Abstract
Low-level replication of hepatitis C virus (HCV) in cultured lymphoblastoid cells inoculated with H77 serum inoculum led to the appearance of new virus variants containing identical substitutions at three sites within the viral 5′ nontranslated RNA (5′NTR): G107→A, C204→A, and G243→A (N. Nakajima, M. Hijikata, H. Yoshikura, and Y. K. Shimizu, J. Virol. 70:3325–3329, 1996). These results suggest that virus with this 5′NTR sequence may have a greater capacity for replication in such cells, possibly due to more efficient cap-independent translation, since these nucleotide substitutions reside within the viral internal ribosome entry site (IRES). To test this hypothesis, we examined the translation of dicistronic RNAs containing upstream and downstream reporter sequences (Renilla and firefly luciferases, respectively) separated by IRES sequences containing different combinations of these substitutions. The activity of the IRES was assessed by determining the relative firefly and Renilla luciferase activities expressed in transfected cells. Compared with the IRES present in the dominant H77 quasispecies, an IRES containing all three nucleotide substitutions had significantly greater translational activity in three of five human lymphoblastoid cell lines (Raji, Bjab, and Molt4 but not Jurkat or HPBMa10-2 cells). In contrast, these substitutions did not enhance IRES activity in cell lines derived from monocytes or granulocytes (HL-60, KG-1, or THP-1) or hepatocytes (Huh-7) or in cell-free translation assays carried out with rabbit reticulocyte lysates. Each of the three substitutions was required for maximally increased translational activity in the lymphoblastoid cells. The 2- to 2.5-fold increase in translation observed with the modified IRES sequence may facilitate the replication of HCV, possibly accounting for differences in quasispecies variants recovered from liver tissue and peripheral blood mononuclear cells of the same patient.
Persons with chronic hepatitis C virus (HCV) infections of the liver are at a significantly increased risk for cirrhosis and hepatocellular carcinoma. Despite a high level of interest in this virus, the mechanisms responsible for viral persistence are poorly understood, as are many other aspects of the biology of this flavivirus (21). One question that is important to both pathogenesis and persistence is whether HCV undergoes replication in cells outside of the liver. Genomic RNA has been detected in peripheral blood mononuclear cells (PBMC) as well as liver tissue and serum or plasma from infected persons by reverse transcription (RT)-PCR (4, 17–19, 22, 30). However, although a considerable number of studies have focused on the possible presence of the virus in PBMC, many of these reports remain controversial because of uncertainty concerning the strand specificity of putative negative-strand-specific RT-PCR assays used for the detection of viral replicative intermediates (16). Nonetheless, several recent studies using well-validated and highly strand-specific RT-PCR assays have demonstrated the presence of negative-strand RNA in PBMC from infected patients (18, 19, 22). These studies suggest the existence of a potentially important extrahepatic site of replication for HCV, although the magnitude of the pool of virus replicating outside of the liver and the exact nature of the cell types in which this replication may occur remain unknown.
Other data that indirectly support PBMC as an extrahepatic site of replication come from in vitro studies, as several lymphoblastoid cell lines appear to be permissive for HCV replication. Shimizu and colleagues (31–34) extensively characterized the replication of the virus in both B-cell (Daudi) and T-cell (HPBMa10-2 and Molt4) lines, while Kato et al. (14) and Nissen et al. (25) also described the replication of HCV in human T-cell lines (MT-2 and H9, respectively). By sequencing the hypervariable region of the E2 coding segment as well as the viral 5′ nontranslated RNA (5′NTR), Nakajima et al. (23) demonstrated a change in the dominant viral quasispecies in both Daudi cells and HPBMa10-2 cells inoculated with the genotype 1a H77 strain of HCV (34). In both cell lines, a new dominant quasispecies emerged in which there were three identical nucleotide substitutions within the 5′NTR: G107→A, C204→A, and G243→A (hereafter referred to as the A-A-A variant). Quasispecies with this 5′NTR sequence were not detected in the original H77 serum inoculum (23). The parallel selection of the A-A-A variant in long-term cultures of HCV in two different lymphoblastoid cell lines suggests the possibility that these nucleotide substitutions may enhance the replication capacity of the virus in such cells. Thus, these 5′NTR substitutions may reflect a viral phenotype that is particularly well adapted to replication in lymphoid cells. This possibility is further suggested by the fact that Shimizu et al. found the A-A-A variant to be dominant in PBMC (but not liver tissue or serum) collected from chimpanzees that were experimentally inoculated with the H77 inoculum (30). Such a hypothesis is consistent with the observations of other investigators who have also noted differences in dominant HCV quasispecies recovered from serum versus PBMC (17, 20, 24).
