Abstract
Objectives
Antimicrobial resistance is a growing concern and claims over 1 million lives per year. The discovery of new antimicrobial drugs is expensive and often generates low profitability, with very low success rates. One way to combat this is by the improvement of known antimicrobials, such as antimicrobial peptides (AMPs). The aim of this study was to improve the antimicrobial activities of two known AMPs, UyCT3 and indolicidin, with the use of peptide libraries and growth curves.
Methods
Peptide permutation libraries were synthesized for two AMPs, indolicidin and UyCT3, which included 520 peptides. These peptides were subsequently tested against MG1655-K12, to which subsequent peptide design was performed, then tested against three clinically Gram-negative relevant drug-resistant isolates. Best-performing candidates were subjected to a haemolysis assay for toxicity validation.
Results
Single amino acid permutations of UyCT3 and indolicidin were sufficient to inhibit growth of MG1655-K12, and subsequent generations of peptide design were able to inhibit growth of clinical isolates at concentrations as low as 5 µM. Our best-performing AMP, UyCT3I5A, W6Y, K10I, F13I, was not seen to be toxic towards sheep RBCs.
Conclusions
The efficacy of the AMPs improved with the use of our peptide library technology, whereby an AMP was found that inhibited bacterial growth of clinical Gram-negative isolates 4-fold better than its WT counterpart.
Introduction
The rise of antimicrobial resistant (AMR) bacteria is a serious global health issue;1 for example, in 2018 in the USA approximately 26% of bacterial infections were resistant to first-line treatments such as amoxicillin or combinatory treatments of amoxicillin/clavulanic acid.2 An estimated 1.27 million deaths were attributed to AMR infections in 2019, with this number projected to rise to 10 million deaths by the year 2050.3 The development of new antibiotics and novel treatment options is an important component of the fight against AMR, but is scientifically challenging and time consuming. Unfortunately, antibiotic drug development has entered a relatively dry discovery pipeline since the 1980s, with fewer antimicrobial agents undergoing clinical trials in the following decades.4,5 As such, there is a desperate need for new biomolecules targeting pathogenic bacteria.
Antimicrobial peptides (AMPs) have emerged as a promising type of new antimicrobials.6 Since their discovery in 1928, AMPs have been increasingly discussed in the literature and subjected to clinical trials as key antimicrobial agents. The first FDA-approved AMP was nisin, a peptide used primarily as a food preservative. However, wide use of AMPs as antimicrobials has been limited by their toxicity to host cells, as well as other drawbacks such as manufacturing costs and time for synthesis.5 Despite these challenges in translating in vivo antimicrobial effects into prescribable therapeutics, seven AMPs have been granted FDA approval: gramicidin, daptomycin, colistin, vancomycin, ortivancin, dalbavancin and telavancin. Moreover, numerous AMPs are currently in clinical trials, underlining interest in these biomolecules as antimicrobials.5
AMPs are produced by plants, animals, fungi and bacteria. These naturally occurring AMPs thus provide a wealth of raw material for the development of new therapeutics. However, since naturally derived AMPs have evolved against the pathogens faced by organisms in their own environments, they may not have optimal activity against infectious agents of concern to humans. Thus, it is likely that naturally occurring AMPs can be further optimized.
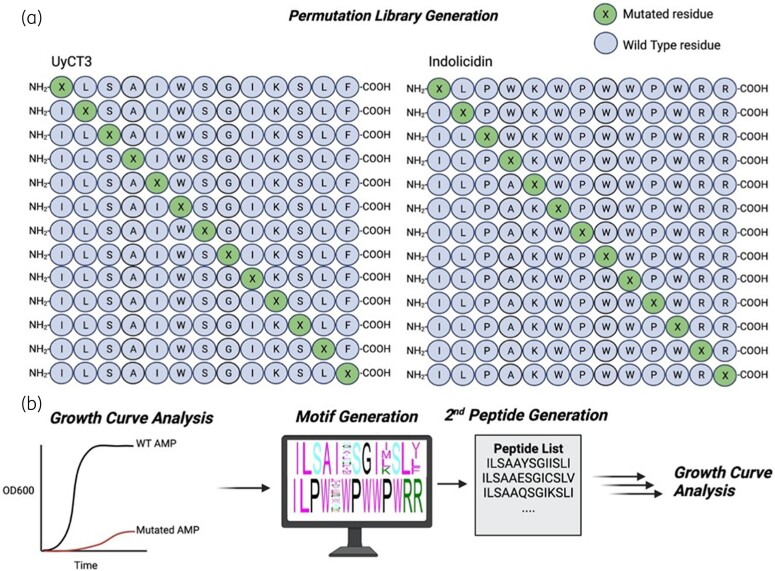
As a new systematic strategy for AMP development, we used in vitro evolution to enhance the activity of two established AMPs, indolicidin and UyCT3, against the opportunistic pathogen Escherichia coli (Figure 1). This method involves systematically permutating amino acids within defined peptide sequences, and selecting substitutions that improve activity.
Figure 1.
