Abstract
Gammaherpesviruses cause important infections of humans, in particular in immunocompromised patients. Recently, murine gammaherpesvirus 68 (MHV-68) infection of mice has been developed as a small animal model of gammaherpesvirus pathogenesis. Efficient generation of mutants of MHV-68 would significantly contribute to the understanding of viral gene functions in virus-host interaction, thereby further enhancing the potential of this model. To this end, we cloned the MHV-68 genome as a bacterial artificial chromosome (BAC) in Escherichia coli. During propagation in E. coli, spontaneous recombination events within the internal and terminal repeats of the cloned MHV-68 genome, affecting the copy number of the repeats, were occasionally observed. The gene for the green fluorescent protein was incorporated into the cloned BAC for identification of infected cells. BAC vector sequences were flanked by loxP sites to allow the excision of these sequences using recombinase Cre and to allow the generation of recombinant viruses with wild-type genome properties. Infectious virus was reconstituted from the BAC-cloned MHV-68. Growth of the BAC-derived virus in cell culture was indistinguishable from that of wild-type MHV-68. To assess the feasibility of mutagenesis of the cloned MHV-68 genome, a mutant virus with a deletion of open reading frame 4 was generated. Genetically modified MHV-68 can now be analyzed in functionally modified mouse strains to assess the role of gammaherpesvirus genes in virus-host interaction and pathogenesis.
Gammaherpesviruses cause important human infections, in particular in immunocompromised patients. In humans, the prototypic gamma-1 herpesvirus, Epstein-Barr virus (EBV), is associated with lymphomas and nasopharyngeal carcinoma (26), and the Kaposi's sarcoma herpesvirus (also called Human herpesvirus 8), a gamma-2 herpesvirus, is associated with lymphoproliferative disorders and Kaposi's sarcoma (12, 28). In vivo studies of gammaherpesvirus pathogenesis have been limited to clinical investigation of the infection because of the restricted host range of these viruses. Useful animal models for the analysis of gammaherpesvirus pathogenesis have been infection of primates with Herpesvirus saimiri (HVS), the prototypic gamma-2 herpesvirus, or EBV infection of marmosets (11, 26). Recently, a mouse model of gammaherpesvirus infection has been established (8, 23, 24, 30, 32, 34). Murine gammaherpesvirus 68 (MHV-68) is a natural pathogen of wild murid rodents (1) and is capable of infecting laboratory mice. Clinically, MHV-68 infection of mice induces a syndrome very similar to EBV in humans (8). Genetically, MHV-68 is similar to EBV but more closely related to HVS and human herpesvirus 8 (35). The host response, in particular the immune response to infection of mice with MHV-68, has been studied by several groups over the past few years and has demonstrated its potential as a model for gammaherpesvirus infections (8, 23, 24, 30, 32, 34). A major advantage of the mouse model over the above-mentioned primate models is the availability of genetically defined mouse strains rendered deficient for specific parameters, e.g., of the immune response, either by gene knockout technology or by depletion of various subsets of immune cells.
The molecular basis for the genetic analysis of this virus has been established by the determination of the complete nucleotide sequence of MHV-68 (35). The genome of MHV-68 encodes genes that are common to other members of the gammaherpesviruses, cellular gene homologues, and MHV-68-specific genes (35). The availability of viral mutants would significantly contribute to the understanding of viral gene functions and to the evaluation of their role in pathogenesis. This was demonstrated recently, for example, by the generation and analysis of recombinant MHV-68 with a deletion of both tRNA-like sequences 1 to 4 and open reading frame (ORF) 1 or of ORF 1 by homologous recombination in eukaryotic cells (5, 29). However, because the frequency of homologous recombination in eukaryotic cells is low and selection against nonrecombinant wild-type (WT) virus is necessary, this technique is often ineffective, laborious, and time-consuming.
Recently, the cloning of several viruses, including mouse cytomegalovirus (MCMV), human cytomegalovirus, herpes simplex virus, pseudorabiesvirus, and EBV, as infectious bacterial artificial chromosomes (BACs) has been described (2, 7, 16, 22, 27, 31, 33). This technique allows the maintenance of viral genomes by a BAC in Escherichia coli and the reconstitution of viral progeny by transfection of the BAC plasmid into eukaryotic cells.
Mutagenesis of the virus genome in E. coli using the bacterial recombination machinery, thereby allowing the generation of mutant viruses, is possible. Another possibility is direct transposon mutagenesis, as has been shown for the MCMV BAC and the pseudorabiesvirus BAC (3, 31). Since insertion of the transposon is random, screening procedures are required to identify selected mutants. Targeted mutagenesis of a BAC-cloned virus genome was first demonstrated for MCMV by mutagenesis of the immediate-early gene 1 (22). However, the mutagenesis method used was still laborious and time-consuming, since it required construction of a recombination plasmid via multiple cloning steps and a two-step replacement strategy in E. coli (22). In addition, cloning of DNA using restriction enzyme-based strategies relies on the favorable disposition of cleavage sites, which has practical limitations. Therefore, for the generation of mutants more efficient procedures are desirable.
In this report, we describe the cloning of MHV-68 as an infectious BAC. A viral mutant with a deletion of ORF 4 was generated by applying a site-specific mutagenesis method using PCR-generated linear DNA fragments (37). Using this method, any genetic modification should be possible, thereby facilitating the analysis of herpesvirus genomes cloned as infectious BACs.
MATERIALS AND METHODS
Virus, cells, and plaque assay.