Interestingly, the three nucleotide substitutions that differentiate the A-A-A variant from the G-C-G variant that dominates in the H77 inoculum are located within the viral internal ribosome entry site (IRES) (Fig. 1). This highly structured RNA element is responsible for directing the cap-independent initiation of translation of the viral polyprotein (27, 35). The activity of the HCV IRES is critically dependent upon a primary nucleotide sequence, as well as secondary and tertiary RNA structures, within the segment extending from about nucleotide (nt) 44 to the initiator AUG codon located at nt 345 of the genome (11, 13, 27). Since cell type-specific variation in IRES activity has been clearly demonstrated among picornaviruses (both hepatitis A virus [HAV] and poliovirus) (15, 29), the fact that the A-A-A substitutions are located within the IRES suggests that they may have a favorable impact on HCV translation in lymphoid cells. To test this hypothesis, we assessed the translational activities of IRES sequences containing one or more of the A-A-A variant 5′NTR substitutions in different human cell lines. Our results indicate that these substitutions do in fact specifically enhance HCV translation in some lymphoblastoid cell lines, a finding that may have broad significance for HCV pathogenesis as well as for the molecular mechanisms that control the internal initiation of translation on the HCV genome.
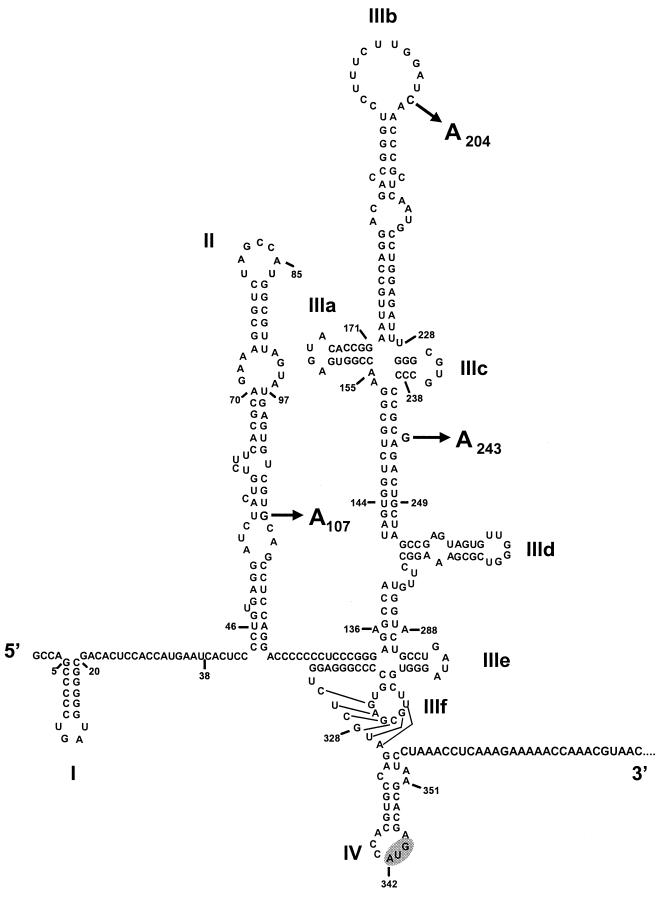
FIG. 1.
Predicted secondary and tertiary RNA structures within the 5′NTR of virus strain H77 (11, 27). The AUG codon at nt 342 (highlighted) is located at the 5′ end of the long open reading frame and is the site at which translation is initiated. The arrows indicate the positions of nucleotide substitutions identified at positions 107, 204, and 243 in HCV sequences amplified from infected Daudi and HPBMa10-2 cells (see Table 1) (23).