Methodology for systematic evolution of antimicrobial peptides. (a) Permutations are performed by the amino acid substitution of single residues on WT peptides, known as the first generation. (b) Said peptides were tested for growth inhibition against MG1655. Second peptide generations were made, and motifs were generated for further growth inhibition testing. Final candidates were tested for haemolytic activity.
Indolicidin is a tridecapeptide (ILPWKWPWWPWRR) that was isolated from the cytoplasmic granules of bovine neutrophils.7 It has broad and potent antimicrobial activity against both Gram-negative and Gram-positive bacteria. It is thought to bind abasic sites of DNA and interfere with the recruitment of DNA machinery, cause DNA filamentation, and to interact with DNA topoisomerase I.8–10 In addition, it has been proposed that indolicidin may also inhibit bacterial proteases.11 This peptide is unusual in that it does not take on the typical α-helical or β structure that is common in other cationic peptides. With its high percentage of tryptophan residues, it adopts a wedge conformation with its hydrophobicity in the bacterial membrane. Although indolicidin has substantial therapeutic potential, it has high toxicity towards mammalian RBCs, yielding a high haemolytic activity.12
UyCT3 is an α-helical peptide derived from the venom of the scorpion Urodacus yaschenkoi.13 The full-length protein comprises 68 amino acids, but the subsequence ILSAIWSGIKSLF is known to confer its antimicrobial activity. It is a member of the non–disulphide-bridged peptide family as well as the small antimicrobial peptide group.14 The mechanism of action for UyCT3 is debated in the literature but it is proposed to create a helical channel in the membrane of the bacterium, causing autolysis and ultimately bacterial death.14
We generated systematic amino acid permutation libraries based on the sequences of indolicidin and UyCT3 to evolve AMPs with increased therapeutic potential. The growth rates of selected AMPs and their analogues were assessed in both laboratory and clinically derived strains of E. coli. We additionally evaluated the haemolytic activity of peptides displaying the highest growth inhibition.
Materials and methods
Strains
The standard laboratory E. coli strain K-12 (MG1655) was used for screening of peptide libraries. Selected AMPs were tested against clinically derived E. coli strains pb3, pb15 and pb35 that were provided by the Zhanel Laboratory from the University of Manitoba and characterized previously.15 The three strains were chosen based on their differing sources rather than their MDR or mutation profiles. Their AMR profiles are shown in Table 1 with relevant MIC values and site-specific mutations obtained from Basra et al.15 These strains were chosen because they originated from differing sources (blood, urine, respiratory). All strains were grown in Mueller–Hinton broth (MHB) purchased from Sigma Aldrich.
Table 1.
Tabulated information on clinical strains
| Strain | Infection source | Drug resistance | Chromosomal mutationsa | |||
|---|---|---|---|---|---|---|
| CIP MIC, ng/μL | CAZ MIC, mg/L | gyrA | gyrB | parC | ||
| pb3 | Urinary tract infection | 15.6 | 4 | D678E, A828S | E185D | D475E |
| pb5 | Respiratory | 32 000 | 0.5 | S83L, D87N, D678E, A828S | S492N, A618T, E656D | S80I, A108V |
| pb15 | Blood | 32 000 | 128 | S83L, D87N, A828S | A618T | S80I, E84V, A192V, A471G, D475E |
CAZ, ceftazidime; CIP, ciproflaxin.
aBolded mutations refer to mutations that lead to resistance phenotype.
Peptide synthesis
UyCT3 and indolicidin permutation libraries were synthesized. Each residue was mutated to one of the remaining 19 amino acids, while keeping the rest of the AMP sequence constant. This led to the synthesis of 520 unique peptides, 260 each for indolicidin and UyCT3.
Peptides were synthesized using solid-phase Fmoc [N-(9-fluorenyl) methoxycarbonyl] chemistry using a ResPep SL peptide synthesizer (Intavis) at a 2 μmol scale using Rink-NH2 resin following procedures described previously.16 All amino acid derivates and activator were purchased from p3Bio Systems. To allow for quantification, peptides were synthesized with a C-terminal tryptophan separated by a 6-aminohecanoic acid (6-ahx) linker. After synthesis, peptides were liberated from the resin, and protecting groups were cleaved using an acidic cleavage solution (95% trifluoroacetic acid, 3% tri-isopropylsilane, 2% water). Cold diethyl ether (−20°C) was used to precipitate and wash the peptides of residual cleavage solution. Once dried, pellets were dissolved in 1X PBS containing 4% acetic acid. Peptides were brought up to pH 7 using 5 M NaOH and quantified by A280 and molar extinction coefficients calculated by ProtParam Expasy.
Growth curves
Overnight cultures of bacteria were grown in MHB with continuous shaking (150 rpm) at 37°C. The overnight cultures were then diluted 1:100 in MHB and experimental peptides were tested at 40 µM in triplicate in a 96-well plate, with the lid left on to prevent evaporation. The OD600 was read every 30 min for 24 h with continuous shaking using Gen5 software with a BioTek plate reader. For serial dilution testing of AMPs, a 2-fold dilution series was performed to test the range of 40 µM to 5 µM in triplicates.