The original stock of MHV-68 (clone G2.4) was obtained from J. Stewart and A. Nash (University of Edinburgh, Edinburgh, United Kingdom). Working stocks of virus were prepared on BHK-21 cells (ATCC CCL10; kindly provided by J. Stewart and A. Nash). BHK-21 cells were maintained in Glasgow modified Eagle's medium (GIBCO) supplemented with 5% newborn calf serum, 5% tryptose-phosphate broth, penicillin (100 U/ml), and streptomycin (100 μg/ml). The BHK-21 cells were infected at a multiplicity of infection (MOI) of 0.1, and virus stocks were prepared when the cytopathic effect (CPE) was complete by the freezing and thawing of the cells two times. Cellular debris was removed by centrifugation, and the supernatants were stored in aliquots at −80°C. Virus titers were determined by plaque assay on BHK-21 cells. Briefly, 10-fold dilutions of virus were adsorbed onto BHK-21 cells. After 90 min of incubation at 37°C, the inoculum was removed and fresh medium containing 1.5% carboxymethylcellulose was added. Cells were stained after 4 to 5 days with 0.1% crystal violet solution to determine the number of plaques. NIH 3T3 cells (ATCC CRL1658) were cultured in Dulbecco's modified Eagle medium supplemented with 10% newborn calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml) and 1% l-glutamine. Rat embryonic fibroblasts stably transfected and expressing recombinase Cre were obtained from W. Burns (Johns Hopkins University School of Medicine, Baltimore, Md.) and were cultured in Dulbecco's modified Eagle medium supplemented with 10% newborn calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 1% l-glutamine in the presence of 300 μg of G418 per ml. To investigate the in vitro growth properties of the ORF 4-deletion mutant RγHV68A98.05, NIH3T3 cells were infected at an MOI of 0.1 for 1 h at 4°C to allow adsorption. For penetration, prewarmed medium was added for a 2-h period of incubation at 37°C. Remaining extracellular virus was inactivated by treatment with low-pH citrate buffer for 1 min (19, 20).
Preparation and analysis of viral DNA and BAC plasmids.
Total cellular DNA was isolated from infected BHK-21 cells. Briefly, cells were harvested and washed once with phosphate-buffered saline. The pellet was resuspended in Tris-EDTA buffer (100 mM Tris-HCl–20 mM EDTA, pH 8.0), and an equal volume of 1% sodium dodecyl sulfate was added. Proteinase K was added to a final concentration of 500 μg/ml, and the suspension incubated at 55°C for at least 3 h. DNA was extracted twice with phenol-chloroform and once with chloroform-isoamylalcohol (24:1) and then precipitated with isopropanol, washed with 70% ethanol, and resuspended in TE buffer (10 mM Tris-HCl–1 mM EDTA, pH 7.5). Circular viral DNA was isolated by the method of Hirt (15) as described previously (22). BAC plasmids were isolated from E. coli cultures using an alkaline lysis procedure (21) and analyzed by restriction enzyme digestion and gel electrophoresis. For Southern blot analysis, the DNA was blotted onto a Hybond N+ membrane (Amersham). Blots were hybridized overnight with digoxigenin (DIG)-labeled probes and developed using an enhanced chemiluminescence system (Boehringer Mannheim) according to the instructions of the manufacturer.
Plasmid construction.
To construct the recombination plasmid pHA2, a 1.5-kbp EcoRI fragment from the left end of the MHV-68 genome (nucleotide positions 50 to 1540) (35) was generated by PCR (forward primer, 5′-TTC AGG GCG GCC GAG AAT TCG ATG CAA ATG-3′; reverse primer, 5′-GAC TTT GGC GTC ATT GGG GAA TTC CAA GAC-3′), using MHV-68 DNA as the template. The resulting fragment was cloned into the EcoRI site of the vector pK18 (25) containing a modified polylinker, providing the following restriction sites: MluI, NotI, AvrII, SgrAI, PacI, SgrAI, EcoRI, ApaLI, and MluI. The BAC vector pKSO-gpt (22) was cloned into the PacI site of this vector. Using an MluI-PstI adapter, a 1.6-kbp NsiI-MluI fragment of the plasmid pEGFP-C1 (Clontech) containing the human cytomegalovirus major immediate-early promoter, the coding sequence for the green fluorescent protein (GFP) and the poly(A) signal of simian virus 40, was cloned into a NsiI site between sequences of the BAC vector and the right loxP site (see Fig. 2A, “recombination plasmid pHA2”).
FIG. 2.
Construction of the MHV-68 BAC genome and structural analysis of reconstituted virus genomes. (A) The BAC cloned genome was generated in eukaryotic cells by homologous recombination of the MHV-68 DNA with the recombination plasmid pHA2. The recombination plasmid contained 1.5 kbp of flanking homologous sequence (shaded box) as well as the BAC vector, the gpt gene, and the gfp gene, flanked by loxP sites. Electroporation of the circular BAC cloned genome RγHV68A98.01 into E. coli generated the MHV-68 BAC-plasmid pHA3. Integration of the BAC vector into the linear recombinant virus genome resulted in a new EcoRI fragment of 7.4 kbp which is indicated by an arrow. An additional EcoRI fragment of approximately 18 kbp in the BAC plasmid resulted from the fusion of the terminal EcoRI fragments (containing the terminal repeats of the virus genome). P, probe. (B) Structural analysis of BAC plasmids and of reconstituted virus genomes by ethidium bromide-stained agarose gel analysis of EcoRI-digested DNA. The lanes show MHV-68 WT DNA isolated from infected cells (lane 1), MHV-68 BAC plasmid pHA3 DNA isolated from E. coli (lane 2), reconstituted MHV-68 BAC virus RγHV68A98.01 DNA isolated from infected cells (lane 3), and reconstituted MHV-68 BAC virus RγHV68A98.02 DNA (with the BAC vector excised by recombinase Cre) isolated from infected cells (lane 4). The upper arrowhead indicates an additional 18-kbp band present only in lane 2, and the lower arrowhead indicates a 7.4-kbp fragment resulting in a double band in lanes 2 and 3. (C) Southern blot analysis of the gel shown in panel B using a DIG-labeled probe (indicated in panel A). Lanes 3 and 4 were from a longer exposure than lanes 1 and 2. The arrowhead indicates the additional 18-kbp band present only in lane 2. Marker (M) sizes (in kilobase pairs) are indicated on the left.
Mutagenesis.
