ABSTRACT
Staphylococcus aureus is a Gram-positive bacterium and one of the most prevalent infectious disease-related causes of morbidity and mortality in adults. This pathogen can trigger a broad spectrum of diseases, from sepsis and pneumonia to severe skin infections that can be fatal. In this review, we will provide an overview of S. aureus and discuss the extensive literature on epidemiology, transmission, genetic diversity, evolution and antibiotic resistance strains, particularly methicillin resistant S. aureus (MRSA). While many different virulence factors that S. aureus produces have been investigated as therapeutic targets, this review examines recent nanotechnology approaches, which employ materials with atomic or molecular dimensions and are being used to diagnose, treat, or eliminate the activity of S. aureus. Finally, having a deeper understanding and clearer grasp of the roles and contributions of S. aureus determinants, antibiotic resistance, and nanotechnology will aid us in developing anti-virulence strategies to combat the growing scarcity of effective antibiotics against S. aureus.
KEYWORDS: Staphylococcus aureus, antibiotic resistance, nanotechnology, mesoporous silica nanoparticles, MDR
1. Introduction to Staphylococcus aureus
Staphylococcus aureus (S. aureus) is a gram-positive bacteria that belongs to the family Micrococcaceae, which includes several species of medical and veterinary importance, including S. aureus, S. epidermidis, S. agnetis, S. pseudintermedius, S. lutrae, S. intermedius, S. hyicus, S. delphini, S. cornubiensis and S. schleiferi subsp [1]. S. aureus is a versatile and adaptable spherical-shaped coccus that is a catalase positive and oxidase negative bacterium commonly found on the skin and in the nasal passages of healthy individuals [2]. It usually acts in a commensal symbiotic relationship with humans but can result into pathogenesis, infecting through wounds or other openings in the skin. S. aureus is the cause of a variety of clinical diseases commonly acquired in either community or hospital settings including skin infections, such as boils and impetigo, as well as more serious infections like pneumonia, toxic shock syndrome, bone and bloodstream infections [3,4]. Because of its ability to produce toxins, S. aureus can also contaminate food resulting in food poisoning [5,6].
In 1880, for the first time, Scottish physician Sir Alexander Ogston isolated staphylococci from surgical abscess fluid [7]. Ogston was the first to discuss post-operative pyogenic infections caused by what he called ‘micrococci’ [4]. He named the bacteria staphylococcus because of its resemblance to grapes as clustered, round shapes. The name comes from the Greek words staphyle, which means ‘bunch of grapes,’ and kokkos, which means, ‘berry’. Staphylococci were distinguished from streptococci, which are arranged in chains and cause post-surgical infections [5,6]. In 1881, Ogston conducted experimental laboratory tests on guinea pigs and mice by injecting staphylococci into their subcutaneous tissues to examine skin-associated illnesses caused by S. aureus [8]. A few years later, in 1884, the German physician Friedrich Rosenbach identified and cultivated staphylococci from individuals. In order to distinguish S. aureus from S. epidermidis (originally known as S. albus), he examined their properties and grouped them according to the creation of golden or whitish colonies, designating the species ‘aureus’ from the Latin term meaning ‘golden’ [8].
Colonies of S. aureus are distinguished by their size, smoothness, elevation, and golden yellow tint [9]. The bacteria’s production of staphyloxanthin, a carotenoid that coats and shields the microorganism from phagocytosis, gives the S. aureus its yellow color [10]. It is worth mentioning that 70% of people are either transiently or not colonized by S. aureus, whereas 20% of healthy individuals are persistently or asymptomatically colonized [11]. On an enriched blood agar, the microorganism typically produces zones around its’ colonies as a result of hemolysis, which is caused by the release of several enzymes known as hemolysins [5,7]. Due to its’ salt tolerance, S. aureus can be distinguished and cultured on a specific medium, such as mannitol salt agar with 7.5–10% sodium chloride [1]. Furthermore, the bacteria ferments mannitol sugar resulting in an acidic environment, thus changing the medium’s color from pink to yellow [12]. This can be used to differentiate S. aureus from S. epidermidis, a non-mannitol fermenter [5].
S. aureus exhibits rapid development of antibiotic resistance, and the emergence of multidrug-resistant strains is a serious issue [13]. According to previous reports, the number of people dying each year from antibiotic-resistant problems has topped 10 million, and by 2050, the mortality rate will surpass those from cancer [14]. Due to the ineffectiveness of conventional antibiotics, the morbidity and mortality outcomes highlight the urgent need to find alternative, effective remedies [15]. Hence, to decrease virulence factors, many approaches have been tested, most notably medication designs based on synthetic analogs. On the other hand, due to toxicity and/or limited bioavailability, these trials have not yet been approved clinically [16]. A new potential anti-staphylococcal alternative is now being tested, with a focus on biological molecules or chemicals that interfere with toxins or toxin-regulator genes [17–22].
This review outlines key points related to S. aureus, such as the structure, health problems, and nanomaterials as new treatment options.
2. Epidemiology of Staphylococcus aureus
S. aureus infections constitute a major public health concern, with an approximately 30% of the population harboring the bacteria colonized in many bodily organs, especially in upper respiratory tracts including the skin, nostrils, axilla, and inguinal regions [23,24]. According to the China Antimicrobial Resistance Surveillance System’s surveillance statistics from 2019, S. aureus has the highest detection rate among gram-positive pathogenic bacteria in China [25]. Nonetheless, certain groups of individuals are at a higher risk, up to 80%, of S. aureus colonization including health care workers, diabetics, people with compromised immune systems, children, elderly individuals, HIV or cystic fibrosis individuals, people who are on indwelling catheter, or have chronic metabolic diseases, or who have previously been infected with Methicillin-Resistant Staphylococcus aureus (MRSA) [5,26]. Indeed, recent S. aureus epidemiology has focused on the rise and dissemination of MRSA in clinical settings and the population [27]. Several studies have attempted to establish the prevalence of community acquired MRSA, CA-MRSA, and colonization amongst patients [28].
CA-MRSA infections have evolved significantly and are now becoming basic causes of nosocomial infections [29–31]. According to a 2012 review of van Hal et al., an annual incidence rate of S. aureus bacteremia range from 20 to 50 cases per 100,000, with 10 to 30% of these patients dying from the infection [32]. Further, according to a 2017 study, the total incidence of deaths in the United States owing to S. aureus bacteremia was 20,000. In this context, Hindy et al. determines the incidence trends of S. aureus bacteremia from population-based studies of 26 geographical populations. The incidence of S. aureus bacteremia in the 11 nations studied is shown to be generally steady and ranges from 9.3 to 65 cases per 100,000 people per year [33]. In a study performed in Ethiopia to assess the prevalence of methicillin-resistant S. aureus and its associated factors among patients with wound infection, data revealed that 34.58% patients suspected with wound had culture-confirmed S. aureus, and of these 28.3% were MRSA [34]. It is important to note the majority of the S. aureus bacteremia data that is currently released originates from high-income, temperate countries. Despite the fact that the majority of the world’s population lives in tropical nations with low and middle-income, there is a dearth of statistics on these regions [35]. According to available data and based on relevant studies found on PubMed search engine published between January 2005 and December 2019, Arab countries in the Middle East and North Africa have a significant prevalence of S. aureus [36]. It is important to note that the number of reported rate showed a wide range. In the Gulf Cooperation Council, infectious S. aureus rates varied from 9% to 38%, in Levant they ranged from 28% to 67%, and in North African regions they ranged from 28% to 57% [36]. Lastly, in Jordan, S. aureus was isolated from 19% of nasal carrier isolates and 57% to 62% of clinical isolates [37, 38].
The prevalence of MRSA nasal carriage among populations has been the subject of numerous studies as well. The percentages vary greatly depending on the regions and populations polled. Nasal carriage rates of MRSA in the US and Europe vary from 0.1% to 2.5%, and with rates going as high as 10.5% recorded in Africa [39,11,12]. A research of S. aureus isolates from pus/wound swab samples in a tertiary care hospital in Nepal revealed that out of 76 isolates, 43 (56.6%) were from outpatients and 33 (43.4%) were from inpatients. MRSA made up 41.9% of the S. aureus isolates from outpatients, while 54.5% of the isolates from inpatients [40,41]. The nasal carriage rates of MRSA found in Middle Eastern countries including KSA, Lebanon, and Palestine ranged from 0–13.2% [10,14]. In Jordan specifically, the transmission rates of MRSA was reported to be 7.5–19% among healthy Jordanians . Another study of 126 Staphylococcus isolates from multiple Jordanian medical sites revealed that 71 (56.3%) of the isolates were S. aureus while 55 (77.5%) were MRSA [37]. Similarly, a further study from Jordan found that 62% of the 232 S. aureus isolates were MRSA which were isolated from various clinical samples (abscesses, wounds, skin lesions, nasal swabs and blood) [42]. Interestingly, a recent study was performed using different published resources between January 1980 and December 2022 found that the prevalence of MRSA colonization in residents of elderly care centers is high across the world and varies by gender and geographic location. This provides important insight into the fundamentals of targeted treatments and screening programs that aim to identify infected individuals, manage risk factors, and lessen the spread of MRSA to elderlies living in elderly care centers [43]. Furthermore, a study documenting the epidemiology of S. aureus in South America using genomics was published in 2019 [44], as Di Gregorio and his colleagues characterized 404 genomes recovered from a prospective observational study of S. aureus bacteremia in 58 hospitals from Argentina, Bolivia, Brazil, Paraguay and Uruguay. The results demonstrate that more than a quarter of S. aureus isolates are resistant to macrolide-lincosamide-streptogramin B (MLSb), whereas 5.2% of S. aureus isolates are phenotypically multi-drug resistant. The genetic diversity of methicillin-susceptible S. aureus (MSSA) was higher than that of MRSA. The prevalence of MRSA and MSSA lineages varied by country even though the most common S. aureus genotypes are high-risk clones that are widespread in South America and lack a distinct phylo-geographical structure that is particular to any one region [44].
3. Clinical manifestations of Staphylococcus aureus
S. aureus has been a significant human pathogen and is currently responsible for the majority of bacterial infections worldwide [30]. It can potentially infect any tissue in the human body and causes mild to life-threatening diseases [1]. The different S. aureus infections can be broadly divided into three groups: toxinoses (food-borne illnesses, scalded skin syndrome, and toxic shock syndrome), systemic and life-threatening disorders (infective endocarditis, osteoarticular, bronchiolitis, meningitis, pleuro-pulmonary, and septicemia) and superficial skin and soft tissue infections [24,45,46].
The severity of an infection depends on several factors including the virulence of the particular strain, the quantity of the inoculum and the person’s immunological state. It is worth noting that pus-filled abscesses and necrotic tissue encircling harmed leukocytes are typical signs of staphylococcal infection [32].
3.1. Staphylococcal food poisoning
Staphylococcal food poisoning (SFP) is a common food-borne disorder that affects people globally, mainly brought on by the consumption of enterotoxigenic strains of coagulase-positive staphylococci called staphylococcal enterotoxins (SEs) [47].
S. aureus can survive in dry conditions and can even form biofilms, contributing to its persistence in food and food-processing environments. However, outbreaks are typically caused by post-process contamination and/or thermal abuse [48]. Depending on eating habits, certain foods are more frequently linked to SFP in different nations. Nonetheless, a number of ways have been suggested, involving physical and/or chemical therapies, the use of natural antimicrobials, and combined hurdles approaches [49].
3.2. Staphylococcal scalded skin syndrome (SSSS)
Staphylococcal scalded skin syndrome (SSSS) is a skin condition caused by bacterial toxins that can be potentially fatal and primarily affects toddlers, but older children and adults can also be affected. S. aureus exotoxins spread hematogenously to the skin and causes SSSS [50]. This syndrome frequently manifests sudden fever. Widespread erythema begins on the head and progress to superficial skin peeling, flaccid bullae, and denuded tender skin, an additional cutaneous symptoms (Figure 1) [23]. However, the two most feared outcomes are sepsis and pneumonia as they can be life-threatening [24].
Figure 1.
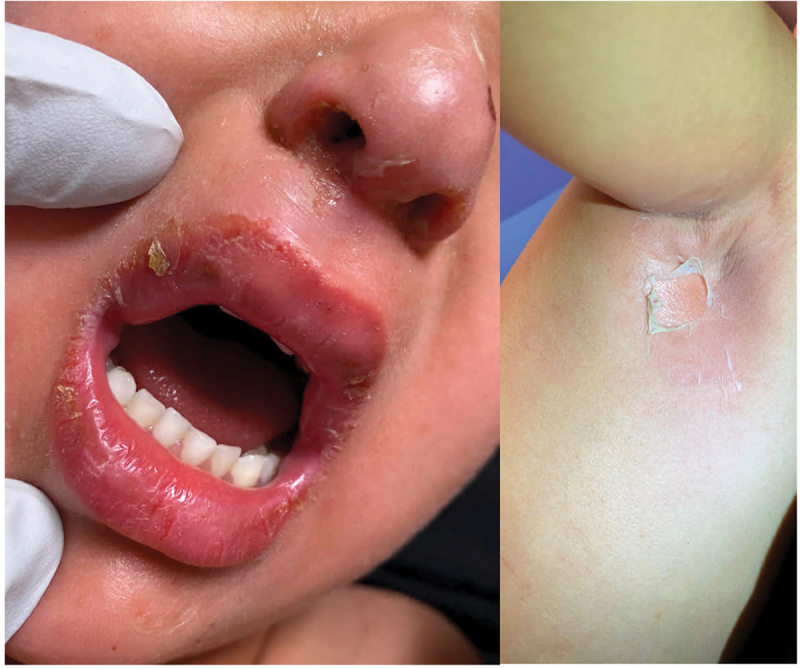
Staphylococcal scalded skin syndrome. superficial blistering on the face (left) and in the axilla (right). Original image provided by Kiran Motaparthi, MD [51].
A skin biopsy sample can verify the diagnosis. According to recent epidemiological research, mortality among adults’ ranges from 40% to 63% but is less than 10% among children. Intravenous immunoglobulin had long been recommended to treat Staphylococcal scalded skin condition despite receiving antimicrobial medication [52,53].
3.3. Staphylococcal toxic shock syndrome
Staphylococcal toxic shock syndrome (TSS) is a rare but serious illness caused by toxins produced through certain strains of S. aureus bacteria. The symptoms of staphylococcal TSS can vary but typically include fever, vomiting, diarrhea, muscle aches and a rash that resembles sunburn. In severe cases, the illness can progress rapidly and lead to organ failure, shock and even death [1].
Staphylococcal TSS can affect anyone, but it is most commonly associated with risk factors such as skin wounds and infections, surgery, childbirth and the use of nasal packings and other medical devices [10]. On another hand, treatment for staphylococcal TSS typically involves hospitalization, intravenous antibiotics to treat the bacterial infection and supportive care to manage symptoms and prevent complications. It is important to seek medical attention immediately if you suspect you may have staphylococcal TSS or have any symptoms suggestive of this illness [1].
