Abstract
Murine gammaherpesvirus 68 (MHV68) is a gammaherpesvirus that was first isolated from murid rodents. MHV68 establishes a latent infection in the spleen and other lymphoid organs. Several gammaherpesviruses, including herpesvirus saimiri, human herpesvirus 8, and MHV68, encode proteins with extensive homology to the D-type cyclins. To study the function of the cyclin homologue, a recombinant MHV68 has been constructed that lacks the cyclin homologue and expresses β-galactosidase as a marker (MHV68cy−). MHV68cy− grows in vitro with kinetics and to titers similar to those of the wild type. BALB/c mice infected with mixtures of equivalent amounts of the wild type and MHV68cy− show deficient growth of the MHV68cy− in an acute infection. Infection of SCID mice with virus mixtures also showed decreased MHV68cy− virus growth, indicating that the deficiency is not mediated by T or B cells. Although mice infected with mixtures containing 100 times as much MHV68cy− had greater splenic titers of the mutant virus than wild-type virus in acute infection, at 28 days postinfection splenocytes from these mice reactivated primarily wild-type virus. Quantitative PCR data indicate that equivalent genomes were present in the latent state. Reinsertion of the cyclin homologue into the cyclin-deleted virus restored the wild-type phenotype. These results indicate that the MHV68 cyclin D homologue mediates important functions in the acute infection and is required for efficient reactivation from latency.
The gammaherpesviruses are an important group of pathogens in humans and animals most notable for their associations with tumor formation in the host and ability to establish latent infections within lymphocytes (18, 31). Herpesvirus saimiri (HVS) causes lymphomas in primates and can transform T lymphocytes (19). Epstein-Barr virus (EBV) is a significant human pathogen causing lymphomas and nasopharyngeal carcinoma as well as transforming lymphocytes in vitro (27; D. H. Crawford, J. A. Thomas, G. Janossy, P. Sweny, O. N. Fernando, J. F. Moorhead, and J. H. Thompson, Letter, Lancet i:1355–1356, 1980). Human herpesvirus 8 (HHV8) is associated with Kaposi's sarcoma, body cavity-based lymphomas, and Castleman's disease in humans (7, 9, 39, 47; P. S. Moore and Y. Chang, Letter, Science 270:15, 1995). Mouse herpesvirus 68 (MHV68) is a gammaherpesvirus that can serve as a model system for these viruses (36, 40).
MHV68 was isolated from wild rodents and infects inbred and outbred rodents (29; D. Blaskovic, M. Stancekova, J. Svobodova, and J. Mistrikova, Letter, Acta Virol. 24:468, 1980). Infection is characterized by hematogenous spread with exudative pneumonia and splenomegaly (29, 48). MHV68 latently infects B lymphocytes as well as other cell types (41, 45, 49, 54, 56) and has also been associated with lymphoproliferative disease (43).
In addition to sharing biological characteristics with the other gammaherpesviruses, there is substantial sequence homology among the gammaherpesviruses (13, 51). Many of the gammaherpesviruses, including HHV8, HVS, and MHV68, encode a homologue to the D-type cyclins (2, 28, 32, 51; Y. Chang, P. S. Moore, S. J. Talbot, C. H. Boshoff, T. Zarkowska, K. Godden, H. Paterson, R. A. Weiss, and S. Mittnacht, Letter, Nature 382:410, 1996). The notable exception is EBV, which upregulates cellular cyclin D2 expression (3, 5, 38). Levels of the cyclin proteins fluctuate throughout the cell cycle and regulate cell cycle progression. The cyclin proteins function by binding to the cyclin-dependent kinases (CDK) which phosphorylate substrates responsible for cell cycle progression. Based on sequence homology, the gammaherpesvirus-encoded cyclins are most similar to the host D-type cyclins. The D-type cyclins (D1, D2, and D3) function in G1 phase to regulate progression into S phase. The host D-type cyclins bind to CDK 4 and 6, which phosphorylate retinoblastoma protein releasing the E2F transcription factors which stimulate progression into S phase (1, 26, 34, 35).
The virally encoded cyclins have been shown to share features with cellular cyclin but have also acquired altered characteristics. Like host D-type cyclins, HHV8 v-cyclin binds to CDK 6 and phosphorylates and inactivates retinoblastoma protein (15). MHV68-encoded cyclin induces cell cycle progression, and transgenic mice expressing MHV68 v-cyclin produce lymphoid tumors as they age (50).
Prior to this work the role of MHV68 v-cyclin in nononcogenic infections of MHV68 had yet to be defined. To explore its role in various aspects of viral infection, we constructed a recombinant MHV68 lacking the cyclin D homologue (MHV68cy−). We demonstrate here that the virus replicates efficiently in cell culture but exhibits a deficiency in replication in acute infection in BALB/c mice. We further show that the cyclin-deficient virus can establish latency but very inefficiently reactivates from the latent state. We repaired the virus by reinserting the cyclin gene and demonstrate that the wild-type phenotype is restored (MHV68rep).
MATERIALS AND METHODS
Viruses and tissue culture.
Our MHV68 is the same strain used by A. Nash and coworkers (44). The complete sequence of this virus is known (51). Virus stocks used for mouse infections were passaged on a previously described line of rat embryo fibroblasts (REFs) until complete cytopathic effect (CPE) was observed (4). The titer of the wild-type virus was 2.5 × 107 PFU/ml, and the recombinant virus titer ranged between 6.0 × 106 and 2.0 × 107 PFU/ml. REF cells were maintained in minimal essential medium (MEM) supplemented with 10% fetal bovine serum, 30 μg of gentamicin per ml, 0.3 μg of amphoterocin B per ml, and 2 mM l-glutamine (complete MEM).
Plasmid construct.
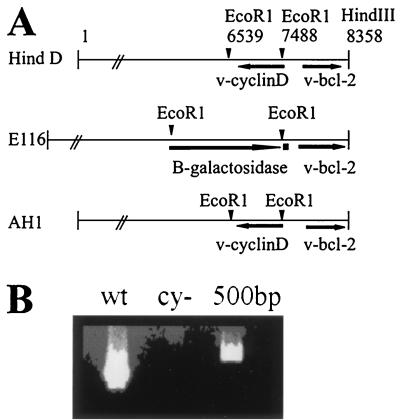
v-cyclin was deleted through homologous recombination with a transfer vector as described in Fig. 1. Virion DNA from MHV68 was digested with HindIII. The 8.35-kb fragment encoding v-cyclin (Hind D) was inserted into pSK (Stratagene, La Jolla, Calif.) lacking an EcoRI restriction site (13). The β-galactosidase (β-Gal) expression cassette in pSVB (Clontech, Palo Alto, Calif.) was excised with EcoRI and inserted into the EcoRI site of the transfer vector, replacing a 949-bp portion of the v-cyclin open reading frame to yield E116. The orientation of β-Gal (Fig. 1) was determined by sequencing the fusion between the 5′ end of β-Gal and the MHV68 genome. To repair the cyclin-deleted recombinant virus, the EcoRI restriction fragment encoding v-cyclin was inserted in place of the β-Gal cassette in the transfer vector to yield AH1.