For mutagenesis of the BAC plasmid pHA3, linear fragments were prepared by PCR using primer pairs that contained 24 nucleotides for amplification of a tetracycline resistance gene from vector pCP16 (4) and an additional 50 nucleotides homologous to the sequences flanking the MHV-68 ORF 4. PCR products were purified by electrophoresis in a 1% low-melting-point agarose gel, extracted with phenol-chloroform, and resuspended in 25 μl of TE buffer. The mutagenesis procedure was performed as described previously (37), with slight modifications. Briefly, 10 μl of the linear, PCR-generated fragments were electroporated into E. coli JC8679 (recBC sbcA) (6) containing the MHV-68 BAC plasmid pHA3. Bacteria were incubated at 37°C for 1 h and plated onto agar plates containing chloramphenicol (17 μg/ml) and tetracycline (10 μg/ml). Plasmid DNA was isolated and analyzed by restriction enzyme digestion, which led to the identification of the mutant BAC plasmid pHA6. To remove the tetracycline resistance gene from the mutant BAC plasmid, pHA6 was retransformed into E. coli DH10B. The Flp expression plasmid pCP20 (4) was electroporated into E. coli DH10B containing the mutant BAC plasmid pHA6. The bacteria were plated on chloramphenicol-ampicillin (100 μg/ml)-containing plates and grown overnight at 30°C. Colonies were replated on chloramphenicol-containing plates and grown overnight at 43°C. Colonies were again replated on both chloramphenicol- and tetracycline-containing plates and grown overnight at 37°C. BAC plasmids from chloramphenicol-positive, tetracycline-negative colonies were analyzed by restriction enzyme analysis for the loss of the tetracycline resistance gene. A revertant BAC was generated by a two-step replacement procedure, as described previously (2, 22). For that purpose, a 5.4-kbp MluI-BglII fragment of MHV-68 (nucleotide positions 7948 to 13310) was cloned into the shuttle plasmid pST76K-SR and electroporated into E. coli strain DH10B, which already contained the mutant BAC plasmid pHA6. pST76K-SR is a derivative of the shuttle plasmid pST76K_SacB (2) and contains, in addition, the recA gene (E.-M. Borst et al., unpublished results). The presence of recA allows recombination to be performed with recA-negative bacteria such as DH10B.
Generation of recombinant viruses.
A total of 5 μg of the recombination plasmid pHA2 was digested with MluI, and the linear fragment (loxP, gpt [guanosine phosphoribosyl transferase gene], BAC vector, gfp, loxP, and MHV-68 in homologous sequence) was cotransfected with about 5 μg of MHV-68 DNA in BHK-21 cells by electroporation (960 μF and 250 V using a Gene Pulser unit [Bio-Rad]). Plaques appeared, and after complete CPE was reached, the supernatant was transferred to new BHK-21 cells and cultured in the presence of 25 μM xanthine and 100 μM mycophenolic acid to select recombinant viruses with an integrated BAC plasmid containing the gpt marker (14). After three rounds of selection, circular DNA was isolated from infected BHK-21 cells and electroporated into E. coli DH10B. Bacteria were plated on agar plates containing chloramphenicol. Plasmid isolation and restriction enzyme analysis led to the identification of an E. coli clone containing the BAC plasmid with the complete genome of MHV-68.
Electroporation of the MHV-68 BAC plasmid pHA3 into BHK-21 cells resulted in plaques expressing the gfp gene. To remove the BAC vector sequences, rat embryonic fibroblasts expressing recombinase Cre were infected with the MHV-68 BAC virus (RγHV68A98.01). A viral clone with the BAC vector sequences deleted (RγHV68A98.02) was purified by limiting dilution using loss of GFP expression as a marker. The absence of the BAC vector sequences was confirmed by Southern blot analysis. Electroporation of DNA of the mutant BAC plasmid pHA6 in BHK-21 cells resulted in the development of plaques. DNA of the mutant virus (RγHV68A98.05) was isolated from infected BHK-21 cells and analyzed by restriction enzyme digestion.
RESULTS
Strategy for cloning and mutagenesis of the MHV-68 genome in E. coli.
The strategy for cloning and mutagenesis of the MHV-68 genome in E. coli is shown in Fig. 1. First, a recombinant virus containing a bacterial vector integrated into the viral genome was constructed by homologous recombination in eukaryotic cells. Using the gpt gene as a selection marker, circular intermediates of recombinant virus were accumulated in infected cells. Circular DNA can be isolated from infected cells and electroporated into E. coli. For mutagenesis, a recently described recombination system in E. coli JC8679 utilizing PCR fragments that provide short homology arms was applied (37). After mutagenesis, the mutated BAC plasmid was retransformed into E. coli DH10B. Since the antibiotic resistance gene was flanked by FRT sites, it can be excised by Flp-mediated recombination. Transfection of mutated BAC plasmids into eukaryotic cells will eventually lead to the reconstitution and release of infectious virus mutants. The BAC vector sequences are flanked by loxP sites, and can be removed by propagation of the recombinant viruses in fibroblasts expressing recombinase Cre.
FIG. 1.
Strategy for cloning and mutagenesis of MHV-68. Viral DNA and the linearized recombination plasmid containing the BAC vector sequences were cotransfected into eukaryotic cells to generate a recombinant virus. Circular DNA of the recombinant virus genome was isolated from cells and electroporated into E. coli. Mutagenesis of the MHV-68 BAC plasmid was performed with E. coli JC8679. The mutated BAC plasmid was retransformed into E. coli DH10B. In E. coli DH10B, the tetracycline resistance gene can be deleted by Flp-mediated recombination. The mutated BAC plasmid was transfected into eukaryotic cells to reconstitute recombinant virus. Propagation of the mutant virus in fibroblasts expressing recombinase Cre results in deletion of the BAC vector sequences. Circled arrows indicate FRT sites. P, loxP site; TR, terminal repeats.
Generation of the MHV-68 BAC plasmid.