3.4. Staphylococcus aureus bacteremia
S. aureus bacteremia (SAB) can colonize and cause difficulties by seeding in almost any part of the body [54]. These problems can lead to serious medical conditions with significant morbidity and mortality [55]. SAB complications are common, with an incidence of 11% to 53%. Due to the anatomic site and the difficulty of rapid diagnosis, some problems require frequent admission to the intensive care unit and may have a poor prognosis [24]. A recent study that was conducted in Brazil show that S. aureus is a common cause of bloodstream infection in neutropenic patients. Indeed, the majority of fatalities occurred after two weeks of febrile neutropenia and MRSA bacteremia were linked to an elevated risk of mortality [56,57].
3.5. Staphylococcal endocarditis
Infectious endocarditis is currently most frequently caused by S. aureus worldwide. S. aureus is a feared pathogen of endocarditis due to its ability to spread serious disease and its frequent drug resistance [31]. Injury to the cardiac endothelium, either directly by trauma, such as injection of drugs-derived particles, or by turbulent blood flow, results in nidus, which causes bacterial colonization and infection, or inflammation resembling degenerative valve disease [58]. Exposure to sub-endothelial cells produces the extracellular matrix proteins and tissue factor, and deposits fibrin and platelets to create sterile vegetations. Bacterial colonization of these thrombotic vegetation can lead to the development of endocarditis [59].
3.6. Staphylococcal osteomyelitis
Osteomyelitis is an inflammatory bone disease that gradually destroys and loses bone mass. The pathogenesis of this disease is a double-edged sword whereby not only can staphylococci utilize bone for colonization, but bone itself can facilitate infection progression . The commensal staphylococci are the most causative agent and infection which originate from either hematogenous (endogenous) or exogenous [60]. Problems like the ability of staphylococci to endure treatment and surgical intervention to remove necrotic and infected bone further exacerbates patient impairment. Different factors that prompt a patient to developing Staphylococcal osteomyelitis, including age, diabetes, peripheral vascular disease, intravenous drug use, surgical implants, and immunodeficiency due to disease or immunosuppressant drugs [61].
3.7. Staphylococcal pneumonia
Staphylococcal pneumonia was first described in 1919 in adults during the influenza pandemic [62]. Although this pulmonary infection is rare, it can afflict people of all ages and manifest itself in many ways [63]. However, a recent meta-analysis review shed light on the fact that S. aureus is a key cause of global pneumonia hospitalization in under-five children [64]. Staphylococcal pneumonia is a disease condition that might have consequences like severe necrotizing pneumonia, bacteremia, or sepsis with or without shock [65,66]. It is associated with airway hemorrhage, epithelial necrosis and high fatality rate in otherwise healthy patients [67]. Subsequent reports confirmed the high fatality rates (40–50%) of Panton – Valentine leukocidin -associated pneumonia in adults [68,69].
4. Virulence factors and pathogenicity of Staphylococcus aureus
S. aureus is a typical component of human mucous membranes and skin, but when given the opportunity to enter the body due to injury or compromised immune systems of the host, illnesses may result [10]. There are numerous different virulence factors involved in the pathogenesis of S. aureus, including surface proteins, enzymes, toxins, and many others. These virulence factors are crucial for S. aureus invasion, colonization, and survival in the hosts to induce staphylococcal infections [1]. S. aureus perceives, reacts to and adapts to the extreme environmental circumstances prevalent in the host during infection. Consequently, this aids in their ability to colonize and survive within the host despite host immune responses and antimicrobial therapy .
4.1. Staphylococcus aureus virulence factors
4.1.1. Genetics of virulence
Bacterial genomes can be generically classified into core and accessory parts [70,71]. All isolates carry the genes known as the core genome, which generally contains essential genetic information related to cellular metabolism and replication [30]. The core of S. aureus’ genome makes up around 75% of the whole genome and is highly conserved between strains [72]. The accessory genome, where mediators of virulence, immune evasion and antibiotic resistance are frequently located, is where a large portion of the genetic diversity of MRSA and other infections occurs [31]. Thus, about 25% of the S. aureus genome is made up of the accessory genes [31].
S. aureus virulence factors are regularly encoded on the pathogen’s accessory genome [1]. The accessory genome contains mobile genetic elements (MGEs) including transposons, plasmids, prophages, insertion sequences, pathogenicity islands (PAIs), and Staphylococcus cassette chromosomes (SCCs) [30]. These MGEs not only include virulence factors but also antibiotic resistance genes [73]. Due to the encoding of the corresponding causative toxins, such as toxic shock syndrome toxin 1 (TSST-1), or even the enterotoxins that cause food poisoning, isolate-specific MGEs are frequently linked to specific diseases, or ‘toxinoses’ [5].
Most of the toxins and other virulence factors produced by S. aureus are found in phage-related and pathogenicity islands [74]. Panton-Valentine leukocidin genes (PVL), the immune evasion proteins CHIPS and SCIN, the Epidermolytic toxins B and A, staphylokinase and a number of enterotoxins are significant S. aureus toxins encoded on prophages [75,76]. The hlb gene, which encodes the β-toxin, has been linked to virulence processes and is crucial for infectious colonization [35]. Enterotoxins and TSST are the main pathogenicity islands’ recognized products [77]. Toxins that are encoded on genomic islands and often only differ in expression among different strains include peptide-spectrum match (PSM), α-toxin, the leukocidin LukDE, SSLs, the lipoprotein-like toxins (LPLs) and various enterotoxins [1].
4.1.2. Virulence regulation
A sophisticated network of genetic and metabolic processes that react to environmental factors such nutrient intake, pH, temperature and host variability controls the virulence of S. aureus [10,78]. The expression of S. aureus virulence factors can be affected by several different regulatory influences [30]. These include the global regulators that control a number of virulence genes and are frequently influenced by specific environmental conditions, as well as locus-specific regulatory factors like the icaR gene adjacent to the ica operon, which is also subject to numerous regulatory impacts, or the PSM-sensing PmtR protein controlling the PSM exporter Pmt operon [31].
The accessory gene regulator (agr) system is one of the major controllers of virulence gene expression in S. aureus . The four genes agrA, agrB, agrC, and agrD that make up this system encode a two component signaling pathway [79]. This agr system is activated by the accumulation of autoinducing peptides (AIPs), a peptide the bacterium produces [80]. Whenever it is active, the agr system suppresses the expression of surface proteins such adhesions and capsule polysaccharides while inducing the expression of virulence genes [81]. This change in gene expression allows the bacteria to more effectively penetrate host tissues and evade the host immune system [82,83].
The staphylococcal accessory regulator (sar) system is another mechanism that controls the expression of virulence genes in S. aureus [84]. The three genes (sarA, sarR, and sarS) that make up the sar system encode a transcriptional regulator as well as two sensor kinases [85]. Surface proteins, adhesions, and toxins are only a few of the virulence factors that the sar system regulates its expression [86].
The ferric uptake regulator (Fur), a global regulator that responds to reduced iron availability, and SrrAB, a regulator that responds to oxygen, are other prominent global regulators triggered with known environmental cues [87]. Fur controls a number of virulence factors, including poisons and immune evasion proteins in addition to iron consumption [82]. The role of Fur is to coordinate the pathogen’s attack on the host, as such iron restriction is assumed to signal entry into the body and the subsequent demand for those components [88]. Meanwhile, SrrAB is a two-part oxygen-sensitive system that depends on redox-sensitive cysteines to provide oxidative stress resistance. In anaerobic environments, it downregulates agr while increasing ica expression, improving resistance to neutrophil invasions as a result [86,89].
The sigma factor, a subunit of the RNA polymerase, is responsible to initiate the transcription in bacteria. The majority of bacteria produce many sigma factors to transcribe their various gene sets [90]. S. aureus also synthesizes four sigma factors (i.e. σA, σB, σH, and σS) to express its diverse array of genes [91]. In depth, the alternative sigma factor B (SigB), σB, is a class of transcription factors that regulates both promoter recognition and RNA polymerase recruitment [25]. SigB is crucial for the expression of their genes when bacteria respond to external stressors [89]. Indeed it has been reported that SigB regulates more than 250 genes [92]. Moreover, a recent study confirmed that sigB is responding to biofilm formation and stress tolerance in S. aureus [25]. Finally, σH is designated as the alternative sigma factors, while σA and σS act as the housekeeping sigma factor and extra cytoplasmic sigma factor, respectively [93].
The global regulators CodY and CcpA, along with the two-component systems Sae and ArlRS, are just a few of the regulatory pathways that regulate the expression of the virulence genes in S. aureus in addition to the main systems of agr and sar [83]. Overall, various genetic and metabolic processes interact to regulate the expression of the virulence genes in S. aureus in a complicated manner [31].
Lastly, S. aureus’ capacity to produce several virulence factors at once and to rapidly control their expression in response to environmental changes is vital to its pathogenic success [94]. Table (1) summarize tools that S. aureus bacteria use to avoid immune responses, which consequently leads to the spread of infection and serious damage throughout the body [80,93–96].
Table 1.
The most important virulence factors of S. aureus and their targets during infection.
| Type of Virulence Factor | Name | Target | Effect |
|---|---|---|---|
| Cell wall-associated factors | Cell wall components – peptidoglycan, teichoic acid, lipoteichoic acid | Immune cells, other tissues | Stimulate immune cell activation and inflammatory response; participate in adhesion and biofilm formation |
| Staphylococcal protein A (SpA) | IgG, IgM, complement | Binds Fc region of IgG and IgM, thus inhibiting opsonization and phagocytosis; activates B cells | |
| Fibronectin-binding proteins (FnBPA, FnBPB) | Fibronectin, fibrinogen, elastin, plasminogen, keratin, complement | Binding to extracellular matrix proteins (ECM), enable adhesion to host tissues and biomaterials; limit phagocytosis and complement activation | |
| Collagen-binding protein (Cna) | Cartilage and collagen-rich tissues, complement | Binding cartilage and collagen, enables adhesion to host tissues; inhibits complement activation | |
| Clumping factors (ClfA, ClfB) | Fibrinogen, blood platelets, complement (ClfA), cytokeratin 10 (ClfB) | Binding to fibrinogen, enables adhesion to host tissues; inhibit complement preventing opsonization and phagocytosis; activate platelets | |
| Serine-aspartate repeat protein E (SdrE) | Complement | Inhibits complement preventing opsonization and phagocytosis | |
| Iron-regulated surface determinant proteins (IsdA, IsdB) | Heme-iron | Heme uptake and iron acquisition contribute to increased pathogenesis, tissue invasion and abscess formation | |
| Polysaccharide intercellular adhesion/polymeric N-acetyl-glucosamine (PIA/PNAG) | Staphylococcal cells, mucous membranes, other tissues, abiotic surfaces | Participates in bacterial aggregation, adhesion and biofilm formation (major component of biofilm matrix); reduces phagocytosis | |
| Capsular polysaccharides | Mucous membranes, other tissues, abiotic surfaces | Reduce phagocytosis; increase the efficiency of colonization and durability on the surface of mucous membranes or biomaterials | |
| Enzymes | Catalase | Hydrogen peroxide | Catalyzes breakdown of hydrogen peroxide into water and oxygen, preventing oxidative stress |
| Coagulase | Prothrombin | Reacts with prothrombin, allowing fibrinogen polymerization and clot formation, thus reducing phagocytosis | |
| Staphylokinase (SAK) | Plasminogen | Converts plasminogen to active serine protease plasmin, which promotes degradation of ECM, complement and IgG | |
| Lipases | Lipids of cell membranes and components of sebum | Decompose lipids, which allows spreading of staphylococci | |
| Nucleases | Nucleic acids | Degrade nucleic acids, thereby releasing them from extracellular traps (ETs) | |
| Proteases, e.g. serine protease V8 (SspA), staphopain A (Scp A) and B (SspB), aureolysin (Aur) | ECM proteins, complement, mucins, pulmonary surfactant | Degrade ECM proteins, mucins and pulmonary surfactant, which allow staphylococcal spread in the host tissues; inhibit chemotaxis and phagocytosis by proteolysis of immune cell receptors; degrade complement preventing opsonization and lysis of bacteria; degrade antimicrobial peptides | |
| Superoxide dismutases | Superoxide | Convert superoxide to hydrogen peroxide and oxygen, thereby preventing oxidative stress | |
| Toxins | Hemolysins (alpha, beta, gamma, delta) | Erythrocytes, platelets, leukocytes | Cause lysis of red blood cells, platelets, leukocytes – evading of host immune response; bacterial spreading |
| Enterotoxins | Enterocytes, lymphocytes T | Cause diarrhea; after translocation into blood, activate lymphocytes T leading to cytokine storm | |
| Exfoliative toxins | Desmosomes between keratinocytes | Cleave the granular layer of the epidermis by damaging desmosomes (staphylococcal scalded skin syndrome) | |
| Panton-–Valentine leukocidin (PVL) | Neutrophils, monocytes, macrophages | Causes lysis of neutrophils, monocytes, macrophages – avoiding innate immune response; development of necrotic changes | |
| Toxic shock syndrome toxin 1 (TSST-1) | Lymphocytes T | Activates lymphocytes T, which causes massive production of cytokines and leads to toxic shock syndrome | |
| Other secreted proteins | Chemotaxis inhibitory protein of Staphylococcus (CHIPS) | Neutrophils | Binds to cell receptors (FPR1 and C5aR) inhibiting neutrophils chemotaxis, thereby preventing phagocytosis |
| Staphylococcal complement inhibitor (SCIN) | Complement (C4, C3b) | Inhibits complement activation, thus preventing bacterial lysis, opsonization and phagocytosis | |
| SSL-5 | Neutrophils, platelets | Binds to cell receptors (PSGL-1 and GPCRs) inhibiting neutrophil diapedesis and activation; activates platelets (aggregate formation) | |
| SSL-7 | IgA, complement (C5) | Binds Fc region of IgA and complement protein C5, thus blocking antibodies and inhibiting complement activation | |
| Extracellular fibrinogen-binding protein (Efb) | Fibrinogen, blood platelets, complement | Binds fibrinogen enabling adhesion and aggregation: interferes with platelet aggregation; inhibits complement activation | |
| Extracellular adherence protein (Eap) | ICAM-1 | Binds ICAM-1 inhibiting neutrophil rolling and migration (diapedesis) |
4.2. Staphylococcus aureus pathogenicity
Since S. aureus is a part of the normal flora of the skin and mucous membranes, it has the potential to infiltrate and cause infection if there is a wound in the skin or if it colonizes people with weakened immune systems. The systematic infection process may be influenced by two potential mechanisms: the synthesis of toxins and colonization leading to tissue invasion and destruction (Figure 2) [1].
Figure 2.