FIG. 1.
Construction of recombinant MHV68cy− and MHV68rep. (A) The cyclin-encoding EcoRI restriction fragment was removed from the 8.35-kb HindIII fragment (Hind D) to generate the transfer vector used to delete v-cyclin (E116). To repair MHV68rep, the EcoRI restriction fragment was reinserted into E116 in place of the β-Gal cassette to generate AH1. (B) PCR amplification of v-cyclin from MHV68cy− (cy−) was compared to MHV68wt (wt).
Transfection and recombinant virus isolation.
Subconfluent REFs were cotransfected with E116 (1 μg) and MHV68-infected cell DNA by the calcium phosphate precipitation method. The repaired virus was generated by cotransfection of AH1 and DNA from cells infected with MHV68cy−. CPE usually occurred within 6 days posttransfection. Supernatants from frozen and thawed cultures were diluted and screened for β-Gal-positive virus. Recombinant virus was isolated by limiting dilution cultures on REF cells in 96-well tissue culture plates. Supernatant from these plates were transferred to empty plates and frozen while the original plates were fixed and stained for β-Gal expression. Wells positive for β-Gal virus were again diluted onto 96-well plates for multiple rounds of limited dilution culture until the recombinant virus was isolated and verified by plaque assay, PCR, restriction analysis, and Southern blotting.
To assess purity of recombinant virus, PCR was performed with cyclin-specific primers. DNA from cells infected with the recombinant virus and wild type were amplified in 50-μl reaction mixtures with Supermix (Gibco BRL). PCR conditions were as follows: a 5-min denaturation at 94°C, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by an extension phase of 10 min at 70°C. PCR products were electophoresed on a 1.3% agarose gel. Primer sequences were as follows: 5′-GCATAGGGCGCAGAAGATT-3′ and 5′-CGCAGCGAAAGAGAACACG-3′.
To examine the possibility that MHV68cy− was reactivating from latency but deleting the β-Gal marker, PCR was performed as above with a cyclin-specific primer, 5′-CCACCCAGTTGGCATACCT-3′, and a primer flanking the cyclin gene, 5′-GTAAGGGAATTCGGTAAATTC-3′.
Southern and Northern analyses of recombinant viruses.
DNA for Southern blot analysis was extracted from infected REFs and digested with EcoRI and HindIII. Electrophoresis, blotting, and hybridization conditions were done by established procedures. A nick translation system (Gibco BRL) was used with [α-32P]ATP (Amersham, Arlington Heights, Ill.) to label the 949-bp v-cyclin-encoding EcoRI restriction fragment. Virion DNA was extracted and was digested with restriction enzymes and electrophoresed on a 0.7% agarose gel for analysis (4). Total cell RNA was extracted from infected REF cells with TRIzol (Gibco BRL) according to manufacturer recommendations. Northern analysis was performed according to standard procedures.
In vitro infections.
REFs or BALB/c 3T3 cells (approximately 5 × 105 per well in a six-well plate) were infected with MHV68wt, MHV68cy−, or MHV68rep at a multiplicity of infection (MOI) of 5 or 0.01. After a 1-h adsorption, the wells were washed with MEM. At various times after infection, the supernatants were frozen and thawed, and the amount of infectious virus in the supernatants was determined by plaque assay on REF cells as described below.
In vivo infections.
Female BALB/c mice (age, 4 to 6 weeks) were obtained from Frederick Cancer Research Labs (Frederick, Md.). Mice were inoculated intraperitoneally with virus in 1 ml of complete MEM. At various times after infection, mice were sacrificed and assayed for infectious virus in spleens by mincing the spleens or isolating splenocytes as described below and sonicating in 1 ml of complete MEM with 2 pulses of 180 W for 3 s/pulse at 4°C using the microprobe from a Branson sonicator. Sonicates were assayed on REF monolayers. After infection for 1 h at 37°C, cells were overlaid with 0.9% methylcellulose in MEM supplemented with 5% fetal bovine serum, 25 mM HEPES, 2 mM l-glutamine, and 30 μg of gentamicin per ml. Six days after infection, monolayers were fixed and stained with methylene blue or X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and plaques were counted. For X-Gal staining, cells were fixed in 2% paraformaldehyde and 0.2% glutaraldehyde in 0.1 M Na2PO4 for 10 min. After rinsing in phosphate-buffered saline, plaques were stained with X-Gal (2 mg/ml) in 1.3 mM MgCl2, 3 mM K4Fe(CN)6, 3 mM K3Fe(CN)6, and 100 mM Na2PO4. For methylene blue staining, cells were fixed in ethanol and then stained with 0.5% methylene blue.
To determine the sensitivity of this assay, an uninfected spleen in 1 ml of complete MEM was spiked with virus of known titer and the spleen was treated as described above to determine the sensitivity of the assay. The titer of the sonicated virus decreased by 80% from the titer of the unsonicated virus which was determined in parallel, reducing the sensitivity of the assay from approximately 2 to 10 PFU/spleen. To assay for latent virus, splenocytes were isolated 28 days after infection. Spleen cells were separated from stromal tissue by teasing with needles. Stroma and spleen cells were resuspended in MEM and allowed to settle for 10 min. Cells remaining in suspension were pelleted and resuspended in 1 ml of distilled water to lyse red blood cells. Ten milliliters of complete MEM was added quickly, and splenocytes were washed in complete MEM. Thirty to 100 times more cells were sonicated and assayed for infectious virus than were explanted to recover reactivated virus.
For treatment of splenocytes with lipopolysaccharide (LPS), splenocytes were isolated as above and resuspended in complete MEM with or without 5 μg of LPS per ml (VWR, Batavia, Ill.). After 48 h, splenocytes were transferred to monolayers of REFs and observed for 2 to 4 weeks for CPE.
Real-time PCR analysis.
Quantitation of viral genomes was performed with real-time PCR using the 7700 Sequence Detection System (PE Biosystems, Foster City, Calif.). This method is based on continuous optical monitoring of a fluorogenic PCR (14, 17). In addition to the two amplification primers, an oligonucleotide homologous to the amplified region is included in the reaction mixture. The oligonucleotide is bound to a fluorescent tag that serves as a reporter and a quencher that suppresses fluorescence. During the extension phase of PCR, the 5′-to-3′ exonuclease activity of Taq cleaves the reporter from the probe, releasing it from the quencher and resulting in an increase in fluorescence emission which was detected by the laser detector of the ABI Prism 7700. After crossing a sequence detection threshold, the PCR amplification results in a fluorescent signal proportional to the amount of PCR product generated. Initial template concentration is derived from the cycle number (CT) at which the fluorescent signal crosses a threshold in the exponential phase of the PCR. Standard graphs of CT values obtained from serially diluted positive controls were used to derive values for unknowns.