MATERIALS AND METHODS
Plasmids.
Using standard techniques, we constructed a plasmid (pRL-HL) containing a dicistronic transcriptional unit under the control of a composite cytomegalovirus (CMV)-T7 promoter (from pRC-CMV; InVitrogen) (12) (Fig. 2). Transcripts produced from this vector contain an upstream cistron that encodes Renilla luciferase and a downstream cistron representing an in-frame fusion of the first 66 nts of the HCV polyprotein-coding region with the firefly luciferase sequence, separated by a sequence corresponding to the 5′NTR of the genotype 1b virus, HCV-N (GenBank accession number AF139594) (3). Thus, the firefly luciferase reporter protein expressed from this transcript is under the translational control of the HCV IRES, while the upstream Renilla luciferase is translated by canonical cap-dependent translation mechanisms. Seven additional plasmids were subsequently generated by introducing into the IRES sequence one or more of the nucleotide substitutions identified within H77 viral RNA amplified from infected lymphoblastoid cells (23) (Table 1). Mutagenesis was accomplished by a PCR-based strategy. The IRES segments of the mutated plasmids were subsequently sequenced to verify that no other mutations had been introduced.
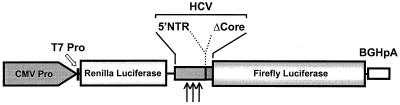
FIG. 2.
Organization of the transcriptional units present in the plasmids used in this study. Transcription is initiated under the control of a composite CMV-T7 promoter (Pro). The upstream cistron encodes Renilla luciferase and is translated by a cap-dependent mechanism in transfected cells, while a downstream cistron encoding firefly luciferase is translated under the control of the HCV IRES. The HCV sequence within the intercistronic space represents the 5′ 407 nts of the HCV genome, corresponding to the entire 5′NTR and 66 nts of the core protein-coding segment (ΔCore) of a genotype 1b virus fused in frame to firefly luciferase. The bovine growth hormone polyadenylation signal (BGHpA) is located downstream of the firefly sequence. Arrows indicate the locations of the nucleotide substitutions introduced into the IRES sequence.
TABLE 1.
Reporter plasmids containing HCV IRES sequences with nucleotide substitutions identified in lymphoblastoid cells infected with H77 virus
| Sequence designation
|
Nucleotide substitution at base position:
|
Quasispecies distributiona
|
|||||
|---|---|---|---|---|---|---|---|
| Plasmid | Nakajima et al. (23)b | 107 | 204 | 243 | H77 inoculum | HPBMa10-2 cells | Daudi cells |
| G-C-Gc | NC1 | G | C | G | 7/8 | ||
| G-A-A | NC6 | A | A | 1/8 | |||
| A-A-A | NC7 | A | A | A | 12/12 | 10/11 | |
| A-C-A | NC9 | A | A | 1/11 | |||
| G-A-G | A | ||||||
| G-C-A | A | ||||||
| A-C-G | A | ||||||
| A-A-G | A | A | |||||
Number of clones with the indicated sequence/number of all clones recovered from the source (23). HPBMa10-2 cells were infected for 193 days and Daudi cells were infected for 308 days prior to RT-PCR amplification and isolation of sequences.
Designation of quasispecies sequence identified in H77 inoculum or in virus recovered from infected lymphoblastoid cells by Nakajima et al. (23).
Dominant wild-type virus quasispecies in H77 inoculum.
In vitro translation.
Plasmids were linearized with ApaI (New England Biolabs), and runoff transcripts were synthesized using bacteriophage T7 RNA polymerase (Promega). One microgram of RNA synthesized from each plasmid was used to program translation in 25 μl of rabbit reticulocyte lysate (Promega). Following separation of the reaction products by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), the amount of 35S-methionine-labeled protein product was quantified by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, Calif.).
Cell lines.
