Abstract
Reverse transcriptase (RT) switches templates frequently during DNA synthesis; the acceptor template can be the same RNA (intramolecular) or the copackaged RNA (intermolecular). Previous results indicated that intramolecular template switching occurred far more frequently than intermolecular template switching. We hypothesized that intermolecular template-switching events (recombination) occurred at a lower efficiency because the copackaged RNA was not accessible to the RT. To test our hypothesis, the murine leukemia virus (MLV) extended packaging signal (Ψ+) containing a dimer linkage structure (DLS) was relocated from the 5′ untranslated region (UTR) to between selectable markers, allowing the two viral RNAs to interact closely in this region. It was found that the overall maximum recombination rates of vectors with Ψ+ in the 5′ UTR or Ψ+ between selectable markers were not drastically different. However, vectors with Ψ+ located between selectable markers reached a plateau of recombination rate at a shorter distance. This suggested a limited enhancement of recombination by Ψ+. The locations of the recombination events were also examined by using restriction enzyme markers. Recombination occurred in all four regions between the selectable markers; the region containing 5′ Ψ+ including DLS did not undergo more recombination than expected from the size of the region. These experiments indicated that although the accessibility of the copackaged RNA was important in recombination, other factors existed to limit the number of viruses that were capable of undergoing intermolecular template switching. In addition, recombinants with multiple template switches were observed at a frequency much higher than expected, indicating the presence of high negative interference in the MLV-based system. This extends our observation with the spleen necrosis virus system and suggests that high negative interference may be a common phenomenon in retroviral recombination.
Retroviruses package two copies of viral RNA into each virion (9, 28). During reverse transcription, both copackaged RNAs can be used as templates to produce a recombinant with a mixture of genetic information from each of the parental RNAs (8, 18). Retroviruses recombine at high rates (6, 15, 23, 24, 26, 29, 30, 46–48). Using murine leukemia virus (MLV)-based vectors with markers separated by 1.0, 1.9, and 7.1 kb, recombination rates of 4.7, 7.4, and 8.2%, respectively, were observed in one round of viral replication (2).
During reverse transcription of the viral genome, the virus-encoded enzyme reverse transcriptase (RT) has to perform two template-switching events (named minus- and plus-strand DNA transfer) to complete the synthesis of viral DNA (12). It has been hypothesized that RT is evolutionarily selected to have a low affinity to the template and low processivity in order to complete the two obligatory template-switching events (44). A consequence of this low processivity is that RT may also perform other nonobligatory template-switching events during reverse transcription. Because two copies of RNA are present in the virion, after dissociation from the template used for DNA synthesis, RT may reassociate with the same RNA or switch to the copackaged RNA, resulting in intramolecular or intermolecular template-switching events, respectively. Between these two events, only intermolecular template switching generates recombination.
The rates of intramolecular and intermolecular template switching were measured in a spleen necrosis virus (SNV)-based system (17). It was found that intramolecular template switching occurred far more frequently than intermolecular template switching. Furthermore, when recombination was selected in one region of the viral genome, intermolecular template switching was more likely to be observed in another region of the genome. In contrast, when recombination was not selected, intermolecular template switching was not observed in a different region of the viral genome; however, frequent intramolecular template switching was still observed. This observation led to the hypothesis that two viral populations exist. In the first population (recombining population), RT could frequently switch templates to the same RNA or the copackaged RNA. In the second population (nonrecombining population), RT could only switch to the same RNA frequently. Because frequent template switching occurred in both populations, the difference seemed to be the ability of the RT to interact with the other template. We hypothesized that if the barrier to intermolecular template switching is the ability to access the other RNA, then it should be possible to increase recombination by bringing the two copackaged RNAs closer together. In our experimental system, recombination is measured by the presence of two selectable markers, one from each parent, in the progeny provirus. Therefore, to test our hypothesis, the region between the two selectable markers from the two copackaged RNAs should be in close proximity to observe any effect on recombination.
Electron microscopy studies demonstrated that the two copackaged retroviral RNAs were dimerized at the 5′ end of the RNA (27, 28, 32, 39). Genetic and biochemical studies defined a region important for dimerization through noncovalent interaction between the two viral RNAs (13, 40, 42). The region important for dimerization of MLV RNAs, the dimer linkage structure (DLS), is part of the packaging signal (Ψ) located within the 5′ untranslated region (UTR) (11, 13, 39, 40, 42). In the current report, we examined the effect of the naturally occurring dimerization between the two copackaged RNAs on recombination. The extended MLV packaging signal (Ψ+), including the DLS, was relocated from the 5′ UTR to the middle of the viral genome between two selectable markers. Recombination rates between vectors with Ψ+ located in the 5′ UTR were directly compared to those from vectors with Ψ+ located between the selectable markers.
In addition, using a forced-recombination experiment, it was previously observed that the DLS was a putative recombination hot spot (31, 34, 35). To determine the general effect of the DLS on the location of template-switching events, a set of vectors that contained five sets of restriction enzyme markers was used to dissect the locations of template-switching events after one round of retroviral replication.
MATERIALS AND METHODS
Plasmid construction.
Vectors JS30 and JA32-1kb have been described previously (2); to avoid confusion, JA32-1kb is abbreviated as JA32 in this report. Vectors pJA33-1.3kb, pJA9, pJA10ΨNeo, pJA11HyΨ, pJA23, pJA19ΨNeo, and pJA20HyΨ were constructed by standard molecular cloning techniques (33). All of the MLV-based vectors were constructed with various derivatives of pLAEN (38) as backbones. To generate pJA33-1.3kb, pJS30 (2) was partially digested with NdeI, treated with calf intestinal phosphatase (CIP), and ligated to a linker (5′-TAACGCGT-3′) to create a unique MluI site and an 8-bp insertion in the hygromycin phosphotransferase B gene (hygro) (14). To generate pJA9, pWH390-Cla was digested with EcoRI and ligated to annealed linkers PL1 (5′-AATTAGGCCTCATATGCTAGCCTCGAG-3′) and PL2 (5′-AATTCTCGAGGCTAGCATATGAGGCCT-3′) to generate pBWB-1. pBWB-1 was digested with ClaI and treated with CIP and Escherichia coli DNA polymerase I large fragment (Klenow) to fill in the recessive ends. This treated pBWB-1 DNA was ligated to a 1.0-kb DNA fragment containing hygro to generate pJA30ΨBWB2. pJA30ΨBWB2 was digested with XhoI plus BclI, treated with Klenow fragment to fill in the recessive ends, and self-ligated to remove the 0.6-kb sequence containing the internal ribosomal entry site (IRES) (1, 19, 20) to form pCN2. pCN2 was digested with XhoI, treated with CIP, and ligated to annealed linkers 5′ neoJA (5′-TCGAGGTCGACGCGGCCGCCA-3′) and 5′oenJA (5′-TCGATGCGGCCGCGTCGACC-3′). The resulting plasmid, pJA5, contained unique XhoI and NotI restriction sites immediately upstream of the neomycin phosphotransferase gene (neo) (22). pJA5 was partially digested with BamHI, treated with Klenow fragment to fill in recessive ends, and self-ligated to generate pJA6, which contains a unique BamHI site immediately downstream of hygro. pJA6 was digested to completion with BamHI, treated with CIP, and ligated to annealed linkers 3′hygroJA (5′-GATCCGTCGACAAGCTT-3′) and 3′ orgyhJA (5′-GATCAAGCTTGTCGACG-3′). The resulting plasmid, pJA7, contained unique BamHI and HindIII restriction sites immediately downstream of hygro. pJA7 was digested with BamHI and HindIII, treated with CIP, and ligated to annealed linkers Jarzts1 (5′-GATCCGAATGCATCGTGTCAAGTTAGGTCTGCGTA-3′) and JA1stzr (5′-AGCTTACGCAGACCTAACTTGACACGATGCATTCG-3′) to generate pJA8. pJA8 was digested with XhoI plus NotI, treated with CIP, and ligated to annealed linkers Jats2 (5′-TCGAGCATGCATATGTCTCTAACTCTTCGATGCCCGTTCTGTCTACTTGTGC-3′) and JA2st (5′-GGCCGCA CAAGTAGACAGAACGGGCATCGAAGAGTTAGAGACATATGCATGC- 3′) to generate pJA9. To generate pJA10ΨNeo, pJA9 was partially digested with SacII, treated with T4 DNA polymerase to remove the protruding 2 bp at the 3′ termini, and self-ligated. The resulting plasmid, pJA10ΨNeo, contained an NaeI site in the inactivated hygro. To generate pJA11HyΨ, pJA9 was partially digested with NarI (an isoschizomer of EheI), treated with Klenow fragment to fill in recessive ends, and self-ligated. These procedures introduced a BssHII site and inactivated neo in pJA11HyΨ. pJA9 was partially digested with FspI, treated with CIP, and ligated to a linker (5′-GGACGTCC-3′); these procedures generated pJA14, which contained an 8-bp insertion and an AatII site in neo. pJA15 was generated by partial digestion of pJA9 with NcoI followed by Klenow fill-in reaction and self-ligation. These procedures introduced a 4-bp insertion to generate an NsiI site and inactivated hygro. pJA16 was generated by complete digestion of pJA14 with BamHI followed by Klenow fill-in reaction and self-ligation; these procedures introduced a ClaI site immediately downstream of hygro. pJA17 was generated by complete digestion of pJA15 with XhoI, followed by Klenow fill-in reaction and self-ligation; these procedures generated a PvuI site immediately upstream of neo. pJA17 was partially digested with AflII, treated with CIP, and ligated to a linker, JAmlu (5′-TTAACGCG-3′), to generate pJA18; these procedures introduced a unique MluI site in the gag-coding region included in the MLV extended packaging signal Ψ+. To generate pJA19ΨNeo, the splice donor site of pJA18 was mutated from AGGTAAG to TCGACAG by overlapping PCR mutagenesis that also created a unique SalI site. To generate pJA20HyΨ, pJA16 was digested with AscI plus EcoRI, treated with CIP, and ligated to the 0.76-kb AscI-EcoRI-digested DNA fragment from pJA19ΨNeo. pJA23 was constructed by digesting pJA9 with AscI plus EcoRI, treating it with CIP, and ligating it to the same 0.76-kb DNA fragment from pJA19ΨNeo. All of the plasmids were characterized by restriction enzyme mapping to confirm the general structures. DNA sequencing was performed to ensure that the PCR-amplified DNA used for cloning did not introduce any inadvertent mutations. In addition, DNA sequencing was also performed in some plasmids to ensure that only one linker was inserted into the plasmids.
