Abstract
We have previously identified a functionally essential bulged stem-loop in the 3′ untranslated region of the positive-stranded RNA genome of mouse hepatitis virus. This 68-nucleotide structure is composed of six stem segments interrupted by five bulges, and its structure, but not its primary sequence, is entirely conserved in the related bovine coronavirus. The functional importance of individual stem segments of this stem-loop was characterized by genetic analysis using targeted RNA recombination. We also examined the effects of stem segment mutations on the replication of mouse hepatitis virus defective interfering RNAs. These studies were complemented by enzymatic and chemical probing of the stem-loop. Taken together, our results confirmed most of the previously proposed structure, but they revealed that the terminal loop and an internal loop are larger than originally thought. Three of the stem segments were found to be essential for viral replication. Further, our results suggest that the stem segment at the base of the stem-loop is an alternative base-pairing structure for part of a downstream, and partially overlapping, RNA pseudoknot that has recently been shown to be necessary for bovine coronavirus replication.
Mouse hepatitis virus (MHV), one of the best-characterized members of the coronavirus family, has a single-stranded, positive-sense RNA genome some 31 kb in length. Upon infection, the first two-thirds of this exceptional molecule is translated into an RNA-dependent RNA polymerase. Coronavirus RNA synthesis then proceeds by a unique and incompletely understood mechanism described by conflicting models (1, 16, 24, 44–46, 54, 55, 60). Initially, the genomic RNA becomes the template for, at the least, a full-length (negative-sense) antigenome. Further events produce a series of smaller, subgenomic RNAs of both polarities. The positive-sense subgenomic RNAs form a 3′ nested set, with each containing a 70-nucleotide (nt) leader that is identical to the 5′ end of the genome and is joined at a downstream site to a stretch of sequence identical to the 3′ end of the genome. The negative-sense subgenomic RNAs form a 5′ nested set and are roughly 1/10 to 1/100 as abundant as their positive-sense counterparts, with each possessing the complement of this arrangement, including a 5′ oligo(U) tract and a 3′ antileader (10, 47).
Many advances in investigating the mechanism of coronavirus RNA synthesis have been enabled by the discovery of defective interfering (DI) RNAs of MHV (29, 30, 52) and of other coronaviruses (5, 34, 39). DI RNAs are extensively deleted genomic remnants that replicate by using the RNA synthesis machinery of a helper virus, often interfering with viral genomic RNA replication. Studies of naturally occurring and artificially constructed DI RNAs, which can be transfected into helper virus-infected cells, have mapped cis-acting sequence elements from the genome that participate in replication and transcription. These deletion analyses have demonstrated that the minimal extent of the 3′ terminus of the MHV genome that is able to sustain DI RNA replication falls between 417 and 463 nt (17, 25, 28, 53). Notably, this includes a portion of the upstream nucleocapsid (N) gene as well as the entire 301-nt 3′ untranslated region (3′ UTR). Given this requirement, it was surprising when further study showed that the minimal tract of template required for negative-strand RNA synthesis is contained within just the last 55 nt at the 3′ end of the genome in addition to an as-yet-undetermined amount of poly(A) tail (26). This suggested that sequences upstream of the negative-strand promoter are required for positive-strand RNA synthesis, the initiation of which may thus require a circularizing interaction between the 5′ and 3′ termini of the viral genome (26).
Although a full-length infectious cDNA clone of MHV has not yet been attained, some avenues into coronavirus genetics have been made possible through the development of site-directed mutagenesis by targeted RNA recombination (20, 22, 31, 40, 42). This technique allows the incorporation of mutations into the coronavirus genome via RNA-RNA recombination between a synthetic donor RNA and the genome of a recipient virus that can be selected against. Recently, in attempting to replace the MHV 3′ UTR with its counterpart from the bovine coronavirus (BCoV) genome by targeted RNA recombination, we found that the two 3′ UTRs were fully interchangeable (12). Moreover, we discovered that a predicted bulged stem-loop secondary structure adjacent to the stop codon of the N gene is essential for viral replication (Fig. 1) (12). This 68-nt structure is composed of six stem segments, interrupted by five bulges, which range from 1 to 4 nt. Sequence comparison suggests that 8 of the 10 nt that are different between MHV and BCoV in this region form 4 covariant bp falling in two of the stem segments. Our previous molecular genetic analysis suggested that the base pairing, but not the primary sequence, of these 4 covariant bp is necessary for replication (12). In this report, we determined the functional significance of each of the putative stem segments, and we probed the RNA secondary structure of this region by chemical modification and enzymatic analysis. Our results point to a modification of the originally proposed stem-loop, and they suggest a relationship between this structure and an immediately downstream pseudoknot that has been recently described for the 3′ UTR of BCoV (56).
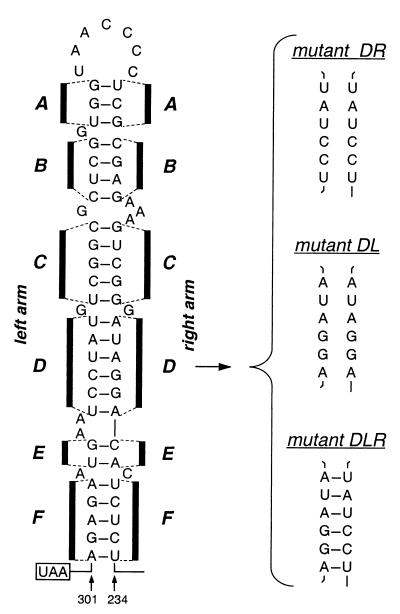
FIG. 1.
Mutational analysis of the proposed bulged stem-loop structure in the MHV 3′ UTR (12). Nucleotide numbering begins at the 3′ end of the genome, excluding the poly(A) tail; the N gene stop codon is boxed. The six stem segments of the structure are designated A through F. Shown at the right are strand replacement mutants for stem segment D.
MATERIALS AND METHODS
Virus and cells.
Growth of all stocks of MHV-A59 wild-type, mutant, and recombinant viruses was carried out in mouse 17 clone 1 (17Cl1) cells. All plaque titrations and plaque purifications were performed with mouse L2 cells. Spinner cultures of L2 cells were maintained for RNA transfection by electroporation, as described previously (32). The interspecies chimeric coronavirus fMHV was grown in feline FCWF cells (22).
Plasmid constructs.
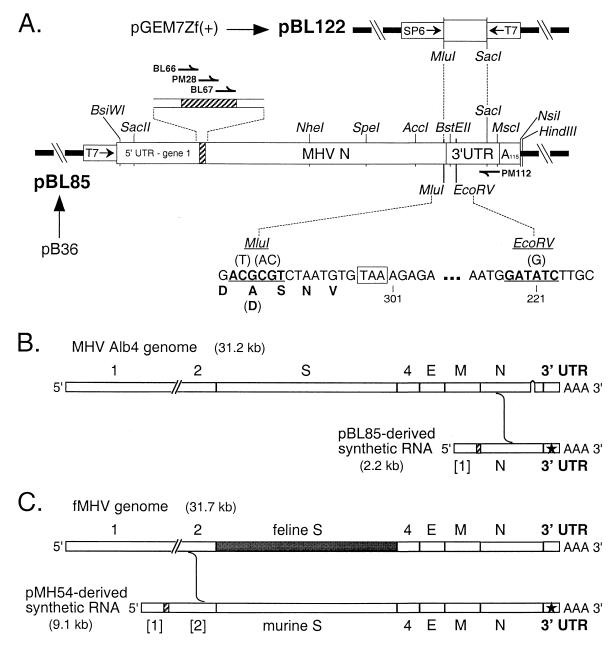
To enable more rapid construction of mutations in the 3′ UTR, convenient restriction sites were incorporated into plasmid pB36, a T7 transcription vector that encodes a replicating MHV DI RNA comprising the 5′ 467 nt of the MHV genome followed by a heterologous 48-nt linker, the entire N gene, the 3′ UTR, and a 115-residue poly(A) tail (32). A unique MluI site was created 9 through 14 nt upstream of the N gene stop codon, and a unique EcoRV site was created 10 through 15 nt downstream of the bulged stem-loop structure (see Fig. 2A). The MluI site was generated by PCR with a mutagenic primer coupled with a primer flanking the upstream AccI site. A second DNA fragment containing both the engineered MluI and EcoRV sites and the downstream SacI site was generated in two steps by splicing overlap extension-PCR (11). The former PCR product was digested with AccI and MluI, the latter was digested with MluI and SacI, and the two resulting fragments were inserted, via a three-way ligation, in place of the corresponding AccI-SacI region of pB36 to yield pBL85 (Fig. 2A).
