Abstract
Background
Kallikrein-related peptidase 7 (KLK7) is a chymotrypsin-like serine protease which is essential for the desquamation of corneocytes and thus plays a pivotal role in maintaining skin homeostasis. In cancer, KLK7 overexpression was suggested to represent a route for metastasis through cleavage of cell junction and extracellular matrix proteins of cancer cells.
Methods
To comprehensively determine KLK7 protein expression in normal and neoplastic tissues, a tissue microarray containing 13,447 samples from 147 different tumor types and subtypes as well as 608 samples of 76 different normal tissue types was analyzed by immunohistochemistry.
Results
KLK7 positivity was found in 64 of 147 tumor categories, including 17 tumor categories with at least one strongly positive case. The highest rate of KLK7 positivity was found in squamous cell carcinomas from various sites of origin (positive in 18.1%-63.8%), ovarian and endometrium cancers (4.8%-56.2%), salivary gland tumors (4.8%-13.7%), bilio-pancreatic adenocarcinomas (20.0%-40.4%), and adenocarcinomas of the upper gastrointestinal tract (3.3%-12.5%). KLK7 positivity was linked to nodal metastasis (p = 0.0005), blood vessel infiltration (p = 0.0037), and lymph vessel infiltration (p < 0.0001) in colorectal adenocarcinoma, nodal metastasis in hepatocellular carcinoma (p = 0.0382), advanced pathological tumor stage in papillary thyroid cancer (p = 0.0132), and low grade of malignancy in a cohort of 719 squamous cell carcinomas from 11 different sites of origin (p < 0.0001).
Conclusions
These data provide a comprehensive overview on KLK7 expression in normal and neoplastic human tissues. The prognostic relevance of KLK7 expression and the possible role of KLK7 as a drug target need to be further investigated.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-024-12552-8.
Keywords: KLK7, Tissue microarray, Immunohistochemistry, Neoplastic human tissues
Introduction
The human kallikrein (KLK, kallikrein-related peptidase) family refers to a group of enzymes that belong to the proteolytic family of serine proteases. To date, there are 15 known members in the human kallikrein family, designated as KLK1-KLK15 [1]. Some kallikreins have been implicated in cancer. For example, KLK3 (prostate specific antigen, PSA) is used as a blood biomarker for prostate cancer [2], and alterations of KLK4, KLK5, and KLK6 have been suggested to play a role for breast cancer aggressiveness [3]. A cancer-relevant role is also suspected for Kallikrein-related peptidase 7. KLK7 is a chymotrypsin-like serine protease with a critical role in the maintenance of skin homeostasis [4–6]. The KLK7 zymogen is secreted by a subset of keratinocytes into the stratum granulosum of the epidermis where it is activated by proteolytic removal of the short N-terminal [4, 7]. Activated KLK7 induces the desquamation of corneocytes from the outer layer of the keratinized squamous epithelium of the epidermis by proteolysis of corneodesmosomes [5, 8]. Dysregulated KLK7 production and secretion occurs in various skin disorders going along with dry, crusty and inflamed skin such as atopic dermatitis, Netherton syndrome, or psoriasis (summarized in [9]).
In normal tissues, KLK7 expression is largely limited to squamous epithelium [4, 5]. In cancer, however, KLK7 expression has been described to occur in a broad range of tumor entities [10–27]. Overexpression of KLK7 has been suggested to promote epithelial-mesenchymal transition (EMT) through the hydrolysis of cell membrane and extracellular matrix molecules (e.g., E-cadherin) and therefore to represent a possible mechanism for metastasis (summarized in [28]). Several studies have described high rates of KLK7 positivity in triple negative breast cancer [17], colorectal cancer [21], ovarian cancer [12–14], adenocarcinomas of the cervix uteri [23, 24], pancreatic cancer [10, 11] and malignant melanoma [19], but the published data on the prevalence of KLK7 immunostaining are discrepant for several tumor entities. For example, KLK7 positivity has been found in 20.0%-92.5% of adenocarcinomas of the endocervix [18, 23, 24] and from 41.0%–100% of squamous cell carcinomas of the oral cavity [20, 22, 25, 26]. Many important tumor entities such as carcinomas of the lung, stomach, esophagus, liver, bile ducts, or the endometrium have not been analyzed for KLK7 expression yet.
To better understand the prevalence and potential role of KLK7 expression in cancer, a comprehensive study analyzing large numbers of neoplastic and non-neoplastic tissues under highly standardized conditions is desirable. For this purpose, KLK7 expression was analyzed in more than 13,000 tumor tissue samples from 147 different tumor types and subtypes as well as 76 non-neoplastic tissue categories by immunohistochemistry (IHC) in a tissue microarray (TMA) format in this study.
Materials and methods
Tissue Microarrays (TMAs)
The tissue microarrays that were utilized in the current study had been described before [29–31]. In brief, our normal tissue TMA contains 76 different normal tissue types, and each tissue is represented by 8 samples from 8 different donors (in total: 608 samples on one slide). Our cancer TMAs were constructed from a total of 13,447 primary tumors obtained from 147 tumor types and subtypes that were distributed across 24 TMA slides. Detailed histopathological data on grade, pathological tumor stage (pT), pathological lymph node status (pN), blood vessel infiltration (V), and lymph vessel infiltration (L) were available from subsets of colon carcinomas (n = 2,351), liver carcinomas (n = 301), endometrium carcinomas (n = 259), ovarian carcinomas (n = 524), pancreas carcinomas (n = 598), papillary thyroid carcinomas (n = 382), gastric carcinomas (n = 398), as well as of 902 squamous cell carcinomas of different sites of origin. All samples were collected from the Institute of Pathology, University Hospital of Hamburg, Germany, the Institute of Pathology, Clinical Center Osnabrueck, Germany, and the Department of Pathology, Academic Hospital Fuerth, Germany. All tissues were formalin-fixed (4% buffered formalin) and subsequently embedded in paraffin. The process of TMA construction has been described earlier [32, 33]. In brief, each tissue is represented by a single 0.6 mm tissue core that was taken from a cancer containing donor block and transferred into an empty recipient paraffin block. The local ethics committee (Ethics commission Hamburg, WF-049/09) has approved the manufacturing and analysis of TMAs from archived remnants of diagnostic tissues. Patient informed consent was not required for this study. Patient data analysis without informed patient consent is covered by local laws (HmbKHG, §12). All work has been carried out in compliance with the Helsinki Declaration.
Immunohistochemistry (IHC)
Freshly cut tissue sections were used for all IHC experiments. All immunohistochemistry experiments were made manually in one day at the Institute of Pathology, University Medical Center Hamburg-Eppendorf. TMA sections of 3µ were cut, deparaffinized in xylol and rehydrated through a descending alcohol series. Immunohistochemistry was performed manually. Sections were placed for 5 min in an autoclave at 121 °C in pH 7.8 DakoTarget Retrieval Solution™ (Agilent, CA, USA) for heat induced antigen retrieval. Endogenous peroxidase was blocked in Dako Peroxidase Blocking Solution™ (Agilent, CA, USA; #52,023). Primary antibody specific for KLK7 protein (mouse monoclonal, MSVA-707M, #4860-707M, MS Validated Antibodies, Hamburg, Germany) was applied at 37 °C for 60 min at a dilution of 1:150. Specificity and cross reactivity of MSVA-707M was checked by Western blotting (Supplementary Fig. 1) and by comparison of the staining pattern with that of a second independent antibody as recommended by The International Working Group for Antibody Validation (IWGAV) [34]. For this purpose, the normal tissue TMA was also analyzed by the rabbit recombinant monoclonal KLK7 antibody EPR22594-203 (Abcam; ab254258) at a dilution of 1:7.5 and an otherwise identical protocol. As controls, skin with stratum granulosum (positive control) and colon mucosa (negative control) were included in the TMA. The EnVision detection Kit™ (Agilent, CA, USA; #K5007) was used to visualize the bound antibody. For counterstain, slides were immersed in haemalaun solution. Staining was membranous and/or cytoplasmic, which fits to the function of KLK7 as a protein that is secreted from the cytoplasm via the cell membrane to the extracellular space [28]. Accordingly, cytoplasmic and membranous staining was scored. Slide scoring was performed as described before [29–31]. In brief, one pathologist scored all slides (SK). In questionable cases, a second pathologist’s opinion was sought. For scoring of tumor tissues, the percentage of KLK7 positive tumor cells was estimated in each spot and the staining intensity was noted in a 4-step scale (0, 1 + , 2 + , 3 +), where 0 indicates no visble staining, 1 + ligth brown staining, 3 + dark brown staining, and 2 + staining between 1 + and 3 + . Examples for the staining intensities are given in Supplementary Fig. 2. The staining results were combined into four groups for statistical analyses: Negative staining: no visible staining, weak staining: staining intensity of 1 + in ≤ 70% or staining intensity of 2 + in ≤ 30% of tumor cells, moderate staining: staining intensity of 1 + in > 70%, staining intensity of 2 + in > 30% but in ≤ 70% or staining intensity of 3 + in ≤ 30% of tumor cells, strong staining: staining intensity of 2 + in > 70% or staining intensity of 3 + in > 30% of tumor cells.
