly with fiber in baculovirus-infected cells. Virology. 1994;202:782–795. doi: 10.1006/viro.1994.1400. [DOI] [PubMed] [Google Scholar]
- 21.Kirby I, Davison E, Beavil A J, Soh C P, Wickham T J, Roelvink P W, Kovesdi I, Sutton B J, Santis G. Mutations in the DG loop of adenovirus type 5 fiber knob protein abolish high-affinity binding to its cellular receptor CAR. J Virol. 1999;73:9508–9514. doi: 10.1128/jvi.73.11.9508-9514.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Gal La Salle G, Robert J J, Berrard S, Ridoux V, Stratford-Perricaudet L D, Perricaudet M, Mallet J. An adenovirus vector for gene transfer into neurons and glia in the brain. Science. 1993;259:988–990. doi: 10.1126/science.8382374. [DOI] [PubMed] [Google Scholar]
- 23.Lemarchand P, Jaffe H A, Danel C, Cid M C, Kleinman H K, Stratford-Perricaudet L D, Perricaudet M, Pavirani A, Lecocq J P, Crystal R G. Adenovirus-mediated transfer of a recombinant human alpha 1-antitrypsin cDNA to human endothelial cells. Proc Natl Acad Sci USA. 1992;89:6482–6486. doi: 10.1073/pnas.89.14.6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leon R P, Hedlund T, Meech S J, Li S, Schaack J, Hunger S P, Duke R C, DeGregori J. Adenoviral-mediated gene transfer in lymphocytes. Proc Natl Acad Sci USA. 1998;95:13159–13164. doi: 10.1073/pnas.95.22.13159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis N, Fender P, Barge A, Kitts P, Chroboczek J. Cell-binding domain of adenovirus serotype 2 fiber. J Virol. 1994;68:4104–4106. doi: 10.1128/jvi.68.6.4104-4106.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maizel J V, Jr, White D O, Scharff M D. The polypeptides of adenovirus. I. Evidence for multiple protein components in the virion and a comparison of types 2, 7A, and 12. Virology. 1968;36:115–125. doi: 10.1016/0042-6822(68)90121-9. [DOI] [PubMed] [Google Scholar]
- 27.Mathias P, Wickham T, Moore M, Nemerow G. Multiple adenovirus serotypes use alpha v integrins for infection. J Virol. 1994;68:6811–6814. doi: 10.1128/jvi.68.10.6811-6814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLeskey S W, Ding I Y, Lippman M E, Kern F G. MDA-MB-134 breast carcinoma cells overexpress fibroblast growth factor (FGF) receptors and are growth-inhibited by FGF ligands. Cancer Res. 1994;54:523–530. [PubMed] [Google Scholar]
- 29.Miller C R, Buchsbaum D J, Reynolds P N, Douglas J T, Gillespie G Y, Mayo M S, Raben D, Curiel D T. Differential susceptibility of primary and established human glioma cells to adenovirus infection: targeting via the epidermal growth factor receptor achieves fiber receptor-independent gene transfer. Cancer Res. 1998;58:5738–5748. [PubMed] [Google Scholar]
- 30.Mittereder N, March K L, Trapnell B C. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nalbantoglu J, Pari G, Karpati G, Holland P C. Expression of the primary coxsackie and adenovirus receptor is downregulated during skeletal muscle maturation and limits the efficacy of adenovirus-mediated gene delivery to muscle cells. Hum Gene Ther. 1999;10:1009–1019. doi: 10.1089/10430349950018409. [DOI] [PubMed] [Google Scholar]
- 32.Philipson S, Stewart P L, Burnett R M. The molecular repertoire of adenovirus I. Berlin, Germany: Springer-Verlag; 1995. [Google Scholar]
- 33.Quantin B, Perricaudet L D, Tajbakhsh S, Mandel J L. Adenovirus as an expression vector in muscle cells in vivo. Proc Natl Acad Sci USA. 1992;89:2581–2584. doi: 10.1073/pnas.89.7.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roelvink P W, Lizonova A, Lee J G, Li Y, Bergelson J M, Finberg R W, Brough D E, Kovesdi I, Wickham T J. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol. 1998;72:7909–7915. doi: 10.1128/jvi.72.10.7909-7915.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roelvink P W, Mi Lee G, Einfeld D A, Kovesdi I, Wickham T J. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science. 1999;286:1568–1571. doi: 10.1126/science.286.5444.1568. [DOI] [PubMed] [Google Scholar]