FIG. 2.
Plasmid construction and schemes for mutant selection. (A) Parent plasmid pBL85, used for construction of vectors for stem segment mutants, encodes a DI RNA composed entirely of MHV components, except for a 48-nt linker region (hatched rectangle). pBL85 was derived from pB36 (32) by creation of an MluI site near the end of the N gene and an EcoRV site in the 3′ UTR. Portions of sequence containing these two sites, as well as the encoded carboxy terminus of the N protein, are shown; the original wild-type nucleotide and amino acid residues are indicated in parentheses. Nucleotide numbering begins at the 3′ end of the genome, excluding the poly(A) tail; the N gene stop codon is boxed. Plasmid pBL122, constructed for generation of the positive-strand transcript used for enzymatic and chemical probing, contains the MluI-SacI fragment derived from pBL85. All restriction sites shown, except for BstEII, are unique in the plasmids in which they appear. The positions of primers BL66, BL67, PM28, and PM112, used for RT-PCR analysis of negative-strand DI RNA, are indicated. (B) Scheme for construction of MHV mutants by targeted recombination using the temperature-sensitive and thermolabile mutant Alb4 as the recipient virus. In this case, recombinants generated by the indicated crossover event can be selected on the basis of having regained wild-type thermal stability. (C) Scheme for targeted recombination using the interspecies chimeric virus fMHV, which grows only in feline cells. In this case, recombinants generated by the indicated crossover event can be selected on the basis of having regained the ability to grow in murine cells. In both panels B and C, the star represents mutations in the 3′ UTR transduced from the synthetic donor RNA into the recipient genome.
A series of 15 vectors containing mutations in the bulged stem-loop, between the MluI and EcoRV sites (Fig. 1 and 3A), were then constructed by cassette mutagenesis. For left-arm mutants MBL, MDL, and MFL, a pair of mutation-containing oligonucleotides bounded by MluI and BstEII sites was ligated with BsiWI-MluI and BstEII-BsiWI fragments from pBL85. For right-arm mutants MAR, MBR, MCR, MDR, MER, and MFR, a pair of mutation-containing oligonucleotides bounded by BstEII and EcoRV sites was ligated with SpeI-BstEII and EcoRV-SpeI fragments from pBL85. For double-arm mutants MALR, MBLR, MCLR, MDLR, MELR, and MFLR, the SpeI-BstEII fragment from a left-arm mutant (or the MluI-BstEII oligonucleotide cassette) and the BstEII-HindIII fragment from its right-arm mutant counterpart were swapped with the corresponding region of pBL85 via a three-way ligation.
FIG. 3.
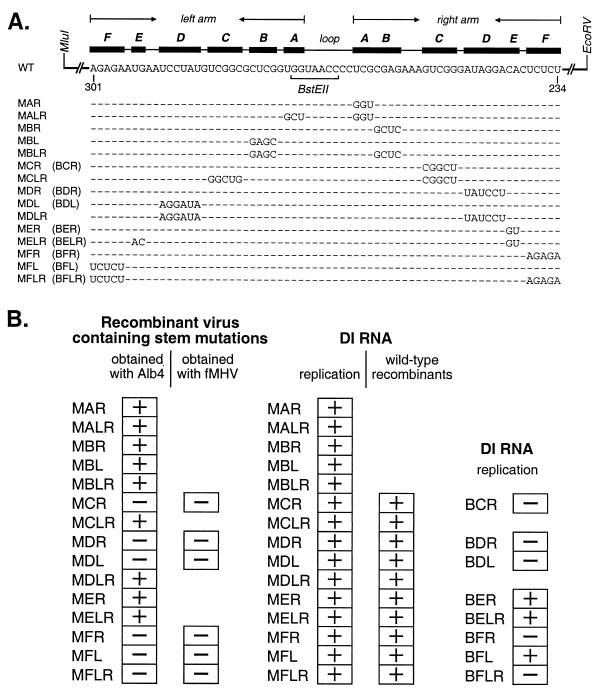
Composition and viability of stem segment mutants. (A) Nucleotide changes in stem segment mutants. The primary sequence of the wild-type (WT) bulged stem-loop structure is shown at the top, with stem segments labeled as in Fig. 1. Nucleotide numbering begins at the 3′ end of the genome, excluding the poly(A) tail. The loci of restriction sites MluI, EcoRV, and BstEII in the plasmid vector are indicated. For each mutant, only those bases that differ from the wild type are shown. The first letter of the name of each mutant (M or B) indicates the origin (MHV or BCoV, respectively) of the remainder of the 3′ UTR downstream of the stem-loop. The second letter (A, B, C, D, E, or F) indicates the stem segment. The final letters (L, R, or both) indicate which arm of the stem segment has been mutagenized. (B) Summary of the replicative ability of stem segment mutants in recombinant viruses or in DI RNAs. On the left are listed those donor RNA mutations for which viable recombinant viruses containing the mutations could (+) or could not (−) be obtained. For each mutation that scored negatively by the Alb4 targeted recombination strategy, an attempt was made to incorporate it into the viral genome by selection with fMHV. In the center are listed DI RNA replication results for stem mutations constructed in the background of the MHV 3′ UTR. For all replication results that were discrepant with the recombinant virus results, wild-type recombinant DI RNAs were detected by negative-strand-specific RT-PCR. On the right are listed DI RNA replication results for stem mutations constructed in the background of the BCoV 3′ UTR.
Another set of bulged stem-loop mutants were constructed in which the sequence downstream of the EcoRV site was derived from the BCoV 3′ UTR rather than the MHV 3′ UTR. This segment of the BCoV 3′ UTR, flanked upstream by a primer-generated EcoRV site and downstream by an MscI site, was produced through PCR amplification of template pBL34, which contains this region of the BCoV 3′ UTR (12). The EcoRV-MscI PCR fragment was inserted in place of the corresponding region of MCR, MFR, and MFLR to generate BCR, BFR, and BFLR, respectively. For the remaining constructs, the EcoRV-HindIII fragment of BFR was used to replace the same region in MDR, MDL, MER, MELR, and MFL, to generate BDR, BDL, BER, BELR, and BFL, respectively.