Cells, DNA transfections, and virus propagations.
All cells were obtained from the American Type Culture Collection. PA317 is a murine cell line that expresses MLV gag-pol and env (36). PG13 is a murine cell line that expresses MLV gag-pol and gibbon ape leukemia virus (GaLV) env (37). D17 is a dog osteosarcoma cell line permissive to infection by MLV (41).
All cells were grown in Dulbecco's modified Eagle's medium supplemented with either 6% (D17) or 10% (PA317 and PG13) calf serum. Cells were maintained in a 37°C incubator with 5% CO2. Hygromycin selection was performed at 120 μg/ml (D17 and PA317) or 300 μg/ml (PG13). Selection with G418, an analog of neomycin, was performed at 400 μg/ml (D17 and PA317) or 600 μg/ml (PG13). Double-drug selection was performed with 96 μg of hygromycin per ml and 320 μg of G418 per ml for D17 cells and 240 μg of hygromycin per ml and 480 μg of G418 per ml for PG13 cells.
DNA transfections were performed by either the dimethyl sulfoxide-Polybrene (25) or the calcium phosphate precipitation method (33). Viral infections were performed immediately following viral harvest. Viruses were collected from helper cells and centrifuged at 3,000 × g for 10 min to remove cellular debris. Ten-fold serial dilutions of each viral stock were generated, and the viral infections were performed in the presence of 50 μg of Polybrene per ml. Viral titers were determined by the number of hygromycin-, G418-, or hygromycin-plus-G418-resistant cells. Each titer shown was from one experiment.
Southern hybridization analysis.
Genomic DNA purification, digestion, and hybridization were performed by standard molecular techniques (33). DNA transfers were performed with a vacuum blotter (Pharmacia). All blots were hybridized with probe generated from either a 1.3-kb MluI-EheI fragment of pJA33-1.3kb, a 1.9-kb NcoI-NcoI fragment of pJS30, or a 1.3-kb NcoI-FspI fragment of pJS30. Probes were generated by the random-priming method with [α-32P]dCTP (specific activity of >109 cpm/μg of DNA) (10). Southern hybridization results were obtained by autoradiography or PhosphorImager analysis (Molecular Dynamics).
Mapping of recombinant proviruses.
DNA lysates were prepared from double-drug-resistant cell clones and used as substrates for PCR (16). Proviral genomes were amplified with hygro- and neo-specific primers JA16-995 (5′-GGATATGTCCTGCGGGTAAATAGC-3′) and 16AJ-3327 (5′-ATCGACAAGACCGGCTTCCATCCG-3′), respectively. All PCRs were carried out in a Hybaid Omnigene thermal cycler for 35 cycles. Amplified DNAs were analyzed with various restriction enzyme digestions, separated by gel electrophoresis, and visualized by ethidium bromide staining. All DNA manipulations were performed by standard procedures (33).
RESULTS
Retroviral vectors used to study the effect of the MLV extended packaging signal on retroviral recombination.
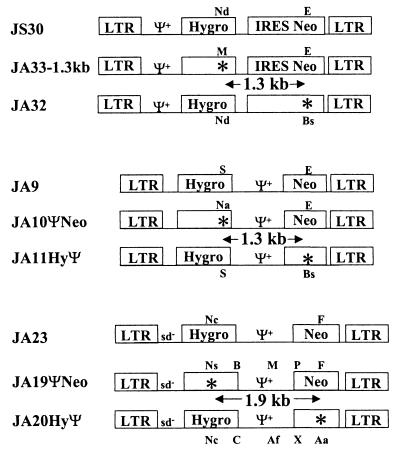
To examine the effect of the MLV Ψ+ on retroviral recombination, we constructed a set of vectors in which Ψ+ was located in the 5′ UTR and another set of vectors in which Ψ+ was located in the middle of the viral genome between two selectable markers. Both sets of vectors were derived from MLV and contained cis-acting elements required for viral replication and gene expression. The first set of vectors (pJS30, pJA33-1.3kb, and pJA32) contained Ψ+ in the 5′ UTR (Fig. 1). These vectors also contained hygro and neo; both genes were expressed by transcripts initiated from the long terminal repeat (LTR). The translation of neo was directed by an IRES from encephalomyocarditis virus (1, 19, 20). These three vectors were highly homologous (>99%) to one another. Vector pJS30 contained functional hygro and neo (14, 22). Vector pJA33-1.3kb contained a functional neo and a nonfunctional hygro with an 8-bp frameshift insertion that destroyed an NdeI site and generated an MluI site. Vector pJA32 (2) contained a functional hygro and a nonfunctional neo with a 4-bp frameshift insertion that destroyed an EheI site and generated a BssHII site. The distance between the two inactivating mutations in hygro and neo was 1.3 kb. Frameshift mutations of more than 1 bp were used to inactivate genes because they exhibit a low reversion rate (2, 18).
FIG. 1.
MLV vectors used to determine the role of the extended packaging signal (Ψ+) in retroviral recombination. sd−, mutated splice donor site; ∗, inactivating frameshift mutation; Nd, NdeI; E, EheI; M, MluI; Bs, BssHII; S, SacII; Na, NaeI; Ns, NsiI; B, BamHI; P, PvuI; F, FspI; Nc, NcoI; C, ClaI; Af, AflII; X, XhoI; Aa, AatII. Translation of Neo in these constructs was directed by IRES from encephalomyocarditis virus or from MLV Ψ+.
The second set of vectors (pJA9, pJA10ΨNeo, and pJA11HyΨ) also contained hygro and neo expressed from U3-regulated transcripts (Fig. 1). However, MLV Ψ+ was moved from the 5′ UTR to between hygro and neo; in these vectors, the translation of neo was directed by the IRES in the MLV packaging signal (4, 45). Vector pJA9 contained functional hygro and neo. Vector pJA10ΨNeo contained a functional neo and a nonfunctional hygro with a 2-bp frameshift deletion that destroyed a SacII site and introduced an NaeI site. Vector pJA11HyΨ contained a functional hygro and a nonfunctional neo with a 2-bp frameshift insertion that destroyed an EheI site and introduced a BssHII site. The distance between the two inactivating mutations in hygro and neo was 1.3 kb.
Experimental protocol used to measure the rates of recombination in one replication cycle.