Three primer-probe concentrations were designed using Primer Express software (PE Biosystems). We utilized a primer and probe set specific to v-cyclin to detect MHV68wt, a set spanning the β-Gal insertion site to detect MHV68cy− and a set specific to the double-copy murine RNase P gene, to which viral genomes were normalized. To generate standard curves for v-cyclin, the EcoRI fragment encoding v-cyclin was cloned into pBluescript SK, and for β-Gal, the previously described plasmid E116 was used. Mouse DNA extracted from spleen cells was used to generate the standard curve for the RNase P control. Standard curves were generated from the CT values from serial dilutions of each template. Copy number for infected and uninfected spleens were calculated from the CT values of samples using standard curves of CT values corresponding to known copy numbers. These calculations were performed utilizing sequence detection systems software (PE Biosystems). Each sample was tested in duplicate, and the mean of the two values was shown as the copy number of the sample. Calculated copy numbers were normalized to RNase P and then adjusted to represent 1 ng of genomic DNA. Correlation coefficients calculated by Sequence Detection Systems software were as follows: 0.984 for cyclin, 0.996 for β-Gal, and 0.966 for RNase P.
Reactions were performed with universal master mix and universal cycling conditions (PE Biosystems). PCR primers were synthesized by PE Biosystems or Life Technologies, Inc. (Gaithersburg, Md). Primers were used at a concentration of 900 nM for each primer and 200 nM of probe in a 25-μl reaction mixture.
Primers sequences were as follows: for v-cyclin, 5′-CCTGTCAGCTACCCACGAGAG-3′ and 5′-CCACCCAGTTGGCATACCT-3′; for B-Gal, 5′-CACATTCCACAGCCAAGCTGTA-3′ and 5′-CAACATTCCACCTTCAACAAACA-3′; and for RNase P, 5′-GATGCCTCCCTCGCCG-3′ and 5′-CTCAGCCATTGAACTCGCAC-3′. Probe sequences were as follows: for cyclin, VIC-TTCCAGAGTCAATAGTTTGTCAGCTGTTGTTG-TAMRA; for β-Gal, VIC-CGAGTCGAATTCTTGACTGGCCATG-TAMRA; and for RNaseP, 6FAM-AGCTTGGAACAGACTCACGGCCAGC-TAMRA.
RNA dot blot.
RNA dot blot assays were performed by adding 3 volumes of denaturing solution (66% formamide, 8% formaldehyde, 0.6× MOPS [morpholinepropanesulfonic acid] buffer, pH 7.0) to 1.5 μg of RNA. The samples were heated for 15 min at 65°C followed by the addition of 2 volumes of cold 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Equivalent amounts of RNA were applied under vacuum (96-well minifold; Schleicher & Schuul, Inc., Keene, N.H.) to BIOTRANS nylon membrane (ICN Biomedicals Inc., East Hills, N.Y.) prewetted with 2× SSC. The filter was then washed three times with 2× SSC, baked at 80°C for 2 h, and prehybridized for 2 h at 42°C in 4 ml of RNA hybridization solution (0.1% Ficoll, 0.1% polyvinylpyrrolidone, 0.1% bovine serum albumin, 5× SSC, 0.1% sodium dodecyl sulfate, 50 mM sodium phosphate [pH 9.5], and 50% deionized formamide). Hybridization was carried out overnight at 42°C in 4 ml of RNA hybridization solution with the addition of 500 ng of cyclin D2 probe (a gift from C. Sherr) labeled as described previously.
RESULTS
Recombinant virus design and purification.
The v-cyclin gene was deleted from the MHV68 genome by homologous recombination with the E116 transfer vector shown in Fig. 1A. To generate the transfer vector used for repair of MHV68cy−, the EcoRI restriction fragment encoding v-cyclin was reinserted in place of the β-Gal cassette (AH1).
To create MHV68cy−, E116 and DNA from cells infected with MHV68wt were cotransfected into REFs. After CPE was detected, viral supernatant was screened for β-Gal expression to detect recombinant virus. The recombinant virus was then isolated from wild-type virus by multiple rounds of limiting-dilution cultures. When dilution cultures yielded all β-Gal-expressing virus, DNA was extracted for analysis. PCR amplification for the cyclin gene from REFs infected with MHV68cy− was negative for cyclin amplification, demonstrating that the v-cyclin gene had been deleted from the MHV68cy− genome (Fig. 1B). DNA isolated from REFs infected with purified MHV68cy− was used in a cotransfection with AH1, and MHV68rep was isolated by limiting dilution for β-Gal-negative virus.
Analysis of recombinant viruses.
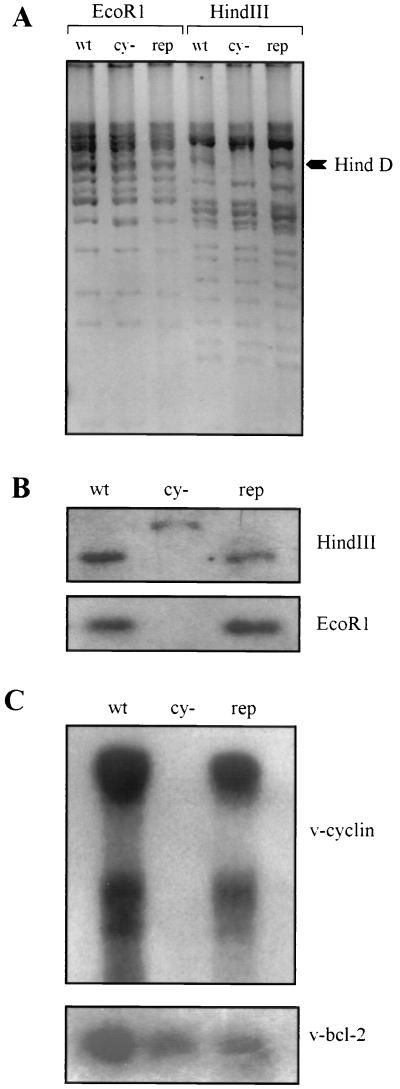
Purified MHV68cy− and MHV68rep were analyzed by restriction digestion (Fig. 2A), Southern hybridization (Fig. 2B), and Northern analysis (Fig. 3C). Virion DNA was digested with EcoRI and HindIII and was electrophoresed on an agarose gel (Fig. 2A). No rearrangements outside of Hind D in the MHV68cy− genome are apparent. The arrow on the right indicates the Hind D restriction fragment that is not visible in MHV68cy− due to the insertion of the β-Gal cassette that adds approximately 3 kb to the size of the restriction fragment. This is more apparent upon hybridization with the Hind D restriction fragment (Fig. 2B). The upper panel shows the altered migration pattern of the HindIII restriction fragment in MHV68cy−. In the lower panel of Fig. 2B, the approximately 1-kb v-cyclin-encoding restriction fragment is absent in MHV68cy− and present in MHV68wt and MHV68rep. To ensure that transcription of v-cyclin from MHV68rep was normal, and that transcription of neighboring bcl-2 homologue has not been altered by deletion of v-cyclin, Northern blot analysis was performed (Fig. 2C). Hybridization with v-cyclin (upper panel) showed an identical expression pattern of two different-sized messages from MHV68wt and MHV68rep and no cyclin expression from MHV68cy−. The same Northern blot was stripped and probed with v-bcl-2. v-bcl-2 expression was similar in all samples, indicating that its transcription was not altered in MHV68cy− and MHV68rep.