To generate growth curves, the OD600 over the 24 h period was plotted. All y-values in the linear portion of the log phase were log2-transformed and plotted against time (min). A LINEST function was used in Microsoft Excel to output the slope and standard error of regression. All growth rates were normalized to the ‘No AMP’ control treatment.
Second-generation peptide clustering
The list of predicted second-generation AMPs was clustered into 100 individual groups that represent the greatest sequence diversity and were used for evaluation. Each set of peptides was ranked in each cluster by varying characteristics: charge, charge density, isoelectric point (pI), instability index, aromaticity, aliphatic index, Boman index and hydrophobicity. The top-ranking peptide in each group was compiled into a new list of 100 AMPs and used for further testing.
Haemolytic assay
A haemolytic assay was performed as described.17 Sheep RBCs (sRBCs) were purchased in a 10% suspension from Fisher Scientific (Cat No. 0855876). UyCT3WT, UyCT3I5A, W6Y, K10I, F13I and UyCT3I5A, W6E, K10C, F13V were prepared at a concentration of 400 µM in PBS buffer. 1% Triton X-100 was used as a positive control, PBS as a negative control, and 0.66% DMSO was used as the vehicle control. We added 75 µL of AMP to 75 µL of sRBCs in a flat, black, clear bottom 96-well plate. The plate was shaken at 37°C for 1 h. The plate was spun at 1000 × g for 15 min to collect the pellet, and supernatant was taken and diluted 1:10 with PBS buffer. The OD414 was read and percentage haemolysis was determined by the following formula: (sample absorbance − average aborbance of negative control)/(average aborbance of positive control − average aborbance of negative control). Absorbances were taken in triplicates.
Pepwheel visualization
Amino acid residue formations for UyCT3WT, UyCT3I5A, W6Y, K10I, F13I and UyCT3I5A, W6E, K10C, F13V were visualized using the EMBOSS Pepwheel tool (EMBOSS; http://emboss.bioinformatics.nl/cgi-bin/emboss/pepwheel), as done previously.18
Statistical analysis
Growth rate data derived from the AMP permutations were manipulated into a 13 × 19 matrix with the WT AMP on the y-axis and amino acid mutations along the x-axis. These two matrices were subjected to peptide specificity analysis (PeSA) to create motifs for a second-generation of AMP synthesis .19 A motif is provided based on a threshold, which is relative to the positive control (WT AMP). A threshold of 2 SDs above the mean ‘WT AMP’ score was set. Mutations that elicit a score that is 2 SDs above the ‘WT AMP’ score were used for motif production to create the second generation of AMPs. Statistical analysis was performed using Graphpad Prism (version 9.0) with independent Student’s t test. Mean values of experimental AMPs were tested against mean WT AMP growth rates. Statistical significance is denoted by asterisks: * denotes P value <0.05, ** denotes P value <0.01, and *** denotes P value <0.001.
Results
Effect of single amino acid mutations on growth inhibition
The peptide sequences of indolicidin and UyCT3 were permutated at each residue, for a total of 520 unique peptides (260 for indolicidin, 260 for UyCT3). Each of these peptides was then tested against E. coli WT strain MG1655 (K12). Each peptide was initially tested at 40 µM, because this is a typical MIC of most AMPs.13 Peptides were deemed stable throughout experimental conditions by monitoring degradation by SDS-PAGE using a Tris-Tricine gel necessary for peptide resolution.20
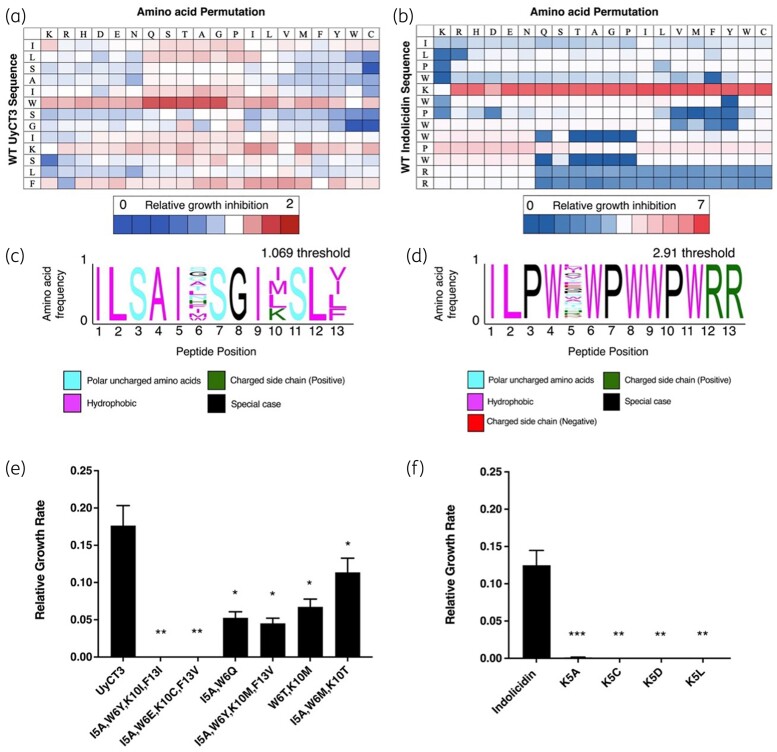
Figure 2 shows data from the initial peptide permutation screens of indolicidin (Figure 2a) and UyCT3 (Figure 2b). From the UyCT3 screen, 118 peptide mutations decreased bacterial growth, whereas 129 peptides increased bacterial growth compared with WT UyCT3. For indolicidin, 125 peptide mutations decreased bacterial growth, whereas 122 peptides increased bacterial growth compared with WT indolicidin.