The protocol used to determine the rate of homologous recombination is illustrated in Fig. 2 with vector set pJA33-1.3kb and pJA32 as an example. Vectors pJA33-1.3kb and pJA32 were separately transfected into PA317 helper cells, selected with either G418 or hygromycin, and the resulting colonies were pooled separately. The pool size for each vector was greater than 160 colonies. Viruses were harvested from these two PA317 cell pools and used to infect PG13 helper cells simultaneously. PG13 cell clones resistant to hygromycin plus G418 were isolated, and the proviral structures in these cell clones were characterized by Southern hybridization analysis. Cell clones containing one intact copy of each of the JA33-1.3kb and JA32 proviruses were selected. Three types of virions can be produced from these dual-infected PG13 cells: virions containing two copies of JA33-1.3kb RNA, virions containing two copies of JA32 RNA, and virions containing one copy of each of the JA33-1.3kb and JA32 RNAs (heterozygotic virions). The first two types of virions generate proviruses containing one functional drug resistance gene that confers resistance to a single drug. Recombination can occur in the heterozygotic virions to generate proviruses that contain two functional drug resistance genes that confer resistance to both drugs (18, 48). Viruses were harvested from the selected PG13 cell clones and used to infect D17 target cells. Infected D17 cells were subjected to single (hygromycin or G418) or double (hygromycin plus G418) drug selection. Virus titers were then determined by counting the numbers of drug-resistant colonies. Recombination rates were calculated from the double- and the single-drug-resistant colony titers (18). In addition, double-drug-resistant D17 cell clones were isolated, and Southern hybridization analysis was performed to confirm the recombinant genotype of the proviruses.
FIG. 2.
Experimental protocol to measure the rates of recombination. Abbreviations and symbols are defined in the legend to Fig. 1.
In this system, the recombination rates were measured in one replication cycle, which was defined by the steps that occurred from the proviral stage in PG13 helper cells to the proviral stage in D17 target cells. Viruses produced from the PG13 cells contained GaLV Env; because PG13 cells do not express the receptor for GaLV Env, reinfection during propagation of the viruses cannot occur. Additionally, viruses cannot be propagated in D17 cells, because they lack helper function. Therefore, this system allowed only a single round of replication to occur.
The control vectors generate similar single- and double-drug-resistant colony titers.
In this system, recombination rates were calculated from single- and double-drug-resistant titers. Therefore, it was important to determine whether the three different drug treatments generated similar viral titers in infected cells.
The control vector pJA9 was structurally similar to the vectors (pJA10ΨNeo and pJA11HyΨ) used to measure the rate of recombination at a 1.3-kb marker distance with Ψ+ located between selectable markers (Fig. 1); however, pJA9 contained a functional hygro and neo. A protocol similar to that shown in Fig. 2 was used to determine the relative titers of JA9 with hygromycin, G418, and hygromycin-plus-G418 drug selection. Double-drug-resistant PG13 cell clones containing JA9 were isolated, and the proviral structures in these cell clones were analyzed by Southern hybridization analysis. Five cell clones were identified to contain one intact copy of JA9 provirus (data not shown). Furthermore, the proviruses in these cell clones were integrated at different sites in the host cell genome, indicating that these five cell clones were generated through independent infection events. Viruses were harvested from these cell clones and were used to infect D17 target cells. The virus titers generated from five JA9-containing cell clones are shown in Table 1. These data indicated that within each cell clone, the single- and double-drug selections resulted in similar numbers of colonies.
TABLE 1.
Virus titers generated from JA9- or JA23-infected PG13 cell clones
| Vector and clone | Titer (105 CFU/ml)
|
||
|---|---|---|---|
| Hygromycin | G418 | Hygromycin + G418 | |
| JA9 | |||
| E4 | 0.23 | 0.17 | 0.17 |
| E5 | 7.6 | 3.9 | 5.9 |
| G1 | 3.1 | 2.9 | 2.8 |
| D4 | 1.6 | 1.2 | 1.2 |
| F4 | 4.0 | 2.7 | 5.0 |
| JA23 | |||
| D3 | 1.6 | 0.75 | 1.2 |
| E3 | 1.0 | 0.28 | 0.62 |
| F1 | 0.4 | 0.12 | 0.24 |
| D2 | 2.2 | 0.64 | 1.7 |
| F6 | 0.68 | 0.092 | 0.34 |
The control vector pJS30 was structurally similar to the vectors used to measure the rate of recombination at a 1.3-kb marker distance with Ψ+ located in the 5′ UTR (JA33-1.3kb and JA32). In a previous study, the viral titers obtained from five independent PG13 cell clones containing an intact copy of JS30 demonstrated that the hygromycin, G418, and hygromycin-plus-G418 titers generated within each cell clone were comparable (2). Therefore, the titers produced by each drug selection directly reflected the amount of cells infected with the control vectors.
Characterization of proviral structures in PG13 cell clones infected with JA33-1.3kb and JA32.
PG13 cell clones containing JA33-1.3kb and JA32 were generated to measure the recombination rate between markers separated by 1.3 kb with Ψ+ located in the 5′ UTR. The proviral structures in these cell clones were characterized by Southern hybridization analysis. Partial restriction enzyme maps of JS30, JA33-1.3kb, and JA32 are shown in Fig. 3A. The JS30 provirus had a unique NdeI site located in hygro and four EheI sites (one in each LTR, one in Ψ+, and one in neo). In JA33-1.3kb, the NdeI site was destroyed to inactivate hygro, whereas in JA32, an EheI site was destroyed to inactivate neo. All three proviruses contained four EcoRV sites, two in each LTR. When genomic DNA from cell clones containing a copy of JA33-1.3kb and a copy of JA32 was digested with EheI, NdeI, and EcoRV and hybridized with probes generated from a 1.3-kb MluI-EheI fragment of JA33-1.3kb, a 1.8- and a 2.4-kb band were expected from the JA33-1.3kb and the JA32 proviruses, respectively (Fig. 3A). A representative Southern blot of three different PG13 cell clones (A2, B1, and E1) is shown in Fig. 3B; each cell clone contained the expected bands. A unique HindIII site was located in JA33-1.3kb and JA32 proviruses at the 5′ end of IRES (Fig. 3A). Therefore, each provirus was expected to generate two bands when digested with HindIII and hybridized with the aforementioned probe. Since retroviruses integrate randomly into the host genome (7), the band sizes should vary according to the site of integration. Therefore, cell clones that contain one copy of JA33-1.3kb and JA32 should produce four bands of different sizes upon HindIII digestion and Southern analysis. A representative Southern analysis with HindIII digestion of genomic DNA from the helper cell clones is shown in Fig. 3B; all cell clones (Fig. 3B and data not shown) exhibited a different band pattern, indicating that they were derived from independent infection events (Fig. 3B, lanes H).
FIG. 3.
Proviral structures of JA33-1.3kb, JA32, and JS30. Recombinants with two functional drug resistance genes have the same structure as JS30. (A) Predicted proviral structures. RV, EcoRV; H, HindIII; zigzag lines, host cell sequences. A 1.3-kb MluI-EheI DNA fragment of JA33-1.3kb was used for a random-priming reaction to generate a probe for Southern hybridization analysis; the asterisk in JA33-1.3kb denotes the MluI site. This probe hybridized to the 3′ end of hygro and the 5′ end of neo and is indicated by the black box. (B) Southern hybridization analysis of the proviral structures in virus-producing and target cell clones. A2, B1, and E1 are PG13 virus-producing cell clones containing a copy of each of the JA33-1.3kb and JA32 proviruses. A2-1a, B1-b, and E1-b are double-drug-resistant D17 cell clones infected with viruses harvested from A2, B1, and E1, respectively. Other abbreviations and symbols are defined in the legend to Fig. 1.
Recombination rate of MLV vectors with Ψ+ located in the 5′ UTR and markers separated by 1.3 kb.
Viruses harvested from eight independent PG13 cell clones that contained a copy of JA33-1.3kb and JA32 were used to infect D17 target cells. Viral titers are shown in Table 2. The hygromycin-resistant colony titers ranged from 0.15 × 105 to 33.0 × 105 CFU/ml, the G418-resistant colony titers ranged from 0.18 × 105 to 31 × 105 CFU/ml, and the hygromycin-plus-G418-resistant colony titers ranged from 0.022 × 104 to 8.9 × 104 CFU/ml.
TABLE 2.
Virus titers generated from PG13 cell clones coinfected with JA33-1.3kb and JA32
| Clone | Titer (CFU/ml)
|
Recombination ratea (%) | ||
|---|---|---|---|---|
| Hygromycin (105) | G418 (105) | Hygromycin + G418 (104) | ||
| D1 | 33.0 | 22.0 | 8.5 | 7.7 |
| L1 | 26.0 | 16.0 | 3.8 | 4.8 |
| J2 | 31.0 | 31.0 | 8.9 | 5.7 |
| C1 | 0.15 | 0.18 | 0.022 | 2.9 |
| B1 | 5.6 | 5.3 | 0.93 | 3.5 |
| A2 | 9.2 | 8.6 | 1.8 | 4.2 |
| G1 | 9.2 | 9.4 | 2.5 | 5.4 |
| E1 | 2.5 | 2.0 | 5.9 | 5.9 |
| Avg ± SE | 5.0 ± 0.5 | |||
Recombination rate = (double-drug-resistant titer/lesser of the single-drug-resistant titers) × 2.