KLK7 RNA expression data source
RNA expression z-scores of KLK7 relative to diploid samples from RNA sequencing (RNA-Seq by Expectation–Maximization (RSEM) V2, normalized from Illumina HiSeq) were downloaded from cBioportal (http://www.cbioportal.org) in July 2023. The data selection included the 32 studies labeled as “TCGA PanCancer Atlas Studies” with a total of 10,976 samples.
Western Blot
Human tissue samples of skin, kidney and liver were shock-frozen in liquid nitrogen, grinded and lysed with 5% SDS. Samples were heated for 5 min at 95 °C and the suspension was centrifuged at 10,000 g for 5 min at 8 °C. Supernatant was further used for protein quantification via Qubit protein quantification kit (Invitrogen, Carlsbad, California, USA). Protein concentration was adjusted with sample buffer (4XLDS, BioRad, Hercules, California, USA) and DTT (1:10) and the sample was heated up to 95 °C for 5 min. 30 µg per human tissue sample was used for separation on a Bis–Tris 4–20% gel. The gel was then blotted on a 0.2 µm nitrocellulose membrane. The membrane was blocked for 1 h in 5% BSA in PBST before the primary murine antibody KLK7 (MSVA-707M, MS Validated Antibodies, GmbH, Hamburg, Germany, diluted 1:1,000 in 5% BSA in PBST) was incubated on the membrane over night at 4 °C. On the next day, the membrane was incubated with the secondary antibody goat-anti-mouse-HRP (1:2,000) for 1 h at room temperature and developed with Pierce ECL Western Blotting Substrate (BioRad). Blots were imaged using a BioRad Chemidoc imager. For loading control, the membrane was incubated with a primary anti-tubulin antibody (anti-rabbit, Abcam, Cambridge, UK, ab4074) using the same protocol.
Statistics
The JMP 16 software package (SAS Institute Inc., NC, USA) was used for statistical analysis. Contingency table analysis and chi2-testing was applied to search for associations between KLK7 immunostaining and tumor phenotype. For statistical adjustment the Bonferroni correction (p = p-value/number of statistical tests) was performed, and p ≤ 0.0018 was considered as statistically significant.
Results
Technical issues
A total of 12,345 (91.8%) of 13,447 tumor samples and at least 4 normal samples per tissue category were interpretable in our TMA analysis. Non-interpretable samples demonstrated lack of unequivocal tumor cells or absence of tissue in the respective TMA spots.
KLK7 in normal tissues
KLK7 immunostaining was predominantly seen in squamous epithelia where the staining preferably occurred in a zone between the middle and the top 20% of the squamous epithelium. This area contains the granular layer in keratinizing squamous epithelium. Staining was predominantly membranous but in case of high staining intensity the cytoplasm was also involved. The extent of KLK7 staining in squamous epithelium also depended on its localization. KLK7 staining was low in the cervix uteri, higher in the esophagus and tonsil surface, and highest in the skin. In the skin, a strong staining was seen in the stratum granulosum and the keratinizing cell layers. KLK7 immunostaining was also strong in the cortex of hair follicles, the luminal area of sebaceous glands, and the central (keratinizing) zone of corpuscles of Hassall’s in the thymus. Few scattered epithelial cells or groups of cells with a moderate KLK7 positivity were occasionally seen in renal tubuli, the fallopian tube, and in salivary glands. Representative images of KLK7 positive normal tissues are shown in Fig. 1. All these findings were observed by both MSVA-707M and EPR22594-203 (Supplementary Fig. 3). An additional cytoplasmic smooth muscle staining and nuclear staining was observed in several organs for EPR22594-203 but not for MSVA-707M and was thus considered an antibody specific cross-reactivity of EPR22594-203.
Fig. 1.
KLK7 immunostaining in normal tissues. The panels show a strong membranous and cytoplasmic KLK7 staining of the granular layer of the epidermis of the skin (A) (inset shows higher magnification of the granular layer), a weak to moderate KLK7 positivity of the upper two thirds of the squamous epithelium of the esophagus (B), a strong KLK7 staining around the keratinizing zone of corpuscles of Hassall‘s of the thymus (C), and a weak to moderate KLK7 positivity in a subset of cells of squamous epithelium of tonsil crypts (D). KLK7 staining is largely absent in non-keratinizing squamous epithelium of the uterine cervix (E) and completely lacking in the mucosa of the colon (F)
KLK7 in cancer tissues
Positive KLK7 immunostaining was detectable in 1,600 (13.0%) of the 12,345 analyzable tumors, including 1,352 (11.0%) with weak, 203 (1.6%) with moderate, and 45 (0.4%) with strong immunostaining. Overall, 64 (43.5%) of 147 tumor categories showed detectable KLK7 expression with 17 (11.6%) tumor categories including at least one case with strong positivity (Table 1). Representative images of KLK7 positive tumors are shown in Fig. 2. The highest rate of KLK7 positivity was found in squamous cell carcinomas from various sites of origin (positive in 18.1%-63.8%), various categories of ovarian and endometrium cancers (4.8%-56.2%), salivary gland tumors (4.8%-13.7%), bilio-pancreatic adenocarcinomas (20.0%-40.4%), and adenocarcinomas of the upper gastrointestinal tract (3.3%-12.5%). Twenty other tumor entities showed—a usually weak—KLK7 positivity in less than 9%. A graphical representation of a ranking order of KLK7 positive and strongly positive cancers is given in Fig. 3. The relationship between KLK7 expression and clinically important histopathological and molecular tumor features in colon, liver, endometrial, ovarian, pancreas, papillary thyroid, and gastric carcinomas is shown in Table 2. Detectable KLK7 immunostaining was linked to pN + (p = 0.0005), V1 (p = 0.0037), and L1 (p < 0.0001) in colorectal adenocarcinoma, pN + in hepatocellular carcinoma (p = 0.0382), advanced pT stage in papillary thyroid cancer (p = 0.0132), and low grade of malignancy in a cohort of 719 squamous cell carcinomas from 11 different sites of origin (p < 0.0001).
Table 1.