Figure 2.
Amino acid permutations show increased antimicrobial activity. (a) Growth rate detection through single amino acid permutation of UyCT3. (b) Growth rate detection through single amino acid permutation of indolicidin. Relative growth inhibition is shown with each respective bar. Blue coloration represents mutations that did not result in any growth inhibition, whereas red represents mutations that performed better than the WT peptide. Any boxes coloured white represent inhibition matching the WT. All points are normalized to the substitution that results in the WT AMP (showed in bolded boxes). Motifs are shown for (c) UyCT3 and for (d) indolicidin. (e, f) Peptides made from motif for UyCT3 and indolicidin, respectively, that demonstrated significant growth inhibition of MG1655 at 40 μM. All growth rates were normalized to ‘No AMP treatment’, and statistical comparisons were with the WT peptide. Error bars show the standard error of regression. *P value <0.05, **P value <0.01, ***P value <0.001.
PeSA software was used to generate a sequence motif that represents all AMP mutations that increased the potency of either UyCT3 (Figure 2c) or indolicidin (Figure 2d) peptides; the threshold cut-off for the motif was based on +2 SDs above the ‘WT AMP’ score (2.91 and 1.069 for indolicidin and UyCT3, respectively). Many of the residues found in the original AMP sequence are held constant in the motifs, but a large array of amino acid families can also be identified.
Clustering of candidate AMPs using sequence motifs
To generate the second generation of peptides, PeSA was used to output every combination of peptide sequences that are encoded within each motif. This resulted in a total of 20 and 1440 peptides for indolicidin and UyCT3, respectively. Given the high number of combinatorial peptides from UyCT3, the 1440 peptides were clustered into 100 discrete groups for testing; these clusters were ranked based on peptide characteristics (charge, charge density, pI, instability, aromaticity, Boman index and hydrophobicity). One hundred peptides were generated from the clustering and tested further.
Growth inhibition by second-generation AMPs
A total of 120 peptides representing our second generation of AMPs were synthesized and tested against MG1655 at 40 µM, and their growth rates monitored. Only four peptides from second-generation indolicidin showed growth inhibition (Figure 2f). For UyCT3, 6 of the 100 predicted AMPs significantly decreased growth compared with UyCT3. Notably, these peptides differed from the first generation of peptides tested (Figure 2e) as they contain at least one or more amino acid mutations and are not derived directly from the WT sequence. The growth rates of the 120 peptides and 20 peptides are shown in Table S1 (available as Supplementary data at JAC-AMR Online) for UyCT3 and Table S2 for indolicidin. The peptides used further in this study are shown in Table 2.
Table 2.
AMP candidates from the second-generation screen
| Peptide name | Sequence |
|---|---|
| IndoK5A | ILPWAWPWWPWRR |
| IndoK5C | ILPWCWPWWPWRR |