A double-drug-resistant D17 cell clone could be generated from dual infection of the two parental viruses or a recombinant provirus that had functional hygro and neo. If the double-drug-resistant D17 cell clones contained one of each parental virus, then Southern analysis would reveal a band pattern identical to that of the PG13 cell clones with the same probe and restriction enzyme digests. In contrast, if D17 cell clones contained a recombinant provirus, Southern analyses would generate a 1.3-kb band when digested with the same three enzymes (EcoRV, NdeI, and EheI) and hybridized with the same probe (Fig. 3A). To verify that the double-drug-resistant colony titers were generated by recombinant proviruses, 16 hygromycin-plus-G418-resistant D17 cell clones were isolated, and the proviral structures were analyzed. A representative Southern analysis of three D17 cell clones (A2-1a, B1-b, and E1-b) is shown in Fig. 3B; each of the cell clones was derived from one of the three PG13 cell clones shown in the same figure. Of the 16 cell clones, 13 had a 1.3-kb band indicating that they contained a recombinant provirus (Fig. 3B and data not shown), and 3 had a genotype consistent with a double infection event. This indicated that most of the double-drug-resistant cell clones contained recombinant proviruses and the hygromycin-plus-G418 titer reflected the titer of the recombinant viruses containing functional hygro and neo.
This assay measured the formation of half of the recombinants—those containing functional hygro and neo. The other half of the recombinants, those containing inactivated hygro and neo, could not be measured in the viral titer assay. Therefore, the recombination rate for viruses generated from each cell clone was calculated by dividing the double-drug-resistant colony titers by the lesser of the two single-drug-resistant colony titers and then multiplying by 2 (Table 2). Therefore, the recombination rates of vectors JA33-1.3kb and JA32 from these eight PG13 cell clones ranged from 2.9 to 7.7%, with an average of 5.0% ± 0.5% (standard error [SE]).
Characterization of proviral DNA structures in PG13 cells infected with JA10ΨNeo and JA11HyΨ.
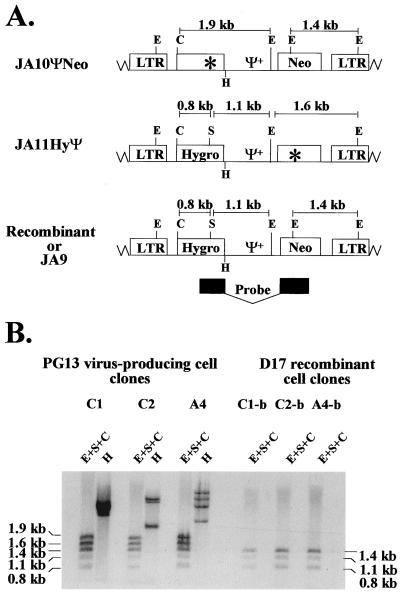
PG13 cell clones containing JA10ΨNeo and JA11HyΨ were generated to examine the effect of Ψ+ located between the selectable markers on the rate of recombination. Southern analyses were performed to examine the proviral structures in coinfected PG13 cell clones. Partial restriction enzyme maps of JA9, JA10ΨNeo, and JA11HyΨ are shown in Fig. 4A. JA9, JA10ΨNeo, and JA11HyΨ proviruses each contained a unique ClaI site between the 5′ LTR and hygro. JA9 proviruses contained four EheI sites: one in each LTR, one in Ψ+, and one in neo. JA10ΨNeo proviruses contained all four EheI sites, whereas JA11HyΨ contained only three EheI sites, because one of the sites was mutated to inactivate neo. A unique SacII site was located in hygro of JA9 and JA11HyΨ; this site was mutated in JA10ΨNeo to inactivate hygro. Genomic DNAs from PG13 cell clones coinfected with JA10ΨNeo and JA11HyΨ were digested with three enzymes: EheI, SacII, and ClaI. Five bands were expected from this digestion when hybridized with a probe containing the 3′ portion of hygro and 5′ portion of neo. A 1.4-kb band and a 1.9-kb band were expected to be generated from the JA10ΨNeo provirus, whereas 0.8-, 1.1-, and 1.6-kb bands were expected to be generated from the JA11HyΨ provirus (Fig. 4A). A representative Southern blot of three different PG13 cell clones (C1, C2, and A4) is shown in Fig. 4B. Each cell clone contained the expected fragments consistent with the predicted structures of JA10ΨNeo and JA11HyΨ. In addition, all DNAs were digested with HindIII to confirm that all of the PG13 cell clones used were generated through independent infection events (Fig. 4B and data not shown).
FIG. 4.
Proviral structures of JA10ΨNeo, JA11HyΨ, and JA9. Recombinants with two functional drug resistance genes have the same structure as JA9. (A) Predicted proviral structures. A 1.9-kb NcoI-NcoI DNA fragment from JS30 was used for a random-priming reaction to generate a probe for Southern hybridization analysis. This probe hybridized to the 3′ end of hygro and the 5′ end of neo and is indicated by the black boxes. (B) Southern hybridization analysis of the proviral structures in virus-producing and target cell clones. C1, C2, and A4 are PG13 virus-producing cell clones infected with JA10ΨNeo and JA11HyΨ. C1-b, C2-b, and A4-b are double-drug-resistant D17 cell clones infected with viruses harvested from C1, C2, and A4, respectively. Other abbreviations and symbols are defined in the legends to Fig. 1 and 3.
Recombination rate of MLV vectors with Ψ+ located between two selectable markers that were 1.3 kb apart.
Six PG13 cell clones coinfected with JA10ΨNeo and JA11HyΨ were identified; viruses were harvested from these cell clones and were used to infect D17 target cells. The infected target cells were placed on hygromycin, G418, or hygromycin-plus-G418 drug selection. Virus titers generated from these six cell clones are shown in Table 3. The hygromycin-resistant colony titers ranged from 1.1 × 105 to 5.4 × 105 CFU/ml, the G418-resistant colony titers ranged from 0.50 × 105 to 1.9 × 105 CFU/ml, and the hygromycin-plus-G418-resistant colony titers ranged from 3.1 × 103 to 13.4 × 103 CFU/ml.
TABLE 3.
Virus titers generated from PG13 cells coinfected with JA10ΨNeo and JA11HyΨ
| Clone | Titer (CFU/ml)
|
Recombination ratea (%) | ||
|---|---|---|---|---|
| Hygromycin (105) | G418 (105) | Hygromycin + G418 (103) | ||
| A3 | 1.1 | 0.50 | 3.1 | 12.4 |
| A4 | 5.4 | 1.9 | 13.4 | 13.7 |
| C1 | 1.4 | 1.0 | 3.9 | 7.8 |
| C2 | 1.6 | 1.0 | 4.1 | 8.2 |
| B2 | 2.7 | 1.2 | 7.4 | 12.3 |
| Y4 | 2.2 | 0.9 | 7.9 | 17.5 |
| Avg ± SE | 12.0 ± 1.5 | |||
Recombination rate = (double-drug-resistant titer/lesser of the single-drug-resistant titers) × 2.
Southern analyses of double-drug-resistant D17 cell clones were performed. Analyses of three D17 cell clones (C1-b, C2-b, and A4-b) are shown in Fig. 4B. If D17 cell clones contained a recombinant provirus, Southern analyses would generate band sizes of 0.8, 1.1, and 1.4 kb when digested with the same three enzymes (EheI, SacII, and ClaI) and hybridized with the same probe used to analyze the PG13 cell clones (Fig. 4A). A total of 12 double-drug-resistant D17 cell clones were examined; 10 contained recombinant proviruses, whereas 2 were the result of double infections (Fig. 4B and data not shown). These results indicated that most of the double-drug-resistant colonies contained recombinant viruses containing functional hygro and neo.
The recombination rates of JA10ΨNeo and JA11HyΨ were calculated from the single- and double-drug-resistant colony titers (Table 3). Based on the titers generated from the six cell clones, recombination rates ranged from 7.8% to 17.5%, with an average of 12.0% ± 1.5% (SE).
Effect of Ψ+ on the rate of recombination when markers were separated by 1.3 kb.
The recombination rate of vectors with markers separated by 1.3 kb when Ψ+ was located in the 5′ UTR or between selectable markers was 5.0 or 12.0%, respectively. These data indicated that the recombination rate was approximately twofold higher when Ψ+ was located between the selectable markers. This increase is statistically significant (P = 0.00002; one-way analysis of variance [ANOVA]). This higher rate of recombination could be caused by an increase in the size of viral population that had the potential to undergo recombination (recombining population). Alternatively, the size of the recombining population might not have changed; however, a larger proportion of the viruses within the recombining population might have undergone recombination between the selectable markers when Ψ+ was relocated to the middle of the genome. We previously observed that when Ψ+ was located in the 5′ UTR, recombination rates increased linearly between marker distances of 1.0 kb (4.7%) and 1.9 kb (7.4%) (2). If the recombining population had been altered, we should observe an increase in the recombination rate when the marker distance was increased. However, if the recombining population had not changed, we should not observe an increase in recombination rates when the marker distance was increased because the observed rate was already close to the maximum rate detected (8.2%) (2). To distinguish between these two possibilities, we measured recombination rates between markers 1.9 kb apart.