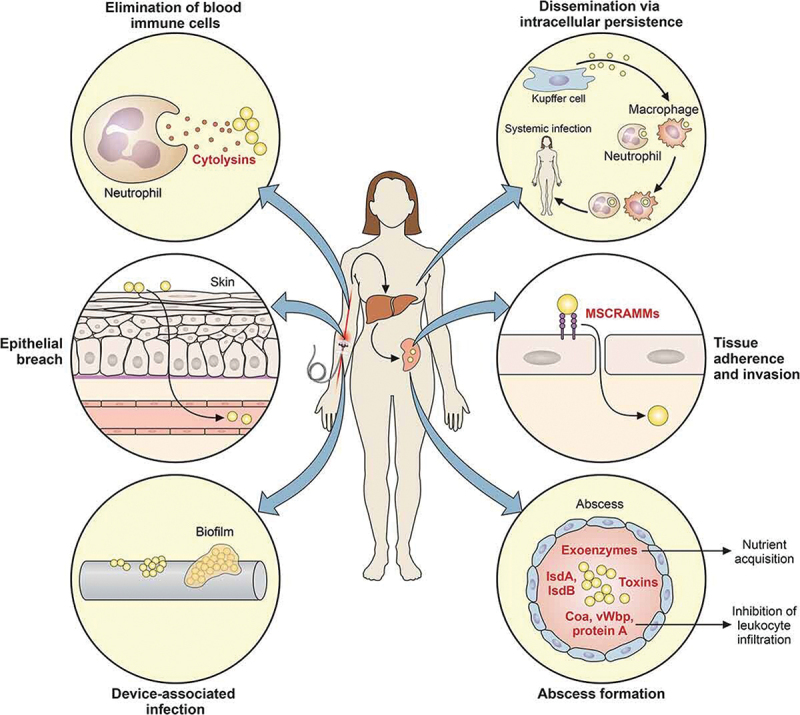
S. aureus mechanisms to initiate systematic infections. When S. aureus penetrates the skin’s natural defenses or spread through a biofilm that might develop on indwelling medical equipment, systemic infection is commonly the result. By actively attacking and killing immune cells like neutrophils in the circulation with cytolytic toxins, the bacteria can also remain in these cells and spread throughout the body, the bacteria can also spread through the bloodstream and invade more cells. Many bacterial factors, including particular surface proteins, toxins, exoenzymes and other compounds, can affect the development of an abscess later on [1]. .
For S. aureus to produce infection, adhesion is a necessary step. S. aureus has a number of surface proteins and other components that enables it to adhere to host cells and tissues. Via the use of these adhesions, the bacteria are able to attach to a range of host tissues and organs, such as skin, nasal mucosa, and heart valves [32,97]. Certain examples of adhesions produced by S. aureus involve Microbial Surface Components Recognizing Adhesive Matrix Molecules (MSCRAMMs). Nonetheless, S. aureus produces different MSCRAMMs, like collagen-binding protein (Can), fibrinogen-binding protein (ClfA) and fibronectin-binding proteins (FnBPs), that allow the bacteria to adhere to extracellular matrix elements of host tissues [1,24]. Moreover, S. aureus produces teichoic acids and lipoteichoic acid, which are acidic polymers that can interact with host proteins and glycoproteins to help the bacterium attach to host cells and enhance bacterial adherence. Last but not least, S. aureus creates a capsule that enables the bacterium to bind to host cells and tissues as well as elude the host’s defenses [5].
After adhering to the tissues of the host, invasion is the next stage of S. aureus pathogenicity. Several strategies are employed by S. aureus enable the cell to penetrate host tissues while evading host defenses including enzyme secretion. In fact, S. aureus produces a number of enzymes, including hyaluronidase and fibrinolysin, that can break down host tissues and allow the bacterium to infiltrate into the host tissue more deeply [4]. In order to overcome host defenses, S. aureus also produces proteases such aureolysin and staphopain that cleave host proteins like immunoglobulins and complement proteins [30]. S. aureus is capable of forming abscesses, which serve as both a barrier between the bacteria and the host’s immune system as well as a source of nourishment for the bacterium [1]. S. aureus is able to create biofilms, which are bacterial populations encased in extracellular matrix. Therefore, because of these biofilms, the host’s defenses, antibiotics, and disinfectants will have no affect the bacteria [98,99].
Immune evasion can be brought on by S. aureus through releasing chemotaxis inhibitory proteins, anti-opsonizing proteins, that stop neutrophils from phagocytosing it. Additionally, S. aureus cell surface protein A exhibits antiphagocytic characteristics. Furthermore, S. aureus secretes Periventricular leukomalacia (PVL), which kills leukocytes, and superantigens (enterotoxin and TSST1) that disrupt the immune system’s usual response by stimulating T cells (receptor -variable specific T cells) and expanding them into polyclonal populations. As a result, these T cells would be suppressed to an anergic condition [24,31,100,101].
5. Antimicrobial resistance
S. aureus’s relatively high virulence enables the cell to adapt to a variety of environmental conditions [102–104]. Unfortunately, S. aureus strains have developed resistance against practically all antimicrobial agents used in therapy, specifically to beta-lactams, glycopeptides and oxazolidinones, which are frequently used to treat Gram-positive infections [1]. Inadequate infection control procedures, the misuse and abuse of antibiotics and S. aureus‘s capacity to acquire resistance genes through horizontal gene transfer are just a few of the causes that lead to the emergence of antibiotic resistance in this species [31]. Inadequate infection control procedures make it easier for resistant strains to spread among patients in hospital settings, while misuse and abuse of antibiotics lead to the development of novel resistant strains by placing a selection pressure on their survival [10].
5.1. Mechanisms of antibiotic resistance
S. aureus has evolved a number of mechanisms for resistance against methicillin and other antibiotics. Among the primary aspects of resistance are, first, the synthesis of beta-lactamases because these enzymes can break down and render beta-lactam drugs, like methicillin, and second, the modification of penicillin-binding proteins (PBPs), which are crucial for bacterial cell wall production [105]. Moreover, certain S. aureus strains can alter their PBPs to make them more resistant to beta-lactam drugs [106].
Some S. aureus strains reach resistance reducing cell membrane permeability, thus compromising beta-lactam antibiotics’ ability to enter cells and reach their target. Changes in membrane permeability can be achieved through increased antimicrobial efflux pumps of the cells. In order to reduce the effectiveness of beta-lactam antibiotics, some strains of S. aureus have developed efflux pumps which can pump antibiotics out removing the drugs from the bacterial cell and making it more difficult to pass through the cell membrane.
Another way S. aureus develops antimicrobial resistance is through the horizontal gene transfer of various MGEs [107]. Based on the size of the plasmids that the bacteria contain, it is possible to predict that each of the aforementioned MGEs could carry genes that are resistant to antibiotics [108]. Small plasmids may include genes resistant to tetracycline and erythromycin, whilst large plasmids carry genes resistant to beta lactams and macrolides [109,110]. Contrarily, larger plasmids have genes that when combined with other MGEs result in resistance to medications such vancomycin, erythromycin, beta lactams, and trimethoprim [107,111]. S. aureus first developed antibiotic resistance during the early stages of antibiotic treatment. In contrast, only a few years after penicillin was first used as a treatment, the first penicillin-resistant S. aureus strains were described in the 1940s [4]. At this context, S. aureus has become resistant to various types of antibiotics over time. One of the most notable cases of S. aureus antibiotic resistance is MRSA. Early in the 1960s, a few years after methicillin was initially used as a treatment, the UK recorded its first cases of MRSA and this epidemic was primarily constrained to Europe [30]. Soon after, MRSA was identified in the US and Australia. So far, MRSA has become a concern to public health, causing a variety of infections, including those that can be serious and affect the skin and soft tissues [99].
It is crucial to note that other groups have become a concern to public health including Vancomycin-intermediate S. aureus (VISA), Vancomycin-resistant S. (VRSA), heterogeneous VISA (hVISA) are staphylococcal bacteria that are less susceptible (VISA) or fully resistant (VRSA) to the antibiotic vancomycin. However, VISA and VRSA are usually susceptible to other antibiotics, and infections caused by these organisms are treatable [112,113]. The first Vancomycin Intermediate S. aureus (VISA) strain with an 8 g/ml Minimum Inhibitory Concentration (MIC) was discovered in Japan in 1997 [114], and the first case of Vancomycin-resistant S. aureus (VRSA) in a diabetic patient in the U.S.A. was reported back in 2002 [115]. Since the prevalence of VRSA, VISA, and hVISA has been on the rice recently (particularly in the Asian and American continents), strict monitoring of vancomycin treatment, its therapeutic response, and the definition of appropriate control guidelines depending on geographical regions is highly advised and essential to stop the further spread of vancomycin-resistant S. aureus [113]. Distribution of VRSA, VISA and hVISA isolates among different countries based on meta-analysis of published original articles were illustrated in Figure 3.
Figure 3.
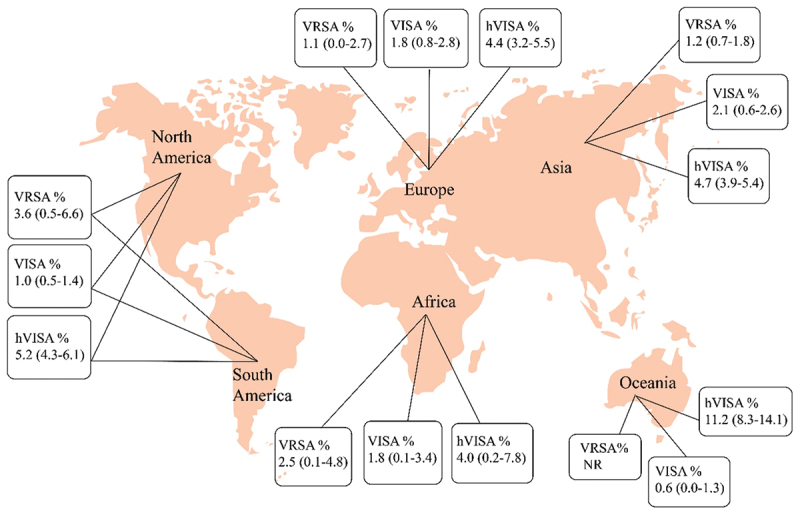
Distribution of VRSA, VISA and hVISA isolates among different countries based on meta-analysis of published original articles [113].
5.1.1. Mechanisms of antibiotic resistance in MRSA
For many years, hospital-acquired MRSA, which is acquired in hospitals, care homes and other healthcare settings, was considered the standard reference point for MRSA strains. Infections in the community, such as those in schools and other public settings, were linked to the emergence of additional MRSA strains in the 1990s, noted as community acquired MRSA (CA-MRSA). Also at the beginning of the 21st century, it has been established that Livestock-associated MRSA, which is present in animals and particularly pigs and cows, can be passed on to humans by contact or consumption of contaminated meat or dairy products [116,117].
The methicillin resistance is mediated by the chromosomally placed mecA gene, which encodes for a modified PBP known as PBP2a. MGEs called staphylococcal cassette chromosome mec (SSCmec) include the mecA gene (Figure 4) [48]. Because of its poor affinity for β-lactams, PBP2a replaces other PBP in the cross-linking of peptidoglycan chains, allowing staphylococci to survive even in high doses of these antibiotics [34,118] .
Figure 4.
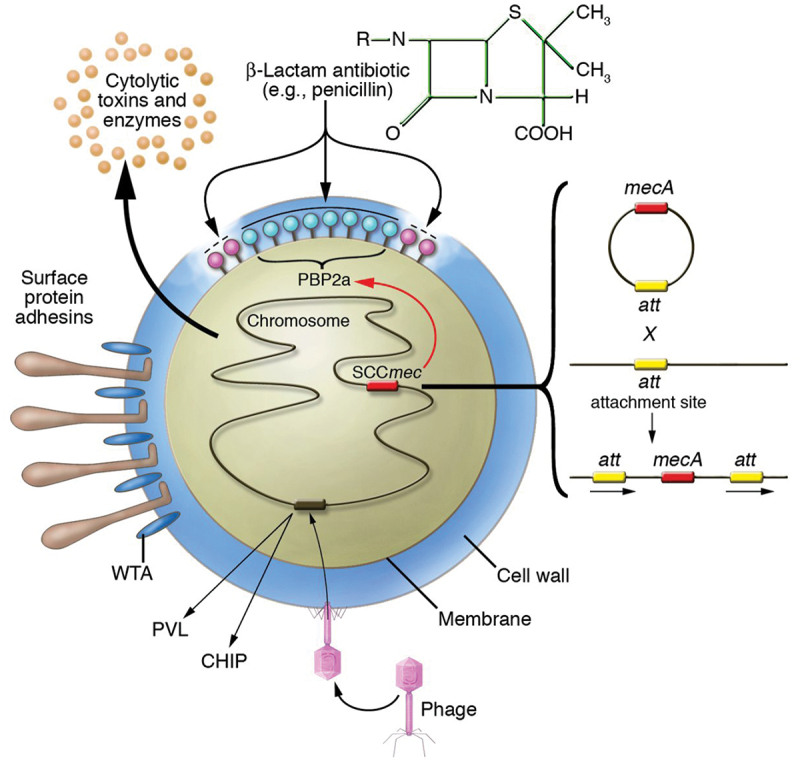
MRSA antibiotic resistance mechanism S. aureus has methicillin resistance. Nasal and skin colonization are influenced by the expression of the surface protein adhesins and the wall teichoic acid. Insertion of the SCCmec horizontally transmitted DNA element results in methicillin resistance. Site-specific recombination allows for the integration of five separate SCCmec components at the same site. The mecA gene encodes PBP2a, a new β-lactam-insensitive penicillin binding protein [119].
The MRSA pathogen, in particular, is resistant to many antibiotics, including all penicillins and cephalosporins as well as most macrolides, tetracyclines, fluoroquinolones and aminoglycosides. The strains of MRSA are often treated with glycopeptides, such as vancomycin, or newer antibiotics, such as daptomycin and linezolid [120]. As a result, standard antibiotics are ineffective against resistant bacteria since they are unable to intracellularly achieve a therapeutic level. Given that present MRSA treatment options have been either unsuccessful or partake severe side effects, there is a critical need for new tactics other than novel medication discovery and reuse of existing drugs [31].
6. The role of nanotechnology in combating multi-drug resistant bacteria
Nanotechnology refers to the identification and manipulation of matter at dimensions ranging from approximately 1 to 100 nm, wherein unique phenomena manifest, leading to the emergence of innovative applications [121,122]. Throughout the industrial revolution, there was a notable increase in the exposure of nanoparticles, which have always existed. The significance of this increase can be attributed to the pioneering work of Richard Zsigmondy, a distinguished chemist who was awarded the Nobel Prize in 1925. Zsigmondy’s contribution to the field of nanotechnology is multifaceted, as he not only conducted groundbreaking research but also coined the term ‘nanometer’ for the first time. His innovative use of a microscope to measure the size of particles, such as gold colloids, marked a significant advancement in the field. Furthermore, Zsigmondy’s introduction of the term ‘nanometer’ to describe particle size has had a lasting impact on the scientific community, cementing his legacy as a pioneer in the field of nanotechnology.