KLK7 immunostaining in human tumors
| Tumor entity | KLK7 immunostaining result | ||||||
|---|---|---|---|---|---|---|---|
| on TMA (n) | analyzable (n) | negative (%) | weak (%) | moderate (%) | strong (%) | ||
| Tumors of the skin | Pilomatrixoma | 35 | 23 | 100.0 | 0.0 | 0.0 | 0.0 |
| Basal cell carcinoma | 89 | 75 | 84.0 | 9.3 | 6.7 | 0.0 | |
| Benign nevus | 29 | 28 | 96.4 | 0.0 | 3.6 | 0.0 | |
| Squamous cell carcinoma of the skin | 145 | 141 | 36.2 | 55.3 | 6.4 | 2.1 | |
| Malignant melanoma | 65 | 62 | 90.3 | 8.1 | 1.6 | 0.0 | |
| Malignant melanoma Lymph node metastasis | 86 | 79 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Merkel cell carcinoma | 48 | 43 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the head and neck | Squamous cell carcinoma of the larynx | 109 | 106 | 49.1 | 48.1 | 2.8 | 0.0 |
| Squamous cell carcinoma of the pharynx | 60 | 58 | 60.3 | 36.2 | 3.4 | 0.0 | |
| Oral squamous cell carcinoma (floor of the mouth) | 130 | 128 | 44.5 | 46.1 | 7.0 | 2.3 | |
| Pleomorphic adenoma of the parotid gland | 50 | 47 | 97.9 | 0.0 | 2.1 | 0.0 | |
| Warthin tumor of the parotid gland | 104 | 103 | 46.6 | 38.8 | 13.6 | 1.0 | |
| Adenocarcinoma, NOS (Papillary Cystadenocarcinoma) | 14 | 12 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Salivary duct carcinoma | 15 | 14 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Acinic cell carcinoma of the salivary gland | 181 | 160 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adenocarcinoma NOS of the salivary gland | 109 | 89 | 94.4 | 5.6 | 0.0 | 0.0 | |
| Adenoid cystic carcinoma of the salivary gland | 180 | 118 | 89.0 | 11.0 | 0.0 | 0.0 | |
| Basal cell adenoma of the salivary gland | 101 | 95 | 86.3 | 13.7 | 0.0 | 0.0 | |
| Basal cell adenocarcinoma of the salivary gland | 25 | 23 | 87.0 | 13.0 | 0.0 | 0.0 | |
| Epithelial-myoepithelial carcinoma of the salivary gland | 53 | 53 | 94.3 | 5.7 | 0.0 | 0.0 | |
| Mucoepidermoid carcinoma of the salivary gland | 343 | 300 | 92.0 | 7.7 | 0.3 | 0.0 | |
| Myoepithelial carcinoma of the salivary gland | 21 | 21 | 95.2 | 4.8 | 0.0 | 0.0 | |
| Myoepithelioma of the salivary gland | 11 | 10 | 90.0 | 10.0 | 0.0 | 0.0 | |
| Oncocytic carcinoma of the salivary gland | 12 | 12 | 91.7 | 8.3 | 0.0 | 0.0 | |
| Polymorphous adenocarcinoma, low grade, of the salivary gland | 41 | 36 | 88.9 | 8.3 | 2.8 | 0.0 | |
| Pleomorphic adenoma of the salivary gland | 53 | 39 | 94.9 | 2.6 | 2.6 | 0.0 | |
| Tumors of the lung, pleura and thymus | Adenocarcinoma of the lung | 196 | 187 | 97.3 | 2.7 | 0.0 | 0.0 |
| Squamous cell carcinoma of the lung | 80 | 72 | 81.9 | 16.7 | 1.4 | 0.0 | |
| Small cell carcinoma of the lung | 16 | 16 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mesothelioma, epitheloid | 40 | 35 | 97.1 | 2.9 | 0.0 | 0.0 | |
| Mesothelioma, other types | 77 | 72 | 98.6 | 0.0 | 1.4 | 0.0 | |
| Thymoma | 29 | 28 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the female genital tract | Squamous cell carcinoma of the vagina | 78 | 73 | 64.4 | 28.8 | 4.1 | 2.7 |
| Squamous cell carcinoma of the vulva | 157 | 145 | 43.4 | 48.3 | 4.8 | 3.4 | |
| Squamous cell carcinoma of the cervix | 136 | 131 | 74.0 | 22.9 | 3.1 | 0.0 | |
| Adenocarcinoma of the cervix | 23 | 23 | 91.3 | 4.3 | 4.3 | 0.0 | |
| Endometrioid endometrial carcinoma | 338 | 289 | 93.1 | 6.6 | 0.3 | 0.0 | |
| Endometrial serous carcinoma | 86 | 73 | 84.9 | 12.3 | 1.4 | 1.4 | |
| Carcinosarcoma of the uterus | 57 | 42 | 97.6 | 2.4 | 0.0 | 0.0 | |
| Endometrial carcinoma, high grade, G3 | 13 | 10 | 80.0 | 20.0 | 0.0 | 0.0 | |
| Endometrial clear cell carcinoma | 9 | 8 | 87.5 | 12.5 | 0.0 | 0.0 | |
| Endometrioid carcinoma of the ovary | 130 | 106 | 76.4 | 18.9 | 3.8 | 0.9 | |
| Serous carcinoma of the ovary | 580 | 527 | 43.8 | 45.5 | 8.5 | 2.1 | |
| Mucinous carcinoma of the ovary | 101 | 80 | 81.3 | 16.3 | 1.3 | 1.3 | |
| Clear cell carcinoma of the ovary | 51 | 42 | 95.2 | 2.4 | 0.0 | 2.4 | |
| Carcinosarcoma of the ovary | 47 | 41 | 90.2 | 9.8 | 0.0 | 0.0 | |
| Granulosa cell tumor of the ovary | 44 | 43 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leydig cell tumor of the ovary | 4 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli cell tumor of the ovary | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli Leydig cell tumor of the ovary | 3 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Steroid cell tumor of the ovary | 3 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Brenner tumor | 41 | 41 | 97.6 | 2.4 | 0.0 | 0.0 | |
| Tumors of the breast | Invasive breast carcinoma of no special type | 80 | 68 | 95.6 | 1.5 | 2.9 | 0.0 |
| Lobular carcinoma of the breast | 122 | 108 | 99.1 | 0.9 | 0.0 | 0.0 | |
| Medullary carcinoma of the breast | 15 | 15 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tubular carcinoma of the breast | 18 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mucinous carcinoma of the breast | 22 | 20 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Phyllodes tumor of the breast | 50 | 49 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the digestive system | Adenomatous polyp, low-grade dysplasia | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 |
| Adenomatous polyp, high-grade dysplasia | 50 | 48 | 97.9 | 2.1 | 0.0 | 0.0 | |
| Adenocarcinoma of the colon and rectum | 2483 | 2354 | 92.3 | 7.2 | 0.5 | 0.0 | |
| Gastric adenocarcinoma, diffuse type | 215 | 209 | 96.7 | 2.9 | 0.5 | 0.0 | |
| Gastric adenocarcinoma, intestinal type | 215 | 205 | 90.2 | 8.8 | 0.5 | 0.5 | |
| Gastric adenocarcinoma, mixed type | 62 | 60 | 88.3 | 11.7 | 0.0 | 0.0 | |
| Adenocarcinoma of the esophagus | 83 | 80 | 87.5 | 10.0 | 0.0 | 2.5 | |
| Squamous cell carcinoma of the esophagus | 76 | 64 | 67.2 | 32.8 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the anal canal | 91 | 88 | 59.1 | 35.2 | 5.7 | 0.0 | |
| Cholangiocarcinoma | 58 | 57 | 91.2 | 7.0 | 0.0 | 1.8 | |
| Gallbladder adenocarcinoma | 51 | 34 | 79.4 | 11.8 | 8.8 | 0.0 | |
| Extrahepatic bile duct carcinoma | 42 | 40 | 80.0 | 17.5 | 2.5 | 0.0 | |
| Hepatocellular carcinoma | 312 | 285 | 95.8 | 3.2 | 0.7 | 0.4 | |
| Ductal adenocarcinoma of the pancreas | 659 | 631 | 62.4 | 30.3 | 6.2 | 1.1 | |
| Pancreatic/Ampullary adenocarcinoma | 98 | 94 | 59.6 | 27.7 | 11.7 | 1.1 | |
| Acinar cell carcinoma of the pancreas | 16 | 15 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Gastrointestinal stromal tumor (GIST) | 62 | 62 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the urinary system | Non-invasive papillary urothelial carcinoma, pTa G3 | 32 | 27 | 100.0 | 0.0 | 0.0 | 0.0 |
| Urothelial carcinoma, pT2-4 G3 | 162 | 136 | 91.9 | 6.6 | 1.5 | 0.0 | |
| Squamous cell carcinoma of the bladder | 22 | 21 | 52.4 | 47.6 | 0.0 | 0.0 | |
| Small cell neuroendocrine carcinoma of the bladder | 22 | 22 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sarcomatoid urothelial carcinoma | 25 | 24 | 95.8 | 0.0 | 4.2 | 0.0 | |
| Urothelial carcinoma of the kidney pelvis | 62 | 59 | 96.6 | 1.7 | 1.7 | 0.0 | |
| Clear cell renal cell carcinoma | 50 | 41 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Papillary renal cell carcinoma | 50 | 31 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Chromophobe renal cell carcinoma | 50 | 34 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Oncocytoma | 50 | 30 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of the male genital organs | Adenocarcinoma of the prostate, Gleason 3 + 3 | 83 | 83 | 100.0 | 0.0 | 0.0 | 0.0 |
| Adenocarcinoma of the prostate, Gleason 4 + 4 | 80 | 76 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adenocarcinoma of the prostate, Gleason 5 + 5 | 85 | 82 | 98.8 | 1.2 | 0.0 | 0.0 | |
| Adenocarcinoma of the prostate (recurrence) | 258 | 246 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Small cell neuroendocrine carcinoma of the prostate | 19 | 18 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Seminoma | 111 | 93 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Embryonal carcinoma of the testis | 54 | 46 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leydig cell tumor of the testis | 31 | 29 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sertoli cell tumor of the testis | 2 | 2 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sex cord stromal tumor of the testis | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Spermatocytic tumor of the testis | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Yolk sac tumor | 53 | 43 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Teratoma | 53 | 27 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Squamous cell carcinoma of the penis | 92 | 84 | 52.4 | 40.5 | 6.0 | 1.2 | |
| Tumors of endocrine organs | Adenoma of the thyroid gland | 113 | 111 | 99.1 | 0.9 | 0.0 | 0.0 |
| Papillary thyroid carcinoma | 391 | 363 | 97.0 | 2.8 | 0.0 | 0.3 | |
| Follicular thyroid carcinoma | 154 | 147 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Medullary thyroid carcinoma | 111 | 109 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Parathyroid gland adenoma | 43 | 34 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Anaplastic thyroid carcinoma | 45 | 42 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adrenal cortical adenoma | 50 | 38 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Adrenal cortical carcinoma | 28 | 28 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Phaeochromocytoma | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Appendix, neuroendocrine tumor (NET) | 3 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Colorectal, neuroendocrine tumor (NET) | 1 | 1 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ileum, neuroendocrine tumor (NET) | 4 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Lung, neuroendocrine tumor (NET) | 10 | 9 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pancreas, neuroendocrine tumor (NET) | 49 | 49 | 93.9 | 4.1 | 2.0 | 0.0 | |
| Colorectal, neuroendocrine carcinoma (NEC) | 2 | 2 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ileum, neuroendocrine carcinoma (NEC) | 8 | 8 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Pancreas, neuroendocrine carcinoma (NEC) | 3 | 3 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of haemotopoetic and lymphoid tissues | Hodgkin Lymphoma | 103 | 101 | 100.0 | 0.0 | 0.0 | 0.0 |
| Small lymphocytic lymphoma, B-cell type (B-SLL/B-CLL) | 50 | 48 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Diffuse large B cell lymphoma (DLBCL) | 113 | 108 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Follicular lymphoma | 88 | 84 | 100.0 | 0.0 | 0.0 | 0.0 | |
| T-cell Non Hodgkin lymphoma | 25 | 25 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Mantle cell lymphoma | 18 | 17 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Marginal zone lymphoma | 16 | 15 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Diffuse large B-cell lymphoma (DLBCL) in the testis | 16 | 16 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Burkitt lymphoma | 5 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Tumors of soft tissue and bone | Tenosynovial giant cell tumor | 45 | 45 | 100.0 | 0.0 | 0.0 | 0.0 |
| Granular cell tumor | 53 | 48 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leiomyoma | 50 | 50 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Leiomyosarcoma | 94 | 93 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Liposarcoma | 145 | 142 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Malignant peripheral nerve sheath tumor (MPNST) | 15 | 15 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Myofibrosarcoma | 26 | 26 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Angiosarcoma | 74 | 68 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Angiomyolipoma | 91 | 91 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Dermatofibrosarcoma protuberans | 21 | 19 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ganglioneuroma | 14 | 13 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Kaposi sarcoma | 8 | 6 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Neurofibroma | 117 | 97 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Sarcoma, not otherwise specified (NOS) | 74 | 72 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Paraganglioma | 41 | 40 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Ewing sarcoma | 23 | 19 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Rhabdomyosarcoma | 7 | 7 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Schwannoma | 122 | 112 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Synovial sarcoma | 12 | 11 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Osteosarcoma | 44 | 38 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Chondrosarcoma | 40 | 29 | 100.0 | 0.0 | 0.0 | 0.0 | |
| Rhabdoid tumor | 5 | 5 | 100.0 | 0.0 | 0.0 | 0.0 | |
Fig. 2.