Vectors used to further study the effect of Ψ+ on the rate and location of the recombination events.
To further characterize the effect of Ψ+ on recombination, a third set of vectors was constructed to measure the recombination rate when markers were separated by 1.9 kb. The vectors pJA23, pJA19ΨNeo, and pJA20HyΨ were very similar in sequence to the second set of vectors, pJA9, pJA10ΨNeo, and pJA11HyΨ, respectively. The structures of these viral vectors are shown in Fig. 1.
It has been proposed that the DLS region within the Ψ is a recombinational hot spot (34, 35). To examine the general effect of DLS on the locations of the intermolecular template switching events, three sets of restriction enzyme markers were placed at the 5′ end, the middle, and the 3′ end of Ψ+ of pJA19ΨNeo and pJA20HyΨ. It was shown that relocation of the packaging signal to the 3′ UTR could result in packaging of spliced RNA (21). Although it was demonstrated that the recombination rates were not affected by the packaging of spliced RNA (21), this could result in a potential bias in the analysis of the locations of the recombination events. To avoid this potential bias, the splice donor sites were destroyed by PCR mutagenesis in all three vectors used in this experiment.
Similar to pJS30 and pJA9, pJA23 is a control vector and contained functional hygro and neo. The vector pJA19ΨNeo contained a functional neo and a nonfunctional hygro with a 4-bp frameshift insertion that destroyed an NcoI site and generated an NsiI site. The vector pJA20HyΨ contained a functional hygro and a nonfunctional neo with an 8-bp frameshift insertion that destroyed an FspI site and generated an AatII site. The distance between the two inactivating mutations in hygro and neo was 1.9 kb. The presence of the NcoI-NsiI and AatII-FspI markers determined the abilities of the provirus to confer drug resistance; therefore, these two sets of markers are referred to as “selectable markers.” In addition to the selectable markers, these two vectors differed at three other restriction enzyme markers. These markers are located between hygro and Ψ+ (BamHI-ClaI), in the middle of Ψ+ (MluI-AflII), and between Ψ+ and neo (PvuI-XhoI). The MluI-AflII markers in Ψ+ were located past the minimum packaging signal and at the 5′ end of the gag containing the mutated AUG. Therefore, it was expected that this mutation would not interfere with the efficiency of RNA packaging. The other two mutations were located between Ψ+ and drug resistance genes and were not expected to interfere with the abilities of these genes to confer drug resistance. The nature of these three sets of markers was 4- to 8-bp insertions.
To determine whether the control vector, pJA23, generated similar single- and double-drug-resistant colony titers, five independent PG13 cell clones containing an intact copy of JA23 were identified by Southern hybridization analysis (data not shown). Viral titers generated from these five cell clones are shown in Table 1. Parallel to JA9 and JS30, the viral titers generated within each cell clone for JA23 were similar; thus, mutation of the SD did not adversely affect expression of either hygro or neo. The viral titers ranged from 0.4 × 105 to 2.2 × 105 CFU/ml for hygromycin, 0.092 × 105 to 0.75 × 105 CFU/ml for G418, and 0.24 × 105 to 1.7 × 105 CFU/ml for hygromycin-plus-G418 drug selection. The results indicated that the titers produced by each drug selection directly reflected the amount of cells infected with the control vectors.
Proviral DNA analysis of PG13 cell clones infected with JA19ΨNeo and JA20HyΨ.
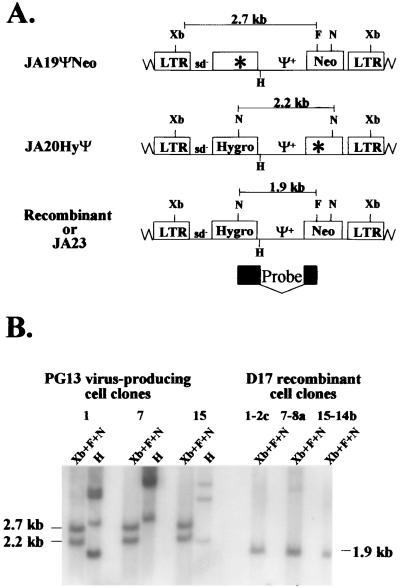
PG13 cell clones containing JA19ΨNeo and JA20HyΨ were generated to further examine the effect of Ψ+ on recombination. Southern hybridization analyses were performed to determine the proviral structures. Partial restriction enzyme maps of JA23, JA19ΨNeo, and JA20HyΨ are shown in Fig. 5A. JA23, which has functional hygro and neo, has an NcoI site located in each drug resistance gene and a unique FspI site located in neo. One of the NcoI sites was destroyed to inactivate hygro in JA19ΨNeo, whereas the FspI site was destroyed to inactivate neo in JA20HyΨ. In addition, all three vectors contained an XbaI site located in each LTR.
FIG. 5.
Proviral structures of JA19ΨNeo, JA20HyΨ, and JA23. Recombinants with two functional drug resistance genes have the same structure as JA23. (A) Predicted proviral structures. Xb, XbaI. A 1.3-kb NcoI-FspI fragment of JS30 was used for a random-priming reaction to generate a probe for Southern hybridization analysis. This probe hybridized to the 3′ end of hygro and the 5′ end of neo and is indicated by the black boxes. (B) Southern hybridization analysis of the proviral structures in virus-producing and target cell clones. Clones 1, 7, and 15 are PG13 virus-producing cell clones infected with JA19ΨNeo and JA20HyΨ. 1-2c, 7-8a, and 15-14b are double-drug-resistant D17 cell clones infected with viruses harvested from clones 1, 7, and 15, respectively. Other abbreviations and symbols are defined in the legends to Fig. 1 and 3.
Genomic DNAs were digested with XbaI, FspI, and NcoI and hybridized with a probe generated from a 1.3-kb NcoI-FspI fragment of JS30 (Fig. 5A). If the cell clones contained a copy of JA19ΨNeo and a copy of JA20HyΨ, 2.7- and 2.2-kb bands were expected (Fig. 5A). A representative Southern blot of three PG13 cell clones (clones 1, 7, and 15) is shown in Fig. 5B; each cell clone contained the expected bands. In addition, all DNAs were digested with HindIII and analyzed by Southern analyses as previously described to confirm that cell clones were generated through independent infection events (Fig. 5B and data not shown).
Recombination rate of MLV vectors with Ψ+ located between selectable markers that were 1.9 kb apart.
Viruses were harvested from five independent PG13 cell clones that contained a copy each of JA19ΨNeo and JA20HyΨ and used to infect D17 target cells. Viral titers are shown in Table 4. The hygromycin-resistant colony titers ranged from 0.50 × 105 to 7.8 × 105 CFU/ml, the G418-resistant colony titers ranged from 0.20 × 105 to 20 × 105 CFU/ml, and the hygromycin-plus-G418-resistant colony titers ranged from 1.2 × 103 to 5.6 × 103 CFU/ml.
TABLE 4.
Virus titers generated from PG13 cells coinfected with JA19ΨNeo and JA20HyΨ
| Clone | Titer (CFU/ml)
|
Recombination ratea (%) | ||
|---|---|---|---|---|
| Hygromycin (105) | G418 (105) | Hygromycin + G418 (103) | ||
| 1 | 2.3 | 1.2 | 3.8 | 6.3 |
| 7 | 0.50 | 0.20 | 1.2 | 12.0 |
| 15 | 1.5 | 0.44 | 2.5 | 11.4 |
| 4 | 7.8 | 20.0 | 3.7 | 9.5 |
| 25 | 1.7 | 0.87 | 5.6 | 12.9 |
| Avg ± SE | 10.4 ± 1.2 | |||
Recombination rate = (double-drug-resistant titer/lesser of the single-drug-resistant titers) × 2.
The proviral structures in double-drug-resistant D17 cells were examined. Figure 5B shows a representative Southern blot of three D17 cell clones (1-2c, 7-8a, and 15-14b). Recombinant proviruses should generate a single 1.9-kb band when digested with XbaI, NcoI, and FspI and hybridized with the probe generated from the 1.3-kb NcoI-FspI fragment of JS30. Alternatively, the genotypes of the proviruses can be examined by amplifying a portion of the proviral sequences, using PCR and subjecting the DNA to restriction enzyme mapping (described in detail below). A total of 53 D17 cell clones were analyzed by Southern blot analysis and/or restriction enzyme mapping; 42 contained a recombinant provirus, whereas 11 were the result of a double-infection event (Fig. 5B and 6B, and data not shown).