Nanoparticles possess unique physical and chemical properties due to their high surface area and nanoscale size. Their optical properties are dependent on size, resulting in different colors due to the absorption in the visible region [124]. The physical properties of nanoparticles, including size, shape, charge, and elasticity, as they play a crucial role in their pharmaceutical functions [125]. Nanoparticles can have surprising optical, physical, and chemical characteristics due to their small size and ability to create quantum effects [126]. The surface energy of nanoparticles is significantly greater than that of microparticles, leading to a non-equilibrium state and the possibility of self-organization [127]. Nanoparticles can be classified based on their structure analysis and property management, and they possess versatile properties that make them suitable for various applications in fields such as catalysis, imaging, and medicine [128]. In the dominion of pharmaceutical industries, the incorporation of nanotechnology has exerted a profound impact on the advancement of medical devices, including imaging probes, drug delivery systems, and diagnostic biosensors. In the field of biomedicine, nanoparticles find utility in a range of biomedical applications including medical diagnostics, targeted drug delivery, tissue engineering, regenerative medicine, and biomedical textiles [129]. Nanoparticles that have been functionalized with biological molecules possess distinct characteristics and are efficacious in medical diagnostics [130]. Their therapeutic potential extends to the development of nanodrug delivery systems and the creation of novel drugs [131]. Moreover, nanoparticles facilitate the coupling of marker-specific antibodies or pertinent antigens, thereby improving early detection methods based on molecular markers [132]. They have also demonstrated antibacterial properties against various pathogens rendering them a viable alternative for antibacterial treatment [133]. Furthermore, progress in nanotechnology holds promise for enhancing disease diagnosis, refining targeted drug delivery, improving imaging of therapeutic responses, regulating cellular and tissue responses, and guiding resection procedures. Presently, nanotechnology exerts a pervasive influence on the everyday lives of individuals. Today, the impact of nanotechnology on human existence is a constant occurrence. An array of possible advantages exists that covers a broad spectrum. However, as a result of the significant exposure of humans to nanoparticles, a considerable amount of apprehension arises regarding the potential risks to both health and the environment [53]. These apprehensions have led to the emergence of new scientific disciplines, namely nanomedicine and nanotoxicology, which delve into the investigation of the detrimental effects that nanoparticles may have on human well-being [79]. While nanoparticles have several disadvantages in the field of medicine, one critical drawback is the difficulty in maintaining therapeutic action in three dimensions inside the human body [134]. Another disadvantage is the limitation to maintain efficacy in the target organ once the magnetic field is removed from the outside [135]. Nanoparticles also bring unique environmental and societal challenges, particularly concerning toxicity [136]. There is a possibility of side effects caused by the action of metal nanoparticles absorbed by organisms [137]. When employing nanoparticles in medicine, several obstacles must be overcome, such as issues with biodegradability and porosity [138].
6.1. Nanoparticles and drug delivery system
The fusion of nanotechnology and medicine has given rise to the relatively new field of nanomedicine, where its basis is the modification of matter at the nanoscale for purposes in the fields of human health. By altering essential drug properties like solubility, diffusivity, bloodstream half-life and drug release and distribution profiles, the usage of materials in this range has greatly advanced pharmacology [8]. In 1986, Matsumura and Maeda discovered that an anticancer protein coupled to polymeric nanoparticles demonstrated larger accumulation in tumor tissues than in healthy ones. As a result, various researchers began to perceive connections between the nanotechnology and medicine fields [51]. Due to the physiology of tumors and the size of nanoparticles (NPs) (200 nm), small size NPs have better tumor penetration. This discovery gave rise to the hypothesis of enhanced permeability and retention (EPR) [139].
In the realm of nanotechnology, it is possible to classify nanoparticles into three distinct categories: organic, inorganic, and carbon-based materials [140]. There exists a plethora of examples that fall under the umbrella of nanoparticles, including but not limited to nano-silver, nano-silica, nano-aluminum, nano-zinc oxide, nano-copper, carbon nanotubes, and nano-titanium dioxide [141]. Metallic nanoparticles, in particular, find widespread application across a variety of fields, with the pharmaceutical industry being no exception. These metallic nanoparticles can be synthesized through various means, namely physical, chemical, and biological methods [142]. It is worth noting that the pharmaceutical industry specifically utilizes a range of metallic nanoparticles, such as gold, silver, iron oxide, zinc oxide, platinum, copper oxide, and palladium nanoparticles [143]. Moreover, it is essential to recognize that nanoparticles can be further categorized based on their size, shape, composition, and functionalities, leading to different platforms that include liposomes, Albumin-bound, Polymeric, Quantum Dots, and Iron Oxide [144]. Researchers have successfully synthesized multi metallic nanoparticles, each possessing distinct configurations like core-shell, three-shell, random alloy, and dumbbell-like, in order to harness their unique and specialized catalytic properties. As shown in Table (2), the combination of nanomaterials with many types of antibiotics were used to combat bacterial resistance [145].
Table 2.
Antibacterial activity of nanomaterials combined with antibiotics (147).
| Nanomaterial | Antibiotics | Target bacteria | References |
|---|---|---|---|
| Ag NPs | Ciprofloxacin Vancomycin Clotrimazole |
VRE MRSA MRSA, S. aureus |
[145] [146] [147] |
| Au NPs | Vancomycin Ampicillin |
MRSA MRSA, P. aeruginosa, Enterobacter aeruginosa, E. coli |
[148] [149] |
| ZnO NPs | Ciprofloxacin, ceftazidime | MDR A. baumannii | [150] |
| Fe3O4 NPs | Ampicillin Ampicillin |
S. aureus E. coli, P. aeruginosa, MRSA |
[151] [152] |
| CuO NPs | Cephalexin | E. coli | [153] |
| SWCNTs | Ciprofloxacin | S. aureus, P. aeruginosa, E.coli | [154] |
| GO | Lincomycin hydrochloride Chloramphenicol Gentamycin sulfate |
E. coli, S. aureus | [155] |
| AMP-NPs | Gentamicin, vancomycin, azithromycin, amoxicillin | S, aureus, E. coli, P. aeruginosa, A. baumanni | [156] |
| Chitosan | Streptomycin Ciprofloxacin |
Listeria monocytogenes Uropathogenic E. coli |
[157] [203] |
6.2. Nanoparticles and bacterial infections
The use of antibacterial therapy based on nanomaterials is extremely important in tackling the various difficulties presented by bacterial infections and antibiotic resistance. The inherent antibacterial properties offered by nanomaterials make them an invaluable resource in the battle against bacterial infections. Furthermore, their ability to act as drug carriers presents a novel and alternative strategy in the fight against bacterial infections. By utilizing nanomaterials as drug carriers, scientists and researchers can explore new avenues and approaches to combat bacterial infections, thereby addressing the growing concern of antibiotic resistance [146,147]. Hybrid bacteria systems, which have been subject to modification with highly versatile nanomaterials, have emerged as a highly promising avenue for enhancing the efficacy of tumor therapeutic interventions [148]. Photodynamic antibacterial therapy (PDAT) using nanomaterials as carriers for photosensitizers has emerged as a non-antibiotic treatment method for bacterial infections [149]. In the current era, characterized by the advancements of the 21st century, the prevalence of infections continues to persist as a significant predicament, necessitating the urgent requirement for novel and alternative approaches to combat bacterial growth and proliferation [150]. The investigation and study of therapeutic treatments that utilize nanomaterials for the purpose of combating bacteria is of utmost significance and relevance, especially when considering the prevailing issue of drug-resistant bacteria, which is an immense challenge and threat to public health [151].
NPs have countless advantages, including more precise targeting, a larger surface-to-volume ratio, more regulated and prolonged drug release [123]. For the convenience of patients, the abovementioned characteristics can reduce the quantity of antibiotic and administration times [152]. One strategy was developed to employ NPs as biosensors that can recognize particular bacterial biomolecules, such as elements of bacterial cell walls or bacterial DNA or RNA [153]. This can be shown, for instance, in the usage of gold NPs that have been functionalized with certain antibodies that can bind to bacterial antigens. The antibodies will bind to the antigens when these nanoparticles come into contact with the sample containing the target bacterium, changing the NPs characteristics like color or optical qualities [154]. Then, this alteration can be discovered using several analytical methods like spectrometry, microscopy, or fluorescence. Also, Mesoporous silica nanoparticles (MSNs) have a significant potential to give sensitive, selective and quick diagnosis of bacterial infections [155]. NPs are made to target infected host cells and get intracellular access for bioactivity in order to pass the cellular barrier [145]. For instance, due to the phagocytic nature of macrophages, nanoparticles locally delivered to the infection sites may be spontaneously taken up by macrophages infected with Mycobacterium TB, Salmonella typhimurium, or MRSA. Moreover, Chlamydia trachomatis-infected mouse fibroblasts and human lung epithelial cells showed enhanced antibiotic efficacy when treated with nanoparticles [156]. Another form of NP, the Au-NPs which have been functionalized with cationic ligands, have antibiofilm action by dissolving the biofilm matrix of S. aureus and P. aeruginosa [157]. Additionally, a complex made of solid-lipid NPs that included the second-generation cephalosporin cefuroxime axetil (CA) was investigated for its antibacterial and antibiofilm properties against S. aureus. Encapsulation made prolonged release possible. This combination has a lower minimum inhibitory concentration (MIC) against S. aureus and generates an inhibitory zone to biofilm formation that is bigger than CA alone (13 mm vs. 9 mm) [61]. Nodes of polymeric PNPs also have been used to deliver antibiotics, particularly hydrophobic medicines. The antibiotic Levofloxacin exhibits antibiofilm activity when encapsulated in a poly lactic-co-glycolic acid (PLGA) nanocapsule, as it eliminates the biofilm and stops the remaining bacteria from proliferating [158].
Noble metal nanoparticles, including gold, silver, and platinum, have demonstrated considerable potential in the field of antimicrobial applications, owing to their inherent bactericidal properties and the ease with which they can be synthesized [159]. Bismuth halide nanomaterials (BiONs) have garnered considerable interest due to their exceptional optoelectronic capabilities and biosafety characteristics, rendering them well-suited for applications in photocatalytic antibacterial processes and photodynamic therapy. Antimicrobial photodynamic therapy (aPDT) exploiting the potential of nanomaterials has demonstrated remarkable efficacy in the eradication of both bacteria and fungi, with the added advantage of circumventing the issue of resistance, which has plagued conventional antimicrobial treatments. However, it is imperative to acknowledge that despite these encouraging findings, additional advancements and refinements are imperative to ensure the successful translation of this technology into clinical application, thus warranting further investigation and exploration [160]. Graphene-based nanomaterials, which are composed of a single layer of carbon atoms arranged in a hexagonal lattice, have exhibited remarkable antibacterial properties that arise from their unique structural and chemical characteristics. These nanomaterials have been found to possess the ability to physically damage bacteria, thereby offering a promising avenue for combating antibiotic-resistant strains. The physical damage-based mechanisms employed by graphene-based nanomaterials involve the piercing and rupturing of bacterial cell membranes, leading to the leakage of cellular contents and ultimately cell death. This novel approach to tackling antibiotic resistance holds great potential for addressing the urgent and increasingly prevalent issue of bacterial infections that are unresponsive to conventional antibiotics. Photothermal therapy (PTT), a therapeutic approach that utilizes nanomaterials as photothermal agents, has demonstrated great potential in the treatment of drug-resistant bacteria and biofilms. This cutting-edge therapy holds promise for targeted delivery and controlled release, which could effectively minimize the occurrence of side effects associated with traditional treatment methods. The utilization of nanomaterials in PTT enables the precise targeting of bacteria and biofilms, allowing for enhanced efficacy and reduced damage to healthy tissues. Furthermore, the controlled release of therapeutic agents through nanomaterials offers the possibility of extended treatment duration and sustained therapeutic effects. These advancements in PTT hold immense potential in revolutionizing the field of antibacterial therapy and could significantly contribute to the development of more effective treatments for drug-resistant bacteria and biofilms [161].
6.2.1. Graphene-based nanomaterials
Graphene-based nanomaterials, which are composed of a single layer of carbon atoms arranged in a hexagonal lattice, have exhibited remarkable antibacterial properties in various studies conducted, thereby highlighting their potential as promising functional agents for the development of antibacterial therapy [162–164]. These nanomaterials demonstrate the ability to inhibit the growth of bacteria through a multitude of mechanisms, which encompass the stimulation of oxidative stress within the bacterial cells, the disruption of their delicate cell membranes, as well as the induction of physical damage to their structural integrity [165,166]. Graphene-based nanomaterials have demonstrated their efficacy in fighting against both Gram-positive and Gram-negative bacteria, which is a significant breakthrough in the realm of antibacterial treatment [167]. The antibacterial activity of these nanomaterials is heavily influenced by their size, morphology, and composition, thus highlighting the criticality of these factors in determining their effectiveness [168]. Furthermore, the potential applications of graphene-based nanomaterials extend beyond their antibacterial properties, as they have also exhibited promising results in wound healing, biofilm removal, and addressing the challenge posed by antimicrobial resistance [169]. Nonetheless, it is imperative to conduct further research in order to fully comprehend the intricate molecular mechanisms that underlie the antibacterial activity of graphene-based nanomaterials [166]. By gaining a deeper understanding of these mechanisms, we can unlock the true potential of these nanomaterials and harness them as valuable tools in the field of antibacterial therapy, thereby addressing the persistent threat posed by bacterial infections [170]. In summary, the advent of graphene-based nanomaterials has paved the way for a new era in antibacterial treatment, offering immense potential for combating bacterial infections and improving patient outcomes.
6.2.2. MXene, a novel 2D nanomaterial
MXene, which is a groundbreaking two-dimensional nanomaterial, has demonstrated immense promise as an exceedingly effective antibacterial agent, thereby showcasing its remarkable capabilities in combating bacterial infections and diseases [171]. MXene, a two-dimensional material that consists of transition metal carbides, nitrides, or carbonitrides, has been found to possess extraordinary antimicrobial properties, thereby making it a highly promising candidate for the development of metal-based bactericides [172]. The size, concentration, and surface charge of MXene nanosheets are three key parameters that play a significant role in determining the antimicrobial efficacy of these nanomaterials [170]. These factors have a direct impact on the interaction between MXene nanosheets and microorganisms, ultimately influencing the effectiveness of their antimicrobial properties [173]. By manipulating the size of the nanosheets, researchers can control their ability to penetrate microbial cells and disrupt vital cellular processes [174]. Furthermore, the concentration of MXene nanosheets in a given solution can dictate the extent of antimicrobial activity, as higher concentrations may lead to more efficient eradication of pathogens. Additionally, the surface charge of the nanosheets can influence their interaction with microbial membranes, with positively charged nanosheets potentially exhibiting enhanced antimicrobial efficacy. Overall, understanding and optimizing the size, concentration, and surface charge of MXene nanosheets is crucial for harnessing their full potential as antimicrobial agents [175]. MXene has been discovered to possess an array of therapeutic properties, including but not limited to its ability to inhibit viral replication, mitigate inflammatory responses, and impede the growth and proliferation of cancerous cells, thereby highlighting its immense potential as a multifaceted remedy for a diverse range of ailments [176]. The potential cytotoxicity of MXene, a class of two-dimensional transition metal carbides and nitrides, has raised concerns within the scientific community [177]. In order to address these concerns and better understand the biocompatibility of MXene, numerous studies have been carried out to evaluate its interactions with living organisms [178]. The biocompatibility and cytotoxicity of MXene are contingent upon various factors, including its size, dose, and surface coating [179]. To comprehensively examine the biological response and cytotoxicity of MXene, both in vitro and in vivo toxicological studies have been conducted. However, it is imperative to conduct further research in order to gain a complete understanding of the potential of MXene as an antibacterial agent [180]. Additionally, addressing the concerns pertaining to its cytotoxicity is of utmost importance [181].