KLK7 immunostaining in cancer. The panels show a focal KLK7 immunostaining (staining intensity (int) 2 + in 10% of tumor cells) in areas of keratinization of an HPV-negative squamous cell carcinoma of the vulva (A), the oral cavity (int 1 + in 10%) (B), and of the urinary bladder (int 1 + in 20%) (C). A focal KLK7 staining is also seen in a urothelial carcinoma of the bladder with focal squamous differentiation (int 2 + in 10%) (D) while KLK7 staining is absent in a poorly differentiated squamous cell carcinoma of the esophagus lacking keratinization (int 0) (E). KLK7 positivity of variable intensity is also seen in samples from a serous high-grade carcinoma of the ovary (int 2 + in 40%) (F) as well as from an intestinal adenocarcinoma of the stomach (int 2 + in 30%) (G) and the pancreas (int 2 + in 30%) (H)
Fig. 3.
Ranking order of KLK7 immunostaining in cancers. Both the percentage of positive cases (blue dots) and the percentage of strongly positive cases (orange dots) are shown
Table 2.
KLK7 immunostaining and tumor phenotype
| KLK7 immunostaining result | ||||||||
|---|---|---|---|---|---|---|---|---|
| n | negative (%) | weak (%) | moderate (%) | strong (%) | p | |||
| Adenocarinoma of the colon and rectum | Tumor stage | pT1 | 88 | 95.5 | 4.5 | 0.0 | 0.0 | 0.6353 |
| pT2 | 442 | 93.7 | 6.1 | 0.2 | 0.0 | |||
| pT3 | 1258 | 91.9 | 7.5 | 0.6 | 0.0 | |||
| pT4 | 451 | 92.0 | 7.5 | 0.4 | 0.0 | |||
| Grade | G1 | 4 | 100.0 | 0.0 | 0.0 | 0.0 | 0.8063 | |
| G2 | 489 | 90.2 | 8.6 | 1.0 | 0.2 | |||
| G3 | 55 | 87.3 | 12.7 | 0.0 | 0.0 | |||
| Lmyph node status | pN0 | 1177 | 94.0 | 5.9 | 0.1 | 0.1 | 0.0005 | |
| pN + | 1057 | 90.4 | 8.7 | 0.9 | 0.0 | |||
| Vascular infiltration status | V0 | 1611 | 92.8 | 7.0 | 0.1 | 0.1 | 0.0037 | |
| V + | 590 | 90.8 | 7.8 | 1.4 | 0.0 | |||
| Lymphatic infiltration status | L0 | 739 | 95.5 | 4.3 | 0.0 | 0.1 | < 0.0001 | |
| L1 | 1474 | 90.6 | 8.6 | 0.7 | 0.0 | |||
| Location | left | 1265 | 94.3 | 5.5 | 0.2 | 0.0 | 0.0088 | |
| right | 459 | 90.0 | 9.4 | 0.7 | 0.0 | |||
| Mismatch repair defective | 88 | 92.0 | 5.7 | 2.3 | 0.0 | |||
| Liver cancer | Tumor stage | pT1 | 72 | 95.8 | 1.4 | 2.8 | 0.0 | 0.1118 |
| pT2 | 109 | 87.2 | 11.0 | 0.9 | 0.9 | |||
| pT3-4 | 76 | 93.4 | 5.3 | 1.3 | 0.0 | |||
| Lmyph node status | pN0 | 93 | 93.5 | 4.3 | 2.2 | 0.0 | 0.0382 | |
| pN + | 62 | 80.6 | 16.1 | 3.2 | 0.0 | |||
| Grade | G 1 | 36 | 97.2 | 2.8 | 0.0 | 0.0 | 0.4041 | |
| G 2 | 157 | 90.4 | 6.4 | 2.5 | 0.6 | |||
| G 3 | 61 | 91.8 | 8.2 | 0.0 | 0.0 | |||
| Endometrioid endometrial carcinoma | Tumor stage | pT1 | 87 | 94.3 | 5.7 | 0.0 | 0.0 | 0.1207 |
| pT2 | 22 | 95.5 | 0.0 | 4.5 | 0.0 | |||
| pT3-4 | 27 | 88.9 | 11.1 | 0.0 | 0.0 | |||
| Lmyph node status | pN0 | 42 | 90.5 | 9.5 | 0.0 | 0.0 | 0.3566 | |
| pN + | 24 | 87.5 | 8.3 | 4.2 | 0.0 | |||
| Endometrioid carcinoma of the ovary | Tumor stage | pT1 | 24 | 83.3 | 16.7 | 0.0 | 0.0 | 0.3779 |
| pT2 | 4 | 75.0 | 25.0 | 0.0 | 0.0 | |||
| pT3 | 5 | 100.0 | 0.0 | 0.0 | 0.0 | |||
| Lmyph node status | pN0 | 19 | 84.2 | 15.8 | 0.0 | 0.0 | 0.6824 | |
| pN1 | 9 | 77.8 | 22.2 | 0.0 | 0.0 | |||
| Serous carcinoma of the ovary | Tumor stage | pT1 | 36 | 38.9 | 52.8 | 8.3 | 0.0 | 0.5420 |
| pT2 | 43 | 44.2 | 48.8 | 4.7 | 2.3 | |||
| pT3 | 261 | 49.4 | 40.2 | 9.6 | 0.8 | |||
| Lmyph node status | pN0 | 83 | 44.6 | 44.6 | 9.6 | 1.2 | 0.4142 | |
| pN1 | 163 | 47.2 | 46.0 | 6.7 | 0.0 | |||
| Pancreas adenocarcinoma | Tumor stage | pT1 | 15 | 73.3 | 26.7 | 0.0 | 0.0 | 0.2744 |
| pT2 | 72 | 63.9 | 29.2 | 6.9 | 0.0 | |||
| pT3 | 398 | 61.8 | 29.9 | 6.8 | 1.5 | |||
| pT4 | 31 | 74.2 | 12.9 | 12.9 | 0.0 | |||
| Grade | 1 | 17 | 70.6 | 23.5 | 0.0 | 5.9 | 0.3752 | |
| 2 | 366 | 62.3 | 29.8 | 7.4 | 0.5 | |||
| 3 | 109 | 64.2 | 27.5 | 6.4 | 1.8 | |||
| Lmyph node status | pN0 | 114 | 63.2 | 29.8 | 5.3 | 1.8 | 0.7658 | |
| pN + | 401 | 63.3 | 28.2 | 7.5 | 1.0 | |||
| Resection margin status | R0 | 261 | 67.0 | 26.4 | 5.7 | 0.8 | 0.2911 | |
| R1 | 215 | 59.5 | 30.7 | 7.9 | 1.9 | |||
| Mismatch repair status | proficient | 464 | 64.0 | 28.9 | 6.3 | 0.9 | 0.3143 | |
| deficient | 4 | 100.0 | 0.0 | 0.0 | 0.0 | |||
| Papillary thyroid carcinoma | ||||||||
| Tumor stage | pT1 | 140 | 98.6 | 1.4 | 0.0 | 0.0 | 0.0132 | |
| pT2 | 75 | 100.0 | 0.0 | 0.0 | 0.0 | |||
| pT3-4 | 98 | 91.8 | 7.1 | 0.0 | 1.0 | |||
| Lmyph node status | pN0 | 85 | 95.3 | 4.7 | 0.0 | 0.0 | 0.2076 | |
| pN + | 120 | 98.3 | 1.7 | 0.0 | 0.0 | |||
| Stomach cancer | Histological type | diffuse | 92 | 96.7 | 2.2 | 1.1 | 0.0 | 0.1035 |
| intestinal | 88 | 94.3 | 5.7 | 0.0 | 0.0 | |||
| mixed | 60 | 88.3 | 11.7 | 0.0 | 0.0 | |||
| Tumor stage | pT1-2 | 59 | 89.8 | 10.2 | 0.0 | 0.0 | 0.4286 | |
| pT3 | 128 | 89.8 | 10.2 | 0.0 | 0.0 | |||
| pT4 | 125 | 95.2 | 4.8 | 0.0 | 0.0 | |||
| Lmyph node status | pN0 | 77 | 93.9 | 6.1 | 0.0 | 0.0 | 0.5413 | |
| pN1 | 208 | 90.8 | 8.7 | 0.0 | 0.4 | |||
| Mismatch repair status | proficient | 255 | 90.2 | 8.6 | 0.8 | 0.4 | 0.8144 | |