The left end of the MHV-68 genome was chosen for the integration of the BAC vector (Fig. 2A). This region, containing ORF 1 and sequences with features of tRNAs, has been recently shown to be dispensable for lytic replication in vitro and for latent infection in vivo. In this study, it was also shown that insertions into the left end of the MHV-68 genome can be achieved by a single crossover event via only one homology region at one side of the recombination plasmid (29). This strategy has been originally described for the generation of HVS recombinants and has been suggested to provide a generally applicable means of gamma-2-herpesvirus mutagenesis (13). Thus, the recombination plasmid pHA2 containing a 1.5-kbp fragment homologous to the left end of the MHV-68 genome and the BAC vector including the gpt and gfp genes was constructed (Fig. 2A, line 2). After cotransfection of both recombination plasmid pHA2 and MHV-68-DNA, virus plaques showing green fluorescence developed, indicating the integration and expression of the gfp gene. Recombinant viruses (Fig. 2A, line 3) were selected using mycophenolic acid and xanthine by utilizing the gpt marker (14). After three rounds of selection, circular viral DNA was isolated from infected cells and electroporated into E. coli. Isolation of plasmids from single E. coli colonies and restriction enzyme analysis led to the identification of a bacterial clone containing a BAC plasmid with the full-length MHV-68 genome (pHA3) (Fig. 2A, line 4). In comparison to MHV-68 WT DNA, the MHV-68 BAC-plasmid pHA3 contains an additional EcoRI fragment of approximately 18 kbp (Fig. 2A and Fig. 2B, lanes 1 and 2). This fragment results from the fusion of the terminal EcoRI fragments, indicating the circular nature of the BAC plasmid. In addition, EcoRI digestion of the BAC vector sequence released a 7.4-kbp fragment (Fig. 2A, line 4, and Fig. 2B, lane 2). Characterization of the BAC plasmid pHA3 with restriction enzymes HindIII, BglII, and BamHI confirmed the successful cloning of the complete genome of MHV-68 in E. coli (data not shown). In addition, Southern blot analysis confirmed the presence of the BAC vector sequence in the MHV-68 BAC plasmid (Fig. 2C, lane 2).
Stability of MHV-68 BAC plasmid pHA3 in E. coli.
The stability of the MHV-68 BAC plasmid pHA3 in E. coli was of interest. Certain DNA sequences, in particular repeats in a genome, provide optimal substrates for recombination. The MHV-68 genome contains a high number of repeated sequences, e.g., the terminal repeats, an internal 40-bp repeat, and an internal 100-bp repeat (35). BAC plasmids are usually propagated in the E. coli strain DH10B that carries a recA mutation in order to minimize the propensity for recombination. Stable maintenance in E. coli DH10B has indeed been shown for several herpesvirus BACs (2, 3, 16, 17, 22). Bacteria with the BAC plasmid pHA3 were grown for three passages of 24 h each and finally plated on agar plates containing chloramphenicol. Overnight cultures were grown from single colonies, and DNA was isolated from the cultures and analyzed by restriction enzyme digestion and gel electrophoresis (Fig. 3). With the exception of one band that varied in size between 4.5 and 5 kbp, all five clones presented in Fig. 3 showed an identical EcoRI restriction pattern, compared to the original BAC plasmid pHA3 (Fig. 2, lane 2). Southern blot analysis with a DIG-labeled probe specific for the EcoRI K fragment, the fragment that contains the 40-bp internal repeat of MHV-68 (9, 35), demonstrated that the shift of the original 5.2-kbp band towards smaller bands was due to a reduced number of 40-bp repeats (data not shown). This was confirmed by digestion with several other restriction enzymes. The band representing the fragment with the 40-bp repeat always shifted to a smaller size (data not shown). In some clones the new band was clearly submolar (as shown, for example, in Fig. 3, lane 3), and the size of the new band varied between clones. This demonstrated that for the cloned genome, a heterogeneous progeny with regard to the number of 40-bp repeats can occur in E. coli.
FIG. 3.
Stability of the MHV-68 BAC plasmid pHA3 in E. coli. The BAC plasmid pHA3 was propagated three times in E. coli DH10B. Afterwards, bacteria were plated on agar plates containing chloramphenicol and plasmid DNA isolated from single colonies was analyzed by EcoRI digestion and gel electrophoresis. The analysis of five clones (lanes 1 to 5) is shown on an ethidium bromide-stained agarose gel. The bands representing the EcoRI K fragment which contains the 40-bp internal repeat of MHV-68 are marked by dots. Marker sizes (in kilobase pairs) are indicated on the left.
Reconstitution of infectious virus from the MHV-68 BAC plasmid pHA3.
Transfection of the MHV-68 BAC plasmid pHA3 into BHK-21 cells led to the development of plaques. Recombinant virus which was named RγHV68A98.01 was harvested, and new BHK-21 cells were infected. EcoRI digestion of DNA isolated from infected cells after reaching complete CPE resulted in a DNA pattern similar to that from digestion of the BAC plasmid (Fig. 2B, lanes 2 and 3). Since DNA isolated from infected cells comprises circular, concatemeric, and linear viral DNA, the amount of the additional EcoRI fragment of approximately 18 kbp caused by fusion of the terminal EcoRI fragments in the circular BAC plasmid was submolar in DNA isolated from infected cells. Southern blot analysis using a BAC vector-specific probe demonstrated the presence of BAC vector sequences in the terminal repeat ladder of the virus (Fig. 2C, lane 3). Thus, infectious recombinant virus (RγHV68A98.01) could be reconstituted from the E. coli-derived BAC plasmid.
The presence of the BAC vector sequences in the BAC-derived viruses will probably not interfere with most analyses in vitro, but it may interfere with analyses performed in vivo. Therefore, as shown before for the MCMV-BAC, the cloned genome was provided with conditions for vector deletion (36). To this end, the BAC vector sequences including the gpt and gfp genes were flanked by loxP sites. MHV-68 BAC virus (RγHV68A98.01) was propagated in rat fibroblasts expressing recombinase Cre. Limiting dilution was performed, and screening for the loss of the gfp marker led to the isolation of clone RγHV68A98.02 devoid of BAC vector sequences. EcoRI digestion resulted in a DNA pattern similar to that from digestion of MHV-68 WT DNA (Fig. 2B, lanes 1 and 4), and Southern blot analysis confirmed the absence of the BAC vector sequences (Fig. 2C, lane 4). WT MHV-68, RγHV68A98.01, and RγHV68A98.02 showed similar growth kinetics in vitro (Fig. 4A, C, and E). Plaque formation by WT MHV-68, RγHV68A98.01, and RγHV68A98.02 was indistinguishable (data not shown). Only RγHV68A98.01 showed green fluorescence due to the presence and expression of the gfp gene (Fig. 4B). The absence of fluorescence in RγHV68A98.02 infected cells indicated the successful deletion of the BAC vector sequences by recombinase Cre (Fig. 4D).
FIG. 4.