FIG. 2.
Analysis of MHV68cy− and MHV68rep. (A) Virion DNA was digested with EcoRI and HindIII, and the restriction pattern of MHV68wt (wt) was compared to those of MHV68cy− (cy−) and MHV68rep (rep). (B) Infected cell DNA was hybridized with Hind D. Cyclin-encoding HindIII restriction fragment is shown in the upper panel, and the EcoRI fragment is shown in the lower panel. (C) Northern analysis of v-cyclin expression is shown in the upper panel and the neighboring gene v-bcl-2 is shown in the lower panel.
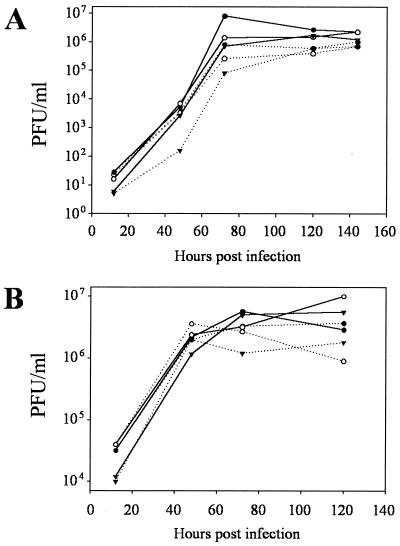
FIG. 3.
Single (A) and multistep (B) growth curves comparing growth of MHV68wt (●), MHV68cy− (○), and MHV68rep (▾) in REF cells (filled lines) and BALB/c 3T3 cells (dashed lines). Supernatants were frozen at given times after infection, and titers were determined on monolayers of REF cells. Error generated in determining supernatant titers is not visible.
Cyclin-deleted MHV68 grows efficiently in cell culture.
The efficiency of replication of MHV68cy− in cell culture was compared to MHV68wt and MHV68rep at MOIs of 5 and 0.01. In the single-step growth curve, in which cells were infected at an MOI of 5, MHV68cy− grew with kinetics and to titers similar to those of MHV68wt and MHV68rep in both REF and BALB/c 3T3 cells (Fig. 3A). In the multistep growth curve, in which cells were infected at an MOI of 0.01, MHV68cy− grew similarly to MHV68wt and MHV68rep (Fig. 3B). All three viruses replicated at about a log higher titer in REF cells than BALB/c 3T3 cells after infection at a low MOI. However, there was no significant difference between MHV68cy− and MHV68wt and MHV68rep in any experiment, indicating that v-cyclin appears to be dispensable for replication in two cell lines.
Cyclin-deleted virus is deficient in acute infections.
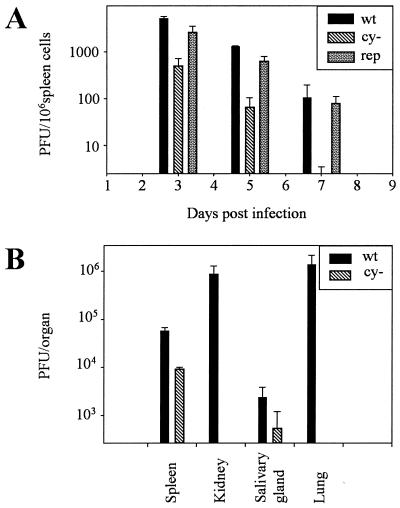
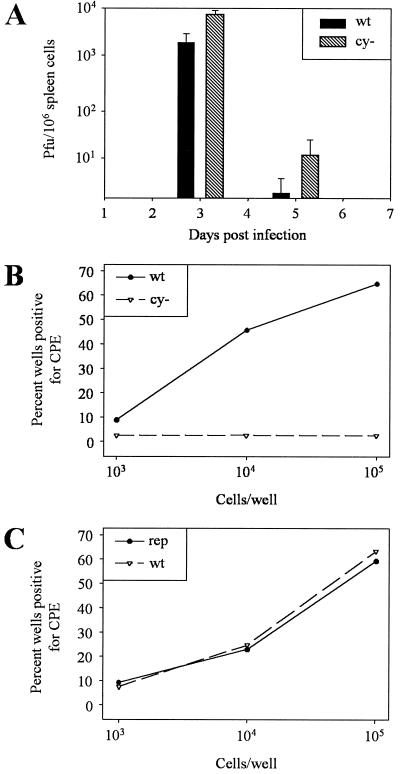
The use of MHV68 as a model system for other gammaherpesviruses allows analysis of the phenotype of the cyclin-deleted virus in vivo. BALB/c mice were infected with 106 PFU of MHV68wt, MHV68cy−, and MHV68rep. Splenic titers of virus were determined at 3, 5, and 7 days after intraperitoneal injection. Splenocytes were disrupted by sonication to release intracellular virus. Sonicates were then assayed on monolayers of REFs, and plaques were stained with X-Gal 6 days after infection. Titers of MHV68cy− drop more quickly than MHV68wt levels, becoming undetectable by 7 days postinfection (Fig. 4A). Repair of the cyclin deletion restores titers to those of MHV68wt, indicating that v-cyclin is required for efficient acute infection.
FIG. 4.
Analysis of MHV68cy− in acute infection. (A) Titers of sonicated spleen cells from BALB/c mice infected with 106 PFU of MHV68wt, MHV68cy−, or MHV68rep were determined at 3, 5, and 7 days postinfection. (B) Titers of sonicated organs from SCID mice infected with 104 PFU of MHV68cy− and MHV68wt in a mixture were determined 7 days postinfection.
One explanation for the deficiency of MHV68cy− in acute infection in light of the efficient replication in cell culture is that the deficiency is immunologically mediated. To address this possibility, we infected SCID mice with MHV68cy− in a mixture of 104 PFU MHV68wt and 104 PFU MHV68cy− viruses (Fig. 4B). After 7 days, splenic titers of MHV68cy− were 13% of titers of MHV68wt. The titer of MHV68cy− in other organs was consistently lower than that of MHV68wt. This provides a qualitative assessment of the nature of the replication deficiency, which supports the observation that MHV68cy− is deficient in acute infections and that this deficiency is inherent to the virus and is not immunologically mediated by mechanisms absent in the SCID mouse, namely T and B cells.