For the subset of the original 15 bulged stem-loop mutants that did not give rise to recombinant viruses, the same mutations were introduced into another vector, pMH54 (22). pMH54 is a T7 transcription vector containing the 5′ 467 nt of the MHV genome connected by a heterologous 72-nt linker to the 3′ 8.6 kb of the MHV genome, beginning at codon 28 of the hemagglutinin (HE) gene (see Fig. 2C) (22). For mutants MCR, MDR, MDL, MFR, MFL, and MFLR, the NheI-SacI fragment of the corresponding pBL85-derived construct was transferred to the larger vector through a three-way ligation with the SacII-NheI and SacI-SacII fragments from pMH54.
DNA manipulations were carried out by standard methods (41). The sequences of all junctions created by ligations and all segments generated by PCR were verified by dideoxy sequencing (43), using a modified T7 DNA polymerase kit (Sequenase; Amersham).
Targeted recombination.
Donor RNAs were transcribed from pBL85-derived plasmids truncated with HindIII. Mutations in these shorter synthetic donor RNAs were incorporated into the MHV genome by targeted recombination with the recipient virus Alb4 exactly as described previously (see Fig. 2B) (12, 20, 32). Negative results that were obtained with a subset of mutant donor RNAs by this method were then confirmed by attempting targeted recombination with donor RNAs transcribed from PacI-truncated vectors derived from pMH54. In this case, the interspecies chimeric coronavirus fMHV was used as the recipient virus (see Fig. 2C) (22). Briefly, confluent feline FCWF cells were infected with fMHV at a multiplicity of approximately 1 PFU per cell for 5 h at 37°C. Infected monolayers were then suspended by trypsin treatment, washed in calcium- and magnesium-free phosphate-buffered saline, and transfected with donor RNA by electroporation with two consecutive pulses at 960 μF and 0.3 kV using a Bio-Rad Gene Pulser. Infected and transfected cells were then plated onto monolayers of murine 17Cl1 cells. After 48 h of incubation at 37°C, progeny viruses were harvested and candidate recombinants were analyzed following two rounds of plaque purification on L2 cells.
Cytoplasmic RNA from infected 17Cl1 cell monolayers was purified either by a Nonidet P-40 gentle lysis method (18) or with Ultraspec reagent (Biotecx) per the manufacturer's instructions. Direct RNA sequencing was performed by a modification of a dideoxy chain termination procedure, using avian myeloblastosis virus reverse transcriptase (Life Sciences) (6, 38).
Radiolabeling of viral RNA and analysis of DI RNA replication.
Metabolic labeling of virus-specific RNA was carried out as previously described (12, 32). In brief, L2 cells in spinner culture were infected with wild-type MHV at a multiplicity of 1 PFU per cell. At 2 h postinfection, DI RNA was introduced into cells by electroporation, and cells were then plated onto a 20-cm2 monolayer of 17Cl1 cells, which was incubated at 37°C until the monolayer developed approximately 50% syncytia. Cells were starved for 2 h in Eagle's minimal essential medium containing 5% dialyzed fetal bovine serum and 1/10 of the normal phosphate concentration. Cells were then labeled for 2 h with [33P]orthophosphate in phosphate-free Eagle's minimal essential medium containing 5% dialyzed fetal bovine serum and 20 μg of actinomycin D (Sigma) per ml. Purified cytoplasmic RNA samples containing equal amounts of radioactivity were analyzed by electrophoresis on 1% agarose gels containing formaldehyde.
To analyze DI RNA-specific negative-strand RNA in infected and transfected cells, positive-sense primers BL66 (5′GGATCCAGATCGATCAGC3′), PM28 (5′TGATAAATGGCTTCCTAT3′), and BL67 (5′CCTATTTACATCCTAGGC3′), all specific for the heterologous (non-MHV) linker of pB36 (Fig. 2A), were used in seminested reverse transcription-PCR (RT-PCR) together with the negative-sense primer PM112 (5′CCATGATCAACTTCATTC3′), which is complementary to nt 18 to 35 of the 3′ UTR.
RNA substrate for structural probing.
An RNA substrate corresponding to the 3′ end of the N gene and almost the entire 3′ UTR was transcribed in vitro from SacI-truncated plasmid pBL122 (Fig. 2A). Synthesis of uncapped RNA was carried out with an SP6 polymerase transcription kit (Ambion) per the manufacturer's instructions. The resulting transcript was 264 nt long, containing 22 vector-derived (non-MHV) nt at its 5′ end followed by the 3′ 17 nt of the N gene and 225 nt of the 3′ UTR. Product RNA was treated with RNase-free DNase I (Ambion) and was purified by extraction twice with phenol-chloroform and twice with chloroform, followed by two precipitations from ethanol in the presence of 2 M ammonium acetate. Prior to enzymatic or chemical probing, RNA (in the relevant reaction buffer) was denatured by incubation at 65°C for 5 min and then allowed to renature by cooling slowly to room temperature in a beaker containing 200 ml of H2O initially at 65°C.
Enzymatic structural probing.
Ten micrograms of synthetic RNA substrate was denatured and renatured in 100 μl of 30 mM Tris HCl (pH 7.5)–20 mM MgCl2–300 mM KCl containing 10 μg of yeast tRNA. Aliquots (20 μl) were then incubated at 25°C for 40 min with 10 μg of yeast tRNA and 1, 5, 10, or 15 U of RNase T1 (Boehringer Mannheim); 0.0001, 0.001, or 0.01 U of RNase A (Boehringer Mannheim); or 0.05, 0.1, 0.3, or 0.5 U of RNase V1 (Pharmacia) (50). Subsequently, the enzyme-cleaved RNA was subjected to phenol-chloroform and chloroform extraction, followed by ethanol precipitation prior to primer extension analysis (9, 51).
Chemical modification.
For modification of RNA with dimethyl sulfate (DMS; Fluka), 10 μg of synthetic RNA was incubated in 200 μl of 80 mM sodium cacodylate (pH 7.2)–100 mM KCl–5 mM MgCl2 containing 0.5, 1.0, 1.5, or 2.0% DMS for 15 min at 25°C (9, 21). Reactions were quenched and precipitated by addition of 20 μl of 3 M NaOAc–10 μg of yeast tRNA–500 μl of ethanol.
For modification of RNA with 1-cyclohexyl-3-[2-morpholinoethyl] carbodiimide metho-p-toluene sulfonate (CMCT; Aldrich), 10 μg of synthetic RNA was incubated in 200 μl of 12.5 mM sodium borate (pH 8.1)–12.5 mM KCl–2.5 mM MgCl2 containing 4.2, 8.4, 12.6, or 16.8 mg of CMCT per ml for 15 min at 25°C (9, 21). Reactions were quenched and precipitated by addition of 20 μl of 3 M NaOAc–10 μg of yeast tRNA–600 μl of ethanol.
Primer extension analysis.
Primer PM165 (5′TCTATCTGTTATGACAGC3′; complementary to nt 199 to 216 of the MHV 3′ UTR) was 5′ end labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs) (21). Purified primer was then annealed to RNase-cleaved or chemically modified RNA at 90°C for 3 min and chilled on ice for 5 min. Primer extension was carried out in 7-μl reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM KCl, 8 mM MgCl2, 2 mM dithiothreitol, 0.85 mM (each) deoxynucleoside triphosphates, and 20 U of avian myeloblastosis virus reverse transcriptase (Life Sciences) at 42°C for 45 min (51). Samples were separated on a standard 6.0% polyacrylamide DNA sequencing gel containing 8 M urea. A sequence ladder was generated directly by dideoxy sequencing of synthetic substrate RNA using the same 5′-end-labeled primer (20).