Comparison of the in vitro growth properties of several recombinant MHV-68 mutants and WT MHV-68. BHK-21 cells were infected at an MOI of 0.1. Cells and supernatants were harvested at the indicated time points, and viral titers were determined by plaque assay. Titers at 0 h represent input inocula. (A) Growth properties of RγHV68A98.01 compared to WT MHV-68; (B) Expression of gfp in NIH3T3 cells infected with RγHV68A98.01; (C) Growth properties of RγHV68A98.02 compared to RγHV68A98.01; (D) Lack of gfp expression in NIH3T3 cells infected with RγHV68A98.02; (E) Growth properties of RγHV68A98.03 and RγHV68A98.04 compared to RγHV68A98.01; (F) Southern blot analysis of EcoRI-digested DNA isolated from cells infected with WT MHV-68 (lane 1), RγHV68A98.01 (lane 2), RγHV68A98.03 (lane 3), and RγHV68A98.04 (lane 4) with a probe specific for the EcoRI K fragment after digestion of the DNA with EcoRI.
To analyze whether the loss of some of the 40-bp internal repeats has an impact on the growth properties of reconstituted viruses in vitro, two viral clones (RγHV68A98.03 and RγHV68A98.04) were reconstituted from BAC plasmids which had lost some of the 40-bp internal repeats (compare Fig. 3). The loss of some of the 40-bp internal repeats was demonstrated by Southern blot analysis of viral DNA using a probe specific for the EcoRI K fragment. Loss of some of the 40-bp internal repeats led to a smaller size of the EcoRI K fragment (Fig. 4F). Both viruses, RγHV68A98.03 and RγHV68A98.04, showed indistinguishable growth kinetics when compared to RγHV68A98.01 (Fig. 4E). Thus, variation in the number of the 40 bp internal repeats had no influence on the in vitro growth properties of these viruses.
Generation of an MHV-68 ORF 4 deletion mutant by site-specific mutagenesis in E. coli.
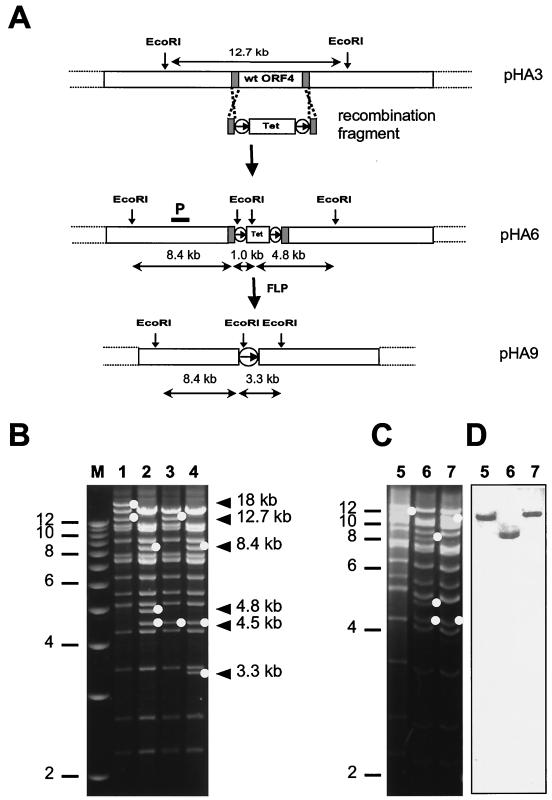
Recently, a new method for DNA recombination in E. coli using short linear DNA fragments has been described (37). This method is based on recombination between linear and circular DNA molecules, and uses the recombination proteins RecE and RecT, and is therefore referred to as “ET cloning” (37). To test the applicability of this method for site-directed mutagenesis of the cloned gamma-2-herpesvirus genome, a deletion mutant was generated. As an example, we deleted ORF 4, which has significant homology to various complement regulatory proteins (35). To delete ORF 4, a linear recombination fragment containing the tetracycline resistance gene flanked by FRT sites and 50-bp regions homologous to MHV-68 sequences was constructed by PCR (Fig. 5A, line 2). This DNA fragment was transferred to the MHV-68 BAC plasmid pHA3 by homologous recombination in the E. coli strain JC8679 (37). The homologous recombination resulted in insertion of the tetracycline resistance gene marker and in deletion of ORF 4 from nucleotide positions 9954 to 10984. The correct insertion of the recombination fragment within the MHV-68 genome was confirmed by sequencing (data not shown). As expected, the mutagenesis resulted in the loss of the 12.7-kbp EcoRI fragment (Fig. 5A, line 1, and Fig. 5B, lanes 1 and 2) and the generation of new 8.4-, 4.8-, and 1.0-kbp fragments (Fig. 5A, line 3, and Fig. 5B, lane 2). Transfection of the mutated BAC plasmid pHA6 into BHK-21 cells led to plaque formation. Viral DNA was isolated from infected cells and analyzed by digestion with EcoRI. Again, the 12.7-kbp fragment was replaced by the expected new bands (Fig. 5C, compare lanes 5 and 6). Thus, the mutation introduced in the MHV-68 BAC plasmid pHA6 was maintained after reconstitution of mutant virus RγHV68A98.05.
FIG. 5.
Construction of the ΔORF4 mutant, structural analysis of the mutated BAC plasmids, and the genomes of reconstituted mutant viruses. (A) Recombination fragment containing the tetracycline resistance gene flanked by FRT sites (circled arrows) and homology regions was generated for mutagenesis in E. coli. Recombination resulted in the deletion of ORF 4 by replacement with the tetracycline resistance gene. Using recombinase Flp, the tetracycline resistance gene was afterwards excised and left one FRT site. (B) Ethidium bromide-stained agarose gel of EcoRI-digested plasmid DNA. MHV-68 BAC plasmid (pHA3) DNA (lane 1), ΔORF4-mutant plasmid (pHA6) DNA containing the tetracycline resistance gene (lane 2), ΔORF4 revertant plasmid (pHA12) DNA (lane 3), ΔORF4 mutant plasmid with the tetracycline resistance gene excised (pHA9) DNA (lane 4). (C) Ethidium bromide-stained agarose gel of EcoRI-digested DNA of reconstituted viruses. BAC MHV-68 (RγHV68A98.01) (lane 5), ORF 4 deletion mutant (RγHV68A98.05) (lane 6), and ORF 4 revertant virus (RγHV68A99.03) (lane 7). (D) Southern blot analysis of the gel shown in panel C with probe P, which is indicated in panel A. As illustrated in panel A, this probe recognizes the 12.7-kbp EcoRI fragment of RγHV68A98.01 and RγHV68A99.03 and the 8.4-kbp EcoRI fragment of RγHV68A98.05. The arrowheads indicate the following bands which are in addition marked by dots: 18-kbp band only present in lane 1; 12.7-kbp band only present in lanes 1 and 3; 8.4-kbp band only present in lanes 2, 4, and 5; 4.8-kbp band present only in lanes 2 and 4; 4.5-kbp band present only in lanes 2, 4 and 5; and 3.3-kbp band present only in lane 5. Marker (M) sizes (in kilobase pairs) are indicated on the left.