| IndoK5D | ILPWDWPWWPWRR |
| IndoK5L | ILPWLWPWWPWRR |
| UyCT3I5A, W6Y. K101, F131 | ILSAAYSGIISLI |
| UyCT3I5A, W6E, K10C, F13V | ILSAAESGICSLV |
| UyCT3I5A, W6Q | ILSAAQSGIKSLF |
| UyCT3I5A, W6Y, K10M, F13V | ILSAAYSGIKSLF |
| UyCT3w6T. K10M | ILSAITSGIMSLF |
| UyCT3I5A W6M, K10T | ILSAAMSGITSLF |
Growth inhibition of clinical strains by second-generation AMPs
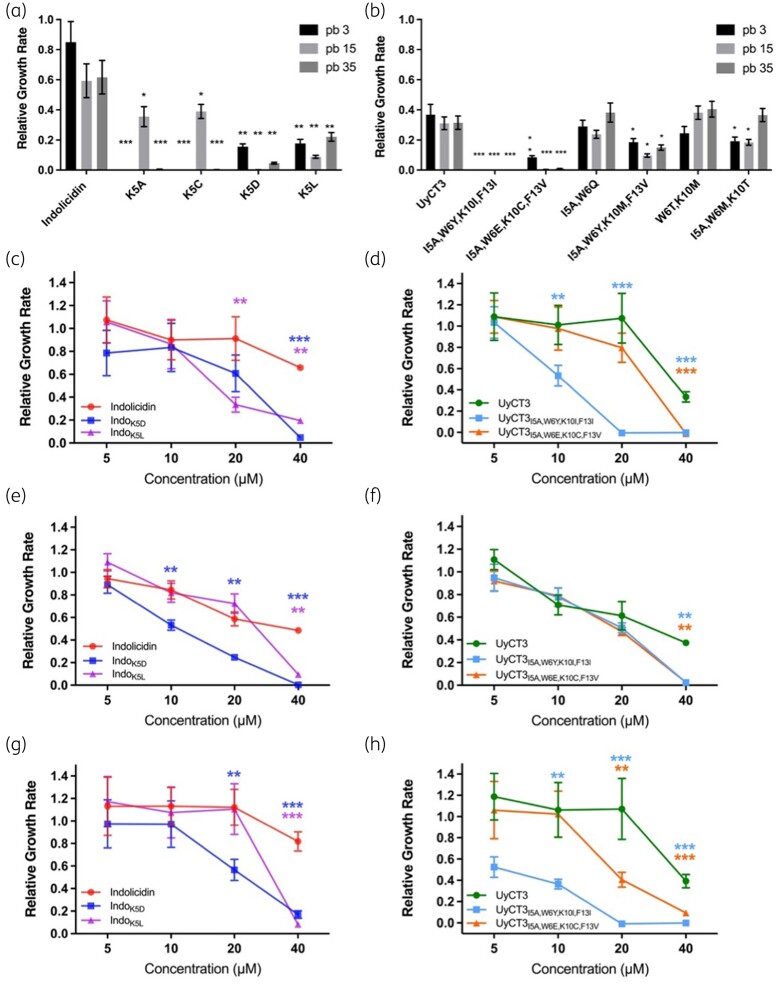
To give insight into the clinical relevance of our chosen peptides, three strains of E. coli were chosen (AMR profiles shown in Table 1). Notably, the indolicidin second-generation AMPs were found to display a broad range of effects on these clinical strains (Figure 3a). Specifically, there was greater effect of IndoK5A and IndoK5C against strains pb3 and pb35, with a reduced but still significant effect on pb15. The indolicidin-derived AMPs IndoK5D and IndoK5L gave an equivalent response across all three strains and were chosen for further characterization. For UyCT3-derived AMPs, only three out of the nine peptides significantly decreased growth across all three strains, with UyCT3I5A, W6Y, K10I, F13I and UyCT3I5A, W6E, K10C, F13V displaying significant growth inhibition (P < 0.01) (Figure 3b).
Figure 3.
AMPs show broad inhibition across three clinical strains. AMPs shown in Table 2 originating from (a) indolicidin and (b) UyCT3 tested against strains pb3 (black), pb15 (light grey) and pb35 (dark grey). All AMPs tested at 40 μM. Serial dilutions were performed for pb35, pb15 and pb3 for indolicidin variants (c, e, g) and UyCT3 variants (d, f, h), respectively. All growth rates were normalized to ‘No AMP treatment’, and statistical comparisions were with the WT peptide. Error bars show the standard error of regression. *P value <0.05, **P value <0.01, ***P value <0.001.
Dose–response of candidate AMPs
Serial dilutions of AMPs were tested for growth inhibition on clinical strains (Figure 3c–h). IndoK5L inhibited pb35 and pb3 strains significantly at 20 µM (Figure 3c and g). In contrast, IndoK5D significantly inhibited bacterial growth at concentrations down to 10 µM in pb15 (Figure 3e) compared with IndoWT. IndoWT inhibited pb35 and pb15 in a concentration-dependent manner, whereas pb3 growth was not inhibited across the concentrations tested. IndoK5D was the most promising derivative since it inhibited strains pb15 at 10 µM and pb3 at 20 µM.
Across all three tested strains, UyCT3 and its derivates displayed antimicrobial activity. UyCT3WT was inhibitory at 40 µM for pb35 (Figure 3d) and pb3 (Figure 3h) but showed a linear range of inhibition for pb15 (Figure 3f). UyCT3I5A, W6Y, K10I, F13I showed significant inhibition of pb35 (Figure 3d) and pb3 (Figure 3h) at concentrations greater than or equal to 10 µM. All three UyCT3 tested peptides had a similar response to pb15 (Figure 3f), with UyCT3I5A, W6Y, K10I, F13I and UyCT3I5A, W6E, K10C, F13V inhibiting bacterial growth significantly at 40 µM. UyCT3I5A, W6Y, K10I, F13I was the most promising derivative due to its ability to inhibit growth at concentrations of 5–10 µM of most strains.