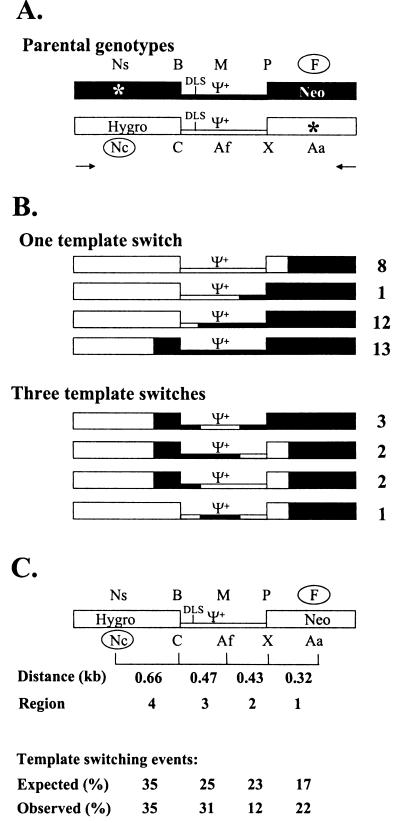
FIG. 6.
Strategy for mapping recombinant proviruses in cell clones. (A) Structures of the internal portions of the parental viruses. JA19ΨNeo is shown in black, and JA20HyΨ is shown in white. Restriction enzyme sites are indicated above or below each parental genotype. The two selectable markers are indicated by the open circles. Arrows, hygro- and neo-specific primers for PCR; other abbreviations and symbols are the same as in Fig. 1. (B) Restriction enzyme maps of 42 recombinant proviruses containing one or three template switches. JA19ΨNeo-derived sequences are shown in black, whereas JA20HyΨ-derived sequences are shown in white. The number to the right of each genotype indicates the number of recombinants with the same genotype observed in the 42 proviruses analyzed. (C) Summary of template-switching events. Restriction enzyme markers corresponding to JA19ΨNeo and JA20HyΨ are shown above and below a generic recombinant structure, respectively. The distances between each set of restriction enzyme markers for four different regions are indicated below recombinant structure. In addition, the expected and observed frequencies of template-switching events are shown beneath the four regions. Expected frequencies were calculated by dividing the distance between each set of markers by the total distance between the selectable markers. Abbreviations are the same as those in Fig. 1 and 6.
The genotype analyses data indicated that most of the double-drug-resistant colonies contained recombinant proviruses. This indicated that the double-drug-resistant colony titers reflected the number of recombinants containing functional hygro and neo and thus could be used to calculate recombination rates. The recombination rates of JA19ΨNeo and JA20HyΨ from five independent cell clones ranged from 6.3% to 12.9%, with an average of 10.4% ± 1.2% (SE). Statistical analysis indicated that there was no significant difference between the rates of recombination at the 1.3- and 1.9-kb marker distances with Ψ+ located between hygro and neo (P = 0.30; one-way ANOVA). These results indicated that the recombination rate reached a plateau by a marker distance of 1.3 kb when Ψ+ was located between hygro and neo.
PCR amplification and restriction enzyme analysis of recombinant proviruses generated from JA19ΨNeo and JA20HyΨ.
The molecular natures of the recombinant proviruses generated from JA19ΨNeo and JA20HyΨ were examined. To ensure that independent recombination events were studied, most of the cell clones were isolated from different cell culture dishes. For cell clones isolated from the same culture dishes, Southern hybridization analyses were performed to ensure that these proviruses had arisen through independent infection events (data not shown). A portion of the proviral genome (2.3 kb) was amplified by PCR with primers specific to hygro and neo (Fig. 6A). Restriction enzyme mapping was performed on PCR products to determine the molecular nature of the recombinant proviruses. For each DNA sample, seven different single-enzyme digestions (NsiI, BamHI, ClaI, MluI, AflII, PvuI, and XhoI) and two double-enzyme digestions (AatII plus SacII and NcoI plus FspI) were performed. The genomes of 42 recombinant proviruses were analyzed and are shown in Fig. 6B. Of the 42 recombinant proviruses analyzed, 34 contained one template switch and 8 contained three template switches in the 2.3-kb region amplified by PCR. Among the 34 proviruses with one template switch, recombination occurred in all four regions between the five sets of restriction enzyme markers. Of these template-switching events, 8 occurred in the 5′ portion of neo between FspI of JA19ΨNeo and XhoI of JA20HyΨ, 1 occurred in the 3′ portion of Ψ+ between PvuI of JA19ΨNeo and AflII of JA20HyΨ, 12 occurred in the 5′ portion of Ψ+ between MluI of JA19ΨNeo and ClaI of JA20HyΨ, and 13 occurred in the 3′ portion of hygro between BamHI of JA19ΨNeo and NcoI of JA20HyΨ (Fig. 6B). Of the proviruses containing three template switches, four patterns were observed. Three proviruses had the first template switch between PvuI of JA19ΨNeo and AflII of JA20HyΨ, the second template switch between AflII of JA20HyΨ and BamHI of JA19ΨNeo, and the third template switch between BamHI of JA19ΨNeo and NcoI of JA20HyΨ. Two proviruses had the first template switch between FspI of JA19ΨNeo and XhoI of JA20HyΨ, the second template switch between XhoI of JA20HyΨ and MluI of JA19ΨNeo, and the third template switch between BamHI of JA19ΨNeo and NcoI of JA20HyΨ. Two proviruses had the first template switch between FspI of JA19ΨNeo and XhoI of JA20HyΨ, the second template switch between AflII of JA20HyΨ and BamHI of JA19ΨNeo, and the third template switch between BamHI of JA19ΨNeo and NcoI of JA20HyΨ. One provirus had the first template switch between FspI of JA19ΨNeo and XhoI of JA20HyΨ, the second template switch between XhoI of JA20HyΨ and MluI of JA19ΨNeo, and the third template switch between MluI of JA19ΨNeo and ClaI of JA20HyΨ.
Distribution of the template-switching events in the recombinant proviruses.
During reverse transcription, recombination had to occur to obtain the NcoI site in hygro and the FspI site in neo to generate recombinants with functional hygro and neo. In the region between these two sets of selectable markers, JA19ΨNeo and JA20HyΨ differed in three other sets of restriction enzyme sites. These five sets of markers divided the 1.9-kb region between the selected markers into four regions (Fig. 6C). Region 4 was located between NsiI-NcoI and BamHI-ClaI markers; this region was 0.66 kb in length and contained the 3′ portion of hygro. Region 3 was located between the BamHI-ClaI and MluI-AflII markers; this region was 0.47 kb in length and contained the 5′ portion of Ψ+, including the DLS. Region 2 was located between the MluI-AflII and PvuI-XhoI markers; this region was 0.43 kb in length and contained the 3′ region of the Ψ+. Region 1 was located between the PvuI-XhoI and FspI-AatII markers; this region was 0.32 kb in length and contained the 5′ portion of neo.
Of the recombinants analyzed, 34 contained a single template switch and 8 contained three template switches; therefore, a total of 58 template switches were observed (Fig. 6B). The total numbers of template switches within regions 4, 3, 2, and 1 were 20, 18, 7, and 13, respectively (Fig. 6C). If the frequencies of the recombination events were proportional to the marker distance, then the expected frequencies of template switches within regions 4, 3, 2, and 1 would be 35, 25, 23, and 17%, respectively. The observed frequencies of template switches within regions 4, 3, 2, and 1 were 35, 31, 12, and 22%, respectively. The differences between the observed and expected frequencies of template switches in regions 4, 3, 2, and 1 were not highly significant (P = 0.93, P = 0.29, P = 0.048, and P = 0.27, respectively; Pearson's chi-square test). Therefore, these results indicated that recombination events occurred in all four regions, and the presence of DLS did not cause an increased number of template-switching events in region 3.
DISCUSSION
Recombination during retroviral replication provides an additional mechanism besides mutation to increase variation in viral populations. Although frequent recombination has been observed in most retroviruses (6, 15, 23, 24, 26, 29, 30, 46–48), many aspects of this phenomenon are still unclear. In this report, we tested whether the rate of recombination could be altered and determined the nature of this potential alteration. Specifically, we examined the effect of the extended packaging signal in recombination and determined the general effect of the DLS on the location of the template-switching events.
Recombination rates of MLV vectors with Ψ+ located in the 5′ UTR.