| deficient | 40 | 90.0 | 10.0 | 0.0 | 0.0 | |||
| Squamous cell carcinoma of different sites* | Tumor stage | pT1 | 278 | 58.6 | 37.1 | 2.2 | 2.2 | 0.4833 |
| pT2 | 279 | 59.9 | 34.8 | 4.7 | 0.7 | |||
| pT3 | 150 | 54.0 | 42.0 | 2.7 | 1.3 | |||
| pT4 | 137 | 54.0 | 40.9 | 4.4 | 0.7 | |||
| Lmyph node status | pN0 | 331 | 58.3 | 36.3 | 3.9 | 1.5 | 0.3838 | |
| pN + | 321 | 60.7 | 36.4 | 2.2 | 0.6 | |||
| Grade | G1 | 34 | 44.1 | 38.2 | 14.7 | 2.9 | < 0.0001 | |
| G2 | 418 | 45.5 | 47.6 | 4.8 | 2.2 | |||
| G3 | 267 | 70.0 | 28.5 | 1.1 | 0.4 | |||
Abbrevations: pT pathological tumor stage, pN pathological lymph node status
*oral, pharynx, larynx, esophagus, lung, cervix, vagina, vulva, penis, skin, and anal canal
Discussion
Our successful analysis of 12,345 tumors from 147 entities identified KLK7 expression in 64 cancer categories and enabled a ranking of tumor types according to their KLK7 positivity rate.
The most commonly KLK7 positive cancers included squamous cell carcinomas from various sites of origin, ovarian carcinomas, pancreatic adenocarcinomas, and salivary gland tumors. This is largely consistent with RNA expression data from The Cancer Genome Atlas Research Network (https://www.cancer.gov/tcga), suggesting highest rates and levels of KLK7 expression in cancers of the head and neck, the ovary and the pancreas (Fig. 4). Our data markedly expand the available data on KLK7 protein expression in cancer. Fifty of the 64 tumor categories with at least occasional KLK7 expression had not been studied by IHC before. Previous IHC studies had analyzed a total of 1,646 samples from 14 different tumor entities (summarized in Fig. 5) and often described substantially higher positivity rates than found in this study. For example, KLK7 positivity rates of ≥ 90% have been described for breast cancers [17], ovarian carcinomas [12–14], oral squamous cell carcinomas [20, 22, 25], adenocarcinomas of the colon [21], adenocarcinoma of the cervix [23], and different subtypes of renal cell carcinomas [27] in individual studies. Most of these discrepancies are likely to be caused by the use of different antibodies, staining protocols and definitions of thresholds to determine KLK7 positivity. Existing RNA data, along with our IHC results, do not suggest a high frequency of high KLK7 expression levels in these tumor entities [35–38]. Other reasons for discrepant findings may include variable expression in a specific molecular background. This might particularly apply for the breast cancers in our study. The general lack of detectable KLK7 expression fits well to earlier studies reporting downregulation of KLK7 mRNA in breast cancers [39, 40], but there are also studies suggesting that KLK7 can be upregulated specifically in hormone receptor negative [41] and triple negative breast cancers [17].
Fig. 4.
KLK7 in the TCGA data set and in the literature. KLK7 mRNA expression in different cancer types. Data plot generated from the cBioPortal [42, 43] database querying 10,976 samples from the “TCGA PanCancer Atlas Studies” sample set including 32 studies with KLK7 mRNA sequencing data
Fig. 5.
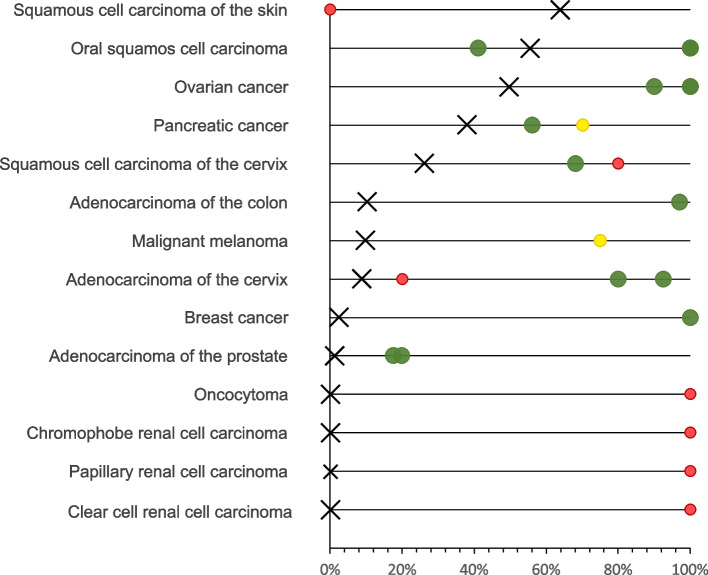
Comparison with previous KLK7 literature. Comparison of the percentage of samples with KLK7 expression (at least weak staining) with the corresponding data from published studies. An “X” indicates the fraction of KLK7 positive cancers in the present study, dots indicate the reported frequencies from the literature for comparison: red dots mark studies with ≤ 10 analyzed tumors, yellow dots mark studies with 11–25 analyzed tumors, and green dots mark studies with > 25 analyzed tumors. All studies are listed in the reference list
A frequent KLK7 expression in squamous cell carcinomas of various sites was expected based on the function of the protein in the desquamation of corneocytes. Desquamation of keratinized cells also occurs in cancers [44] and explains the expression of KLK7 in keratinized squamous cell carcinomas. Keratinization (or leucoplakia) is considered as a precancerous process in squamous epithelium [45–47] and defines a low histologic grade of malignancy in squamous cell carcinomas [48]. A significant association between KLK7 positivity and low grade could therefore be expected in these cancers. Cytokeratin 10, another protein that is specifically expressed in cornifying squamous epithelium, has also been found to be preferentially expressed in low grade squamous cell carcinomas [49]. In agreement with our observation, Leusink et al. found a link between strong KLK7 immunostaining and a favorable prognosis in a cohort of 83 squamous cell carcinomas of the oral cavity [26]. However, an association between high KLK7 protein levels and adverse pathological features or poor clinical outcome of squamous cell carcinomas has been described in two other studies on 30 and 80 cancers of the oral cavity [20, 25].