As for the BAC vector sequences, the presence of the antibiotic resistance gene in the recombinant viruses may interfere with analyses performed in vivo. In order to provide the possibility of deleting the antibiotic resistance gene by Flp-mediated recombination, the gene was flanked by FRT sites. An Flp-expressing plasmid was transformed into E. coli which already contained the mutant MHV-68 BAC plasmid pHA6. As expected, expression of the Flp recombinase led to the excision of the tetracycline resistance cassette, the loss of the 4.8- and 1.0-kbp EcoRI fragments, and the generation of a new 3.3-kbp fragment (Fig. 5A, line 4, and Fig. 5B, lane 4).
Interestingly, the band representing the additional EcoRI fragment of approximately 18 kbp present in the MHV-68 BAC plasmid pHA3 was absent in the mutant BAC plasmid pHA6, in the revertant BAC plasmid pHA12, and in pHA9 (Fig. 5B, lanes 1 to 4). Southern blot analysis demonstrated that this band shifted to a size of approximately 15 kbp (data not shown). This observation suggested that the absolute number of terminal repeats in the MHV-68 BAC plasmids does vary and has no influence on the reconstitution of infectious virus from the mutant plasmid. Furthermore, a new EcoRI fragment of about 4.5 kbp, due to a reduced copy number of the 40-bp internal repeats, appeared in both the mutant plasmid pHA6 and the mutant virus RγHV68A98.05 (Fig. 5B, lanes 2 and 4, and Fig. 5C, lane 6).
On the basis of the ΔORF4 mutant BAC plasmid pHA6, a revertant BAC plasmid was constructed in E. coli strain DH10B using the two-step replacement procedure described in Materials and Methods. The revertant BAC plasmid pHA12 was analyzed by restriction enzyme analysis. As expected, the revertant BAC plasmid pHA12 displayed restriction patterns like the parental BAC plasmid pHA3, with the exception that it contained a smaller number of the terminal as well as of the 40-bp internal repeats as described above for the ΔORF4 mutant BAC plasmid pHA6 (Fig. 5B, compare lanes 1 and 3). Transfection of the revertant BAC plasmid pHA12 into BHK-21 cells led to plaque formation and the generation of the revertant virus RγHV68A98.03. Southern blot analysis of the reconstituted viruses with probe P, indicated in Fig. 5A, line 3, which detects a 12.7-kbp EcoRI fragment both in RγHV68A98.01 and RγHV68A98.03 and an 8.4-kbp fragment in RγHV68A98.05, confirmed the successful mutagenesis (Fig. 5D, lanes 5 to 7).
Growth properties of the ORF 4 deletion mutant RγHV68A98.05.
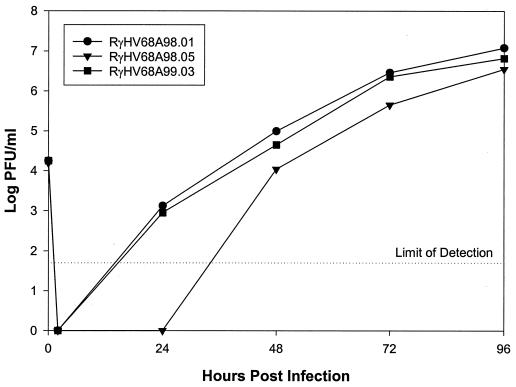
Transfection of the mutant BAC plasmid pHA6 into BHK-21 cells led to the development of plaques, already indicating that ORF 4 is dispensable for lytic replication in vitro. To determine the growth kinetics of the mutant virus RγHV68A98.05, NIH3T3 cells were infected at an MOI of 0.1. Cells and supernatants were harvested at different time points after infection, and viral titers were determined by plaque assay. The mutant virus RγHV68A98.05 showed a delayed growth compared to the parental virus RγHV68A98.01 (Fig. 6). The revertant virus RγHV68A99.03 displayed growth identical to that of the parental virus RγHV68A98.01, indicating that the delayed growth of the mutant virus RγHV68A98.05 was due to the deletion of ORF 4.
FIG. 6.
In vitro growth properties of BAC MHV-68 RγHV68A98.01, of ORF 4 deletion mutant RγHV68A98.05, and of revertant virus RγHV68A99.03. NIH3T3 cells were infected at an MOI of 0.1 for 1 h at 4°C to allow adsorption. For penetration, prewarmed medium was added for a 2-h period of incubation at 37°C. Remaining extracellular virus was inactivated by treatment with low-pH citrate buffer for 1 min. Cells and supernatants were harvested at the indicated time points, and viral titers were determined by plaque assay.
DISCUSSION
In this report, we describe the cloning and mutagenesis of the MHV-68 genome as an infectious BAC in E. coli. Following transfection of the MHV-68 BAC plasmid into BHK-21 cells, infectious virus could be reconstituted which showed growth in cell culture identical to that of WT MHV-68. Since the gfp gene was incorporated into the cloned BAC, infected cells could be easily detected. A selected viral gene (encoding ORF 4) was disrupted by site-specific mutagenesis using PCR-generated linear fragments.
For the generation of a recombinant MHV-68 containing the BAC vector sequences, the strategy described by Grassmann and Fleckenstein (13) was applied. Thus, insertion of the BAC vector was achieved by a single crossover event via only one homology region at the left end of the unique sequence of the MHV-68 genome. This strategy may also be applicable for the cloning of other gammaherpesviruses.