Previously, we determined that in one round of replication, the homologous recombination rates with markers separated by 1.0, 1.9, and 7.1 kb were 4.7, 7.4, and 8.2%, respectively (2). In all of these vectors, Ψ+ was located in the 5′ UTR. It was found that the recombination rates reached a plateau when markers were separated by 1.9 kb and did not increase significantly even when the markers were further apart (7.1 kb). However, recombination rates increased as the distance between markers increased from 1.0 to 1.9 kb (4.7 and 7.4%, respectively). These data suggested that recombination rates increased in linear proportion when the distances between markers ranged from 1.0 to 1.9 kb. However, this relationship could not be determined because only two data points were obtained in the previous study. In the current study, a recombination rate of 5.0% ± 0.5% (SE) was determined with markers 1.3 kb apart. This rate is within the expected range calculated from the previous measured recombination rates when markers were separated by 1.0 and 1.9 kb (4.7%÷1.0 kb × 1.3 kb = 6.1%; 7.4%÷1.9 kb × 1.3 kb = 5.1%). Together with the previous published recombination rates, these data suggested that when the distances between the selectable markers were in the range of 1.0 to 1.9 kb, recombination rates increased linearly in proportion to the distance between markers. This is the first set of evidence to support that within a limited range, a linear relationship exists between recombination rates and the distances between markers.
The effect of the locations of Ψ+ on the recombination rates of MLV vectors.
It was previously observed that the entire viral population was capable of undergoing frequent template switching (17). However, only a subpopulation of viruses underwent recombination (intermolecular template switching). We hypothesized that the barrier to intermolecular template switching is the accessibility of the copackaged RNA. We suggested that the recombination rate may be altered by bringing the two copackaged RNAs closer together between the selectable markers by using the dimerization signal in Ψ+. However, it was not clear whether the distance between the two copackaged RNAs was the only factor separating the viral subpopulation that could undergo recombination from the rest of the viruses. If the distance between the copackaged RNAs was the only factor, then the size of the subpopulation would be changed by relocating Ψ+. In contrast, if the distance between the two RNAs was not the only factor, then it was quite possible that the size of the recombining subpopulation would not be drastically altered. To test our hypothesis, the recombination rates of MLV vectors with Ψ+ located in the 5′ UTR or between the selectable markers were compared. When selectable markers were separated by 1.3 kb, vectors with Ψ+ located between hygro and neo had an approximately twofold-higher recombination rate than those from vectors with Ψ+ located in the 5′ UTR. This enhancement of recombination was less pronounced when the selectable markers were separated by 1.9 kb. The recombination rates of vectors with selectable markers separated by 1.3 and 1.9 kb were not significantly different when Ψ+ was located between hygro and neo (12.0 and 10.4%, respectively; P = 0.30; one-way ANOVA). These data also suggested that the rate of recombination reached a plateau when the markers were separated by 1.3 kb. This was in sharp contrast with the observation that when Ψ+ was located in the 5′ UTR and markers were 1.0 to 1.9 kb apart, recombination rates were in linear proportion to the distances between the selectable markers.
Taken together, our data indicated that with the relocation of the Ψ+, recombination rates reached a plateau at a shorter distance between the markers. However, the overall maximum recombination rates were not drastically different between vectors with Ψ+ in the 5′ UTR and Ψ+ between the two selectable markers: 7.4 to 8.2% and 10.4 to 12%, respectively. The maximum observed recombination rates approximated the size of the recombining subpopulation (2). Therefore, these data indicated that relocating Ψ+ to between the two selectable markers produced a minor alteration in the size of the recombining subpopulation. This also suggested that factors other than the distance between the two RNAs were involved in separating the recombining and nonrecombining populations.
Our study demonstrated that when markers were separated by 1.3 kb, the recombination rate was altered by placing Ψ+ between the selectable markers. A previous study using an SNV-based system had shown that relocating the packaging signal to the 3′ end of the viral RNA did not alter the recombination rate when markers were separated by 1.0 kb (21). The different effects of packaging signal relocation on recombination rates in these two studies could be easily explained by the positions of the packaging signal. In the previous study, the packaging signal was moved from the 5′ UTR to the 3′ UTR; this would not affect the relative distance of the RNAs between the two selected markers. In our study, however, the packaging signal was placed between the selectable markers; this affected the relative distance of the RNAs between the two selectable markers and, as a consequence, altered the recombination rate.
Recombination in MLV exhibits high negative interference.
High negative interference described a phenomenon in which a greater-than-expected probability of multiple recombination events was observed (3, 5, 17, 49). Using the observed recombination rate of 10.4% with Ψ+ between markers separated by 1.9 kb, it was estimated that one in five proviruses should contain a single recombination event (see reference 17 for a detailed calculation). Since all of the proviruses analyzed in these experiments were recombinants, all proviruses contained at least one recombination event. Therefore, 1 in 5 and 1 in 25 recombinants would be expected to contain two and three recombination events, respectively. It was expected that recombinants with two template switches between the 1.9-kb region would contain a parental genotype and would not survive double-drug selection for functional hygro and neo. However, recombinants with three template-switching events could contain functional hygro and neo and should be observed. Analysis of 42 recombinant proviruses indicated that 34 contained one template switch, whereas 8 contained three template switches (Fig. 6B). Approximately 19% of the recombinant proviruses contained three template switches; this was fivefold greater than expected. Therefore, similar to the observations made with SNV (3, 17), homologous recombination in MLV also exhibited high negative interference. In addition, recombinants with multiple template switches were also frequently observed in human immunodeficiency virus type 1 (6, 50). Taken together, we propose that high negative interference is an inherent property of retroviral recombination.
The general effect of packaging signal on the locations of the template-switching events.
Using a system selecting recombination in the MLV packaging signal, it was shown that template-switching events occurred frequently within a 33-nucleotide region that coincided with the DLS (31, 34, 35). These data suggested that DLS is a hot spot for recombination.
In our study, recombination can occur within any of the four regions defined by the restriction enzyme sites between the two selectable markers (Fig. 6). We found that the region containing the 5′ Ψ+ sequences including the DLS did not experience significantly more template switching than expected. This region is 0.47 kb in length, and 25% of the recombination events were expected to occur in this region (0.47 kb/1.9 kb = 25%) if the recombination events occurred randomly. We observed that 31% of the recombinants had a template switch in this region (Fig. 6C). Therefore, the presence of DLS did not cause an increase in recombination events in this general region. This result was in agreement with our previous study using SNV-based vectors (3), in which the locations of the template-switching events throughout the entire vector genomes were determined. It was found that the 0.24-kb region containing the DLS did not experience increased template-switching events. In both the SNV and the MLV studies, we examined the frequencies of template-switching events within the general region. The exact location of the cross-overs in the recombinants could not be determined due to the lack of markers flanking the DLS. We elected not to place mutations flanking the DLS in these studies to avoid the potential impact on the frequencies of RNA heterodimer formation that could influence other aspects of our studies.
Currently, there are several series of studies pointing at different roles of DLS in recombination. It was shown that two HIV-1 clones with different DLS could undergo efficient recombination (43). Our studies indicated that the presence of DLS did not increase the recombination rate within a 0.47- or 0.24-kb region. However, forced recombination studies indicated a cluster of crossovers occurring in the DLS region (31, 34, 35). The data generated from these studies are not necessarily conflicting, because distinct selection pressures were applied in each system and different aspects of recombination were examined. In addition, there were many differences in the experimental systems. For example, the vectors used in the human immunodeficiency virus type 1 study and our studies contained much longer stretches of homology, whereas the vectors used in the forced-recombination systems mainly have homologies in the LTRs and the packaging signals. These differences could create factors that impact recombination events. For example, it has been demonstrated that the length of the 3′ homology can significantly affect the point of template switching (8a). When two vectors have high homology throughout the entire viral genome, the effect of the 3′ homology would not be as strong as that with vectors that only contain a patch of homology. The 3′ homology is only one example of the factors that can generate the differences between these two series of studies, and other factors are likely to also play important roles. Other studies are currently in progress in our laboratories to further explore the role of the DLS in recombination.
ACKNOWLEDGMENTS
We thank Gerry Hobbs, Department of Computer Sciences and Department of Community Medicine, West Virginia University, for performing the statistical analyses. We thank B. Beasley, S. Cheslock, Q. Dang, K. Delviks, E. Halvas, and C. Hwang for critical reading of the manuscript. We also thank J. Coffin and V. KewalRamani for discussions and suggestions for the manuscript.
This work was supported by research grants from the American Cancer Society (MBC-97322 to W.-S.H. and VM-84706 to V.K.P.), by a research grant from NIH (PHS CA58875 to V.K.P.), and by the HIV Drug Resistance Program, National Cancer Institute. J.A.A. is supported by the West Virginia University Medical Scientist Training Program.