Human cell lines were obtained from the American Type Culture Collection. These included Huh-7, a cell line derived from a hepatocellular carcinoma; Bjab and Raji, B-cell lines derived from Burkitt's lymphoma; Molt4 and Jurkat, T-cell lines derived from acute lymphoblastic leukemia and T-cell leukemia, respectively; KG-1, a myeloblastic cell line; HL-60, a promyelocytic cell line; and THP-1, a monocytic cell line. HPBMa10-2 cells were obtained from the Laboratory of Infectious Diseases, National Institutes of Health, Bethesda, Md. With the exception of Huh-7, these cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, and 1% glutamine at 37°C in a 5% CO2 atmosphere. Cells were passed twice weekly at the appropriate dilution for exponential growth. Huh-7 cells were maintained in minimal essential medium supplemented with 10% fetal calf serum, glutamine, and 1% penicillin-streptomycin at 37°C in a 5% CO2 atmosphere. Medium components were purchased from Gibco BRL.
DNA transfection.
Suspension cell cultures were transfected with plasmid DNA by electroporation with a Gene Pulser II (Bio-Rad, Hercules, Calif.) apparatus equipped with a capacitance extender and a pulse controller. Conditions were optimized for each suspension cell line. For electroporation, 20 μg of DNA was incubated with cells in a 0.4-cm cuvette for 10 min at room temperature. Bjab, HPBMa10-2, and THP-1 cells were resuspended at 5 × 106 to 7.5 × 106 cells/300 μl of complete medium and pulsed once with 300 V and 950 μF. Raji cells, resuspended at 10 × 106 cells/300 μl of complete medium, and Jurkat cells, resuspended at 5 × 106 cells/300 μl, were pulsed once with 260 V. Molt4 cells, resuspended at 10 × 106 cells/300 μl of HEPES buffer (10 mM HEPES [pH 7.2], 150 mM NaCl, 5 mM CaCl2), were pulsed once with 400 V and 950 μF. KG-1 cells were resuspended at 10 × 106 cells/500 μl of complete medium and pulsed once with 300 V. HL-60 cells were resuspended at 10 × 106 cells/800 μl of complete medium and pulsed once with 500 V. After electroporation, cells were allowed to recover for 5 min at room temperature, diluted into 6 ml of complete medium, and kept at 37°C in a 5% CO2 atmosphere for 48 h.
Monolayer cultures of Huh-7 cells were transfected with a cationic lipid-DNA complex. Cells were grown in six-well plates until 80 to 90% confluent. One microgram of DNA was added to 100 μl of Opti-MEM (Gibco BRL), mixed with 15 μl of Lipofectin (Gibco BRL) diluted in 100 μl of Opti-MEM, and kept for 15 min at room temperature. The cells were washed twice and overlaid with 0.8 ml of Opti-MEM and then with the DNA-Lipofectin mixture. The cells were incubated at 37°C (5% CO2) for approximately 24 h. The transfection mixture was removed and replaced with 2 ml of complete medium, and the cells were cultured for an additional 24 h.
Luciferase assays.
The enzymatic activity of reporter proteins was quantified using a dual-luciferase reporter assay system (Promega) and a TD 20/20 luminometer (Turner Designs). Briefly, 48 h following transfection, cells were collected by centrifugation and lysed in 50 to 100 μl of 2× passive lysis buffer (Promega). A 5- to 20-μl aliquot of this lysate was placed in the luminometer, which was programmed to deliver sequentially 100 μl of substrate specific for each luciferase: beetle luciferin with ATP and magnesium, or coelenterazine for the firefly and Renilla enzymes, respectively. Light emission was quantified 3 s after injection and integrated over a 12-s interval. The light emission background was determined with mock-transfected cells.
RESULTS
Cell culture passage-related nucleotide substitutions within the IRES do not alter translation efficiency in rabbit reticulocyte lysates.