Following transformation of the circular MHV-68 BAC genomes into E. coli, we obtained BAC plasmids that contained the MHV-68 genome with a defined number of terminal repeats. Transfection of the MHV-68 BAC plasmids into BHK-21 cells reproducibly led to reconstitution of the desired virus genomes and recombinant viruses. As expected from previous work by others (9), in the recombinant virus genomes terminal fragments form a submolar ladder. This is due to the genomic structure of MHV-68 representing a unique stretch of DNA flanked by variable numbers of a terminal repeat unit (9, 35). To generate virus clones with as little foreign genetic material as possible, we flanked the BAC vector sequences by loxP sites and demonstrated targeted excision, allowing the generation of a recombinant virus genome with WT features.
To assess the stability of the MHV-68 BAC genome, the BAC plasmid was propagated in the E. coli strain DH10B. This strain carries a mutation in the recA gene and is therefore severely impaired in its ability to perform homologous recombination. Recombination between direct repeats causes the deletion of intervening sequences and a reduction in the copy number of the repeated sequences. The MHV-68 genome contains a number of repeats, i.e., the terminal repeats, an internal 100-bp repeat, and an internal 40-bp repeat (35), which may be prone to recombination events. We did indeed observe changes in some of the repetitive sequences of MHV-68 but not in other regions of the MHV-68 BAC plasmid. The fragments containing the 40-bp internal repeat varied in size after extended propagation of the BAC plasmid in E. coli DH10B. This was most likely due to the loss of some of the 40-bp repeat units and is probably mediated by a recA-independent mechanism. This observation is supported by the report of Virgin, IV, et al. (35) on the inability to stably clone the 40-bp repeat region. All clones which were recovered by these authors showed deletions in the repeat (35). It is therefore likely that recombination events in repeat structures may occur in other BAC genomes of gammaherpesviruses as well. Size variation due to alterations in repeat structures also occurs in other gammaherpesviruses. For example, the molecular masses of some EBV nuclear antigen proteins vary considerably between virus isolates due to variation in the number of the repetitive sequences (10). Whether the number of repeats in the MHV-68 WT genome remains constant under physiological conditions is not known (9). Obviously, the loss of some of the 40-bp repeats has no influence on the reconstitution of infectious virus and on the in vitro growth pattern. A size variation of the fragment that contains the terminal repeats was noted in the ΔORF4 mutant BAC plasmid after propagation in the recombination-proficient E. coli strain JC8679 due to the loss of terminal repeat units. Notably, this band representing a constant number of terminal repeats is present in the BAC plasmid but not in the reconstituted virus genome where the submolar ladder pattern is observed (see above). The variability of terminal repeat units in the mutant BAC plasmid had no influence on the reconstitution of infectious virus. Finally, changes within the fragments containing the 100-bp internal repeat seemed to be very rare and were only sporadically observed. Further experiments are required to determine whether the variability of the MHV-68 repeat structures has any biological effect in vivo.
For mutagenesis of the cloned MHV-68 genome, a recently described method for site-specific recombination in E. coli using short, PCR-generated linear DNA-fragments (37) was applied for the first time to a cloned herpesvirus. This technique utilizes the E. coli strain JC8679, a recombination-proficient strain which is recBC- and sbcA-negative and expresses the recombination proteins RecE and RecT (6). The mutagenesis is based on recombination between linear and circular DNA molecules (37). For recombination, a linear recombination fragment containing an antibiotic resistance gene was generated by PCR using oligonucleotides consisting of 50 nucleotides of homology to the chosen region in the BAC plasmid and 24 nucleotides for amplification of the antibiotic resistance gene (compare Fig. 5A, line 2). The target gene was disrupted and replaced by the tetracycline resistance gene. As for the BAC vector sequences, targeted excision of the tetracycline resistance gene is possible. This mutagenesis method offers advantages since it does not require construction of a recombination plasmid via multiple cloning steps, and is therefore not dependent on the disposition of restriction enzyme cleavage sites. Because of the short homology required, the complete modified region can be sequenced in a single step to determine whether recombination had occurred as planned. This method may be in particular useful for mutagenesis of BAC-cloned virus genomes for which only limited sequence information is available. Knowledge of short (50-bp) homology regions suffices for the generation of the recombination fragment.
ORF 4 of MHV-68 has been predicted to encode a complement regulatory protein (35). Both the supernatant of MHV-68 infected cells and a recombinant MHV-68 complement regulatory protein inhibited complement activation, as measured by inhibition of C3 deposition on zymosan (18). Furthermore, it was suggested that the protein encoded by ORF 4 may have additional functions independent of complement regulation, e.g., the induction of intracellular signals (18). In our study we generated a recombinant virus with a deletion of ORF 4 and the corresponding revertant. Deletion of ORF 4 had no influence on the generation of infectious virus, demonstrating that this gene is not essential for replication in cell culture. Interestingly, the ORF 4 deletion mutant displayed a delayed growth in vitro. This finding is consistent with the above-mentioned hypothesis, i.e., that the protein might have additional functions beside complement regulation. Construction of a revertant virus clearly demonstrated that the delayed growth of the mutant virus was due to the specific mutation and not to changes elsewhere in the genome of MHV-68.
In conclusion, we have cloned the MHV-68 genome as an infectious BAC and have introduced efficient site-specific mutagenesis procedures for the MHV-68 BAC plasmid. These techniques may be of use for the subjection of gammaherpesvirus and other herpesvirus genomes to fast molecular genetic analysis. For MHV-68, this technique will considerably speed up the construction of mutants, thereby allowing assessment of the role of viral genes in host-virus interaction and improving its value as a small rodent model of gamma-2-herpesvirus pathogenesis.
ACKNOWLEDGMENTS
We thank J. Stewart and A. Nash for providing MHV-68 and BHK-21 cells, W. Burns for providing REF-Cre cells, G. Posfai for providing the shuttle plasmid pST76KSR, A. J. Clarke for providing the E. coli strain JC8679, and K. C. Murphy and A. F. Stewart for useful discussions of recombination procedures. We are grateful to R. Cardin for technical advice, to C. Burgmeier for excellent technical assistance, and to B. Adler for critical reading of the manuscript.
This work was supported by grants from the Bundesministerium für Bildung und Forschung (BMBF), Stipendienprogramm Infektionsforschung, to H.A.; from the BMBF FKZ 01K1960612 to U.H.K.; and from the Deutsche Forschungsgemeinschaft (DFG) (SFB 455) to M.M. and U.H.K.