- 36.Rogers B E, Douglas J T, Ahlem C, Buchsbaum D J, Frincke J, Curiel D T. Use of a novel cross-linking method to modify adenovirus tropism. Gene Ther. 1997;4:1387–1392. doi: 10.1038/sj.gt.3300541. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg S A, Blaese R M, Brenner M K, Deisseroth A B, Ledley F D, Lotze M T, Wilson J M, Nabel G J, Cornetta K, Economou J S, Freeman S M, Riddell S R, Oldfield E, Gansbacher B, Dunbar C, Walker R E, Schuening F G, Roth J A, Crystal R G, Welsh M J, Culver K, Heslop H E, Simons J, Wilmott R W, Habib N A, et al. Human gene marker/therapy clinical protocols. Hum Gene Ther. 1999;10:3067–3123. doi: 10.1089/10430349950016465. [DOI] [PubMed] [Google Scholar]
- 38.Rosenfeld M A, Yoshimura K, Trapnell B C, Yoneyama K, Rosenthal E R, Dalemans W, Fukayama M, Bargon J, Stier L E, Stratford-Perricaudet L, et al. In vivo transfer of the human cystic fibrosis transmembrane conductance regulator gene to the airway epithelium. Cell. 1992;68:143–155. doi: 10.1016/0092-8674(92)90213-v. [DOI] [PubMed] [Google Scholar]
- 39.Stevenson S C, Rollence M, White B, Weaver L, McClelland A. Human adenovirus serotypes 3 and 5 bind to two different cellular receptors via the fiber head domain. J Virol. 1995;69:2850–2857. doi: 10.1128/jvi.69.5.2850-2857.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Straus S E. Adenovirus infections in humans. In: Ginsberg H S, editor. The adenoviruses. New York, N.Y: Plenum Press; 1984. pp. 451–496. [Google Scholar]
- 41.Tillman B W, de Gruijl T D, Luykx-de Bakker S A, Scheper R J, Pinedo H M, Curiel T J, Gerritsen W R, Curiel D T. Maturation of dendritic cells accompanies high-efficiency gene transfer by a CD40-targeted adenoviral vector. J Immunol. 1999;162:6378–6383. [PubMed] [Google Scholar]
- 42.Tomko R P, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walters R W, Grunst T, Bergelson J M, Finberg R W, Welsh M J, Zabner J. Basolateral localization of fiber receptors limits adenovirus infection from the apical surface of airway epithelia. J Biol Chem. 1999;274:10219–10226. doi: 10.1074/jbc.274.15.10219. [DOI] [PubMed] [Google Scholar]
- 44.Wang X, Bergelson J M. Coxsackievirus and adenovirus receptor cytoplasmic and transmembrane domains are not essential for coxsackievirus and adenovirus infection. J Virol. 1999;73:2559–2562. doi: 10.1128/jvi.73.3.2559-2562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watkins S J, Mesyanzhinov V V, Kurochkina L P, Hawkins R E. The 'adenobody' approach to viral targeting: specific and enhanced adenoviral gene delivery. Gene Ther. 1997;4:1004–1012. doi: 10.1038/sj.gt.3300511. [DOI] [PubMed] [Google Scholar]
- 46.Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637–643. doi: 10.1016/s1357-2725(99)00015-1. [DOI] [PubMed] [Google Scholar]
- 47.West M A, Bretscher M S, Watts C. Distinct endocytotic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J Cell Biol. 1989;109:2731–2739. doi: 10.1083/jcb.109.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickham T J, Carrion M E, Kovesdi I. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 1995;2:750–756. [PubMed] [Google Scholar]
- 49.Wickham T J, Lee G M, Titus J A, Sconocchia G, Bakacs T, Kovesdi I, Segal D M. Targeted adenovirus-mediated gene delivery to T cells via CD3. J Virol. 1997;71:7663–7669. doi: 10.1128/jvi.71.10.7663-7669.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wickham T J, Mathias P, Cheresh D A, Nemerow G R. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell. 1993;73:309–319. doi: 10.1016/0092-8674(93)90231-e. [DOI] [PubMed] [Google Scholar]
- 51.Wickham T J, Roelvink P W, Brough D E, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 52.Wickham T J, Segal D M, Roelvink P W, Carrion M E, Lizonova A, Lee G M, Kovesdi I. Targeted adenovirus gene transfer to endothelial and smooth muscle cells by using bispecific antibodies. J Virol. 1996;70:6831–6838. doi: 10.1128/jvi.70.10.6831-6838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickham T J, Tzeng E, Shears II L L, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeh P, Perricaudet M. Advances in adenoviral vectors: from genetic engineering to their biology. FASEB J. 1997;11:615–623. doi: 10.1096/fasebj.11.8.9240963. [DOI] [PubMed] [Google Scholar]
- 55.Zabner J, Freimuth P, Puga A, Fabrega A, Welsh M J. Lack of high affinity fiber receptor activity explains the resistance of ciliated airway epithelia to adenovirus infection. J Clin Investig. 1997;100:1144–1149. doi: 10.1172/JCI119625. [DOI] [PMC free article] [PubMed] [Google Scholar]