REFERENCES
- 1.Adam M A, Ramesh N, Miller A D, Osborne W R A. Internal initiation of translation in retroviral vectors carrying picornavirus 5′ nontranslated regions. J Virol. 1991;65:4985–4990. doi: 10.1128/jvi.65.9.4985-4990.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson J A, Bowman E H, Hu W-S. Retroviral recombination rates do not increase linearly with marker distance and are limited by the size of the recombining subpopulation. J Virol. 1998;72:1195–1202. doi: 10.1128/jvi.72.2.1195-1202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson J A, Teufel II R J, Yin P D, Hu W-S. Correlated template-switching events during minus-strand DNA synthesis: a mechanism for high negative interference during retroviral recombination. J Virol. 1998;72:1186–1194. doi: 10.1128/jvi.72.2.1186-1194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berlioz C, Darlix J-L. An internal ribosomal entry mechanism promotes translation of murine leukemia virus gag polyprotein precursors. J Virol. 1995;69:2214–2222. doi: 10.1128/jvi.69.4.2214-2222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chase M, Doermann A. High negative interference over short segments of the genetic structure of bacteriophage T4. Genetics. 1958;43:332–353. doi: 10.1093/genetics/43.3.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clavel F, Hoggan M D, Willey R L, Strebel K, Martin M A, Repaske R. Genetic recombination of human immunodeficiency virus. J Virol. 1989;63:1455–1459. doi: 10.1128/jvi.63.3.1455-1459.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin J M. Retroviridae: the viruses and their replication. Vol. 3. New York, N.Y: Raven Press; 1996. [Google Scholar]
- 8.Coffin J M. Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J Gen Virol. 1979;42:1–26. doi: 10.1099/0022-1317-42-1-1. [DOI] [PubMed] [Google Scholar]
- 8a.Delviks K A, Pathak V K. Effect of distance between homologous sequences and 3′ homology on the frequency of retroviral reverse transcriptase template switching. J Virol. 1999;73:7923–7930. doi: 10.1128/jvi.73.10.7923-7932.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duesberg P H. Physical properties of Rous sarcoma virus RNA. Proc Natl Acad Sci USA. 1968;60:1511–1518. doi: 10.1073/pnas.60.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y-X, Fu W, Winter A J, Levin J G, Rein A. Multiple regions of Harvey sarcoma virus RNA can dimerize in vitro. J Virol. 1995;69:2486–2490. doi: 10.1128/jvi.69.4.2486-2490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilboa E, Mitra S W, Goff S, Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979;18:93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- 13.Girard P M, Bonnet-Mathoniere B, Muriaux D, Paoletti J. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- 14.Gritz L, Davies J. Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 15.Gu Z, Gao Q, Faust E A, Wainberg M A. Possible involvement of cell fusion and viral recombination in generation of human immunodeficiency virus variants that display dual resistance to AZT and 3TC. J Gen Virol. 1995;76:2601–2605. doi: 10.1099/0022-1317-76-10-2601. [DOI] [PubMed] [Google Scholar]
- 16.Higuchi R. Simple and rapid preparation of samples for PCR. New York, N.Y: Stockton Press; 1989. [Google Scholar]
- 17.Hu W-S, Bowman E H, Delviks K A, Pathak V K. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J Virol. 1997;71:6028–6036. doi: 10.1128/jvi.71.8.6028-6036.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu W S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang S K, Krausslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones J S, Allan R W, Temin H M. Alteration of location of dimer linkage sequence in retroviral RNA: little effect on replication or homologous recombination. J Virol. 1993;67:3151–3158. doi: 10.1128/jvi.67.6.3151-3158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorgensen R A, Rothstein S J, Reznikoff W S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177:65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- 23.Katz R A, Skalka A M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- 24.Kawai S, Hanafusa H. Genetic recombination with avian tumor virus. Virology. 1972;49:37–44. doi: 10.1016/s0042-6822(72)80005-9. [DOI] [PubMed] [Google Scholar]
- 25.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellam P, Larder B A. Retroviral recombination can lead to linkage of reverse transcriptase mutations that confer increased zidovudine resistance. J Virol. 1995;69:669–674. doi: 10.1128/jvi.69.2.669-674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kung H J, Hu S, Bender W, Bailey J M, Davidson N, Nicolson M O, McAllister R M. RD-114, baboon, and woolly monkey viral RNA's compared in size and structure. Cell. 1976;7:609–620. doi: 10.1016/0092-8674(76)90211-7. [DOI] [PubMed] [Google Scholar]
- 28.Kung H-J, Bailey J M, Davidson N, Vogt P K, Nicolson M O, McAllister R M. Electron microscope studies of tumor virus RNA. Cold Spring Harbor Symp Quant Biol. 1975;39:827–834. doi: 10.1101/sqb.1974.039.01.096. [DOI] [PubMed] [Google Scholar]
- 29.Linial M, Blair D. Genetics of retroviruses. New York, N.Y: Cold Spring Harbor Laboratory; 1985. [Google Scholar]
- 30.Linial M, Brown S. High-frequency recombination within the gag gene of Rous sarcoma virus. J Virol. 1979;31:257–260. doi: 10.1128/jvi.31.1.257-260.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lund A H, Mikkelsen J G, Schmidt J, Duch M, Pedersen F S. The kissing-loop motif is a preferred site of 5′ leader recombination during replication of SL3-3 murine leukemia viruses in mice. J Virol. 1999;73:9614–9618. doi: 10.1128/jvi.73.11.9614-9618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangel W F, Delius H, Duesberg P H. Structure and molecular weight of the 60-70S RNA and the 30-40S RNA of the Rous sarcoma virus. Proc Natl Acad Sci USA. 1974;71:4541–4545. doi: 10.1073/pnas.71.11.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 34.Mikkelsen J G, Lund A H, Duch M, Pedersen F S. Recombination in the 5′ leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization region. J Virol. 1998;72:6967–6978. doi: 10.1128/jvi.72.9.6967-6978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikkelsen J G, Lund A H, Kristensen K D, Duch M, Sørensen M S, Jørgensen P, Pedersen F S. A preferred region for recombinational patch repair in the 5′ untranslated region of primer binding site-impaired murine leukemia virus vectors. J Virol. 1996;70:1439–1447. doi: 10.1128/jvi.70.3.1439-1447.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–982. , 984–986, 989–990. [PMC free article] [PubMed] [Google Scholar]
- 39.Murti K G, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prats A-C, Roy C, Wang P, Erard M, Housset V, Gabus C, Paoletti C, Darlix J-L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riggs J L, McAllister R M, Lennette E H. Immunofluorescent studies of RD-114 virus replication in cell culture. J Gen Virol. 1974;25:21–29. doi: 10.1099/0022-1317-25-1-21. [DOI] [PubMed] [Google Scholar]
- 42.Roy C, Tounekti N, Mougel M, Darlix J L, Paoletti C, Ehresmann C, Ehresmann B, Paoletti J. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Res. 1990;18:7287–7292. doi: 10.1093/nar/18.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.St. Louis D C, Gotte D, Sanders-Buell E, Ritchey D W, Salminen M O, Carr J K, McCutchan F E. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J Virol. 1998;72:3991–3998. doi: 10.1128/jvi.72.5.3991-3998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vagner S, Waysbort A, Marenda M, Gensac M C, Amalric F, Prats A C. Alternative translation initiation of the Moloney murine leukemia virus mRNA controlled by internal ribosome entry involving the p57/PTB splicing factor. J Biol Chem. 1995;270:20376–20383. doi: 10.1074/jbc.270.35.20376. [DOI] [PubMed] [Google Scholar]
- 46.Varmus H, Swanstrom R. Replication of retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1985. [DOI] [PubMed] [Google Scholar]
- 47.Vogt P K. Genetically stable reassortment of markers during mixed infection with avian tumor viruses. Virology. 1971;46:947–952. doi: 10.1016/0042-6822(71)90093-6. [DOI] [PubMed] [Google Scholar]
- 48.Weiss R A, Mason W S, Vogt P K. Genetic recombinants and heterozygotes derived from endogenous and exogenous avian RNA tumor viruses. Virology. 1973;52:535–552. doi: 10.1016/0042-6822(73)90349-8. [DOI] [PubMed] [Google Scholar]
- 49.White R L, Fox M S. On the molecular basis of high negative interference. Proc Natl Acad Sci USA. 1974;71:1544–1548. doi: 10.1073/pnas.71.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu H, Jetzt A E, Ron Y, Preston B D, Dougherty J P. The nature of human immunodeficiency virus type 1 strand transfers. J Biol Chem. 1998;273:28384–28391. doi: 10.1074/jbc.273.43.28384. [DOI] [PubMed] [Google Scholar]