6.2.3. Chitosan and metal/metal oxides
Functional materials, including but not limited to chitosan and various metal oxides, have been extensively explored due to their immense potential in the realm of antibacterial therapy [182]. The utilization of these materials has garnered significant attention from researchers worldwide, who have persistently strived to devise innovative strategies to effectively combat bacterial infections [183]. In their tireless pursuit, scientists have made remarkable progress in the development of cutting-edge antibacterial agents, with a particular focus on harnessing the power of metal oxide-based polymers [184]. The integration of metal oxides within polymer frameworks has proven to be a promising approach, enabling enhanced antibacterial efficacy and exhibiting the ability to overcome the challenges posed by bacterial resistance [185]. Through ingenious design and meticulous engineering, these novel antibacterial agents hold immense promise in revolutionizing the field of antibacterial therapy, potentially leading to breakthroughs in the fight against bacterial infections and the associated health complications [186]. Chitosan, a polymer composite material that is derived from chitin, a natural polysaccharide found in the exoskeleton of crustaceans, such as shrimp and crab, has been ingeniously combined with metal organic frameworks (MOFs), a class of highly porous materials composed of metal ions or clusters coordinated to organic ligands, in order to not only amplify but also augment its inherent antibacterial ability, thereby revolutionizing the field of antimicrobial materials and opening up new possibilities for various applications in medicine, biotechnology, and environmental science [187]. Chitosan films incorporating nano-metal oxides, including but not limited to zinc oxide, magnesium oxide, titanium dioxide, copper oxide, iron oxide, silicon dioxide, and silica, have been extensively employed in the realm of food packaging with the primary objective of enhancing the antimicrobial characteristics, thereby effectively inhibiting the growth of various microorganisms and ensuring the safety and quality of the packaged food products [188]. Chitosan-based materials incorporated with metallic nanoparticles and metal oxides have also been fabricated and demonstrated to possess antimicrobial properties [189]. These discoveries underscore the promise of functional substances, more specifically chitosan and metal/metal oxides, in the context of antibacterial therapy and their utilization in diverse domains, such as medicine and the food industries.
6.2.4. Mesoporous silica nanoparticles (MSNs)
Mesoporous silica nanoparticles (MSNs) are one of the most promising nanocarriers for drug delivery due to their distinct qualities, which include a high loading capacity, biocompatibility, simplicity in manufacturing, and customizable pore diameters and volumes [190]. Additionally, by utilizing several methods, MSNs can be manufactured on a massive scale with a variety of sizes, morphologies and surface functionalities [191]. Due to these crucial characteristics, MSNs provide good Nano platforms for assembling various multifunctionalities for the treatment of bacterial infections, emphasizing targeted and stimuli-responsive antimicrobial administration (Figure 5) [65]
Figure 5.
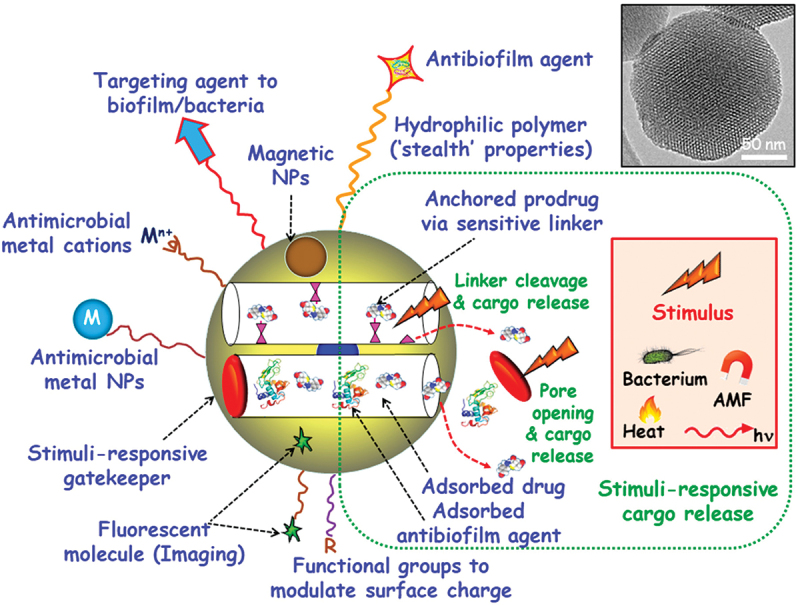
Multifunctionalities of MSNs. MSNs have multiple uses in the treatment of bacterial infections. On the top surface, agents that target bacteria and/or biofilm can be grafted. Adsorbed or grafted antimicrobial medications and/or antibiofilm substances are both possible for MSNs. To stop cargo leakage, stimuli-responsive gatekeepers can be included into blocking nanocaps. Pore uncapping and cargo release are triggered by exposure to internal (bacteria, pH, redox potential) or external (heat, light, alternating magnetic fields (AMF)) stimuli. MSNs can have antimicrobial metal NPs (M) and ions (Mn+) implanted within the mesoporous structure or coated externally. To add “stealth” qualities, biocompatible hydrophilic polymers can be grafted to the surface. To alter surface charge, different organic groups (R) can be externally functionalized. It is also possible to integrate fluorescent compounds and magnetic NPs [192].
The safety dose or toxicity concerns associated with silica nanoparticles have garnered extensive examination and remain a topic of ongoing research and discussion [193,194]. Hollow silica nanoparticles have exhibited promising outcomes in animal studies concerning the controlled release of chemical compounds, demonstrating a low toxicity profile even at higher doses or repeated administrations. However, other research posits that the biocompatibility of silica nanoparticles is contingent upon the dosage, with toxicity only emerging at elevated doses [195]. It is imperative to persist in investigating the safety and biocompatibility of silica nanoparticles, particularly within the realm of biomedical applications, to ensure their responsible and efficacious utilization. This is of particular significance as certain studies have indicated the absence of observable organ damage or inflammation following in vivo toxicity assessments [196]. Numerous studies have showcased that silica nanoparticles possess negligible levels of toxicity, with some even demonstrating no disparity in toxicity when compared to larger particles. However, it should be noted that they can become toxic at high doses [197]. The safety and effectiveness of hollow mesoporous silica nanoparticles, renowned for their aptitude as delivery systems for the controlled release of chemicals within live cells, have been substantiated through both in vitro and in vivo investigations, thus suggesting their potential for biomedical applications [198]. In an in vitro study, Natsheh et al. and their colleagues have illustrated that a high viability rate above 70% was achieved against normal fibroblast cells. Furthermore, the biocompatibility of these nanoparticles in living organisms has also been documented, with evidence indicating the absence of bioaccumulation or biopersistence and the presence of rapid physiological excretion mechanisms [147,199].
6.3. Nanoparticles and staphylococcus aureus
The traits of S. aureus as an multi-drug resistance (MDR) bacterium, such as biofilm formation, facultative intracellular survival and increasing resistance, present a significant problem [66]. Finding effective alternate methods for the eradication of drug-resistant bacterial agents seems to be a pressing necessity given the costs, side effects, and time required for the production of innovative medications. The use of nanoparticles is seen to be a possible solution to the S. aureus resistance-related concerns in the treatment of infections [152]. NPs, especially those created utilizing green synthesis technique which aim to reduce or eliminate the use of hazardous chemicals and energy-intensive processes typically associated with traditional nanoparticle synthesis. These techniques often utilize sustainable starting materials and environmentally benign reaction conditions like using plant extracts, microorganisms and Microwave-assisted synthesis, have demonstrated antibacterial effects [200]. As well, MRSA can be diagnosed with NPs. The primary mechanisms of NP antimicrobial actions include bacterial integrity or metabolism impairment (CuNPs), interruption of transcription and replication, inhibition of membrane-bound enzymes and biofilm, protein denaturation (AgNPs) and formation of reactive oxygen species (ROS) [201]. Many NPs, including gold, silver and the less expensive NPs like nickel, titanium, zinc oxide, silica and bismuth, have been found to effectively eradicate MRSA bacteria in both in vitro and in vivo tests [191].
One study showed that cefotax loaded on barium ferrite NPs had a potential efficacy in controlling MRSA by boosting the drug’s bactericidal power and increasing its penetration through bacterial cells [201]. An MSN-based nanosystem with the possibility of targeting bone through utilization of eight repeating aspartate sequences was proposed in a subsequent study. Eight repeating aspartate sequences have been used as a targeting element to bone to functionalize MSNs because of their affinity for hydroxyapatite. This nanocarrier specifically delivered the peptide into infected bone tissue in vivo, replicating the effects shown in vitro, and lowered both S. aureus bacterial viability and osteoclasticity in vitro [202]. By studying the synergistic bactericidal activity of loaded chlorhexidine and silver-decorated MSNPs against S. aureus and E. coli, Lu et al. took things a step further in 2017 by an easy and sustainable process; the monodisperse MSN nanospheres were successfully created as an appropriate carrier for the delivery of Chlorhexidine (CHX) and nanosilver. The CHX-loaded, silver-decorated MSNs demonstrated simultaneous CHX and silver ion release in response to pH, which had a synergistic antibacterial impact against both gram-positive S. aureus and gram-negative Escherichia coli [139]. To improve the pharmacologic activity against sensitive and resistant S. aureus in the normal phenotypic states and action times, as well as to decrease the drug’s side effects, several antimicrobial drugs are now integrated into or conjugated with NPs. Consequently, NPs medication delivery systems proved to be the best tool for overcoming the problems we had with S. aureus infection [78].
7. Conclusion
The emergence and growth of S. aureus and its MDR problem have shown to be a serious health concern that must be managed on a global scale. To treat persistent MRSA, nanomaterials offer a novel, ‘outside the box’ strategy. Optimization of their physical characteristics, particularly size and surface charge, is essential to maximize their curative potential and decrease host hazard. Clinical use is currently limited by issues concerning the long-term effects of nanoparticles on the patient and its systemic safety. Additionally, the exact and detailed mechanism of interaction of NPs with biological systems should be explained in future trials to develop nanomaterials with favorable physicochemical properties that will allow them to be more responsive to varied biological settings for therapeutic advantages while having no negative effect. The field of nanoparticles promise, in the near future, to be the next-generation of medicines to combat MDR bacteria.
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Authors’ contributions
RB and ROA wrote the first version, IN and RB edited the manuscript, and OA wrote, edit, and supervised the overall project.
Data availability statement
Data files are available upon request.
References
- [1].Cheung GYC, Bae JS, Otto M.. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 2021;12(1):547–569. doi: 10.1080/21505594.2021.1878688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Javid F, Taku A, Bhat MA, et al. Molecular typing of Staphylococcus aureus based on coagulase gene. Vet World. 2018. Apr;11(4):423–430. doi: 10.14202/vetworld.2018.423-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Boswihi SS, Udo EE. Methicillin-resistant Staphylococcus aureus: an update on the epidemiology, treatment options and infection control. Curr Med Res Pract. 2018;8(1):18–24. InternetAvailable from: https://www.sciencedirect.com/science/article/pii/S2352081717301708. doi: 10.1016/j.cmrp.2018.01.001 [DOI] [Google Scholar]
- [4].Linz MS, Mattappallil A, Finkel D, et al. Clinical impact of Staphylococcus aureus skin and soft tissue infections. Antibiotics. 2023;12(3):1–27. doi: 10.3390/antibiotics12030557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kayan T. Staphylococcus aureus Secreted Toxins & Extracellular Enzymes. Physiol Behav. 2017;176(3):139–148. doi: 10.1016/j.physbeh.2017.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Obee P, Griffith CJ, Cooper RA, et al. An evaluation of different methods for the recovery of meticillin-resistant Staphylococcus aureus from environmental surfaces. J Hosp Infect. 2007;65(1):35–41. doi: 10.1016/j.jhin.2006.09.010 [DOI] [PubMed] [Google Scholar]
- [7].Adhikari RP. Staphylococcal infections: host and pathogenic factors. Vol. 9. Microorganisms. Switzerland: MDPI AG; 2021. doi: 10.3390/microorganisms9051080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ivanova N, Gugleva V, Dobreva M, et al. We are IntechOpen, the world ’ s leading publisher of open access books built by scientists, for scientists TOP 1 %. Intech. 2016;i(tourism):13. [Google Scholar]
- [9].Missiakas DM, Schneewind O. Growth and laboratory maintenance of Staphylococcus aureus. CP Microbiol. 2013. Feb;28(1):Unit 9C.1. doi: 10.1002/9780471729259.mc09c01s28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reddy PN, Srirama K, Dirisala VR. An update on clinical Burden, diagnostic tools, and therapeutic options of Staphylococcus aureus. Infect Dis Res Treat. 2017;10:117991611770399. doi: 10.1177/1179916117703999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Otto M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol. 2010. Apr;5(2):183–195. doi: 10.1586/edm.10.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Luo Z, Yue S, Chen T, et al. Reduced growth of Staphylococcus aureus under high glucose conditions is associated with decreased pentaglycine expression. Front Microbiol. 2020;11:537290. doi: 10.3389/fmicb.2020.537290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Guo Y, Song G, Sun M, et al. Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front Cell Infect Microbiol. 2020;10:107. doi: 10.3389/fcimb.2020.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Shankar P. Book review: tackling drug-resistant infections globally. Arch Pharma Pract. 2016;7(3):110. doi: 10.4103/2045-080X.186181 [DOI] [Google Scholar]
- [15].Chinemerem Nwobodo D, Ugwu MC, Oliseloke Anie C, et al. Antibiotic resistance: the challenges and some emerging strategies for tackling a global menace. J Clin Lab Anal. 2022. Sep;36(9):e24655. doi: 10.1002/jcla.24655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ahmad-Mansour N, Loubet P, Pouget C, et al. Staphylococcus aureus toxins: an update on their pathogenic properties and potential treatments. Toxins (Basel). 2021;13(10):1–22. doi: 10.3390/toxins13100677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Martínez OF, Cardoso MH, Ribeiro SM, et al. Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition. Front Cell Infect Microbiol. 2019;9(APR). doi: 10.3389/fcimb.2019.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim M-K. Staphylococcus aureus toxins: from their pathogenic roles to anti-virulence therapy using natural products. Biotechnol Bioprocess Eng. 2019;24(3):424–435. InternetAvailable from. doi: 10.1007/s12257-019-0059-9 [DOI] [Google Scholar]