A poor prognosis of KLK7 expressing cancers could potentially be explained by in vitro and in vivo studies suggesting a relevant role of KLK7 in tumorigenesis, progression, and metastasis (summarized in [28]). The KLK7 zymogen is activated in the extracellular space to hydrolyze various extracellular matrix substrates (e.g. fibronectin 1/FN1 [50], thrombospondin 1/TSP1 [50]), cell membrane proteins (e.g. E-cadherin [11], desmoglein/DSG 1 and 2 [51]), secreted substrates (e.g. insulin growth factor binding protein/IGFBP 3 and 6 [50]), and other substrates (e.g. midkine [52], pro-matrix metalloprotease/MMP10 [50]). Studies suggested a direct or indirect effect of KLK7 on cell proliferation [19, 52], cell–cell adhesion [50, 51, 53], cell shedding [50], cell migration [50, 52, 54], cell invasion [11, 19, 52, 54], extracellular matrix organization [50, 55], EMT [11, 54], angiogenesis [28, 50], and metabolic reprogramming [28, 50] through the activation or inactivation of KLK7 substrates. For example, KLK7-mediated inactivation of E-cadherin – a hallmark of EMT [56]—and DSG1 and 2 promotes metastasis by inducing cell invasion and migration as well as reducing cell aggregation [11, 51]. KLK7-mediated inactivation of TSP1 may lead to cell proliferation, cell migration, cell invasion, and angiogenesis through the activation of the PI3K/Akt/mTOR pathway and VEGFR2 signaling [50, 57, 58]. KLK7-mediated inactivation of IGFBP3 and 6 may lead to cell proliferation through the activation of the PIK3/Akt, MEK/ERK, and Wnt/ß-Catenin pathways [50, 59] and changes in glycolysis, glycogen synthesis, and gluconeogenesis through the PI3K/Akt/mTOR pathway [50, 59, 60]. KLK7-mediated activation of proMMP10 and proMMP9 may leads to cell invasion and cell migration through the MMP-mediated hydrolysis of extracellular matrix and basement membrane components [50, 61, 62]. Furthermore, KLK7 overexpression resulted in a switch from a proliferative to an invasive phenotype in melanoma cells [19]. However, our own analyses of cancer entities with data on histomorphological features of malignancy did not provide evidence for a paramount prognostic impact of KLK7 expression on cancer malignancy. Only 7 of 28 analyses comparing KLK7 immunostaining and prognostic features resulted in significant associations. Because the number of statistical analyses was so large in our study (n = 28) that a statistical adjustment (Bonferroni correction: p ≤ 0.0018) invalidated almost all significant results, it was conspicuous that 4 of the 7 significant data involved nodal metastasis or tumor infiltration into blood or lymph vessels. These observations would indeed be consistent with a role of KLK7 associated cell dissociation in the process of metastasis.
It is an advantage of our study that only a single 0.6 mm tissue core was analyzed per cancer. The one punch per cancer approach allows for maximal standardization of the analysis because the same amount of tissue (approx. 0.28 mm2) is studied from each tumor. In contrast, the use of multiple punches per tumor almost inevitably leads to statistical bias because virtually never all samples from each tumor are interpretable. Others and we have earlier shown that one 0.6 mm core per cancer is sufficient to find clinically relevant associations between molecular markers and tumor phenotype (reviewed in [63]). For example, Torhorst et al. [64] analyzed 1–4 TMA cores each for p53, ER and PR and demonstrated that known associations between these markers and patient prognosis can be found irrespective of whether four tissue cores per tumor were analyzed separately, or a combined result was generated from the four cores.
As KLK7 is only expressed in a limited number of normal tissues and de-novo expression can occur in cancer, KLK7 may represent a suitable drug target for the treatment of cancer. Several studies have indeed demonstrated successful KLK7 inhibition by protein inhibitors and small molecule inhibitors in preclinical studies (summarized in [28]). In addition, some studies have already shown an antitumor effect of KLK7 inhibition. For example, small molecules inhibiting KLK7 have efficiently inhibited the proliferation, migration and invasion of pancreatic cancer cells [65], and cell shedding of plantar stratum corneum cells [6]. Our data suggest that ovarian, endometrial, pancreatic, and gallbladder carcinomas might represent suitable tumor entities for clinical trials once anti-KLK7 drugs will move towards testing in human patients.
Considering the large scale of our study, our assay was extensively validated by comparing our IHC findings in normal tissues with data obtained by another independent anti-KLK7 antibody and RNA data derived from three different publicly accessible databases [35–38]. To ensure an as broad as possible range of proteins to be tested for a possible cross-reactivity, 76 different normal tissues categories were included in this analysis. Validity of our assay was supported by the detection of significant KLK7 immunostaining in organs with documented KLK7 RNA expression (esophagus, salivary gland, kidney, vagina, cervix, fallopian tube, tonsil, and the skin) with the only exception of breast tissue which always stained KLK7 negative both on TMAs and on large sections. As KLK7 RNA was not detected in the FANTOM5 dataset [35, 36], it appears possible that some breast samples from other databases were contaminated by KLK7 RNAs derived from the skin covering breast tissues. KLK7 positivity in a fraction of cells in corpuscles of Hassall’s was the only IHC finding which was not matched by RNA data. This staining is conceivable, however, because of the squamous epithelial nature of corpuscles of Hassall’s. As for all other normal tissue results, KLK7 positivity of corpuscles of Hassall’s was confirmed by the use of a second independent antibody. In the thymus, the KLK7 positive cells constitute such a small fraction of the total number of cells that KLK7 RNA may not occur at detectable quantities in RNAs extracted from whole organ tissue samples. Independence of the two KLK7 antibodies used in this study is documented by cytoplasmic smooth muscle staining and nuclear staining in several organs (often predominating on nuclear membranes) which was seen by EPR22594-203 but not by MSVA-707M.
Conclusion
Our data provide a comprehensive overview on KLK7 expression in normal and neoplastic human tissues. KLK7 expression predominates in squamous cell carcinomas of different organs but is also common in ovarian and biliopancreatic adenocarcinomas as well as adenocarcinomas of the upper gastrointestinal tract. The prognostic relevance of KLK7 expression and the possible role of KLK7 as a drug target needs to be further investigated.
Supplementary Information
Acknowledgements
We are grateful to Melanie Witt, Inge Brandt, Maren Eisenberg, Laura Behm, and Sünje Seekamp for excellent technical assistance.
Abbreviations
- IHC
Immunohistochemistry
- KLK7
Kallikrein-related peptidase 7
- pT
Pathological tumor stage
- pN
Pathological lymph node status
- TMA
Tissue microarray
Authors’ contributions
SK, FJ, LSH, CPC, MK, RS, GS contributed to conception, design, data collection, data analysis, and manuscript writing. RiS, NG, ML, SDR, SK, VR, FV, CB, FB, TSC, CF, AH, FJ, WW, AM, TK, PL, SS, EB, SM, and AHM participated in pathology data analysis, data interpretation, and collection of samples. RS, MK, CPC, CHM: data analysis. RS, GS, FJ: study supervision. All authors agree to be accountable for the content of the work.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approvaland consent to participate
The local ethics committee (Ethics Commission Hamburg, WF-049/09) has approved the manufacturing and analysis of TMAs from archived remnants of diagnostic tissues. Patients data analysis without informed patient consent is covered by local laws (HmbKHG, §12).
The use of archived remnants of diagnostic tissues for manufacturing of TMAs and their analysis for research purposes as well as patient data analysis has been approved by local laws (HmbKHG, §12) and by the local ethics committee (Ethics Commission Hamburg, WF-049/09). All work has been carried out in compliance with the Helsinki.
Consent for publication
Not applicable.