REFERENCES
- 1.Blaskovic D, Stancekova M, Svobodova J, Mistrikova J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980;24:468. [PubMed] [Google Scholar]
- 2.Borst E-M, Hahn G, Koszinowski U H, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J Virol. 1999;73:8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brune W, Menard C, Hobom U, Odenbreit S, Messerle M, Koszinowski U H. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat Biotechnol. 1999;17:360–364. doi: 10.1038/7914. [DOI] [PubMed] [Google Scholar]
- 4.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 5.Clambey E T, Virgin H W, IV, Speck S H. Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency. J Virol. 2000;74:1973–1984. doi: 10.1128/jvi.74.4.1973-1984.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark A J, Sandler S J, Willis D K, Chu C C, Blanar M A, Lovett S T. Genes of the RecE and RecF pathways of conjugational recombination in Escherichia coli. Cold Spring Harbor Symp Quant Biol. 1984;49:453–462. doi: 10.1101/sqb.1984.049.01.051. [DOI] [PubMed] [Google Scholar]
- 7.Delecluse H-J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doherty P C, Tripp R A, Hamilton-Easton A-M, Cardin R D, Woodland D L, Blackman M A. Tuning into immunological dissonance: an experimental model for infectious mononucleosis. Curr Opin Immunol. 1997;9:477–483. doi: 10.1016/s0952-7915(97)80098-2. [DOI] [PubMed] [Google Scholar]
- 9.Efstathiou S, Ho Y M, Minson A C. Cloning and molecular characterization of the murine herpesvirus 68 genome. J Gen Virol. 1990;71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- 10.Falk K, Gratama J W, Rowe M, Zou J Z, Khanim F, Young L S, Oosterveer M A P, Ernberg I. The role of repetitive DNA sequences in the size variation of Epstein-Barr virus (EBV) nuclear antigens, and the identification of different EBV isolates using RFLP and PCR analysis. J Gen Virol. 1995;76:779–790. doi: 10.1099/0022-1317-76-4-779. [DOI] [PubMed] [Google Scholar]
- 11.Fleckenstein B, Desrosiers R C. Herpesvirus saimiri and herpesvirus ateles. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Press; 1982. pp. 253–332. [Google Scholar]
- 12.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 13.Grassmann R, Fleckenstein B. Selectable recombinant herpesvirus saimiri is capable of persisting in a human T-cell line. J Virol. 1989;63:1818–1821. doi: 10.1128/jvi.63.4.1818-1821.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaves R F, Brown J M, Vieira J, Mocarski E S. Selectable insertion and deletion mutagenesis of the human cytomegalovirus genome using Escherichia coli guanosine phosphoribosyl transferase (gpt) gene. J Gen Virol. 1995;76:2151–2160. doi: 10.1099/0022-1317-76-9-2151. [DOI] [PubMed] [Google Scholar]
- 15.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 16.Horsburgh B C, Hubinette M M, Qiang D, MacDonald M L, Tufaro F. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 1999;6:922–930. doi: 10.1038/sj.gt.3300887. [DOI] [PubMed] [Google Scholar]
- 17.Horsburgh B C, Hubinette M M, Tufaro F. Genetic manipulation of herpes simplex virus using bacterial artificial chromosomes. Methods Enzymol. 1999;306:337–352. doi: 10.1016/s0076-6879(99)06022-x. [DOI] [PubMed] [Google Scholar]
- 18.Kapadia S B, Molina H, van Berkel V, Speck S H, Virgin H W., IV Murine gammaherpesvirus 68 encodes a functional regulator of complement activation. J Virol. 1999;73:7658–7670. doi: 10.1128/jvi.73.9.7658-7670.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karger A, Schmidt J, Mettenleiter T C. Infectivity of a pseudorabies virus mutant lacking attachment glycoproteins C and D. J Virol. 1998;72:7341–7348. doi: 10.1128/jvi.72.9.7341-7348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klupp B G, Baumeister J, Dietz P, Granzow H, Mettenleiter T C. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J Virol. 1998;72:1949–1958. doi: 10.1128/jvi.72.3.1949-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nash A A, Sunil-Chandra N P. Interactions of the murine gammaherpesvirus with the immune system. Curr Biol. 1994;6:560–563. doi: 10.1016/0952-7915(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 24.Nash A A, Usherwood E J, Stewart J P. Immunological features of murine gammaherpesvirus infection. Semin Virol. 1996;7:125–130. [Google Scholar]
- 25.Pridmore R D. New and versatile cloning vectors with kanamycin-resistance marker. Gene. 1987;56:309–312. doi: 10.1016/0378-1119(87)90149-1. [DOI] [PubMed] [Google Scholar]
- 26.Rickinson A B, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 27.Saeki Y, Ichikawa T, Saeki A, Chiocca E A, Tobler K, Ackermann M, Breakefield X O, Fraefel C. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum Gene Ther. 1998;9:2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 28.Schulz T F. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 29.Simas J P, Bowden R J, Paige V, Efstathiou S. Four tRNA-like sequences and a serpin homologue encoded by murine gammaherpesvirus 68 are dispensable for lytic replication in vitro and latency in vivo. J Gen Virol. 1998;79:149–153. doi: 10.1099/0022-1317-79-1-149. [DOI] [PubMed] [Google Scholar]
- 30.Simas J P, Efstathiou S. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 1998;6:276–282. doi: 10.1016/s0966-842x(98)01306-7. [DOI] [PubMed] [Google Scholar]
- 31.Smith G A, Enquist L. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J Virol. 1999;73:6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Speck S H, Virgin H W., IV Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr Opin Microbiol. 1999;2:403–409. doi: 10.1016/s1369-5274(99)80071-x. [DOI] [PubMed] [Google Scholar]
- 33.Stavropoulos T A, Strathdee C A. An enhanced packaging system for helper-dependent herpes simplex virus vectors. J Virol. 1998;72:7137–7143. doi: 10.1128/jvi.72.9.7137-7143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Virgin H W, IV, Speck S H. Unraveling immunity to γ-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr Opin Immunol. 1999;11:371–379. doi: 10.1016/s0952-7915(99)80063-6. [DOI] [PubMed] [Google Scholar]
- 35.Virgin H W, IV, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner M, Jonjic S, Koszinowski U H, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Buchholz F, Muyrers J P P, Stewart A F. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]