- [19].Bennett MR, Thomsen IP. Epidemiological and clinical evidence for the role of toxins in S. aureus human disease. Toxins (Basel). 2020;12(6):1–20. doi: 10.3390/toxins12060408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Vlaeminck J, Raafat D, Surmann K, et al. Exploring Virulence Factors and Alternative Therapies against Staphylococcus aureus Pneumonia. Toxins (Basel). 2020;12(11):721. doi: 10.3390/toxins12110721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ford CA, Hurford IM, Cassat JE. Antivirulence strategies for the treatment of Staphylococcus aureus infections: a mini review. Front Microbiol. 2021;11(January):1–10. doi: 10.3389/fmicb.2020.632706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ahmad AL-Fawares RWA O, Abdul F, Salah F, et al. Probiotic therapy: a survey of Middle eastern healthcare providers’ attitudes, Beliefs, and practice patterns. J Appl Pharm Sci. 2023;13(10):1–9. doi: 10.7324/JAPS.2023.143603 [DOI] [Google Scholar]
- [23].Gnanamani A, Hariharan P, Paul-Satyaseela M. Staphylococcus aureus: Overview of Bacteriology, Clinical Diseases, Epidemiology, Antibiotic Resistance and Therapeutic Approach. Front Staphylococcus Aureus. 2017;4. doi: 10.5772/67338 [DOI] [Google Scholar]
- [24].Tong SYC, Davis JS, Eichenberger E, et al. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28(3):603–661. doi: 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang H, Hou X, Shen J, et al. Alternative sigma factor B reduces biofilm formation and stress response in milk-derived Staphylococcus aureus. Lwt. 2022;162(October 2021):113515. InternetAvailable from. doi: 10.1016/j.lwt.2022.113515 [DOI] [Google Scholar]
- [26].Sharma S, Mohler J, Mahajan SD, et al. Microbial biofilm: a review on formation, infection, antibiotic resistance, control measures, and innovative treatment. Microorganisms. 2023;11(6):1614. doi: 10.3390/microorganisms11061614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Shoaib M, Aqib AI, Muzammil I, et al. MRSA compendium of epidemiology, transmission, pathophysiology, treatment, and prevention within one health framework. Front Microbiol. 2023;13(January): doi: 10.3389/fmicb.2022.1067284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Amir N, Rossney A, Veale J, et al. Spread of community-acquired meticillin-resistant Staphylococcus aureus skin and soft-tissue infection within a family: implications for antibiotic therapy and prevention. J Med Microbiol. 2010 Apr 1;59(4):489–492. doi: 10.1099/jmm.0.015925-0 [DOI] [PubMed] [Google Scholar]
- [29].O’Riordan K, Lee JC. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev. 2004;17(1):218–234. doi: 10.1128/CMR.17.1.218-234.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Prosperi M, Azarian T, Johnson JA, et al. Unexpected predictors of antibiotic resistance in housekeeping genes of Staphylococcus aureus. ACM-BCB 2019 - Proc 10th ACM Int Conf Bioinformatics, Comput Biol Heal Informatics. 2019;259–268. doi: 10.1145/3307339.3342138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Turner NA, Sharma-Kuinkel BK, Maskarinec SA, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019;17(4):203–218. doi: 10.1038/s41579-018-0147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].van Hal SJ, Jensen SO, Vaska VL, et al. Predictors of mortality in staphylococcus aureus bacteremia. Clin Microbiol Rev. 2012;25(2):362–386. doi: 10.1128/CMR.05022-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hindy J-R, Quintero-Martinez JA, Lee AT, et al. Incidence trends and epidemiology of Staphylococcus aureus bacteremia: a systematic review of population-based studies. Cureus. 2022;14(5): doi: 10.7759/cureus.25460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tsige Y, Tadesse S, G/Eyesus T, et al. Prevalence of methicillin-resistant Staphylococcus aureus and associated risk factors among patients with wound infection at referral hospital, Northeast Ethiopia. J Pathog. 2020;2020:1–7. doi: 10.1155/2020/3168325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Loftus MJ, Young-Sharma TEMW, Wati S, et al. Epidemiology, antimicrobial resistance and outcomes of Staphylococcus aureus bacteraemia in a tertiary hospital in Fiji: a prospective cohort study. Lancet Reg Health West Pac InternetAvailable from. 2022;22:100438. doi: 10.1016/j.lanwpc.2022.100438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tabaja H, Hindy JR, Kanj SS. EPIDEMIOLOGY OF METHICILLIN RESISTANT STAPHYLOCOCCUS AUREUS IN ARAB COUNTRIES OF THE MIDDLE EAST AND NORTH AFRICAN REGION. Mediterr J Hematol Infect Dis. 2021;13(1):e2021050. doi: 10.4084/MJHID.2021.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jarajreh D, Aqel A, Alzoubi H, et al. Prevalence of inducible clindamycin resistance in methicillin-resistant Staphylococcus aureus: the first study in Jordan. J Infect Dev Ctries. 2017;11(4):350–354. doi: 10.3855/jidc.8316 [DOI] [PubMed] [Google Scholar]
- [38].Moyo SJ, Aboud S, Blomberg B, et al. High nasal carriage of methicillin-resistant Staphylococcus aureus among healthy Tanzanian under-5 children. Microb Drug Resist. 2014;20(1):82–88. doi: 10.1089/mdr.2013.0016 [DOI] [PubMed] [Google Scholar]
- [39].Harbarth S, François P, Schrenzel J, et al. Community-associated methicillin-resistant Staphylococcus aureus , Switzerland. Emerg Infect Dis. 2005;11(6):962–965. doi: 10.3201/eid1106.041308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Belbase A, Pant ND, Nepal K, et al. Antibiotic resistance and biofilm production among the strains of Staphylococcus aureus isolated from pus/wound swab samples in a tertiary care hospital in Nepal. Ann Clin Microbiol Antimicrob. 2017;16(1):1–5. doi: 10.1186/s12941-017-0194-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Islam SI, Moore C. Prevalence of methicillin-resistant Staphyloccocus aureus and associated risk factors on admission to a specialist care eye hospital. Ann Saudi Med. 2002;22(3–4):153–157. doi: 10.5144/0256-4947.2002.153 [DOI] [PubMed] [Google Scholar]
- [42].Aqel AA, Ibrahim A, Shehabi A. Rare occurrence of mupirocin resistance among clinical Staphylococcus isolates in Jordan. Acta Microbiol Immunol Hung. 2012;59(2):239–247. doi: 10.1556/amicr.59.2012.2.8 [DOI] [PubMed] [Google Scholar]
- [43].Hasanpour AH, Sepidarkish M, Mollalo A, et al. The global prevalence of methicillin-resistant Staphylococcus aureus colonization in residents of elderly care centers: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2023;12(1):4. InternetAvailable from. doi: 10.1186/s13756-023-01210-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Di Gregorio S, Vielma J, Haim MS, et al. Genomic epidemiology of Staphylococcus aureus isolated from bloodstream infections in South America during 2019 supports regional surveillance. Microb Genomics. 2023;9(5): doi: 10.1099/mgen.0.001020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Abdolmaleki Z, Mashak Z, Safarpoor Dehkordi F. Phenotypic and genotypic characterization of antibiotic resistance in the methicillin-resistant Staphylococcus aureus strains isolated from hospital cockroaches. Antimicrob Resist Infect Control. 2019;8(1):1–14. doi: 10.1186/s13756-019-0505-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Vinacke HM, Vinacke HM . International journal. Pacific Affairs. 1946;19(2):222. doi: 10.2307/2752507 [DOI] [Google Scholar]
- [47].Pal M, Berhanu G, Megersa L, et al. Epidemiology, pathogenicity, animal infections, antibiotic resistance, public health significance, and economic impact of Staphylococcus aureus: a comprehensive review. Am J Public Heal Res. 2020;8(1):14–21. [Google Scholar]
- [48].Hennekinne JA, De Buyser ML, Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol Rev. 2012;36(4):815–836. doi: 10.1111/j.1574-6976.2011.00311.x [DOI] [PubMed] [Google Scholar]
- [49].Akineden O, Hassan AA, Schneider E, et al. Enterotoxigenic properties of Staphylococcus aureus isolated from goats’ milk cheese. International Journal Of Food Microbiology. 2008;124(2):211–216. doi: 10.1016/j.ijfoodmicro.2008.03.027 [DOI] [PubMed] [Google Scholar]
- [50].Leung AKC, Barankin B, Leong KF. Staphylococcal-scalded skin syndrome: evaluation, diagnosis, and management. World J Pediatr. 2018;14(2):116–120. doi: 10.1007/s12519-018-0150-x [DOI] [PubMed] [Google Scholar]
- [51].Brazel M, Desai A, Are A, et al. Staphylococcal scalded skin syndrome and bullous impetigo. Medicina. 2021;57(11):1157. doi: 10.3390/medicina57111157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Handler MZ, Schwartz RA. Staphylococcal scalded skin syndrome: diagnosis and management in children and adults. J Eur Acad Dermatol Venereol. 2014;28(11):1418–1423. doi: 10.1111/jdv.12541 [DOI] [PubMed] [Google Scholar]
- [53].Tsujimoto M, Makiguchi T, Nakamura H, et al. Staphylococcal scalded skin syndrome caused by burn wound infection in an infant: a case report. Burns Open. 2018;2(3):139–143. doi: 10.1016/j.burnso.2018.05.003 [DOI] [Google Scholar]
- [54].Poolman JT, Anderson AS. Escherichia coli and Staphylococcus aureus: leading bacterial pathogens of healthcare associated infections and bacteremia in older-age populations. Expert Rev Vaccines. 2018;17(7):607–618. doi: 10.1080/14760584.2018.1488590 [DOI] [PubMed] [Google Scholar]
- [55].Nandhini P, Kumar P, Mickymaray S, et al. Recent developments in methicillin-resistant Staphylococcus aureus (MRSA) treatment: a review. Antibiotics. 2022;11(5):1–21. doi: 10.3390/antibiotics11050606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Guaraná M, Nucci M, Nouér SA. EPIDEMIOLOGY, PREDISPOSING FACTORS and OUTCOME of STAPHYLOCOCCUS AUREUS BACTEREMIA in NEUTROPENIC PATIENTS. Hematol Transfus Cell Ther. 2022;44:S70–1. InternetAvailable from: https://www.sciencedirect.com/science/article/pii/S2531137922002334 [Google Scholar]
- [57].Nappi F, Singh SSA. Molecular pathways in Staphylococcus aureus endocarditis: a systematic review. Int. J. Mol. Sci. 2023;24(13):11068 doi: 10.3390/ijms241311068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liesenborghs L, Meyers S, Lox M, et al. Staphylococcus aureus endocarditis: distinct mechanisms of bacterial adhesion to damaged and inflamed heart valves. Eur Heart J. 2019;40(39):3248–3259. doi: 10.1093/eurheartj/ehz175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Peña-Moreno A, Torres-Soblechero L, López-Blázquez M, et al. Fatal Staphylococcus aureus endocarditis misdiagnosed as multisystem inflammatory syndrome in children. Pediatr Infect Dis J. 2022;41(2):e58–e59. doi: 10.1097/INF.0000000000003417 [DOI] [PubMed] [Google Scholar]
- [60].Oryan A, Alidadi S, Moshiri A, et al. Bone regenerative medicine: classic options, novel strategies, and future directions. J Orthop Surg Res. 2014. Mar;9(1):18. doi: 10.1186/1749-799X-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Calhoun JH, Manring MM, Shirtliff M. Osteomyelitis of the long bones. Semin Plast Surg. 2009. May;23(2):059–072. doi: 10.1055/s-0029-1214158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gillet Y, Tristan A, Rasigade JP, et al. Prognostic factors of severe community-acquired staphylococcal pneumonia in France. Eur Respir J. 2021;58(5):1–13. InternetAvailable from. doi: 10.1183/13993003.04445-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Cone S, Mistretta V, Lambert N, et al. Community-acquired Staphylococcus aureus pneumonia. Revue Medicale de Liege Belgium. 2021;76:595–597. [PubMed] [Google Scholar]
- [64].Kulkarni D, Wang X, Sharland E, et al. The global burden of hospitalisation due to pneumonia caused by Staphylococcus aureus in the under-5 years children: a systematic review and meta-analysis. EClinicalMedicine. 2022. Feb;44:101267. doi: 10.1016/j.eclinm.2021.101267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Vraa EP. Atypical presentation of sepsis from community-acquired Staphylococcus aureus pneumonia in a previously healthy 47-year-old male: case report. J Emerg Crit Care Med. 2021;5:1–5. doi: 10.21037/jeccm-21-1 [DOI] [Google Scholar]
- [66].Clark SB, Hicks MA. Staphylococcal Pneumonia. Treasure Island (FL). 2023. [Google Scholar]
- [67].Gillet Y, Issartel B, Vanhems P, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet (London, England). 2002. Mar;359(9308):753–759. doi: 10.1016/S0140-6736(02)07877-7 [DOI] [PubMed] [Google Scholar]
- [68].Mandell LA, Wunderink R. Methicillin-resistant Staphylococcus aureus and community-acquired pneumonia: an evolving relationship. Clin Infect Dis. 54(8): InternetAvailable from:1134–1136. 2012 Apr 15. doi: 10.1093/cid/cis045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang Y, Sibaii F, Lee K, et al. NOTE: this preprint reports new research that has not been certified by peer review and should not be used to guide clinical practice. 1. medRxiv. 2021;1(165):1–13. [Google Scholar]
- [70].Zhou Z, Charlesworth J, Achtman M. Accurate reconstruction of bacterial pan- and core genomes with PEPPAN. Genome Res. 2020. Nov;30(11):1667–1679. doi: 10.1101/gr.260828.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Seder N, Rayyan WA, Al-Fawares O, et al. Pseudomonas aeruginosa virulence factors and Antivirulence mechanisms to combat drug resistance. A Systematic Review. 2022;56(December):1–23. [Google Scholar]
- [72].Kläui AJ, Boss R, Graber HU, et al. Characterization and comparative analysis of the Staphylococcus aureus genomic island vSaβ: an in silico approach. J Bacteriol. 2019. Nov;201(22). doi: 10.1128/JB.00777-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Patel H, Rawat S. A genetic regulatory see-saw of biofilm and virulence in MRSA pathogenesis. Front Microbiol. 2023;14:14. doi: 10.3389/fmicb.2023.1204428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cervera-Alamar M, Guzmán-Markevitch K, Žiemytė M, et al. Mobilisation mechanism of pathogenicity islands by endogenous phages in Staphylococcus aureus clinical strains. Sci Rep. 2018;8(1):1–13. doi: 10.1038/s41598-018-34918-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kong C, Neoh H, Nathan S. Targeting Staphylococcus aureus toxins: a potential form of anti-virulence therapy. Vol. 8. Toxins: Multidisciplinary Digital Publishing Institute (MDPI); 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Missiakas DM, Schneewind O. Growth and laboratory maintenance of Staphylococcus aureus. Chapter 9. Curr Protoc Microbiol . 2013. p.Unit 9C.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chen Q, Zhao G, Yang W. Investigation into the prevalence of enterotoxin genes and genetic background of Staphylococcus aureus isolates from retain foods in Hangzhou, China. BMC Microbiol. 2023;23(1):1–19. doi: 10.1186/s12866-023-03027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bernardos A, Piacenza E, Sancenón F, et al. Mesoporous silica-based materials with bactericidal properties. Small. 2019;15(24):1–34. doi: 10.1002/smll.201900669 [DOI] [PubMed] [Google Scholar]