The apparent selection of HCV variants with modified 5′NTR sequences in cultured lymphoid cells (23), coupled with the demonstration of identical 5′NTR sequences in PBMC collected from infected chimpanzees (30), suggests that virus with these nucleotide substitutions may have an enhanced capacity for replication in these types of cells. Furthermore, the location of these nucleotide substitutions within the IRES (Fig. 1) (27) suggests that they may enhance the internal initiation of translation on the viral RNA within these cells. To test this hypothesis, we constructed a series of plasmids containing dicistronic transcriptional units under the control of a composite CMV-T7 promoter. The RNA transcripts produced from these plasmids contained two reporter protein sequences (Renilla and firefly luciferases) separated by an HCV sequence representing the 5′NTR and the first 66 nts of the core protein-coding sequence (Fig. 2). Different constructs were created that contained the base composition of the wild-type 5′NTR sequence of the dominant quasispecies in the H77 inoculum (23) or various combinations of the nucleotide substitutions identified in virus recovered from infected lymphoblastoid cells at nts 107, 204, and 243 of the HCV genome (Fig. 1). For simplicity, these constructs were labeled according to their base composition at these loci. Thus, the dominant wild-type sequence in the H77 inoculum, termed “NC1” by Nakajima et al. (23), is represented by the G-C-G construct in Table 1.
Of the eight plasmids constructed, four represent IRES sequences that were identified in various quasispecies from either the H77 inoculum or lymphoblastoid cell-passaged virus in the studies by Nakajima et al. (23) (G-C-G, G-A-A, A-A-A, and A-C-A) (Table 1). The remaining four constructs contain combinations of these nucleotide substitutions that were not observed in these cell culture studies (G-A-G, G-C-A, A-C-G, and A-A-G). The background 5′NTR sequence in each of these constructs was that of the genotype 1b virus, HCV-N (3). This sequence differs from that of the dominant genotype 1a H77 variant (13, 23) by a total of 5 nts (nts 11 to 13 and nts 34 and 35), all of which are situated 5′ of the IRES (13, 27). Although the substitutions at nts 34 and 35 do have an impact on translation efficiency, this effect is due to a long-range RNA interaction outside of the IRES. These substitutions have no influence on translation unless the downstream RNA contains the nearly complete core protein-coding sequence (13). The sequences of the minimal functional IRES domains (nts 44 to 345) are identical in HCV-N and the dominant H77 quasispecies.
Rabbit reticulocyte lysates were programmed for translation with runoff T7 transcripts prepared from these plasmids as described in Materials and Methods. The products of these reactions were separated by SDS-PAGE and subjected to PhosphorImager analysis (Fig. 3A). Both the 61-kDa firefly luciferase protein and the smaller, 36-kDa Renilla luciferase protein were readily apparent in the products of each reaction. Moreover, the quantity of the firefly protein produced from each RNA transcript appeared to be relatively constant in relation to the amount of Renilla protein produced. These results suggest that the inclusion of the nucleotide substitutions shown in Table 1 had no dramatic effect on either quantitative or qualitative aspects of IRES-directed translation. This conclusion was confirmed by quantifying the reporter protein activities expressed from the upstream cistron and downstream, IRES-controlled, cistron of these dicistronic transcripts using specific enzyme assays (Fig. 3B). For this analysis, the proportional abundance of the firefly luciferase and Renilla luciferase activities (i.e., the quantity of firefly luciferase per Renilla luciferase light unit) produced by the G-C-G (wild-type) transcript was arbitrarily assigned a value of 1.0 to facilitate comparisons between the different IRES sequences. The results confirmed that there was little difference in the translational activities of these IRES sequences in reticulocyte lysates. We conclude from this experiment that the nucleotide substitutions that are selected for during passage of virus in lymphoblastoid cells do not enhance the efficiency of viral translation in rabbit reticulocyte lysates, when present either as single nucleotide substitutions or in combination with each other. In replicate experiments, only G-A-G displayed a statistically significant difference in translational activity (−35% ± 6% standard error [SE]) compared with G-C-G (P < 0.05; Student's t test). Notably, none of the quasispecies identified in infected lymphoblastoid cells contained the IRES sequence represented by the G-A-G construct (Table 1) (23).
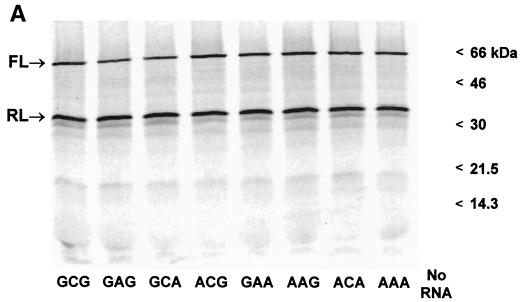
FIG. 3.