α-Helical mapping of UyCT3 and derivatives
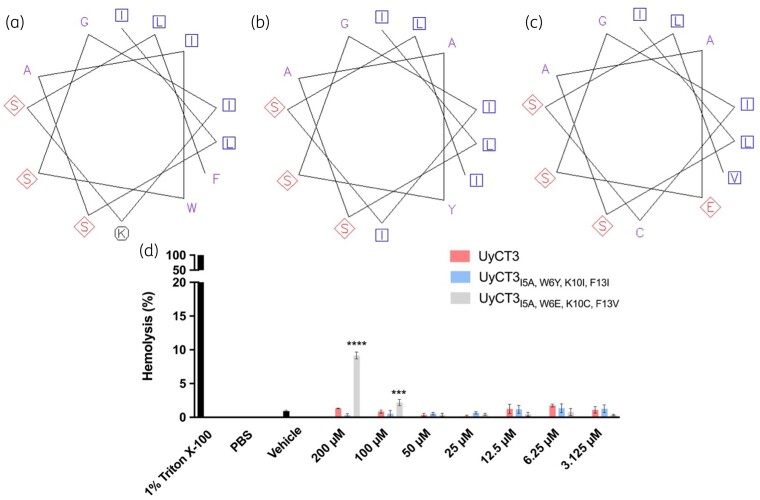
UyCT3WT, UyCT3I5A, W6Y, K10I, F13I and UyCT3I5A, W6E, K10C, F13V were further investigated to explore why these peptide derivatives display a high level of inhibition of bacterial growth compared with UyCT3WT. The EMBOSS Pepwheel was used to visualize the formation of amino acids in our peptide sequence to identify regions of hydrophobicity and hydrophilicity. Aliphatic residues are marked with blue squares, hydrophilic residues are shown in red diamonds, positively charged residues with black octagons, and hydrophobic residues in purple font. With the mutations made to create the UyCT3 derivative peptides, residues are replaced with those that have aliphatic characteristics with branched side groups (Figure 4a–c).
Figure 4.
UyCT3I5A, W6Y, K10I, F13I results in a more amphipathic peptide than UyCT3WT, with similar haemolysis properties. Pepwheels are shown for (a) UyCT3WT, (b) UyCT3I5A, W6Y, K10I, F13I and (c) UyCT3I5A, W6E, K10C, F13V. Aliphatic residues are marked with blue squares, hydrophilic residues are marked in red diamonds, positively charged residues with black octagons, and hydrophobic residues in purple font. Images generated using EMBOSS Pepwheel software. (d) Haemolysis of sRBCs was conducted with UyCT3WT and its variants.
Haemolytic activity of UyCT3 and its variants
As a preliminary screen for cytotoxicity, haemolytic activity of UyCT3, UyCT3I5A, W6Y, K10I, F13I and UyCT3I5A, W6E, K10C, F13V was tested against sRBCs (Figure 4d). HC50 values (i.e., the concentration of peptide that lyses 50% red blood cells) were found for UyCT3 against human erythrocytes at 20 μM21 but to cast a wide net, concentrations of 200 μM to 3.125 μM were used for the assay across all three peptides. At 200 μM, UyCT3I5A, W6E, K10C, F13V showed close to 10% haemolysis activity, whereas UyCT3 and UyCT3I5A, W6Y, K10I, F13I showed less than 5%.
Discussion
Although there have been many advances in antimicrobial therapeutics and combatting AMR in the last decade, there is still a need to find new therapeutics active against AMR pathogens. The process of finding new antibiotics is costly, as most are found through natural sources (plants, fungi, etc.).22 Here, we explored the use of peptide permutation arrays to optimize naturally occurring AMPs. Through permutation of two known AMPs, indolicidin and UyCT3, we developed four AMPs that inhibited the growth of E. coli with more potency than their WT counterparts.
Overnight cultures were grown and diluted 1:100 to allow for the bacterium to grow from lag phase into log phase. Growth rate, which is determined from the log phase, was the metric used throughout all growth curves to interpret effects due to added peptides. Had more bacteria been present at the start of the growth curves, the cells would have already been entering log phase or already in it, which would not allow for the best quantification of growth rate.
Mutating position K5 of indolicidin to any other amino acid showed an increase in antimicrobial activity (Figure 2b). Often, mutations of lysine to arginine do not cause drastic effects on peptide activity because both amino acids carry a positive charge.23 Though the nature of most AMPs is cationic, there is a preference for guanidinium groups in arginine rather than the amine group observed in lysine.23 Another study performed permutations on indolicidin and found that IndoK5A resulted in a reduced MIC, but increased haemolytic activity.24
UyCT3 permutation analysis showed important residues for bacterial growth inhibition. Of particular importance, position W6 had the most leniency for mutations. When mutating this position to either glutamine, serine, threonine, alanine or glycine, growth inhibition increased (Figure 2a). These amino acid mutations result in more amphipathic AMPs, which might perform better at penetrating bacterial membranes.
UyCT3I5A, W6Y, K10I, F13I is the most promising derivative of UyCT3 due to its ability to inhibit growth at concentrations of 5–10 µM of most strains (Figure 3). IndoK5D is the most promising derivative of indocilidin because it inhibits strains pb15 at 10 µM and pb3 at 20 µM (Figure 3). To investigate why UyCT3I5A, W6Y, K10I, F13I might be inhibiting bacterial growth better than WT UyCT3, we plotted the α-helix conformation using the EMBOSS Pepwheel (Figure 4a–c). EMBOSS Pepwheel visualization allows us also to look at the distribution of amino acid residues in the context of peptide structure.25 Interestingly, mutations in UyCT3I5A, W6Y, K10I, F13I and UyCT3I5A, W6Y, K10I, F13I result in a more hydrophobic region on one side versus the WT UyCT3 peptide. Accompanied by the abundance of serine residues on the opposite side, this leads to the creation of a more amphipathic AMP. Because UyCT3 is proposed to create a helical membrane in the channel of the bacterium, such mutations may encourage channel formation leading to loss of cell rigidity.14,21
It is also important to look at the impact of permutation on the oligomerization of AMPs in solution. Figure 4(a) shows UyCT3WT, which has a solvent-exposed lysine residue. Figure 4(b) shows the mutated AMP, UyCT3I5A, W6Y, K10I, F13I, which loses the lysine residue. This loss of positive charge might allow for better oligomerization due to no positive charge repulsions between peptide structures in pore formation. This is termed coulomb repulsion, which is the repulsive force between two positive or two negative species.26 Coulomb repulsion is a driving force for the determination of biological peptide activity.