RESULTS
Mutational analysis of each stem of the stem-loop structure.
To facilitate the construction of mutations in the bulged stem-loop in the 3′ UTR, restriction sites adjacent to this structure were designed in pB36, the plasmid used to generate donor RNA for targeted recombination (12, 32). Near the upstream boundary, we altered codons 450 and 451 of the N gene to create a unique MluI site (Fig. 2A). The first of these changes was coding silent, but the second mutated residue 451 of the N protein from aspartate to alanine. Based on previous mutagenesis studies, this change was expected to be phenotypically silent (B. Hsue and P. S. Masters, unpublished results). Beyond the downstream boundary of the stem-loop, a single base change was introduced into the same vector, creating a unique EcoRV site at nt 219 to 224 of the 3′ UTR (Fig. 2A). [Here and elsewhere in this paper, the coordinates used for the MHV 3′ UTR begin at the 3′ end of the genome, excluding the poly(A) tail.] The locus of this change was specifically chosen to not disrupt a proposed downstream pseudoknot structure (56). The resulting plasmid was designated pBL85.
To test whether the mutations in pBL85 would have any impact on the replication of MHV, targeted RNA recombination was carried out to transduce these mutations into the viral genome (Fig. 2B). Donor RNA transcribed from pBL85 was electroporated into cells infected with the thermolabile mutant Alb4, and recombinant viruses were selected as those able to form large plaques at 39°C. Two independent recombinants, Alb167 and Alb168, were isolated, and we confirmed by RNA sequencing that each contained the MluI and EcoRV mutations from the donor RNA as well as having replaced the 87 nt that were deleted in the Alb4 N gene (data not shown). Since Alb167 and Alb168 had completely wild-type phenotypes, this indicated that the mutations created for the two new restriction sites did not exert any obvious effect on the growth of MHV, and thus pBL85 would be an appropriate vector for construction of stem-loop mutants.
In a stepwise manner, we next genetically analyzed the functional role of each stem segment of the bulged stem-loop, designated A through F (Fig. 1). To explore whether base pairing or primary sequence of each stem segment plays a role in replication, we constructed single-arm mutants (MAR, MBR, MBL, MCR, MDR, MDL, MER, MFR, and MFL) in which each nucleotide of one arm of a stem segment was changed to its complement, thereby disrupting base pairing (Fig. 1 and 3A). To examine whether secondary structure, but not primary sequence, was important, we constructed double-arm mutants (MALR, MBLR, MCLR, MDLR, MELR, and MFLR) in which the two arms of a stem segment were exchanged, thereby restoring the base pairing (Fig. 1 and 3A). In the notation used for these mutants, the first letter (M or B) indicates the origin (MHV or BCoV) of the remainder of the 3′ UTR downstream of the stem-loop, the second letter (A through F) indicates the stem segment, and the final letters (L, R, or both) indicate which arm of the stem segment was replaced.
The functionality of each of these 15 stem segment mutations was tested by determining whether it was able to be incorporated into the MHV genome. In several independent targeted RNA recombination experiments with Alb4 as the recipient virus, at least one recombinant virus was selected, plaque purified, and characterized using MAR, MALR, MBR, MBL, MBLR, MCLR, MDLR, MER, or MELR donor RNA (Fig. 3B). Each of the resulting stem segment mutant viruses formed wild-type-sized plaques at 39°C. In all cases, the presence of the specific stem segment mutations and the repair of the Alb4 deletion were confirmed by direct RNA sequencing of cytoplasmic RNA isolated from cells infected with recombinant virus. Thus, the presence of any one of these nine sets of mutations had no apparent effect on the replication of MHV. In contrast, for MCR, MDR, MDL, MFR, MFL, and MFLR donor RNAs, no recombinant viruses could be selected in repeated trials using the Alb4 selection procedure (Fig. 3B). From this, we concluded that each of these sets of mutations is lethal to the virus, or at least, it results in a virus that is less fit than Alb4 (31).
To further extend this latter result, the mutations of MCR, MDR, MDL, MFR, MFL, and MFLR were transferred to a much larger donor RNA vector, pMH54, which encodes a pseudo-DI RNA containing the 3′ 8.6 kb of the MHV genome, beginning near the 5′ end of the HE gene (Fig. 2C) (22). This allowed us to carry out targeted recombination with the recipient virus fMHV, a recombinant coronavirus that harbors a chimeric MHV-feline infectious peritonitis virus spike gene and can infect feline, but not murine, cell lines (22). We have found that the use of fMHV as the recipient virus with pMH54-derived donor RNAs enables us to isolate extremely defective MHV mutants by selection for reacquisition of the ability to grow in murine cells (Fig. 2C) (L. Kuo and P. S. Masters, unpublished results). Using this selection with donor RNAs containing the six previously negative sets of mutations, we obtained a number of progeny viruses capable of growth in murine cells. However, analysis of these revealed that all contained the wild-type MHV 3′ UTR (Fig. 3B), indicating that they had been formed by double recombination events that incorporated the murine S gene but excluded the mutant 3′ UTRs from progeny genomes. This strongly implies that the MCR, MDR, MDL, MFR, MFL, and MFLR mutations are lethal to MHV.
In sum, the results from this genetic analysis indicated that stems A, B, and E are not necessary for viral replication, since mutations in these segments that disrupted primary sequence, secondary structure, or both did not impede viral growth. In contrast, the secondary structures of stems C and D, as well as both the primary sequence and the base pairing of stem F, appear to be essential to the role of the bulged stem-loop in viral replication.
Replication of DI RNAs containing stem segment mutations.
To seek corroborative support for the genetic results above, we metabolically labeled RNA in wild-type MHV-infected cells that were transfected with each of the mutant DI RNAs. Unexpectedly, cells transfected with all mutant DI RNAs, including MCR, MDR, MDL, MFR, MFL, and MFLR, synthesized, at least to some extent, an extra RNA species identical in size to the positive-control DI RNA (data not shown; summarized in Fig. 3B). This result, by itself, suggested that no portion of the bulged stem-loop was essential for DI RNA replication, in clear contradiction of the viral genetic data as well as of all our previous work (12).
To examine the apparently replicating DI RNA species of mutants in stem segments C through F, we carried out RT-PCR with primers specific for the heterologous linker sequence that is found in pB36 and its derivatives but not in the MHV genome. Negative-strand, rather than positive-strand, DI RNA was analyzed to reduce the background created by the large amount of RNA that was transfected into MHV-infected cells. A replicating DI RNA species would give rise to multiple rounds of negative-strand RNA synthesis, whereas a nonreplicating transfected RNA would be expected to yield, at most, a single round of negative-strand RNA synthesis.
First-strand cDNA from negative-strand RNA was thus reverse transcribed using the linker-specific primer BL66 (Fig. 2A). This was then amplified by seminested PCR with primer pair PM28 and PM112 followed by primer pair BL67 and PM112 (Fig. 2A). Analysis of the resulting PCR products showed that a large fraction of each could not be digested by EcoRV. This was consistent with the interpretation that the observed replicating DI RNA species did not correspond to the original transfected RNA (containing the engineered EcoRV site) but had arisen by acquisition of the wild-type MHV 3′ UTR by homologous recombination with helper virus. A similarly high rate of reversion via recombination with helper virus has been documented by Yu and Leibowitz (58) for DI RNAs containing mutations constructed in another region of the 3′ UTR. The basis for this phenomenon is currently unknown.