In vitro translation of synthetic dicistronic RNAs containing HCV IRES variants. (A) PhosphorImager analysis after SDS-PAGE of representative products of cell-free translation reactions carried out with micrococcal nuclease-treated rabbit reticulocyte lysates. Products are identified on the left, and the positions of molecular mass markers are shown on the right. FL, firefly luciferase; RL, Renilla luciferase. No RNA, lysate programmed with no RNA. (B) Quantitation of the enzymatic activities of reporter proteins produced in cell-free translation reactions. For each transcript, the relative activity of the HCV IRES was calculated by determining the ratio of the firefly luciferase activity produced in a reticulocyte lysate (translated under the control of the IRES) to Renilla luciferase activity (translated from the upstream cistron of the same RNA molecule). The results are plotted for each transcript as the percent change from the ratio obtained with the wild-type H77 IRES (G-C-G variant). Results shown represent means obtained in four separate experiments (each involving two replicate reactions for each transcript) ± SE.
Nucleotide substitutions within the IRES enhance translation in human lymphoblastoid cell lines.
To determine whether the nucleotide substitutions might enhance translation in some lymphoblastoid cells, we compared the translational activities of the wild-type G-C-G IRES and the dominant cell-passaged A-A-A IRES in a variety of human cell types transfected with plasmid DNAs. Since there are significant differences in the efficiency with which these various cell lines can be transfected (data not shown), we determined the relative translational efficiencies of these IRES elements by comparing the proportional abundance of the firefly luciferase and Renilla luciferase activities expressed by each within individual transfected cell cultures. This approach corrects for potential variation in transcript abundance and is identical to the approach used to compare IRES activities in the cell-free translation reactions described above (Fig. 3B).
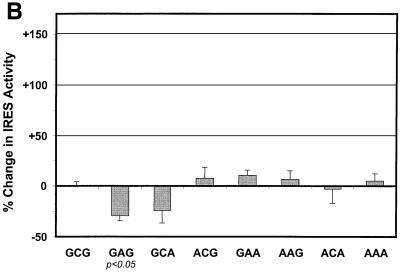
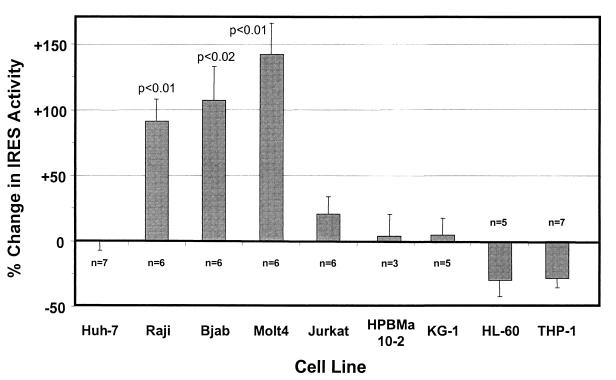
As shown in Fig. 4, these experiments involved a total of nine different cells lines, including two B-cell lines (Bjab and Raji, both derived from human Burkitt's lymphomas), three T-cell lines (Molt4, Jurkat, and HPBMa10-2), a myeloblastic cell line (KG-1), a promyelocytic cell line (HL-60), and a monocytic cell line (THP-1), in addition to Huh-7 cells, which are derived from a hepatocellular carcinoma. We found significant differences in the activity of the A-A-A IRES relative to the wild-type G-C-G IRES in two of two B-cell lines and in one of three T-cell lines but not in any of the other four cell lines that we studied. The activity of the A-A-A IRES was increased approximately twofold in the B-cell lines Bjab (+107% ± 28%; P < 0.02) and Raji (+91% ± 17%; P < 0.01) and slightly more than twofold in the T-cell line Molt4 (+143% ± 24%; P < 0.01). Although of a relatively small magnitude, these differences were both reproducible in multiple experiments and statistically significant. There was only a minimal increase in the activity of the A-A-A IRES in Jurkat cells (+21% ± 13%), which was not statistically significant, and there was no increase in HPBMa10-2 cells, even though both cell lines are derived from T cells. None of the four nonlymphoblastoid cell lines demonstrated any significant increase in the activity of the A-A-A IRES (Fig. 4). Translational activity was slightly reduced in HL-60 and THP-1 cells, but the difference between A-A-A and G-C-G did not reach statistical significance, and there was no difference in Huh-7 cells (−1% ± 6%).