Cyclin-deleted virus cannot efficiently reactivate ex vivo.
We wished to study the effect of the absence of the v-cyclin gene on establishment and reactivation from latency. Based on analogy to studies of latency with murine cytomegalovirus, we first needed to establish an equivalent acute infection (30). We infected mice with 100 times more MHV68cy− than MHV68wt (a mixture of 5 × 106 PFU of MHV68cy− and 5 × 104 PFU of MHV68wt). Three days postinfection there was an average of 104 PFU/106 spleen cells of MHV68cy− and 3.0 × 103 PFU of MHV68wt virus/106 spleen cells. By 5 days, the titers had dropped significantly but the titers of MHV68cy− remained higher (Fig. 5A). We were then prepared to analyze the ability of MHV68cy− to establish and reactivate from a latent infection.
FIG. 5.
Analysis of MHV68cy− reactivation from latency. (A) After infection with a mixture of 5 × 106 PFU of MHV68cy− and 5 × 104 PFU of MHV68wt, acute infection titers were similar. (B) Splenocytes were isolated from spleens of BALB/c mice 28 days after infection. Cells were serially diluted as 103, 104, and 105 cells/well in 96-well plates over monolayers of REFs. After 3 weeks, titers of supernatants from CPE-positive wells were then determined, and supernatants were stained with X-Gal to differentiate between MHV68wt and MHV68cy−. Plates were then scored for percent wells positive for MHV68wt and MHV68cy−. (C) To compare MHV68rep to MHV68wt, mice were infected with 106 PFU of MHV68rep and MHV68wt per ml separately, and spleen cells were explanted and scored for percent wells positive for CPE.
The operational definition of latency requires the absence of preformed infectious virus in an assay of high sensitivity along with the ability to reactivate virus from explanted cells. To exclude the possibility of persisting infectious virus, we analyzed spleens at 28 days after infection and tested all samples containing putative latent virus for infectious virus in an assay of high sensitivity. Since ruptured cells cannot reactivate latent virus, disruption of cells allows determination of preexisting infectious virus. The process of sonication employed reduces the titer of the introduced infectious virus by 80%. In order to compensate for this, we sonicated between 30 and 100 times more spleen cells than were examined for reactivation from latency. In all cases, we and others detected no infectious virus in spleens at 28 days after infection (55). CPE observed from spleen cell explants was therefore thought to be attributable to viral reactivation.
In order to assay for latency, spleen cells from mice were explanted over monolayers of REFs. Spleens from mice infected with a mixture of 5 × 106 PFU of MHV68cy− and 5 × 104 PFU of MHV68wt were analyzed. Spleen cells were plated in 10-fold dilutions of 103, 104, and 105 cells per well. Titers of supernatants from wells showing CPE were determined, and plaques were stained for β-Gal expression (Fig. 5B). MHV68wt and MHV68rep reactivate in a cell dose-response manner while reactivation of MHV68cy− was rarely detected at any dose of spleen cells. In four independent experiments, a total of 250 wells positive for CPE were tested for the presence of MHV68cy−. While 99.2% of the wells containing reactivatable virus were MHV68wt, only 2 wells (0.8%) reactivated MHV68cy−. Mice infected separately with MHV68wt and MHV68cy− yielded similar results (data not shown), indicating that complementation between the viruses was not occurring. Furthermore, complementation would have resulted in more reactivation of MHV68cy−, which was not seen. To determine whether the repair of cyclin would restore the ability of the virus to reactivate from latency, mice were infected with 106 PFU of MHV68wt and 106 PFU of MHV68rep. Spleen cells were examined 28 days after infection, and reactivation was assessed as described above. MHV68rep reactivated in a dose-response manner at a similar frequency as spleen cells from mice infected with MHV68wt (Fig. 5C).
To address the possibility that reactivating virus may represent MHV68cy− virus that had deleted the β-Gal gene, we amplified v-cyclin with PCR on DNA extracted from 19 pools of reactivating virus from mice infected with 5 × 106 PFU of MHV68cy− and 5 × 104 PFU of MHV68wt. v-cyclin was amplified from all samples tested, but not from cells infected with MHV68cy−, indicating that if deletion of β-Gal occurs during reactivation, it is a rare event (data not shown). The cyclin homologue appears to be required for efficient reactivation from latency.
Cyclin-deleted virus can establish latency.
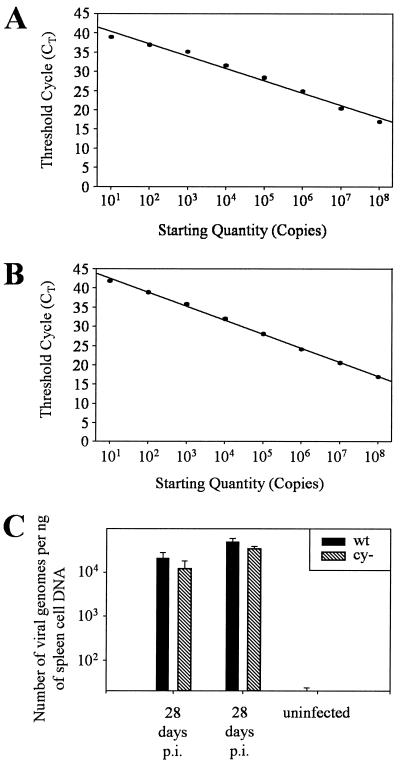
The diminished frequency of reactivation of MHV68cy− could be explained by a deficiency in the establishment of latency or reactivation from latency. To determine if latency was established equivalently by MHV68wt and MHV68cy− under the conditions we used to achieve equivalent acute infections, we employed a real-time PCR method. This method employs flourescently labeled oligonucleotides attached to a quencher that suppresses fluorescence when hybridized to DNA. When the probe is specifically bound to the template it will be released by the exonuclease activity associated with polymerase. Fluorescence can then be quantitatively detected so that the intensity of fluorescence corresponds to the quantity of template, and quantities can be interpolated from a standard curve. To quantitate the viral genomes we utilized a primer and probe set specific to v-cyclin to detect MHV68wt, a set specific to the β-Gal gene to detect MHV68cy−, and a set specific to the double-copy murine RNase P gene, to which viral genomes can be normalized. Standard curves were generated for v-cyclin (Fig. 6A) and β-Gal (Fig. 6B) with known quantities of DNA. The efficiency of amplification of both templates appears to be comparable based on the similar slope of the standard curves. Values for unknowns were then derived from these standard curves. Absolute viral genome copy number was normalized to that of the host genome to generate data shown in Fig. 6C as viral genomes per nanogram of cellular DNA. PCR was performed on DNA extracted from spleen cells from mice 28 days after infection with a mixture containing 100 times as much MHV68cy− as MHV68wt and compared to uninfected spleen cell DNA. At 28 days postinfection, the number of MHV68wt and MHV68cy− genomes was approximately equivalent in two independent experiments. These numbers reflect the acute splenic infection titer data well and predict that with increased MHV68cy− input, MHV68cy− can efficiently establish latency but has a defect in reactivation from latency.