Competing interests
The monoclonal mouse KLK7 antibody, clone MSVA-707M was provided from MS Validated Antibodies GmbH (owned by a family member of GS). All other Authors have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Clements J, Hooper J, Dong Y, Harvey T. The expanded human kallikrein (KLK) gene family: genomic organisation, tissue-specific expression and potential functions. Biol Chem. 2001;382(1):5–14. doi: 10.1515/BC.2001.002. [DOI] [PubMed] [Google Scholar]
- 2.Hong SK. Kallikreins as biomarkers for prostate cancer. Biomed Res Int. 2014;2014:526341. doi: 10.1155/2014/526341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avgeris M, Mavridis K, Scorilas A. Kallikrein-related peptidases in prostate, breast, and ovarian cancers: from pathobiology to clinical relevance. Biol Chem. 2012;393(5):301–317. doi: 10.1515/hsz-2011-0260. [DOI] [PubMed] [Google Scholar]
- 4.Yousef GM, Scorilas A, Magklara A, Soosaipillai A, Diamandis EP. The KLK7 (PRSS6) gene, encoding for the stratum corneum chymotryptic enzyme is a new member of the human kallikrein gene family - genomic characterization, mapping, tissue expression and hormonal regulation. Gene. 2000;254(1–2):119–128. doi: 10.1016/S0378-1119(00)00280-8. [DOI] [PubMed] [Google Scholar]
- 5.Caubet C, Jonca N, Brattsand M, Guerrin M, Bernard D, Schmidt R, Egelrud T, Simon M, Serre G. Degradation of corneodesmosome proteins by two serine proteases of the kallikrein family, SCTE/KLK5/hK5 and SCCE/KLK7/hK7. J Invest Dermatol. 2004;122(5):1235–1244. doi: 10.1111/j.0022-202X.2004.22512.x. [DOI] [PubMed] [Google Scholar]
- 6.de Veer SJ, Furio L, Swedberg JE, Munro CA, Brattsand M, Clements JA, Hovnanian A, Harris JM. Selective Substrates and Inhibitors for Kallikrein-Related Peptidase 7 (KLK7) Shed Light on KLK Proteolytic Activity in the Stratum Corneum. J Invest Dermatol. 2017;137(2):430–439. doi: 10.1016/j.jid.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Hansson L, Strömqvist M, Bäckman A, Wallbrandt P, Carlstein A, Egelrud T. Cloning, expression, and characterization of stratum corneum chymotryptic enzyme. A skin-specific human serine proteinase. J Biol Chem. 1994;269(30):19420–19426. doi: 10.1016/S0021-9258(17)32185-3. [DOI] [PubMed] [Google Scholar]
- 8.Levi K, Baxter J, Meldrum H, Misra M, Pashkovski E, Dauskardt RH. Effect of corneodesmosome degradation on the intercellular delamination of human stratum corneum. J Invest Dermatol. 2008;128(9):2345–2347. doi: 10.1038/jid.2008.107. [DOI] [PubMed] [Google Scholar]
- 9.Kishibe M. Physiological and pathological roles of kallikrein-related peptidases in the epidermis. J Dermatol Sci. 2019;95(2):50–55. doi: 10.1016/j.jdermsci.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Iakovlev V, Siegel ER, Tsao MS, Haun RS. Expression of kallikrein-related peptidase 7 predicts poor prognosis in patients with unresectable pancreatic ductal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2012;21(7):1135–1142. doi: 10.1158/1055-9965.EPI-11-1079. [DOI] [PubMed] [Google Scholar]
- 11.Johnson SK, Ramani VC, Hennings L, Haun RS. Kallikrein 7 enhances pancreatic cancer cell invasion by shedding E-cadherin. Cancer. 2007;109(9):1811–1820. doi: 10.1002/cncr.22606. [DOI] [PubMed] [Google Scholar]
- 12.Shigemasa K, Tanimoto H, Underwood LJ, Parmley TH, Arihiro K, Ohama K, O'Brien TJ. Expression of the protease inhibitor antileukoprotease and the serine protease stratum corneum chymotryptic enzyme (SCCE) is coordinated in ovarian tumors. Int J Gynecol Cancer. 2001;11(6):454–461. doi: 10.1136/ijgc-00009577-200111000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Wang P, Magdolen V, Seidl C, Dorn J, Drecoll E, Kotzsch M, Yang F, Schmitt M, Schilling O, Rockstroh A, et al. Kallikrein-related peptidases 4, 5, 6 and 7 regulate tumour-associated factors in serous ovarian cancer. Br J Cancer. 2018;119(7):1–9. doi: 10.1038/s41416-018-0260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorn J, Gkazepis A, Kotzsch M, Kremer M, Propping C, Mayer K, Mengele K, Diamandis EP, Kiechle M, Magdolen V, et al. Clinical value of protein expression of kallikrein-related peptidase 7 (KLK7) in ovarian cancer. Biol Chem. 2014;395(1):95–107. doi: 10.1515/hsz-2013-0172. [DOI] [PubMed] [Google Scholar]
- 15.Xuan Q, Yang X, Mo L, Huang F, Pang Y, Qin M, Chen Z, He M, Wang Q, Mo ZN. Expression of the serine protease kallikrein 7 and its inhibitor antileukoprotease is decreased in prostate cancer. Arch Pathol Lab Med. 2008;132(11):1796–1801. doi: 10.5858/132.11.1796. [DOI] [PubMed] [Google Scholar]
- 16.Zhang CY, Zhu Y, Rui WB, Dai J, Shen ZJ. Expression of kallikrein-related peptidase 7 is decreased in prostate cancer. Asian J Androl. 2015;17(1):106–110. doi: 10.1002/asia.201402962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng X, Babayeva L, Walch A, Aubele M, Groß E, Kiechle M, Bronger H, Dreyer T, Magdolen V, Dorn J. High levels of KLK7 protein expression are related to a favorable prognosis in triple-negative breast cancer patients. Am J Cancer Res. 2020;10(6):1785–1792. [PMC free article] [PubMed] [Google Scholar]
- 18.Santin AD, Cane' S, Bellone S, Bignotti E, Palmieri M, De Las Casas LE, Roman JJ, Anfossi S, O'Brien T, Pecorelli S. The serine protease stratum corneum chymotryptic enzyme (kallikrein 7) is highly overexpressed in squamous cervical cancer cells. Gynecol Oncol. 2004;94(2):283–288. doi: 10.1016/j.ygyno.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 19.Delaunay T, Deschamps L, Haddada M, Walker F, Soosaipillai A, Soualmia F, El Amri C, Diamandis EP, Brattsand M, Magdolen V, et al. Aberrant expression of kallikrein-related peptidase 7 is correlated with human melanoma aggressiveness by stimulating cell migration and invasion. Mol Oncol. 2017;11(10):1330–1347. doi: 10.1002/1878-0261.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar DV, Sivaranjani Y, Rao GV. Immunohistochemical expression of kallikrein 7 in oral squamous cell carcinoma. J Oral Maxillofac Pathol. 2020;24(3):580. doi: 10.4103/jomfp.JOMFP_244_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker F, Nicole P, Jallane A, Soosaipillai A, Mosbach V, Oikonomopoulou K, Diamandis EP, Magdolen V, Darmoul D. Kallikrein-related peptidase 7 (KLK7) is a proliferative factor that is aberrantly expressed in human colon cancer. Biol Chem. 2014;395(9):1075–1086. doi: 10.1515/hsz-2014-0142. [DOI] [PubMed] [Google Scholar]
- 22.Pettus JR, Johnson JJ, Shi Z, Davis JW, Koblinski J, Ghosh S, Liu Y, Ravosa MJ, Frazier S, Stack MS. Multiple kallikrein (KLK 5, 7, 8, and 10) expression in squamous cell carcinoma of the oral cavity. Histol Histopathol. 2009;24(2):197–207. doi: 10.14670/hh-24.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Termini L, Maciag PC, Soares FA, Nonogaki S, Pereira SM, Alves VA, Longatto-Filho A, Villa LL. Analysis of human kallikrein 7 expression as a potential biomarker in cervical neoplasia. Int J Cancer. 2010;127(2):485–490. doi: 10.1002/ijc.25046. [DOI] [PubMed] [Google Scholar]
- 24.Tian X, Shigemasa K, Hirata E, Gu L, Uebaba Y, Nagai N, O'Brien TJ, Ohama K. Expression of human kallikrein 7 (hK7/SCCE) and its inhibitor antileukoprotease (ALP/SLPI) in uterine endocervical glands and in cervical adenocarcinomas. Oncol Rep. 2004;12(5):1001–1006. [PubMed] [Google Scholar]
- 25.Zhao H, Dong Y, Quan J, Smith R, Lam A, Weinstein S, Clements J, Johnson NW, Gao J. Correlation of the expression of human kallikrein-related peptidases 4 and 7 with the prognosis in oral squamous cell carcinoma. Head Neck. 2011;33(4):566–572. doi: 10.1002/hed.21496. [DOI] [PubMed] [Google Scholar]
- 26.Leusink FK, van Diest PJ, Frank MH, Broekhuizen R, Braunius W, van Hooff SR, Willems SM, Koole R. The Co-Expression of Kallikrein 5 and Kallikrein 7 Associates with Poor Survival in Non-HPV Oral Squamous-Cell Carcinoma. Pathobiology. 2015;82(2):58–67. doi: 10.1159/000381904. [DOI] [PubMed] [Google Scholar]
- 27.Gabril M, White NM, Moussa M, Chow TF, Metias SM, Fatoohi E, Yousef GM. Immunohistochemical analysis of kallikrein-related peptidases in the normal kidney and renal tumors: potential clinical implications. Biol Chem. 2010;391(4):403–409. doi: 10.1515/bc.2010.025. [DOI] [PubMed] [Google Scholar]
- 28.Xiang F, Wang Y, Cao C, Li Q, Deng H, Zheng J, Liu X, Tan X. The Role of Kallikrein 7 in Tumorigenesis. Curr Med Chem. 2022;29(15):2617–2631. doi: 10.2174/0929867328666210915104537. [DOI] [PubMed] [Google Scholar]