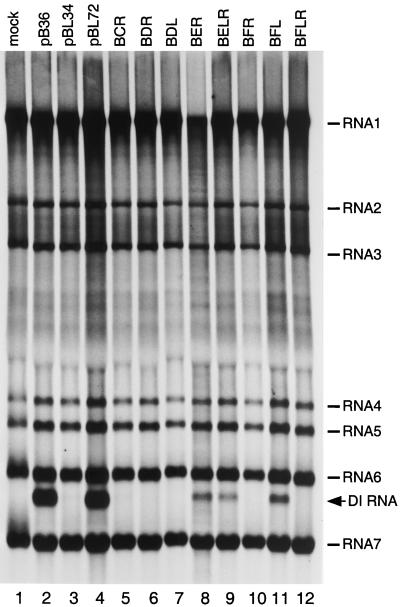
In previous work, we did not observe reversion of nonreplicating DI RNAs that contained mutations in the bulged stem-loop and in which the remainder of the 3′ UTR was from BCoV (12). Therefore, we constructed a series of vectors, BCR, BDR, BDL, BER, BELR, BFR, BFL, and BFLR, that retained the indicated stem segment mutations but replaced the MHV 3′ UTR sequence downstream of the EcoRV site with its BCoV counterpart. Metabolic labeling of MHV-infected cells that were transfected with DI RNAs transcribed from these vectors produced results more in accord with the viral genetic experiments. Compared to the appropriate positive and negative controls, all mutant chimeric DI RNAs except BER, BELR, and BFL failed to replicate (Fig. 4, lanes 5 to 12; summarized in Fig. 3B). These results corroborated the genetic conclusions that disruption of the base pairing of stem C, D, or F is lethal to viral replication but that the composition of stem E is not critical. One anomaly remained: the DI RNA BFL, harboring a mutant left arm of stem F, was capable of replication, despite the fact that we could never obtain a virus with the same stem segment disruption. Possible reasons for this discrepancy are discussed below.
FIG. 4.
Replicative ability of DI RNA stem-loop mutants constructed with the background of the BCoV 3′ UTR. Mouse 17Cl1 cells infected with wild-type MHV were mock transfected (lane 1) or transfected with each indicated DI RNA and then labeled with 134 μCi of [33P]orthophosphate per ml in the presence of 20 μg of actinomycin D per ml from 10 to 12 h postinfection (12). Purified cytoplasmic RNA was denatured with formaldehyde and formamide, separated by electrophoresis through 1% agarose containing formaldehyde, and visualized by autoradiography. Control DI RNAs were pB36 (lane 2) (32), the original wild-type MHV DI progenitor of all DI RNAs in this study; pBL34 (lane 3), the original nonreplicating DI containing a chimeric MHV-BCoV stem-loop in the 3′ UTR (12); and pBL72 (lane 4), a DI RNA with the wild-type MHV stem-loop in the background of the BCoV 3′ UTR (12).
Enzymatic and chemical probing of the bulged stem-loop structure.
We next sought structural evidence to complement the information about the bulged stem-loop derived from our genetic experiments. To accomplish this, a 264-nt synthetic RNA comprising the 5′ 229 nt of the MHV 3′ UTR was transcribed with SP6 RNA polymerase from a subclone of the MluI-SacI fragment of pBL85 (Fig. 2A). The folding of this molecule was probed enzymatically with RNases A, T1, and V1 and chemically with CMCT and DMS. Sites of RNA cleavage or modification by these agents were located by primer extension (9, 21, 51) from a 5′-end-labeled primer complementary to a region of the 3′ UTR beginning 35 nt downstream of the stem-loop structure. Natural reverse transcription stops were detected in control samples, which contained uncleaved, unmodified RNA.
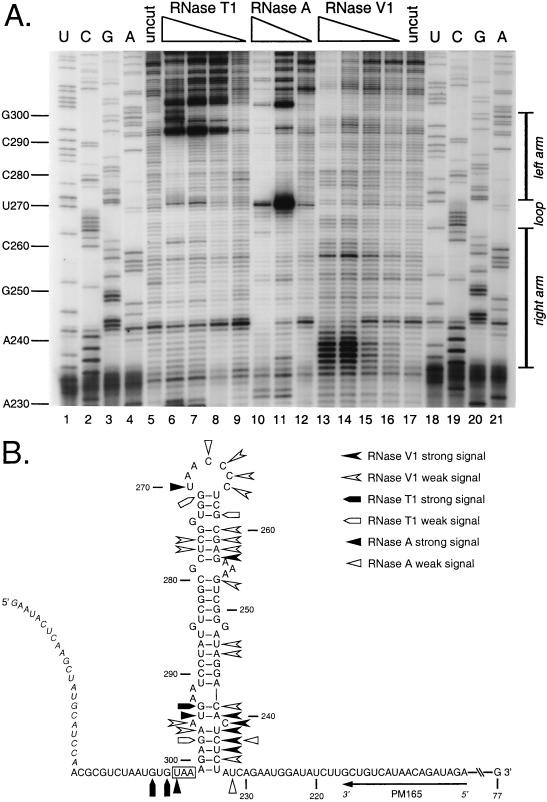
Results for digestion of the synthetic RNA over a range of concentrations of RNase T1 (Fig. 5A, lanes 6 to 9), RNase A (lanes 10 to 12), and RNase V1 (lanes 13 to 16) are summarized in Fig. 5B. RNase T1 and RNase A are single-strand-specific RNases (19). The former cleaves after unpaired guanosines to leave 3′ phosphates, and the latter cleaves after unpaired pyrimidines to leave 3′ phosphates. RNase V1 cleaves double-stranded regions of RNA, leaving 3′ hydroxyl groups, and does not show any base sequence preference (19). RNase V1 also recognizes regions of stacked bases within RNA structures.
FIG. 5.
Enzymatic structural probing of the bulged stem-loop structure. A 264-nt RNA encompassing the stem-loop region of the MHV 3′ UTR was transcribed in vitro from plasmid pBL122 (Fig. 2A), purified, renatured, and digested with various concentrations of RNases. Positions of cleavage sites were determined by primer extension with a 5′-end-labeled primer, PM165, complementary to nt 199 to 216. (A) Enzymatic cleavage sites generated by single-stranded nucleases RNase T1 (G specific) and RNase A (U and C specific) and double-stranded nuclease RNase V1. Lanes 1 to 4 and 18 to 21, sequencing ladders generated with end-labeled PM165 and terminated with ddATP, ddGTP, ddCTP, or ddTTP, respectively; lanes 5 and 17, undigested RNA; lanes 6 to 9, RNA digested with 15, 10, 5.0, and 1.0 U of RNase T1, respectively; lanes 10 to 12, RNA digested with 0.01, 0.001, and 0.0001 U of RNase A, respectively; lanes 13 to 16, RNA digested with 0.5, 0.3, 0.1, and 0.05 U of RNase V1, respectively. Ten-nucleotide intervals and the position of the stem-loop are indicated to the left and right of the autoradiogram, respectively. Each primer extension product from nuclease-digested RNA terminates one base downstream of the corresponding nucleotide in the sequencing ladder because all three RNases cut 3′ to their target bases. (B) Summary of observed enzymatic cleavage sites superimposed on the originally proposed stem-loop structure. Nucleotide numbering begins at the 3′ end of the genome, excluding the poly(A) tail; the N gene stop codon is boxed, and the position of the primer PM165 is shown. Bases indicated in italics at the 5′ end of the synthetic RNA are those derived from the polylinker of the transcription vector. Artifactual RNase T1 signals at A293 and U295 are not included in this diagram.