- [79].Choudhary KS, Mih N, Monk J, et al. The Staphylococcus aureus two-component system AgrAC displays four distinct genomic arrangements that delineate genomic virulence factor signatures. Front Microbiol. 2018;9:1082. doi: 10.3389/fmicb.2018.01082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Joo HS, Chatterjee SS, Villaruz AE, et al. Mechanism of gene regulation by a staphylococcus aureus toxin. MBio. 2016;7(5):2016–2018. doi: 10.1128/mBio.01579-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Jenul C, Horswill AR, Fischetti VA, et al. Regulation of Staphylococcus aureus Virulence. Microbiol Spectr. 2019. Apr;7(2). doi: 10.1128/microbiolspec.GPP3-0031-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Abdelnour A, Arvidson S, Bremell T, et al. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect Immun. 1993;61(9):3879–3885. doi: 10.1128/iai.61.9.3879-3885.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Cheung GYC, Wang R, Khan BA, et al. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun. 2011;79(5):1927–1935. doi: 10.1128/IAI.00046-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Al AA, Alsulami J, Aubee JI, et al. Staphylococcus aureus SigS induces expression of a regulatory protein pair that modulate its mRNA stability J. Bacteriol. 2022;205:e00392–22 doi: 10.1128/jb.00392-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Cheung AL, Schmidt K, Bateman B, et al. SarS, a SarA homolog repressible by agr, is an activator of protein a synthesis in Staphylococcus aureus. Infect Immun. 2001. Apr;69(4):2448–2455. doi: 10.1128/IAI.69.4.2448-2455.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Tiwari N, López-Redondo M, Miguel-Romero L, et al. The SrrAB two-component system regulates Staphylococcus aureus pathogenicity through redox sensitive cysteines. Proc Natl Acad Sci. 2020;117(20):10989–10999. doi: 10.1073/pnas.1921307117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Balasubramanian D, Harper L, Shopsin B, et al. Staphylococcus aureus pathogenesis in diverse host environments. Pathog Dis. 2017 Feb 1;75(1): ftx005. InternetAvailable from. 10.1093/femspd/ftx005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Johnson M, Sengupta M, Purves J, et al. Fur is required for the activation of virulence gene expression through the induction of the sae regulatory system in Staphylococcus aureus. Int J Med Microbiol. 2011 Jan 31;301(1):44–52. doi: 10.1016/j.ijmm.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kinkel TL, Roux CM, Dunman PM, et al. The Staphylococcus aureus SrrAB two-component system promotes resistance to nitrosative stress and hypoxia. MBio. 2013;4(6):1–9. doi: 10.1128/mBio.00696-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu Rev Microbiol. 2003;57(1):441–466. doi: 10.1146/annurev.micro.57.030502.090913 [DOI] [PubMed] [Google Scholar]
- [91].Shaw LN, Lindholm C, Prajsnar TK, et al. Identification and characterization of σS, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One. 2008 Dec 3;3(12): e3844. InternetAvailable from. 10.1371/journal.pone.0003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bischoff M, Dunman P, Kormanec J, et al. Microarray-based analysis of the Staphylococcus aureus σB regulon. J Bacteriol. 2004;186(13):4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sinha D, Sinha D, Dutta A, et al. Alternative sigma factor of Staphylococcus aureus interacts with the cognate antisigma factor primarily using its domain 3. Biochemistry. 2021;60(2):135–151. doi: 10.1021/acs.biochem.0c00881 [DOI] [PubMed] [Google Scholar]
- [94].Wójcik-Bojek U, Różalska B, Sadowska B. Staphylococcus aureus—A known opponent against host defense mechanisms and vaccine development—do we still have a chance to win? Int J Mol Sci. 2022;23(2):948. doi: 10.3390/ijms23020948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Flannagan RS, Heit B, Heinrichs DE. Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens. 2015;4(4):826–868. InternetAvailable from. doi: 10.3390/pathogens4040826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Shettigar K, Murali TS. Virulence factors and clonal diversity of Staphylococcus aureus in colonization and wound infection with emphasis on diabetic foot infection. InternetAvailable from Eur J Clin Microbiol Infect Dis. 2020;39(12):2235–2246. doi: 10.1007/s10096-020-03984-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Al-Bakri AG, Al-Hadithi H, Kasabri V, et al. The epidemiology and molecular characterization of methicillin-resistant staphylococci sampled from a healthy Jordanian population. Epidemiol Infect. 2013;141(11):2384–2391. doi: 10.1017/S0950268813000010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mansour Albalbaki1 M. O’la AL-Fawares1*, Walid Aburayyan1, Nesrin Seder2 OMA-S, Lamya AL-Tahrawe3 MNS. The correlation of gene mutation of coagulopathy cascade with elevated D-dimer levels in COVID-19 patients. J Appl Pharm Sci. 2024;14(1). [Google Scholar]
- [99].Mlynarczyk-Bonikowska B, Kowalewski C, Krolak-Ulinska A, et al. Molecular Mechanisms of Drug Resistance in Staphylococcus aureus. Int J Mol Sci. 2022;23(15):8088. doi: 10.3390/ijms23158088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Durand G, Bes M, Meugnier H, et al. Erratum: detection of new methicillin-resistant Staphylococcus aureus clones containing the toxic shock syndrome toxin 1 gene responsible for hospital- and community-acquired infections in France (journal of clinical Microbiology (2006) 44, 3 (847-853)). J Clin Microbiol. 2006;44(8):3053. doi: 10.1128/JCM.00871-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Silversides JA, Lappin E, Ferguson AJ. Staphylococcal toxic shock syndrome: mechanisms and management. Curr Infect Dis Rep. 2010;12(5):392–400. doi: 10.1007/s11908-010-0119-y [DOI] [PubMed] [Google Scholar]
- [102].Mitevska E, Wong B, Surewaard BGJ, et al. The prevalence, risk, and management of methicillin-resistant Staphylococcus aureus infection in diverse populations across Canada: a systematic review. Pathogens. 2021;10(4):393. doi: 10.3390/pathogens10040393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].AL-Fawares1* O, Al-Khresieh1 RO. Walid Aburayyan1, Nesrin Seder2, Hala I al-Daghistani3, Nasser El-Banna1 and EMA-T. Comparison of preservation enrichment media for long storage duration of Campylobacter jejuni §. Korean J Microbiol. 2023;59(3):192–196. [Google Scholar]
- [104].Al-Khreshieh RO, Al-Fawares O, Em A. Campylobacter jejuni infections: epidemiology, pathophysiology, clinical manifestations and management. J Biomed Res Environ Sci. 2023;4(2):258–269. InternetAvailable from. doi: 10.37871/jbres1670 [DOI] [Google Scholar]
- [105].Brawley DN, Sauer DB, Li J, et al. Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus. Nat Chem Biol. 2022;18(7):706–712. doi: 10.1038/s41589-022-00994-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Lade H, Joo HS, Kim JS. Molecular Basis of Non-β-Lactam Antibiotics Resistance in Staphylococcus aureus. Antibiotics. 2022;11(10):1–28. doi: 10.3390/antibiotics11101378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Bitrus AA, Zunita Z, Bejo SK, et al. In vitro transfer of methicillin resistance determinants mecA from methicillin resistant Staphylococcus aureus (MRSA) to methicillin susceptible Staphylococcus aureus (MSSA). BMC Microbiol. 2017;17(1):1–10. doi: 10.1186/s12866-017-0994-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].de Smalen AW, Ghorab H, Abd El Ghany M, et al. Refugees and antimicrobial resistance: a systematic review. Travel Med Infect Dis. 2017;15:23–28. doi: 10.1016/j.tmaid.2016.12.001 [DOI] [PubMed] [Google Scholar]
- [109].Malachowa N, DeLeo FR. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci. 2010. Sep;67(18):3057–3071. doi: 10.1007/s00018-010-0389-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Jian Z, Zeng L, Xu T, et al. Antibiotic resistance genes in bacteria: occurrence, spread, and control. J Basic Microbiol. 2021;61(12):1049–1070. doi: 10.1002/jobm.202100201 [DOI] [PubMed] [Google Scholar]
- [111].Rasheed NA, Hussein NR. Staphylococcus aureus: An overview of discovery, characteristics, epidemiology, virulence factors and antimicrobial sensitivity. Eur J Mol Clin Med. 2021;8(03):1160–1183. [Google Scholar]
- [112].Conly JM, Johnston BL. VISA, hetero-VISA and VRSA: the end of the vancomycin era? Can J Infect Dis. 2002;13(5):282–284. doi: 10.1155/2002/245109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Shariati A, Dadashi M, Moghadam MT, et al. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta-analysis. Sci Rep. 2020;10(1):1–16. InternetAvailable from. doi: 10.1038/s41598-020-69058-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Gomez-Lus ML, Aguilar L, Martin M, et al. Intracellular and extracellular killing of a penicillin-resistant, serotype-9 strain of streptococcus pneumoniae by polymorphonuclear leucocytes in the presence of sub-inhibitory concentrations of clavulanic acid. J Antimicrob Chemother. 1997;40(1):142–144. doi: 10.1093/jac/40.1.142 [DOI] [PubMed] [Google Scholar]
- [115].Goldrick B. First reported case of VRSA in the United States: an alarming development in microbial resistance. AJN Am J Nurs. 2002;102(11):17. doi: 10.1097/00000446-200211000-00015 [DOI] [PubMed] [Google Scholar]
- [116].Crespo-Piazuelo D, Lawlor PG. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) prevalence in humans in close contact with animals and measures to reduce on-farm colonisation. Ir Vet J. 2021;74(1):1–12. doi: 10.1186/s13620-021-00200-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Khairullah AR, Ramandinianto SC, Effendi MH. A review of Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) on Bovine Mastitis. Syst Rev Pharm. 2020;11(7):172–183. [Google Scholar]
- [118].Lakhundi S, Zhang K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clinical Microbiology Reviews. 2018;31(4):1–103. doi: 10.1128/CMR.00020-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Tu J, PM G. Evolutionary GEM: the evolution of methicillin-resistant Staphylococcus aureus. WURJ: HNS. 2017;8(1):8–10. doi: 10.5206/wurjhns.2017-18.22 [DOI] [Google Scholar]
- [120].Elnasser ZA, Obeidat HM, Bani-Salem ME, et al. Is methicillin-resistant Staphylococcus aureus a common pathogen in ventilation-associated pneumonia?: the experience of a tertiary teaching hospital in Jordan. Med (United States). 2021;100(20):E26069. doi: 10.1097/MD.0000000000026069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Hulla JE, Sahu SC, AW H. Nanotechnology: history and future. Hum Exp Toxicol. 2015;34(12):1318–1321. doi: 10.1177/0960327115603588 [DOI] [PubMed] [Google Scholar]
- [122].Singh S, Numan A, Somaily HH, et al. Nano-enabled strategies to combat methicillin-resistant Staphylococcus aureus. Mater Sci Eng C. 2021;129(August):112384. doi: 10.1016/j.msec.2021.112384 [DOI] [PubMed] [Google Scholar]
- [123].Berk DR, Bayliss SJ. MRSA, staphylococcal scalded skin syndrome, and other cutaneous bacterial emergencies. Pediatr Ann. 2010;39(10):627–633. doi: 10.3928/00904481-20100922-02 [DOI] [PubMed] [Google Scholar]
- [124].Joudeh N, Linke D. Nanoparticle classification, physicochemical properties, characterization, and applications: a comprehensive review for biologists. InternetAvailable from J Nanobiotechnology. 2022;20(1):262. doi: 10.1186/s12951-022-01477-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Miller S. Nanoparticles: properties and applications. ChemXpress. 2021;13(3):143. [Google Scholar]
- [126].Kiio TM, Park S. Physical properties of nanoparticles do matter. J Pharm Investig. 2021;51(1):35–51. InternetAvailable from. doi: 10.1007/s40005-020-00504-w [DOI] [Google Scholar]
- [127].Khan I, Saeed K, Khan I. Nanoparticles: properties, applications and toxicities. Arab J Chem. 2019;12(7):908–931. InternetAvailable from. doi: 10.1016/j.arabjc.2017.05.011 [DOI] [Google Scholar]
- [128].Oake A, Bhatt P, Pathak Y. Understanding surface characteristics of nanoparticles. In. 2019;1–17. [Google Scholar]
- [129].Gisbert-Garzarán M, Vallet-Regí M. Nanoparticles for bio-medical applications. Nanomaterials. 2022;12(7):10–12. doi: 10.3390/nano12071189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Patil MD, Dhas SD, Moholkar AV. Green synthesis of nanocomposites. IGI Global; 2021. p. 77–100. doi: 10.4018/978-1-7998-8936-6.ch004 [DOI] [Google Scholar]
- [131].Khan I, Saeed K, Khan I, et al. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019;12(7):908–931. doi: 10.1016/j.arabjc.2017.05.011 [DOI] [Google Scholar]
- [132].Kanwal A, Bhutta ZA, Ashar A, et al. Antimicrobial applications of nanoparticles. In: Garg R, Garg R, and Eddy N, editors. Handbook of research on green synthesis and applications of nanomaterials. IGI Global. 2022;269–288. [Google Scholar]
- [133].Hoque Apu E, Nafiujjaman M, Sandeep S, et al. Biomedical applications of multifunctional magnetoelectric nanoparticles. Mater Chem Front. 2022;6(11):1368–1390. doi: 10.1039/D2QM00093H InternetAvailable from. [DOI] [Google Scholar]
- [134].Arcos D. Nanomaterials in Biomedicine 2022. IJMS. 2023;24(10):9026. Switzerland. doi: 10.3390/ijms24109026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Schneider-Futschik EK, Reyes-Ortega F. Advantages and disadvantages of using magnetic nanoparticles for the treatment of complicated ocular disorders. Pharmaceutics. 2021;13(8):1–16. doi: 10.3390/pharmaceutics13081157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Joseph TM, Kar Mahapatra D, Esmaeili A, et al. Nanoparticles: Taking a Unique Position in Medicine. Nanomaterials. 2023. Jan;13(3):574. doi: 10.3390/nano13030574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Tedla N, Ruiz J, Mody V. Synthesis, Pharmacokinetics, and Toxicity of Nano-Drug Carriers. In: Shah N, editor. Nanocarriers: Drug Delivery System. Singapore: Springer; 2021. doi: 10.1007/978-981-33-4497-6_3 [DOI] [Google Scholar]
- [138].Shukla AK. Nanoparticles in Medicine. Nanoparticles Med. 2019;10(8):1–220. [Google Scholar]
- [139].Martínez-Carmona M, Gun’ko YK, Vallet-Regí M. Mesoporous silica materials as drug delivery: “the nightmare” of bacterial infection. Pharmaceutics. 2018;10(4):1–29. doi: 10.3390/pharmaceutics10040279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Baazaoui N, Sghaier-Hammami B, Hammami S, et al. A handbook guide to better use of nanoparticles in plants. Commun Soil Sci Plant Anal. 2021 Jan 7;52(4):287–321. doi: 10.1080/00103624.2020.1836198 [DOI] [Google Scholar]
- [141].Maity D, Sahoo SR, Saha S. Synthesis and characterization of nanomaterials for electrochemical sensors. ACS Symp Ser. 2023;1437:193–222. [Google Scholar]