- 29.Rico SD, Mahnken M, Buscheck F, Dum D, Luebke AM, Kluth M, Hube-Magg C, Hinsch A, Hoflmayer D, Moller-Koop C, et al. MUC5AC Expression in Various Tumor Types and Nonneoplastic Tissue: A Tissue Microarray Study on 10 399 Tissue Samples. Technol Cancer Res Treat. 2021;20:15330338211043328. doi: 10.1177/15330338211043328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steurer S, Riemann C, Buscheck F, Luebke AM, Kluth M, Hube-Magg C, Hinsch A, Hoflmayer D, Weidemann S, Fraune C, et al. p63 expression in human tumors and normal tissues: a tissue microarray study on 10,200 tumors. Biomark Res. 2021;9(1):7. doi: 10.1186/s40364-021-00260-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Uhlig R, Gunther K, Broker N, Gorbokon N, Lennartz M, Dwertmann Rico S, Reiswich V, Viehweger F, Buscheck F, Kluth M, et al. Diagnostic and prognostic role of pancreatic secretory granule membrane major glycoprotein 2 (GP2) immunohistochemistry: A TMA study on 27,681 tumors. Pathol Res Pract. 2022;238:154123. doi: 10.1016/j.prp.2022.154123. [DOI] [PubMed] [Google Scholar]
- 32.Bubendorf L, Nocito A, Moch H, Sauter G. Tissue microarray (TMA) technology: miniaturized pathology archives for high-throughput in situ studies. J Pathol. 2001;195(1):72–79. doi: 10.1002/path.893. [DOI] [PubMed] [Google Scholar]
- 33.Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4(7):844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 34.Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E, Rimm DL, Rodriguez H, Hiltke T, Snyder M, et al. A proposal for validation of antibodies. Nat Methods. 2016;13(10):823–827. doi: 10.1038/nmeth.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lizio M, Harshbarger J, Shimoji H, Severin J, Kasukawa T, Sahin S, Abugessaisa I, Fukuda S, Hori F, Ishikawa-Kato S, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lizio M, Harshbarger J, Abugessaisa I, Noguchi S, Kondo A, Severin J, Mungall C, Arenillas D, Mathelier A, Medvedeva YA, et al. Update of the FANTOM web resource: high resolution transcriptome of diverse cell types in mammals. Nucleic Acids Res. 2017;45(D1):D737–D743. doi: 10.1093/nar/gkw995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium G The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, Sjöstedt E, Butler L, Odeberg J, Dusart P, et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7(31):eabh2169. doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yousef GM, Yacoub GM, Polymeris ME, Popalis C, Soosaipillai A, Diamandis EP. Kallikrein gene downregulation in breast cancer. Br J Cancer. 2004;90(1):167–172. doi: 10.1038/sj.bjc.6601451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, Liu J, Wang Y, Zhang L, Ning L, Feng Y. Parallel underexpression of kallikrein 5 and kallikrein 7 mRNA in breast malignancies. Cancer Sci. 2009;100(4):601–607. doi: 10.1111/j.1349-7006.2009.01090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talieri M, Diamandis EP, Gourgiotis D, Mathioudaki K, Scorilas A. Expression analysis of the human kallikrein 7 (KLK7) in breast tumors: a new potential biomarker for prognosis of breast carcinoma. Thromb Haemost. 2004;91(1):180–186. doi: 10.1160/TH03-05-0261. [DOI] [PubMed] [Google Scholar]
- 42.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okegawa T, Pong RC, Li Y, Hsieh JT. The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim Pol. 2004;51(2):445–457. doi: 10.18388/abp.2004_3583. [DOI] [PubMed] [Google Scholar]
- 45.Kramer IR, Lucas RB, Pindborg JJ, Sobin LH. Definition of leukoplakia and related lesions: an aid to studies on oral precancer. Oral Surg Oral Med Oral Pathol. 1978;46(4):518–539. doi: 10.1016/0030-4220(78)90383-3. [DOI] [PubMed] [Google Scholar]
- 46.van Harten AM, Brakenhoff RH. Targeted Treatment of Head and Neck (Pre) Cancer: Preclinical Target Identification and Development of Novel Therapeutic Applications. Cancers (Basel) 2021;13(11):2774. doi: 10.3390/cancers13112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taggart MW, Rashid A, Ross WA, Abraham SC. Oesophageal hyperkeratosis: clinicopathological associations. Histopathology. 2013;63(4):463–473. doi: 10.1111/his.12195. [DOI] [PubMed] [Google Scholar]
- 48.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol. 2015;10(9):1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 49.Uhlig R, Abboud M, Gorbokon N, Lennartz M, Dwertmann Rico S, Kind S, Reiswich V, Viehweger F, Kluth M, Hube-Magg C, et al. Cytokeratin 10 (CK10) expression in cancer: A tissue microarray study on 11,021 tumors. Ann Diagn Pathol. 2022;60:152029. doi: 10.1016/j.anndiagpath.2022.152029. [DOI] [PubMed] [Google Scholar]
- 50.Silva LM, Kryza T, Stoll T, Hoogland C, Dong Y, Stephens CR, Hastie ML, Magdolen V, Kleifeld O, Gorman JJ, et al. Integration of Two In-depth Quantitative Proteomics Approaches Determines the Kallikrein-related Peptidase 7 (KLK7) Degradome in Ovarian Cancer Cell Secretome. Mol Cell Proteomics. 2019;18(5):818–836. doi: 10.1074/mcp.RA118.001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramani VC, Hennings L, Haun RS. Desmoglein 2 is a substrate of kallikrein 7 in pancreatic cancer. BMC Cancer. 2008;8:373. doi: 10.1186/1471-2407-8-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu Y, Prassas I, Dimitromanolakis A, Diamandis EP. Novel Biological Substrates of Human Kallikrein 7 Identified through Degradomics. J Biol Chem. 2015;290(29):17762–17775. doi: 10.1074/jbc.M115.643551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramani VC, Haun RS. Expression of kallikrein 7 diminishes pancreatic cancer cell adhesion to vitronectin and enhances urokinase-type plasminogen activator receptor shedding. Pancreas. 2008;37(4):399–404. doi: 10.1097/MPA.0b013e31817f76f7. [DOI] [PubMed] [Google Scholar]
- 54.Mo L, Zhang J, Shi J, Xuan Q, Yang X, Qin M, Lee C, Klocker H, Li QQ, Mo Z. Human kallikrein 7 induces epithelial-mesenchymal transition-like changes in prostate carcinoma cells: a role in prostate cancer invasion and progression. Anticancer Res. 2010;30(9):3413–3420. [PubMed] [Google Scholar]
- 55.Ramani VC, Haun RS. The extracellular matrix protein fibronectin is a substrate for kallikrein 7. Biochem Biophys Res Commun. 2008;369(4):1169–1173. doi: 10.1016/j.bbrc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 56.Wong SHM, Fang CM, Chuah LH, Leong CO, Ngai SC. E-cadherin: Its dysregulation in carcinogenesis and clinical implications. Crit Rev Oncol Hematol. 2018;121:11–22. doi: 10.1016/j.critrevonc.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 57.Bazzazi H, Zhang Y, Jafarnejad M, Isenberg JS, Annex BH, Popel AS. Computer Simulation of TSP1 Inhibition of VEGF-Akt-eNOS: An Angiogenesis Triple Threat. Front Physiol. 2018;9:644. doi: 10.3389/fphys.2018.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bussolati B, Assenzio B, Deregibus MC, Camussi G. The proangiogenic phenotype of human tumor-derived endothelial cells depends on thrombospondin-1 downregulation via phosphatidylinositol 3-kinase/Akt pathway. J Mol Med (Berl) 2006;84(10):852–863. doi: 10.1007/s00109-006-0075-z. [DOI] [PubMed] [Google Scholar]
- 59.Bach LA. What Happened to the IGF Binding Proteins? Endocrinology. 2018;159(2):570–578. doi: 10.1210/en.2017-00908. [DOI] [PubMed] [Google Scholar]
- 60.Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20(2):74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramani VC, Kaushal GP, Haun RS. Proteolytic action of kallikrein-related peptidase 7 produces unique active matrix metalloproteinase-9 lacking the C-terminal hemopexin domains. Biochim Biophys Acta. 2011;1813(8):1525–1531. doi: 10.1016/j.bbamcr.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davidson B, Reich R, Risberg B, Nesland JM. The biological role and regulation of matrix metalloproteinases (MMP) in cancer. Arkh Patol. 2002;64(3):47–53. [PubMed] [Google Scholar]
- 63.Sauter G. Representativity of TMA studies. Methods Mol Biol. 2010;664:27–35. doi: 10.1007/978-1-60761-806-5_3. [DOI] [PubMed] [Google Scholar]
- 64.Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, Kochli OR, Mross F, Dieterich H, Moch H, Mihatsch M, et al. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol. 2001;159(6):2249–2256. doi: 10.1016/S0002-9440(10)63075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Du JP, Li L, Zheng J, Zhang D, Liu W, Zheng WH, Li XS, Yao RC, Wang F, Liu S, et al. Kallikrein-related peptidase 7 is a potential target for the treatment of pancreatic cancer. Oncotarget. 2018;9(16):12894–12906. doi: 10.18632/oncotarget.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.