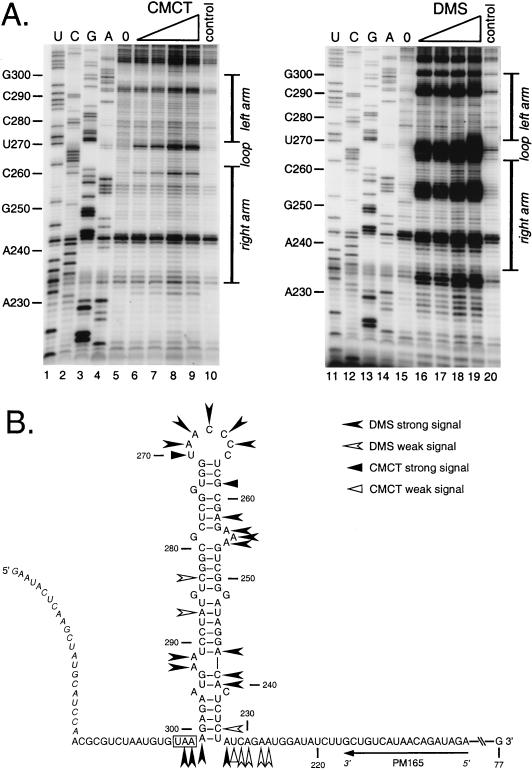
Results for chemical modification of the synthetic RNA with various concentrations of CMCT (Fig. 6A, lanes 6 to 9) and DMS (lanes 16 to 19) are summarized in Fig. 6B. CMCT alkylates the N-3 position of unpaired uridines and the N-1 position of unpaired guanosines. DMS alkylates the N-1 position of unpaired adenosines and the N-3 position of unpaired cytidines (9, 21). Each of these modifications causes termination of reverse transcription. These experiments included chemical modification stop control reactions, wherein the highest concentration of each reagent was added to unmodified RNA following addition of the reaction-quenching and precipitation solutions.
FIG. 6.
Chemical structural probing of the bulged stem-loop structure. A 264-nt RNA encompassing the stem-loop region of the MHV 3′ UTR was transcribed in vitro from plasmid pBL122 (Fig. 2A), purified, renatured, and reacted with various concentrations of chemical reagents. Positions of base modification were determined by primer extension as for Fig. 5. (A) Modification sites generated by single-stranded RNA-specific reagents CMCT (G and U specific) and DMS (A and C specific). Lanes 1 to 4 and 11 to 14, sequencing ladders generated with end-labeled PM165 and terminated with ddATP, ddGTP, ddCTP, or ddTTP, respectively; lanes 5 and 15, unmodified RNA; lanes 6 to 9, RNA modified with 4.2, 8.4, 12.6, and 16.8 mg of CMCT per ml, respectively; lanes 16 to 19, RNA modified with 0.5, 1.0, 1.5, and 2.0% DMS, respectively; lanes 10 and 20, control reactions in which the highest concentration of each reagent was added to RNA after addition of quenching reagents. Ten-nucleotide intervals and the position of the stem-loop are indicated to the left and right of each autoradiogram, respectively. Each primer extension product from chemically modified RNA is positioned one base downstream of the corresponding nucleotide in the sequencing ladder because primer extension terminates at the nucleotide immediately 3′ to the modified base. (B) Summary of observed chemical modification sites superimposed on the originally proposed stem-loop structure. Nucleotide numbering begins at the 3′ end of the genome, excluding the poly(A) tail; the N gene stop codon is boxed, and the position of the primer PM165 is shown. Bases indicated in italics at the 5′ end of the synthetic RNA are those derived from the polylinker of the transcription vector. Artifactual DMS signals at U234, G257, U270, G271, and G294 are not included in this diagram.
The combined enzymatic and chemical probing results revealed that the putative loop region was highly accessible to single-strand-specific reagents (Fig. 5B and 6B). Strong hits by DMS and CMCT at C265 through U270, and weak and strong RNase A cleavage at C267 and U270, respectively, all support a single-stranded structure for this region. Residues C264 through C266 within this region were also digested by RNase V1. This potentially indicates base stacking within the loop or that the loop participates in a tertiary structural interaction. Contrary to our previous model (12) but in accord with the genetic data above, putative stem A was available to both CMCT and RNase T1. Thus, this region is probably not double stranded, and nt 261 through 274 form a larger loop than originally proposed.
For the region of stem segments B, C, and D, the enzymatic evidence is consistent with the double-stranded structure previously proposed. There were multiple RNase V1 hits in this region, particularly in stem B, as well as a complete absence of signals from the single-strand-specific nucleases. A single minor hit from the chemical reagent DMS was detected in each of these stems, but the strongest DMS signals occurred in the bulge of A254 to A256 and at the bases of stems B and D, possibly indicating breathing of the helices at these points.
The region previously designated stem E actually appears to be single stranded, as evidenced by strong RNase A and T1 cleavages at G294 and U295, respectively, and strong DMS signals for A240 and C241. Thus, these bases likely form an open structure that includes A292 and A293, which are also strongly reactive with DMS. This picture is not entirely unambiguous, however, since strong RNase V1 cleavages were detected at C239 and A240, perhaps indicating participation of these bases in an as-yet-uncharacterized tertiary interaction.
Finally, putative stem F was observed to be the region of the RNA substrate most strongly reactive with the double-strand-specific RNase V1. However, weak signals for DMS, RNase A, and RNase T1 were found at C235, C237, and G298, respectively. Thus, it is possible that both helical and open structures are coexistent here, as discussed below. Upstream and downstream of the base of stem F, strong signals from single-strand-specific reagents were exclusively detected. Overall, the data from the enzymatic and chemical probing reinforced the secondary structure model deduced from the genetic experiments.
DISCUSSION
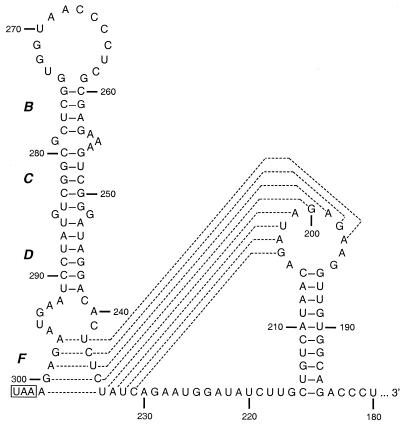
The mutational and structural analyses presented in this work lead us to refine our model of the RNA secondary structure of the 5′ 68 nt of the MHV 3′ UTR, as shown in Fig. 7. Both types of evidence support the notion that the loop of this structure is larger than originally envisioned, encompassing the nucleotides previously designated as stem A. This finding is consistent with our earlier results showing that a 5-nt (20) or 8-nt (12) insertion into the loop had no detectable effect on viral replication, although each had the propensity to contribute to the formation of a different stem adjacent to the loop. Similarly, the region formerly predicted to constitute stem E (12) also appears, by both structural and genetic criteria, to be unpaired, resulting in a larger internal loop between the adjacent helices D and F.
FIG. 7.
Refined model of the stem-loop structure based on genetic and structural experiments. Nucleotide numbering begins at the 3′ end of the genome, excluding the poly(A) tail, and the N gene stop codon is boxed. Dashed lines indicate alternative participation of nt 231 to 238 either in stem F or in one stem of a highly conserved pseudoknot that has been demonstrated to be essential in the 3′ UTR of BCoV (56).