- [142].Pawar S, Takke A. Regulatory aspects, types and bioapplications of metallic nanoparticles: a review. Curr Drug Deliv. 2023;20(7):857–883. doi: 10.2174/1567201819666220817110025 [DOI] [PubMed] [Google Scholar]
- [143].Haroon Anwar S. A brief review on nanoparticles: types of platforms, biological synthesis and applications. Res Rev J Mat Sci. 2018;6(02):109–116. doi: 10.4172/2321-6212.1000222 [DOI] [Google Scholar]
- [144].Akbarzadeh H, Mehrjouei E, Abbaspour M, et al. Thermal behavior of different types of Au–pt–pd nanoparticles: dumbbell-like, three-shell, core-shell, and random-alloy. Mater Chem Phys. 2023 Oct 1;294:126955. doi: 10.1016/j.matchemphys.2022.126955 [DOI] [Google Scholar]
- [145].Naskar A, Kim KS. Nanomaterials as delivery vehicles and components of new strategies to combat bacterial infections: advantages and limitations. Microorganisms. 2019;7(9):356. doi: 10.3390/microorganisms7090356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Geng Z, Cao Z, Liu J. Recent advances in targeted antibacterial therapy basing on nanomaterials. Exploration. 2023;3(1):20210117. doi: 10.1002/EXP.20210117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Natsheh IY, Elkhader MT, Al-Bakheit AA, et al. Inhibition of Acinetobacterbaumannii biofilm formation using different treatments of silica nanoparticles. Antibiotics. 2023;12(9):1365. doi: 10.3390/antibiotics12091365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Zhang Q, Ma X, Zhao X. Hybrid bacteria system with nanomaterials for tumor therapy. Adv NanoBiomed Res. 2023;3(7):2300003. doi: 10.1002/anbr.202300003 [DOI] [Google Scholar]
- [149].Liu H, Jiang Y, Wang Z, et al. Nanomaterials as carriers to improve the photodynamic antibacterial therapy. Front Chem. 2022;10(November):1–8. doi: 10.3389/fchem.2022.1044627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Vasilev K. Antibacterial applications of nanomaterials. Nanomaterials. 2023;13(9):1530. InternetAvailable from. doi: 10.3390/nano13091530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Bai X, Yang Y, Zheng W, et al. Synergistic photothermal antibacterial therapy enabled by multifunctional nanomaterials: progress and perspectives. Mater Chem Front. 2023;7(3):355–380. doi: 10.1039/D2QM01141G InternetAvailable from. [DOI] [Google Scholar]
- [152].Vallet-Regí M, Schüth F, Lozano D, et al. Engineering mesoporous silica nanoparticles for drug delivery: where are we after two decades? Chem Soc Rev. 2022;51(13):5365–5451. doi: 10.1039/D1CS00659B [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Li Z, Zhang Y, Feng N. Mesoporous silica nanoparticles: synthesis, classification, drug loading, pharmacokinetics, biocompatibility, and application in drug delivery. Expert Opin Drug Deliv. 2019;16(3):219–237. doi: 10.1080/17425247.2019.1575806 [DOI] [PubMed] [Google Scholar]
- [154].Yuan J, Wu S, Duan N, et al. A sensitive gold nanoparticle-based colorimetric aptasensor for Staphylococcus aureus. Talanta. 2014;127:163–168. doi: 10.1016/j.talanta.2014.04.013 [DOI] [PubMed] [Google Scholar]
- [155].Karaman DŞ, Pamukçu A, Karakaplan MB, et al. Recent advances in the use of mesoporous silica nanoparticles for the diagnosis of bacterial infections. Int J Nanomedicine. 2021;16:6575–6591. doi: 10.2147/IJN.S273062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Miramoth NS, Di Meo C, Zouhiri F, et al. Self-assembled squalenoylated penicillin bioconjugates: an original approach for the treatment of intracellular infections. ACS Nano. 2012;6(5):3820–3831. doi: 10.1021/nn204928v [DOI] [PubMed] [Google Scholar]
- [157].Sahli C, Moya SE, Lomas JS, et al. Recent advances in nanotechnology for eradicating bacterial biofilm. Theranostics. 2022;12(5):2383–2405. doi: 10.7150/thno.67296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Cheow WS, Chang MW, Hadinoto K. Antibacterial efficacy of inhalable antibiotic-encapsulated biodegradable polymeric nanoparticles against E. coli biofilm cells. J Biomed Nanotechnol. 2010;6(4):391–403. doi: 10.1166/jbn.2010.1116 [DOI] [PubMed] [Google Scholar]
- [159].Ye L, Cao Z, Liu X, et al. Noble metal-based nanomaterials as antibacterial agents. J Alloys Compd InternetAvailable from: https://www.sciencedirect.com/science/article/pii/S0925838822004820. 2022;904:164091. doi: 10.1016/j.jallcom.2022.164091 [DOI] [Google Scholar]
- [160].Awad M, Thomas N, Barnes TJ, et al. Nanomaterials enabling clinical translation of antimicrobial photodynamic therapy. J Control Release Off J Control Release Soc. 2022. Jun;346:300–316. [DOI] [PubMed] [Google Scholar]
- [161].Chen Y, Gao Y, Chen Y, et al. Nanomaterials-based photothermal therapy and its potentials in antibacterial treatment. J Control Release Off J Control Release Soc. 2020. Dec;328:251–262. [DOI] [PubMed] [Google Scholar]
- [162].Edrisi F, Baheiraei N, Razavi M, et al. Potential of graphene-based nanomaterials for cardiac tissue engineering. J Mater Chem B. 2023;11(31):7280–7299. InternetAvailable from. doi: 10.1039/D3TB00654A [DOI] [PubMed] [Google Scholar]
- [163].Gungordu Er S, Edirisinghe M, Tabish TA. Graphene-based nanocomposites as antibacterial, antiviral and antifungal agents. Adv Healthc Mater. 2023;12(6):1–20. doi: 10.1002/adhm.202201523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Chidre P, Chavan A, Mallikarjunaiah HN, et al. Nanomaterials: Potential Broad Spectrum Antimicrobial Agents [Internet]. CNM. 2023;8(4):319–327. Available from: http://www.eurekaselect.com/article/128256 [Google Scholar]
- [165].Wang Y, Li J, Li X, et al. Graphene-based nanomaterials for cancer therapy and anti-infections. Bioact Mater. 2022;14:335–349. InternetAvailable from: https://www.sciencedirect.com/science/article/pii/S2452199X22000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Zhou H, Zou F, Koh K, et al. Antibacterial activity of graphene-based nanomaterials. Adv Exp Med Biol. 2022;1351:233–250. [DOI] [PubMed] [Google Scholar]
- [167].Marković ZM, Kepić DP, Matijašević DM, et al. Ambient light induced antibacterial action of curcumin/graphene nanomesh hybrids. RSC Adv. 2017;7(57):36081–36092. doi: 10.1039/C7RA05027E [DOI] [Google Scholar]
- [168].Zhou X, Hao Y, Yaxin L, et al. MXenes: An emergent materials for packaging platforms and looking beyond. Nano Select. 2022;3(7):1123–1147. doi: 10.1002/nano.202200023 [DOI] [Google Scholar]
- [169].Yaragalla S, Bhavitha KB, Athanassiou A. A review on graphene based materials and their antimicrobial properties. Coatings. 2021;11(10):1–18. doi: 10.3390/coatings11101197 [DOI] [Google Scholar]
- [170].Dwivedi N, Dhand C, Kumar P, et al. Emergent 2D materials for combating infectious diseases: the potential of MXenes and MXene–graphene composites to fight against pandemics. Mater Adv. 2021;2(9):2892–2905. doi: 10.1039/D1MA00003A [DOI] [Google Scholar]
- [171].Amara U, Hussain I, Ahmad M, et al. 2D MXene‐Based Biosensing: A Review. Small. 2022;19(2). Nov 1. doi: 10.1002/smll.202205249 [DOI] [PubMed] [Google Scholar]
- [172].Rizi KS. Mxene nanosheets as a novel nanomaterial with antimicrobial applications: a literature review. J Mol Struct. 2022;1262:132958. InternetAvailable from: https://www.sciencedirect.com/science/article/pii/S0022286022006275 [Google Scholar]
- [173].Li H, Fan R, Zou B, et al. Roles of MXenes in biomedical applications: recent developments and prospects. J Nanobiotechnology. 2023;21(1):1–39. InternetAvailable from. doi: 10.1186/s12951-023-01809-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Xu Z, Zhang Y, Liu M, et al. Two-dimensional titanium carbide MXene produced by ternary cations intercalation via structural control with angstrom-level precision. iScience. 2022;25(12):105562. InternetAvailable from: https://www.sciencedirect.com/science/article/pii/S258900422201834X [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Rosenkranz A, Righi MC, Sumant AV, et al. Perspectives of 2D MXene tribology. Adv Mater. 2023;35(5). doi: 10.1002/adma.202207757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Ramezani Farani M, Nourmohammadi Khiarak B, Tao R, et al. 2D MXene nanocomposites: electrochemical and biomedical applications. Environ Sci Nano. 2022 Jan 1;9(11):4038–4068. doi: 10.1039/D2EN00527A [DOI] [Google Scholar]
- [177].Marchwiany ME, Birowska M, Popielski M, et al. Surface-related features responsible for cytotoxic behavior of mxenes layered materials predicted with machine learning approach. Materials. 2020;13(14):1–17. doi: 10.3390/ma13143083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Lim GP, Soon CF, Ma NL, et al. Cytotoxicity of MXene-based nanomaterials for biomedical applications: a mini review. Environ Res InternetAvailable from. 2021;201:111592. doi: 10.1016/j.envres.2021.111592 [DOI] [PubMed] [Google Scholar]
- [179].Iravani S, Varma RS. Smart MXene quantum dot-based nanosystems for biomedical applications. Nanomaterials. 2022;12(7):1–16. doi: 10.3390/nano12071200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Cheah YJ, Buyong MR, Mohd Yunus MH. Wound healing with electrical stimulation technologies: a review. Polymers. 2021;13(21):1–21. doi: 10.3390/polym13213790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Jang JH, Lee EJ. Influence of MXene particles with a stacked-lamellar structure on osteogenic differentiation of human mesenchymal stem cells. Materials. 2021;14(16):4453. doi: 10.3390/ma14164453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Qiang M, Wu J, Zhang H, et al. Ag/cu-chitosan composite improves laundry hygiene and reduces silver emission in washing machines. Polymers. 2023;15(3):695. doi: 10.3390/polym15030695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Li Q, Wang S, Jin X, et al. The application of polysaccharides and their derivatives in pigment, barrier, and functional paper coatings. Polymers. 2020;12(8):1837. doi: 10.3390/polym12081837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Hadidi N, Mohebbi M. Anti-infective and toxicity properties of carbon based materials: graphene and functionalized carbon nanotubes. Microorganisms. 2022;10(12):2439. doi: 10.3390/microorganisms10122439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Smirnova VV, Chausov DN, Serov DA, et al. A novel biodegradable composite polymer material based on plga and silver oxide nanoparticles with unique physicochemical properties and biocompatibility with mammalian cells. Materials. 2021;14(22):6915. doi: 10.3390/ma14226915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Mizwari Z, Oladipo A, Yilmaz E. Chitosan/Metal oxide nanocomposites: synthesis, characterization, and antibacterial activity. Int J Polym Mater. 2021 Feb 22;70(6):383–391. doi: 10.1080/00914037.2020.1725753 [DOI] [Google Scholar]
- [187].Cai J, Liu R. Introduction to antibacterial biomaterials. Biomater Sci. 2020;8(24):6812–6813. InternetAvailable from. doi: 10.1039/D0BM90100H [DOI] [PubMed] [Google Scholar]
- [188].Shahbazi Y, Shavisi N. Current advancements in applications of chitosan based nano-metal oxides as food preservative materials. Nanomed Res J. 2019;4(3):122–129. doi: 10.22034/NMRJ.2019.03.001 [DOI] [Google Scholar]
- [189].Shahbazi Y, Shavisi N. Current advancements in applications of chitosan based nano-metal oxides as food preservative materials. Nanomedicine Res J. 2019;4(3):122–129. InternetAvailable from. http://www.nanomedicine-rj.com/article_36898.html [Google Scholar]
- [190].Lu MM, Wang QJ, Chang ZM, et al. Synergistic bactericidal activity of chlorhexidine-loaded, silver-decorated mesoporous silica nanoparticles. Int J Nanomedicine. 2017;12:3577–3589. doi: 10.2147/IJN.S133846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Narayan R, Nayak UY, Raichur AM, et al. Mesoporous silica nanoparticles: a comprehensive review on synthesis and recent advances. Pharmaceutics. 2018;10(3):1–49. doi: 10.3390/pharmaceutics10030118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Colilla M, Vallet-Regí M. Targeted stimuli-responsive mesoporous silica nanoparticles for bacterial infection treatment. Int J Mol Sci. 2020;21(22):8605–8632. doi: 10.3390/ijms21228605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Borodina T, Kostyushev D, Zamyatnin AA, et al. Nanomedicine for treating diabetic retinopathy vascular degeneration. IJTM. 2021;1(3):306–322. doi: 10.3390/ijtm1030018 [DOI] [Google Scholar]
- [194].Grunberger JW, Ghandehari H. Layer-by-layer Hollow Mesoporous silica nanoparticles with tunable degradation profile. Pharmaceutics. 2023;15(3):832. doi: 10.3390/pharmaceutics15030832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Sharma SK, Sharma AR, Pamidimarri SDVN, et al. Bacterial compatibility/toxicity of biogenic silica (B-SiO2) nanoparticles synthesized from biomass rice husk ash. Nanomaterials. 2019;9(10):1–10. doi: 10.3390/nano9101440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Kang SH, Lee YK, Park IS, et al. Biomimetic gold nanoshell-loaded macrophage for photothermal biomedicine. Bio Med Res Int. 2020;2020:1–14. doi: 10.1155/2020/5869235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Danylenko S, Marynchenko L, Bortnyk V, et al. Use of highly dispersed silica in biotechnology of complex probiotic product based on bifidobacteria. Innov Biosyst Bioeng [Internet]. 2022; Available from: http://ibb.kpi.ua/article/view/256179
- [198].Zakiyyah SN, Rakhmawaty Eddy D, Firdaus M, et al. Application of gold silica nanocomposites in electrochemical biosensors: a review. pendipa jurnal pendik sains. 2021;5(2):122–132. doi: 10.33369/pendipa.5.2.122-132 [DOI] [Google Scholar]
- [199].Sprenger S. Nanosilica-toughened epoxy resins. Switzerland: MDPI AG; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Antimicrobial E, Activities P, Al-Askar AA, et al. Green biosynthesis of zinc oxide nanoparticles using pluchea. Molecules. 2023;28:4679. doi: 10.3390/molecules28124679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Mohamed MBED, Abo El-Ela FI, Mahmoud RK, et al. Cefotax-magnetic nanoparticles as an alternative approach to control methicillin-resistant Staphylococcus aureus (MRSA) from different sources. Sci Rep. 2022;12(1):1–10. doi: 10.1038/s41598-021-04160-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Fan F, Saha S, Hanjaya-Putra D. Biomimetic hydrogels to promote wound healing. Front Bioeng Biotechnol. 2021;9(September):1–24. doi: 10.3389/fbioe.2021.718377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Singh B, Vuddanda PR, V MR, et al. Cefuroxime axetil loaded solid lipid nanoparticles for enhanced activity against S. aureus biofilm. Colloids Surf B Biointerfaces. 2014;121:92–98. doi: 10.1016/j.colsurfb.2014.03.046 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data files are available upon request.