FIG. 6.
Equivalent numbers of MHV68wt and MHV68cy− genomes are present in latently infected spleen cells. Real-time PCR was used to generate a standard curve from CT values of serially diluted plasmids for detection of MHV68wt (v-cyclin [A]) and MHV68cy− (β-Gal [B]). DNA was extracted from spleen cells of mice 28 days postinfection with 5 × 106 PFU of MHV68cy− and 5 × 104 PFU of MHV68wt and from uninfected mice. Copy number was determined from standard curves and normalized to the quantity of genomic DNA determined by amplification of RNase P and then represented as the number of genomes per nanogram of spleen cell DNA. Data shown were generated from two independent experiments.
Stimulation of cellular cyclin D2 expression cannot substitute for the absence of MHV68 cyclin in reactivation from latency.
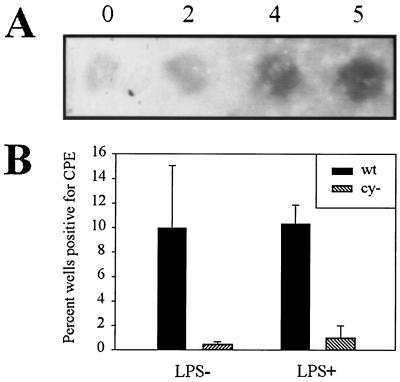
The virus may encode the cellular cyclin homologue to regulate the timing of cyclin expression. Alternatively, the function or properties of v-cyclin may be qualitatively different than those of cellular cyclin, as has been described for the cyclin encoded by HHV8 (15, 46). To address this question, we asked whether appropriately timed cellular cyclin expression would stimulate MHV68cy− reactivation. Since it has been shown previously that LPS stimulates expression of cyclin D2 in B cells (47) and MHV68 latently infects primarily B cells in the spleen (49; our unpublished observations), we stimulated cellular cyclin D2 expression by culturing latently infected spleen cells in the presence of LPS. A volume of 106 spleen cells obtained 28 days postinfection with a mixture of 5 × 106 PFU of MHV68cy− and 5 × 104 PFU of MHV68wt were cultured in the presence of LPS. Cellular cyclin D2 expression was shown to increase after treatment with LPS (Fig. 7A). After CPE was observed, the titer of the virus from the supernatant was determined, and the plaques were stained with X-Gal. Reactivation of MHV68cy− was virtually absent in the presence or absence of LPS. Reactivation of MHV68wt was not substantially influenced by LPS stimulation of splenocytes. This provides a preliminary indication that the function of v-cyclin cannot be substituted by appropriately timed cellular cyclin D2 expression. It is still possible that v-cyclin can be substituted by cyclin D1 or D3.
FIG. 7.
Effect of LPS stimulation on MHV68cy− reactivation. Spleen cells from mice infected with a mixture of 5 × 106 PFU of MHV68cy− and of 5 × 104 PFU MHV68wt were cultured in the presence or absence of LPS. RNA from spleen cells was extracted at the designated numbers of days after explantation. (A) An RNA dot blot from these samples was probed with murine cyclin D2. (B) The effect of LPS on reactivation was determined by scoring percent of wells positive for CPE attributable to MHV68wt or MHV68cy−.
DISCUSSION
Most gammaherpesviruses, with the notable exception of EBV (which upregulates cellular cyclin) encode homologues to cyclin D (2, 28, 32, 51; Chang et al., Letter, Nature). This observation indicates that D-type cyclins may have an important role in the biology of the virus that the host cyclin D cannot provide. There are several possible roles for v-cyclin that can be envisioned. Cyclin may be important in the lytic infection by stimulating progression into S phase, which would increase expression of host proteins required for viral replication. Our data indicate clearly that in tissue culture infection this is not the case. Furthermore, data from our group and others have shown that v-cyclin is expressed with leaky-late kinetics in lytic viral replication (50). If the function of v-cyclin expression is to increase expression of proteins that function in the S phase of cell cycle to optimize viral replication, late expression would seem inappropriate unless the v-cyclin is a virion protein that acts upon entry into the cell. This raises the possibility of roles for v-cyclin in transformation or latency. Many studies have focused on the role of viral cyclins in transformation by the gammaherpesviruses (8, 12, 20, 50). MHV68 v-cyclin expressed in transgenic mice has been shown to function as an oncogene and transform lymphocytes (50). Although interesting, this observation sheds little light on the biologically relevant role of v-cyclin. Another possibility is that v-cyclin plays a role in latency. Our experiments were designed to examine which of these portions of the life cycle of MHV68 require v-cyclin.
We deleted the v-cyclin gene from MHV68 and studied the phenotype of the mutant virus. We confirmed the deletion by PCR, restriction pattern, and Southern and Northern analyses. Expression of the neighboring bcl-2 homologue was not altered in the recombinant virus. The recombinant virus grew efficiently in cell culture but was deficient in replication in vivo. We infected mice with a mixture with MHV68cy− and MHV68wt to provide a control for each animal. This deficiency appeared to be inherent to the virus and not immunologically mediated at least by T or B lymphocytes since MHV68cy− replicated deficiently in the immunologically deficient SCID mouse.
Work with murine cytomegalovirus indicated that the number of latently infected genomes correlates with the titer of the virus in acute infection (30). In order to create equivalent acute infections, we adjusted the input virus to 100 times more MHV68cy− than MHV68wt. We were thus able to establish an acute infection with MHV68cy− that was comparable to wild-type infection. In order to investigate the role of v-cyclin in reactivation, it was critical to establish that equivalent latent genomes were present. We used quantitative real-time PCR to establish that there were equivalent numbers of latent MHV68cy− as MHV68wt genomes present in latently infected spleen cells. Under these conditions, we observed a dramatic deficiency in the ability of MHV68cy− to reactivate from latency. This phenotype was corrected by repair of the cyclin-deleted virus.
What would be the advantage to the virus to encode cyclin, when the cells it infects contain a functional cyclin? Viral cyclin may be qualitatively different from cellular cyclin. HHV8 and HVS v-cyclins preferentially bind CDK 6 and have extended substrate specificity which includes phosphorylation of a histone protein (15, 22). More recent investigations have demonstrated that HHV8- and HVS-encoded cyclins are resistant to inhibition by the CDK inhibitors, p16, p21, and p27 and that HHV8 cyclin can downregulate p27 (25, 46). Furthermore, HHV8 cyclin activates cyclin A expression in a manner that is distinct from cellular cyclin D or E (12). The usefulness of these alterations for the virus is not clear.