The present work fully corroborated the original model of stems B, C, D, and F, and we now know more about the functional significance of each. The presence of stem B was strongly supported by the structural probing experiments, but surprisingly, the genetic analysis showed that both its primary sequence and base pairing could be disrupted without consequence to viral replication. In contrast, the base pairing of stems C and D was found to be essential for the virus, although the left and right arms of each helix could be exchanged. This suggests that the double-stranded structures, but not the primary sequences, of these are functionally relevant. More-extensive mutagenesis will be necessary before we can rule out possible requirements for specific bases at given positions in this region. In addition, we do not currently know whether G248 and G285 constitute two single-base bulges between stems C and D or if they form a non-Watson-Crick base pair in what is actually a single, longer helix. Finally, our evidence leads us to conclude that not only is the base pairing of stem F functionally essential but at least part of the primary sequence of this region will not tolerate alteration, since we could not exchange the left and right arms of this helix.
The nonessential nature of stem B was unexpected. Based on phylogenetic evidence, coupled with a preliminary mutational analysis, we originally concluded that the pairing, but not the primary sequence, of 4 bp in stems B and D that are covariant between MHV and BCoV was critical for the function of the bulged stem-loop structure (12). We have now learned that the replicative inability of structures with mixed pairs of MHV-BCoV or BCoV-MHV stem arms must have been due solely to disruption of the G242-U291 or the G244-C289 base pair in stem D (12). Thus, it is not clear why the base pairings in stem B are conserved among MHV, BCoV, human coronavirus OC43, and bovine enteric coronavirus (13). This may suggest that stem B can promote the folding or overall stability of the structure. It is possible that in recombinant viruses harboring the stem B disruptions (MBR and MBL) there exist subtle changes in growth properties that we were unable to detect.
In our genetic analysis, we observed a nearly complete correspondence between the ability of a given mutation to be incorporated into the viral genome and its ability to allow DI RNA replication (Fig. 3B and 4). However, it was necessary that the DI RNA construct contained a chimeric MHV-BCoV 3′ UTR, rather than an entirely MHV 3′ UTR, since the latter resulted in the rapid selection for wild-type DI RNAs formed by recombination with the genome of the helper virus. Similar high-frequency homologous recombination was observed by Yu and Leibowitz in their study of a conserved 11-nt motif, further downstream in the MHV 3′ UTR, that is required for host protein binding and DI RNA replication (58). In the present study, for reasons we do not yet understand, false-positive DI replication results were eliminated if stem-loop mutations were constructed in the context of the BCoV 3′ UTR. One discrepancy persisted, however. We were never able to obtain a viable viral recombinant with mutations in either the left or right arm or both arms of stem F, even with the powerful selection permitted by use of the host range mutant fMHV as the recipient virus (22). By contrast, the left-arm stem F mutant DI RNA (BFL) was capable of replication (Fig. 4). This may reflect a more complex role for the 3′ UTR in the intact viral genome than in DI RNA, since the former must be competent in replication, transcription, and translation, while survival of the latter depends exclusively on replication.
The higher sensitivity of stem F to mutagenesis may also reflect requirements imposed by its participation in another RNA structure. While this work was in progress, Williams et al. (56) presented compelling evidence for a phylogenetically conserved pseudoknot in the 3′ UTR of the BCoV genome that is essential for DI RNA replication. This structure is immediately adjacent to, and partially overlaps, the bulged stem-loop that we have described (Fig. 7). The upstream stem of the pseudoknot is formed, in part, by the right arm of stem F. In retrospect, it can be seen that compensatory pairs of stem mutations made by Williams et al. to establish the structure of the pseudoknot would have disrupted only a single, terminal base pair of stem F, U234-A301. Thus, none of the genetic or structural results of that study are incompatible with the bulged stem-loop. Conversely, none of our results above are incompatible with the pseudoknot. Although the exact relationship of the two structures remains to be delineated, the two cannot exist simultaneously. This raises the interesting possibility that they are alternative conformers of the same region of RNA and that one is a switch regulating the other, perhaps governing transition between different steps of viral replication. Such an RNA conformational switch has recently been proposed to operate in the 3′ UTR of alfalfa mosaic virus (37).
RNA secondary structures have been discovered in the 3′ UTRs of a number of positive-strand RNA viruses, including picornaviruses (15, 33, 35), togaviruses (36), and flaviviruses (4, 48, 59). In some cases, these have been demonstrated to specifically interact with viral or host cellular proteins (2, 8, 23, 36), a subset of which have been identified (3, 7, 49). It is thought that some of these protein-RNA interaction sites serve as assembly platforms for the viral RNA polymerase machinery to initiate negative-strand RNA synthesis. Such a role is unlikely, however, for the MHV 3′ UTR bulged stem-loop and pseudoknot, since these are distant from the last 55 nt of the MHV genome, which have been shown by Lin et al. to contain all information sufficient for initiation of negative-strand RNA synthesis (26). In addition, no host or viral proteins have yet been shown to specifically bind to this upstream region of the MHV 3′ UTR, although a set of host proteins have been detected that bind to tracts of nt 26 to 36 and nt 129 to 139 (27, 57, 58), and polypyrimidine tract-binding protein has been reported to bind to negative-strand RNA complementary to nt 53 to 149 (14). Thus, considerable work remains to be done to determine why the bulged stem-loop and pseudoknot are essential to the virus. We are currently seeking to better define the relationship between these two structures, and we are attempting to incorporate into them destabilizing, but nonlethal, mutations that may provide evidence of their function.
ACKNOWLEDGMENTS
We thank Matthew Shudt and Tim Moran of the Molecular Genetics Core Facility of the Wadsworth Center for the synthesis of oligonucleotides.
This work was supported by Public Health Service grant AI 39544 from the National Institutes of Health.