A haemolytic assay was performed to measure toxicity towards mammalian cells, shown in Figure 4(d). Haemolysis assays are implemented as a tool for the viability of AMPs as therapeutics, as it allows for the screening of toxic compounds against mammalian cells.27 At 200 µM, UyCT3I5A, W6E, K10C, F13V showed close to 10% haemolysis activity, whereas UyCT3 and UyCT3I5A, W6Y, K10I, F13I showed less than 5%. At clinically relevant concentrations, UyCT3 and UyCT3I5A, W6Y, K10I, F13I showed the least toxicity towards sRBCs. Even though UyCT3I5A, W6Y, K10I, F13I takes on a more amphipathic structure, it appears to have low affinity for mammalian membranes.
We have focused our efforts on activity against a Gram-negative bacterium, E. coli. It will also be interesting to investigate the effects of our novel AMPs against Gram-positive pathogens such as MRSA. Gram-positive bacteria have a substantially thicker peptidoglycan layer than Gram-negative bacteria, with a more negative charge overall.28 As previously discussed, most AMPs interact with negatively charged bacterial cell walls as an initial mechanism of action, but our best-acting AMP has a neutral charge.
We have optimized an AMP that is more potent than its WT counterpart at a 3-fold concentration difference, including against MDR clinical strains. This systematic AMP design was accomplished using a permutation library of a known AMP sequence. With the success of this approach in vitro, this application can be implemented to optimize additional AMPs, examining how systematic permutating and in vitro evolution of existing AMP sequences might lead to better antimicrobial activity.
Supplementary Material
Contributor Information
Ali Shukri, Institute of Biochemistry and Department of Biology, Carleton University, Ottawa, Ontario, Canada K1S 5B6.
Amanda C Carroll, Institute of Biochemistry and Department of Biology, Carleton University, Ottawa, Ontario, Canada K1S 5B6.
Ryan Collins, Institute of Biochemistry and Department of Biology, Carleton University, Ottawa, Ontario, Canada K1S 5B6.
Francois Charih, Department of Systems and Computer Engineering, Carleton University, Ottawa, Ontario, Canada K1S 5B6.
Alex Wong, Institute of Biochemistry and Department of Biology, Carleton University, Ottawa, Ontario, Canada K1S 5B6.
Kyle K Biggar, Institute of Biochemistry and Department of Biology, Carleton University, Ottawa, Ontario, Canada K1S 5B6.
Funding
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant to K.B. (grant no. RGPIN-2016-06151).
Transparency declarations
None to declare.
Supplementary data
Tables S1 and S2 are available as Supplementary data at JAC-AMR Online.
References
- 1. Murray CJL, Ikuta KS, Sharara F et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 2022; 399: 629–55. 10.1016/S0140-6736(21)02724-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anon . Adult outpatient treatment recommendations. 2017. https://www.cdc.gov/antibiotic-use/hcp/clinical-care/adult-outpatient.html
- 3. Salam M, Al-Amin M, Salam MT et al. Antimicrobial resistance: a growing serious threat for global public health. Healthcare (Basel) 2023; 11: 1946. 10.3390/healthcare11131946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen CH, Lu TK. Development and challenges of antimicrobial peptides for therapeutic applications. Antibiotics (Basel) 2020; 9: 24. 10.3390/antibiotics9010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dijksteel GS, Ulrich MMW, Middelkoop E et al. Review: lessons learned from clinical trials using antimicrobial peptides (AMPs). Front Microbiol 2021; 12: 616979. 10.3389/fmicb.2021.616979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Santos C, Rodrigues GR, Lima LF et al. Advances and perspectives for antimicrobial peptide and combinatory therapies. Front Bioeng Biotechnol 2022; 10: 1051456. 10.3389/fbioe.2022.1051456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Subbalakshmi C, Sitaram N. Mechanism of antimicrobial action of indolicidin. FEMS Microbiol Lett 1998; 160: 91–6. 10.1111/j.1574-6968.1998.tb12896.x [DOI] [PubMed] [Google Scholar]