It is possible that MHV68-encoded cyclin may have evolved similar qualities distinct from cellular cyclin. Other possibilities might include an altered pattern of degradation through ubiquitination, or it may interact with proteins which modify its function in unexpected ways. Alternatively, the viral cyclin could be encoded by the virus so that the virus may control the timing of expression. This might be particularly true for reactivation if this event were to occur in quiescent cells unprepared for DNA replication. Our observation that the efficiency of MHV68cy− reactivation is only slightly increased with LPS stimulation doesn't support this hypothesis, at least with respect to cyclin D2. To address this question more precisely, experiments are underway to place the open reading frame of the cellular D-type cyclins under the regulatory elements of v-cyclin in the viral genome.
The timing of cyclin expression is consistent with a role in reactivation. Results from us and others indicate that MHV68 v-cyclin is not expressed during latency and is expressed in lytic infection (37, 52). This contrasts with the expression pattern in HHV8, in which cyclin is expressed in a cell line derived from a latently infected primary effusion lymphoma (11). v-cyclin may have a different expression pattern in tumor cells than in cells in which the virus is normally latent. Genes like cyclin that have described roles in transformation may or may not have been selected in evolution for their transformative properties. If not, they have been selected for a different function in the life cycle of the virus and lead to transformation as a secondary effect or as a result of dysregulation of viral genes.
The role of cell cycle regulatory elements in herpes viral reactivation is not without precedent. Cell cycle proteins may regulate herpesvirus latency and reactivation through distinct mechanisms. The alphaherpesvirus bovine herpesvirus expresses a single RNA during latency, the latency-related gene product. This RNA has been shown to bind to cyclin A and prevent cell cycle progression (33). In HSV, the immediate-early protein Vmw110 is required for efficient reactivation from latency and has also been shown to inhibit progression of transfected cells though mitosis and at the G1/S phase border (16, 24, 57). Apparently paradoxically, it has also been shown to stabilize a D-type cyclin (21). The EBV-encoded gene product Zta, which contributes to the switch from latent to lytic gene expression, inhibits cell cycle progression prior to S phase (6, 10, 23). The apparent discrepancy in the stimulation and arrest of cell cycle could be explained in many cases by the possibility that genes expressed late in G1 could be important for viral replication and reactivation. In any case, cell cycle positioning is an important element in regulating latency and reactivation from latency.
If latency is to have any value as a survival strategy the viral genome must persist intact so that at some later time a productive acute infection can the initiated. Gene products in EBV and HHV8 that trigger reactivation have been identified by utilizing latently infected cell lines (10, 42). While latently infected cell lines are useful for identification of proteins that trigger reactivation, they allow limited conclusions to be made about the role of these proteins in vivo. The observation that cyclin is required for reactivation in MHV68 will allow further investigation into the mechanism of reactivation of the gammaherpesviruses in vivo.
ACKNOWLEDGMENTS
We thank J. Headrick and M. Stanley for excellent technical assistance and helpful discussions.
REFERENCES
- 1.Ajchenbaum F, Ando K, DeCaprio J A, Griffin J D. Independent regulation of human D-type cyclin gene expression during G1 phase in primary human T lymphocytes. J Biol Chem. 1993;268:4113–4119. [PubMed] [Google Scholar]
- 2.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvanitakis L, Yaseen N, Sharma S. Latent membrane protein-1 induces cyclin D2 expression, pRb hyperphosphorylation, and loss of TGF-beta 1-mediated growth inhibition in EBV-positive B cells. J Immunol. 1995;155:1047–1056. [PubMed] [Google Scholar]
- 4.Burns W H, Barbour G M, Sandford G R. Molecular cloning and mapping of rat cytomegalovirus DNA. Virology. 1988;166:140–148. doi: 10.1016/0042-6822(88)90155-9. [DOI] [PubMed] [Google Scholar]
- 5.Cannell E J, Farrell P J, Sinclair A J. Epstein-Barr virus exploits the normal cell pathway to regulate Rb activity during the immortalisation of primary B-cells. Oncogene. 1996;13:1413–1421. [PubMed] [Google Scholar]
- 6.Cayrol C, Flemington E K. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Cesarman E, Nador R G, Bai F, Bohenzky R A, Russo J J, Moore P S, Chang Y, Knowles D M. Kaposi's sarcoma-associated herpesvirus contains G protein-coupled receptor and cyclin D homologs which are expressed in Kaposi's sarcoma and malignant lymphoma. J Virol. 1996;70:8218–8223. doi: 10.1128/jvi.70.11.8218-8223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 10.Countryman J, Jenson H, Seibl R, Wolf H, Miller G. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J Virol. 1987;61:3672–3679. doi: 10.1128/jvi.61.12.3672-3679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duro D, Schulze A, Vogt B, Bartek J, Mittnacht S, Jansen-Durr P. Activation of cyclin A gene expression by the cyclin encoded by human herpesvirus-8. J Gen Virol. 1999;80:549–555. doi: 10.1099/0022-1317-80-3-549. [DOI] [PubMed] [Google Scholar]
- 13.Efstathiou S, Ho Y M, Minson A C. Cloning and molecular characterization of the murine herpesvirus 68 genome. J Gen Virol. 1990;71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- 14.Gibson U E, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 15.Godden-Kent D, Talbot S J, Boshoff C, Chang Y, Moore P, Weiss R A, Mittnacht S. The cyclin encoded by Kaposi's sarcoma-associated herpesvirus stimulates cdk6 to phosphorylate the retinoblastoma protein and histone H1. J Virol. 1997;71:4193–4198. doi: 10.1128/jvi.71.6.4193-4198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris R A, Everett R D, Zhu X X, Silverstein S, Preston C M. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J Virol. 1989;63:3513–3515. doi: 10.1128/jvi.63.8.3513-3515.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 18.Honess R W. Herpes simplex and ‘the herpes complex’: diverse observations and a unifying hypothesis. The eighth Fleming lecture. J Gen Virol. 1984;65:2077–2107. doi: 10.1099/0022-1317-65-12-2077. [DOI] [PubMed] [Google Scholar]
- 19.Jung J U, Choi J K, Ensser A, Biesinger B. Herpesvirus saimiri as a model for gammaherpesvirus oncogenesis. Semin Cancer Biol. 1999;9:231–239. doi: 10.1006/scbi.1998.0115. [DOI] [PubMed] [Google Scholar]