REFERENCES
- 1.An S, Maeda A, Makino S. Coronavirus transcription early in infection. J Virol. 1998;72:8517–8524. doi: 10.1128/jvi.72.11.8517-8524.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwell J L, Brinton M A. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J Virol. 1995;69:5650–5658. doi: 10.1128/jvi.69.9.5650-5658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell J L, Brinton M A. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J Virol. 1997;71:6433–6444. doi: 10.1128/jvi.71.9.6433-6444.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight K J, Rice C M. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang R-Y, Hofmann M A, Sethna P B, Brian D A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fichot O, Girard M. An improved method for sequencing of RNA templates. Nucleic Acids Res. 1990;18:6162. doi: 10.1093/nar/18.20.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gontarek R R, Gutshall L L, Herold K M, Tsai J, Sathe G M, Mao J, Prescott C, Del Vecchio A M. hnRNP C and polypyrimidine tract-binding protein specifically interact with the pyrimidine-rich region within the 3′NTR of the HCV RNA genome. Nucleic Acids Res. 1999;27:1457–1463. doi: 10.1093/nar/27.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris K S, Xiang W, Alexander L, Lane W S, Paul A V, Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J Biol Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 9.Hartshorne T, Agabian N. A common core structure for U3 small nucleolar RNAs. Nucleic Acids Res. 1994;22:3354–3364. doi: 10.1093/nar/22.16.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann M A, Brian D A. The 5′ end of coronavirus minus-strand RNAs contains a short poly(U) tract. J Virol. 1991;65:6331–6333. doi: 10.1128/jvi.65.11.6331-6333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horton R M, Pease L R. Recombination and mutagenesis of DNA sequences using PCR. In: McPherson M J, editor. Directed mutagenesis, a practical approach. New York, N.Y: IRL Press; 1991. pp. 217–247. [Google Scholar]
- 12.Hsue B, Masters P S. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsue B, Masters P S. An essential secondary structure in the 3′ untranslated region of the mouse hepatitis virus genome. Adv Exp Med Biol. 1998;440:297–302. doi: 10.1007/978-1-4615-5331-1_39. [DOI] [PubMed] [Google Scholar]
- 14.Huang P, Lai M M C. Polypyrimidine tract-binding protein binds to the complementary strand of the mouse hepatitis virus 3′ untranslated region, thereby altering conformation. J Virol. 1999;73:9110–9116. doi: 10.1128/jvi.73.11.9110-9116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobson S J, Konings D A M, Sarnow P. Biochemical and genetic evidence for a pseudoknot structure at the 3′ terminus of the poliovirus RNA genome and its role in viral RNA amplification. J Virol. 1993;67:2961–2971. doi: 10.1128/jvi.67.6.2961-2971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong Y S, Makino S. Evidence for coronavirus discontinuous transcription. J Virol. 1994;68:2615–2623. doi: 10.1128/jvi.68.4.2615-2623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim Y-N, Jeong Y S, Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingsman S M, Samuel C E. Mechanism of interferon action. Interferon-mediated inhibition of simian virus-40 early RNA accumulation. Virology. 1980;101:458–465. doi: 10.1016/0042-6822(80)90459-6. [DOI] [PubMed] [Google Scholar]
- 19.Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- 20.Koetzner C A, Parker M M, Ricard C S, Sturman L S, Masters P S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krol A, Carbon P. A guide for probing native small nuclear RNA and ribonucleoprotein structures. Methods Enzymol. 1989;180:212–227. doi: 10.1016/0076-6879(89)80103-x. [DOI] [PubMed] [Google Scholar]
- 22.Kuo L, Godeke G-J, Raamsman M J B, Masters P S, Rottier P J M. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusov Y, Weitz M, Dollenmeier G, Gauss-Muller V, Siegl G. RNA-protein interactions at the 3′ end of the hepatitis A virus RNA. J Virol. 1996;70:1890–1897. doi: 10.1128/jvi.70.3.1890-1897.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y-J, Lai M M C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Y-J, Liao C-L, Lai M M C. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J Virol. 1994;68:8131–8140. doi: 10.1128/jvi.68.12.8131-8140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Yu W, Leibowitz J L. A specific host cellular protein binding element near the 3′ end of mouse hepatitis virus genomic RNA. Virology. 1997;232:74–85. doi: 10.1006/viro.1997.8553. [DOI] [PubMed] [Google Scholar]
- 28.Luytjes W, Gerritsma H, Spaan W J M. Replication of synthetic interfering RNAs derived from coronavirus mouse hepatitis virus-A59. Virology. 1996;216:174–183. doi: 10.1006/viro.1996.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makino S, Fujioka N, Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985;54:329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino S, Shieh C-K, Keck J G, Lai M M C. Defective interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988;163:104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masters P S. Reverse genetics of the largest RNA viruses. Adv Virus Res. 1999;53:245–264. doi: 10.1016/S0065-3527(08)60351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Masters P S, Koetzner C A, Kerr C A, Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J Virol. 1994;68:328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melchers W J, Hoenderop J G, Bruins Slot H J, Pleij C W, Pilipenko E V, Agol V I, Galama J M. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′ untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Méndez A, Smerdou C, Izeta A, Gebauer F, Enjuanes L. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology. 1996;217:495–507. doi: 10.1006/viro.1996.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirmomeni M H, Hughes P J, Stanway G. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J Virol. 1997;71:2363–2370. doi: 10.1128/jvi.71.3.2363-2370.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakhasi H L, Cao X-Q, Rouault T A, Liu T-H. Specific binding of host cell proteins to the 3′-terminal stem-loop structure of rubella virus negative-strand RNA. J Virol. 1991;65:5961–5967. doi: 10.1128/jvi.65.11.5961-5967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsthoorn R C L, Mertens S, Brederode F T, Bol J F. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 1999;18:4856–4864. doi: 10.1093/emboj/18.17.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng D, Koetzner C A, Masters P S. Analysis of second-site revertants of a murine coronavirus nucleocapsid protein deletion mutant and construction of nucleocapsid protein mutants by targeted RNA recombination. J Virol. 1995;69:3449–3457. doi: 10.1128/jvi.69.6.3449-3457.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penzes Z, Tibbles K, Shaw K, Britton P, Brown T D K, Cavanaugh D. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology. 1994;203:286–293. doi: 10.1006/viro.1994.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips J J, Chua M M, Lavi E, Weiss S R. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J Virol. 1999;73:7752–7760. doi: 10.1128/jvi.73.9.7752-7760.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Sanchez C M, Izeta A, Sanchez-Morgado J M, Alonso S, Sola I, Balasch M, Plana-Duran J, Enjuanes L. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J Virol. 1999;73:7607–7618. doi: 10.1128/jvi.73.9.7607-7618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sawicki S G, Sawicki D L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawicki S G, Sawicki D L. A new model for coronavirus transcription. Adv Exp Med Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- 46.Schaad M C, Baric R S. Genetics of mouse hepatitis virus transcription: evidence that subgenomic negative strands are functional templates. J Virol. 1994;68:8169–8179. doi: 10.1128/jvi.68.12.8169-8179.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sethna P B, Hofmann M A, Brian D A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991;65:320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi P-Y, Brinton M A, Veal J M, Zhong Y Y, Wilson W D. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry. 1996;35:4222–4230. doi: 10.1021/bi952398v. [DOI] [PubMed] [Google Scholar]
- 49.Singh N K, Atreya C D, Nakhasi H L. Identification of calreticulin as a rubella virus RNA binding protein. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skinner M A, Racaniello V R, Dunn G, Cooper J, Minor P D, Almond J W. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J Mol Biol. 1989;207:379–392. doi: 10.1016/0022-2836(89)90261-1. [DOI] [PubMed] [Google Scholar]
- 51.Stern S, Moazed D, Noller H F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 52.van der Most R G, Bredenbeek P J, Spaan W J M. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van der Most R G, Luytjes W, Rutjes S, Spaan W J M. Translation but not the encoded sequence is essential for the efficient propogation of defective interfering RNAs of the coronavirus mouse hepatitis virus. J Virol. 1995;69:3744–3751. doi: 10.1128/jvi.69.6.3744-3751.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Most R G, Spaan W J M. Coronavirus replication, transcription, and RNA recombination. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 11–31. [Google Scholar]
- 55.van Marle G, Dobbe J C, Gultyaev A P, Luytjes W, Spaan W J M, Snijder E J. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams G D, Chang R Y, Brian D A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu W, Leibowitz J L. Specific binding of host cellular proteins to multiple sites within the 3′ end of mouse hepatitis virus genomic RNA. J Virol. 1995;69:2016–2023. doi: 10.1128/jvi.69.4.2016-2023.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu W, Leibowitz J L. A conserved motif at the 3′ end of mouse hepatitis virus genomic RNA required for host protein binding and viral RNA replication. Virology. 1995;214:128–138. doi: 10.1006/viro.1995.9947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng L, Falgout B, Markoff L. Identification of specific nucleotide sequences within the conserved 3′-SL in the dengue type 2 virus genome required for replication. J Virol. 1998;72:7510–7522. doi: 10.1128/jvi.72.9.7510-7522.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, Liao C-L, Lai M M C. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J Virol. 1994;68:4738–4746. doi: 10.1128/jvi.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]