- 8. Hsu C-H, Chen C, Jou M-L et al. Structural and DNA-binding studies on the bovine antimicrobial peptide, indolicidin: evidence for multiple conformations involved in binding to membranes and DNA. Nucleic Acids Res 2005; 33: 4053–64. 10.1093/nar/gki725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang K-Y, Lee T-Y, Kao H-J et al. dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res 2019; 47: D298–308. 10.1093/nar/gky1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marchand C, Krajewski K, Lee H-F et al. Covalent binding of the natural antimicrobial peptide indolicidin to DNA abasic sites. Nucleic Acids Res 2006; 34: 5157–65. 10.1093/nar/gkl667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huan Y, Kong Q, Mou H et al. Antimicrobial peptides: classification, design, application and research progress in multiple fields. Front Microbiol 2020; 11: 582779. 10.3389/fmicb.2020.582779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nan YH, Bang J-K, Shin SY. Design of novel indolicidin-derived antimicrobial peptides with enhanced cell specificity and potent anti-inflammatory activity. Peptides 2009; 30: 832–8. 10.1016/j.peptides.2009.01.015 [DOI] [PubMed] [Google Scholar]
- 13. Luna-Ramírez K, Quintero-Hernández V, Vargas-Jaimes L et al. Characterization of the venom from the Australian scorpion Urodacus yaschenkoi: molecular mass analysis of components, cDNA sequences and peptides with antimicrobial activity. Toxicon 2013; 63: 44–54. 10.1016/j.toxicon.2012.11.017 [DOI] [PubMed] [Google Scholar]
- 14. Almaaytah A, Albalas Q. Scorpion venom peptides with no disulfide bridges: a review. Peptides 2014; 51: 35–45. 10.1016/j.peptides.2013.10.021 [DOI] [PubMed] [Google Scholar]
- 15. Basra P, Alsaadi A, Bernal-Astrain G et al. Fitness tradeoffs of antibiotic resistance in extraintestinal pathogenic Escherichia coli. Genome Biol Evol 2018; 10: 667–79. 10.1093/gbe/evy030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wei R, Kaneko T, Liu X et al. Interactome mapping uncovers a general role for numb in protein kinase regulation. Mol Cell Proteomics 2018; 17: 2216–28. 10.1074/mcp.RA117.000114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Darnowski MG, Lanosky TD, Labana P et al. Armeniaspirol analogues with more potent Gram-positive antibiotic activity show enhanced inhibition of the ATP-dependent proteases ClpXP and ClpYQ. RSC Med Chem 2022; 13: 436–44. 10.1039/D1MD00355K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Biggar KK, Kotani E, Furusawa T et al. Expression of freeze-responsive proteins, Fr10 and Li16, from freeze-tolerant frogs enhances freezing survival of BmN insect cells. FASEB J 2013; 27: 3376–83. 10.1096/fj.13-230573 [DOI] [PubMed] [Google Scholar]
- 19. Topcu E, Biggar KK. PeSA: a software tool for peptide specificity analysis. Comput Biol Chem 2019; 83: 107145. 10.1016/j.compbiolchem.2019.107145 [DOI] [PubMed] [Google Scholar]
- 20. Schägger H. Tricine-SDS-PAGE. Nat Protoc 2006; 1: 16–22. 10.1038/nprot.2006.4 [DOI] [PubMed] [Google Scholar]
- 21. Luna-Ramírez K, Sani M-A, Silva-Sanchez J et al. Membrane interactions and biological activity of antimicrobial peptides from Australian scorpion. Biochim Biophys Acta 2014; 1838: 2140–8. 10.1016/j.bbamem.2013.10.022 [DOI] [PubMed] [Google Scholar]
- 22. Dutescu IA, Hillier SA. Encouraging the development of new antibiotics: are financial incentives the right way forward? A systematic review and case study. Infect Drug Resist 2021; 14: 415–34. 10.2147/IDR.S287792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cutrona KJ, Kaufman BA, Figueroa DM et al. Role of arginine and lysine in the antimicrobial mechanism of histone-derived antimicrobial peptides. FEBS Lett 2015; 589: 3915–20. 10.1016/j.febslet.2015.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smirnova MP, Kolodkin NI, Kolobov AA et al. Indolicidin analogs with broad-spectrum antimicrobial activity and low hemolytic activity. Peptides 2020; 132: 170356. 10.1016/j.peptides.2020.170356 [DOI] [PubMed] [Google Scholar]
- 25. Lamprecht A-L, Naujokat S, Margaria T et al. Semantics-based composition of EMBOSS services. J Biomed Semantics 2011; 2: S5. 10.1186/2041-1480-2-S1-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Norouzy A, Assaf KI, Zhang S et al. Coulomb repulsion in short polypeptides. J Phys Chem B 2015; 119: 33–43. 10.1021/jp508263a [DOI] [PubMed] [Google Scholar]
- 27. Greco I, Molchanova N, Holmedal E et al. Correlation between hemolytic activity, cytotoxicity and systemic in vivo toxicity of synthetic antimicrobial peptides. Sci Rep 2020; 10: 13206. 10.1038/s41598-020-69995-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cremin K, Jones B, Teahan J et al. Scanning ion conductance microscopy reveals differences in the ionic environments of gram positive and negative bacteria. Anal Chem 2020; 92: 16024–32. 10.1021/acs.analchem.0c03653 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.