- 20.Jung J U, Stager M, Desrosiers R C. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman P M, Hardwick J M, Sample J, Hayward G S, Hayward S D. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J Virol. 1990;64:1143–1155. doi: 10.1128/jvi.64.3.1143-1155.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomonte P, Everett R D. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G(1) into S phase of the cell cycle. J Virol. 1999;73:9456–9467. doi: 10.1128/jvi.73.11.9456-9467.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann D J, Child E S, Swanton C, Laman H, Jones N. Modulation of p27(Kip1) levels by the cyclin encoded by Kaposi's sarcoma-associated herpesvirus. EMBO J. 1999;18:654–663. doi: 10.1093/emboj/18.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsushime H, Roussel M F, Sherr C J. Novel mammalian cyclins (CYL genes) expressed during G1. Cold Spring Harbor Symp Quant Biol. 1991;56:69–74. doi: 10.1101/sqb.1991.056.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Miller G. Epstein-Barr virus: biology, pathogenesis, and medical aspects. In: Fields B N, Knipe D M, editors. Fields virology. Vol. 2. New York, N.Y: Raven Press, Ltd.; 1990. pp. 1921–1958. [Google Scholar]
- 28.Nicholas J, Cameron K R, Honess R W. Herpesvirus saimiri encodes homologues of G protein-coupled receptors and cyclins. Nature. 1992;355:362–365. doi: 10.1038/355362a0. [DOI] [PubMed] [Google Scholar]
- 29.Rajcani J, Blaskovic D, Svobodova J, Ciampor F, Huckova D, Stanekova D. Pathogenesis of acute and persistent murine herpesvirus infection in mice. Acta Virol. 1985;29:51–60. [PubMed] [Google Scholar]
- 30.Reddehase M J, Balthesen M, Rapp M, Jonjic S, Pavic I, Koszinowski U H. The conditions of primary infection define the load of latent viral genome in organs and the risk of recurrent cytomegalovirus disease. J Exp Med. 1994;179:185–193. doi: 10.1084/jem.179.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roizman B, Carmichael L E, Deinhardt F, de The G, Nahmias A J, Plowright W, Rapp F, Sheldrick P, Takahashi M, Wolf K. Herpesviridae. Definition, provisional nomenclature, and taxonomy. The Herpesvirus Study Group, the International Committee on Taxonomy of Viruses. Intervirology. 1981;16:201–217. doi: 10.1159/000149269. [DOI] [PubMed] [Google Scholar]
- 32.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schang L M, Hossain A, Jones C. The latency-related gene of bovine herpesvirus 1 encodes a product which inhibits cell cycle progression. J Virol. 1996;70:3807–3814. doi: 10.1128/jvi.70.6.3807-3814.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherr C J. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 35.Sherr C J. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 36.Simas J P, Efstathiou S. Murine gammaherpesvirus 68: a model for the study of gammaherpesvirus pathogenesis. Trends Microbiol. 1998;6:276–282. doi: 10.1016/s0966-842x(98)01306-7. [DOI] [PubMed] [Google Scholar]
- 37.Simas J P, Swann D, Bowden R, Efstathiou S. Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J Gen Virol. 1999;80:75–82. doi: 10.1099/0022-1317-80-1-75. [DOI] [PubMed] [Google Scholar]
- 38.Sinclair A J, Palmero I, Holder A, Peters G, Farrell P J. Expression of cyclin D2 in Epstein-Barr virus-positive Burkitt's lymphoma cell lines is related to methylation status of the gene. J Virol. 1995;69:1292–1295. doi: 10.1128/jvi.69.2.1292-1295.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 40.Speck S H, Virgin H W. Host and viral genetics of chronic infection: a mouse model of gamma-herpesvirus pathogenesis. Curr Opin Microbiol. 1999;2:403–409. doi: 10.1016/s1369-5274(99)80071-x. [DOI] [PubMed] [Google Scholar]
- 41.Stewart J P, Usherwood E J, Ross A, Dyson H, Nash T. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J Exp Med. 1998;187:1941–1951. doi: 10.1084/jem.187.12.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sunil-Chandra N P, Arno J, Fazakerley J, Nash A A. Lymphoproliferative disease in mice infected with murine gammaherpesvirus 68. Am J Pathol. 1994;145:818–826. [PMC free article] [PubMed] [Google Scholar]
- 44.Sunil-Chandra N P, Efstathiou S, Arno J, Nash A A. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J Gen Virol. 1992;73:2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 45.Sunil-Chandra N P, Efstathiou S, Nash A A. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol. 1992;73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 46.Swanton C, Mann D J, Fleckenstein B, Neipel F, Peters G, Jones N. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature. 1997;390:184–187. doi: 10.1038/36606. [DOI] [PubMed] [Google Scholar]
- 47.Tanguay D A, Chiles T C. Regulation of the catalytic subunit (p34PSK-J3/cdk4) for the major D-type cyclin in mature B lymphocytes. J Immunol. 1996;156:539–548. [PubMed] [Google Scholar]
- 48.Usherwood E J, Ross A J, Allen D J, Nash A A. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J Gen Virol. 1996;77:627–630. doi: 10.1099/0022-1317-77-4-627. [DOI] [PubMed] [Google Scholar]
- 49.Usherwood E J, Stewart J P, Robertson K, Allen D J, Nash A A. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J Gen Virol. 1996;77:2819–2825. doi: 10.1099/0022-1317-77-11-2819. [DOI] [PubMed] [Google Scholar]
- 50.van Dyk L F, Hess J L, Katz J D, Jacoby M, Speck S H, Virgin H W., IV The murine gammaherpesvirus 68 v-cyclin gene is an oncogene that promotes cell cycle progression in primary lymphocytes. J Virol. 1999;73:5110–5122. doi: 10.1128/jvi.73.6.5110-5122.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Virgin H W, Presti R M, Li X Y, Liu C, Speck S H. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J Virol. 1999;73:2321–2332. doi: 10.1128/jvi.73.3.2321-2332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virgin H W, Speck S H. Unraveling immunity to gamma-herpesviruses: a new model for understanding the role of immunity in chronic virus infection. Curr Opin Immunol. 1999;11:371–379. doi: 10.1016/s0952-7915(99)80063-6. [DOI] [PubMed] [Google Scholar]
- 54.Weck K E, Barkon M L, Yoo L I, Speck S H, Virgin H W., IV Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol. 1996;70:6775–6780. doi: 10.1128/jvi.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weck K E, Kim S S, Virgin IV H W, Speck S H. B cells regulate murine gammaherpesvirus 68 latency. J Virol. 1999;73:4651–4661. doi: 10.1128/jvi.73.6.4651-4661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weck K E, Kim S S, Virgin H W, IV, Speck S H. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J Virol. 1999;73:3273–3283. doi: 10.1128/jvi.73.4.3273-3283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu X X, Chen J X, Young C S, Silverstein S. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J Virol. 1990;64:4489–4498. doi: 10.1128/jvi.64.9.4489-4498.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]