FIG. 4.
Translational activities of HCV IRES variants in different human cell lines. Duplicate cultures of each cell type were electroporated with plasmids expressing dicistronic RNAs containing the G-C-G IRES or the A-A-A IRES. At 48 h after transfection, cells were harvested and luciferase activities were measured. The activity of the IRES in each transcript was determined by comparing firefly luciferase activity with Renilla luciferase activity as described in the legend to Fig. 3B. The results are plotted as the percent change in this ratio in cells transfected with the A-A-A plasmid versus those transfected with the wild-type H77 G-C-G plasmid. Results shown represent the means obtained in five to seven replicate paired cultures for each cell type ± SE.
We conclude from these results that the A-A-A substitutions result in a modest but significant enhancement of the translational activity of the HCV IRES in some lymphoblastoid cell lines. These results support the hypothesis that these nucleotide substitutions may be selected during passage of the virus in cultured B or T cells because they enhance IRES-directed translation in these cells. However, it is important to note that we were unable to demonstrate a translational advantage conferred by the A-A-A substitutions in HPBMa10-2 (Fig. 4), one of the two cell lines used for the propagation of HCV by Nakajima et al. (23) (see Discussion). We were unable to obtain a transfection efficiency sufficient for this experiment with the other cell line, Daudi.
Each of the three cell passage-related nucleotide substitutions contributes to enhanced IRES activity in B cells.
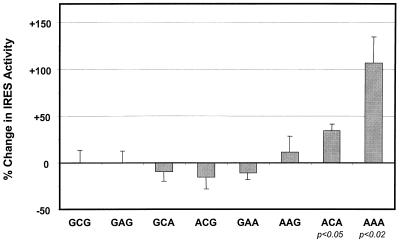
To determine which of the three substitutions in the A-A-A IRES is responsible for its increased translational activity in lymphoblastoid cells, we transfected Bjab cells with each of the eight plasmids shown in Table 1. These plasmids direct the transcription of RNAs that contain various permutations of the IRES substitutions. Only two of the constructs, A-C-A and A-A-A, showed a statistically significant increase in IRES efficiency (+33% ± 6% SE [P < 0.05] and +107% ± 28% SE [P < 0.02], respectively) compared to the wild-type G-C-G construct (Fig. 5). This result indicates that the increase in IRES activity that we observed with Bjab cells (Fig. 4) requires the presence of each of the three substitutions: A107, A204, and A243. There was no statistically significant increase in IRES activity in the absence of both adenosine substitutions at nts 107 and 243; the additional adenosine substitution at nt 204 was required for maximal efficiency in these cells. These results are in contrast to those obtained with rabbit reticulocyte lysates, in which the same nucleotide substitutions conferred no increase in translational activity (Fig. 3), and indicate that these substitutions have a cooperative effect on translation. Interestingly, the A-C-A and A-A-A constructs contained the combinations of substitutions that were identified most often among the HCV quasispecies present in infected lymphoblastoid cell lines (Table 1), with the A-A-A sequence being dominant (22 of 23 clones examined at 193 to 308 days after infection of Daudi cells) (23).
FIG. 5.
Effect of individual nucleotide substitutions and combinations of substitutions in Bjab cells. See the legends to Fig. 3B and 4 for details. The results shown represent the means obtained in three separate experiments (each involving two replicate cultures transfected with each clone, except for A-C-A [n = 4] and A-A-A [n = 6]) ± standard errors.
DISCUSSION
Previous studies indicated that certain established human lymphoblastoid cell lines are permissive for low-level replication of HCV (14, 25, 31–34). Among the most compelling data for HCV replication in two such cell lines, Daudi and HPBMa10-2, is the observation that the quasispecies diversity of the original HCV inoculum (H77 serum from patient H) was significantly altered during the passage of virus in these cells (Table 1) (23). The parallel selection of virus containing identical nucleotide substitutions within the IRES (G107→A, C204→A, and G243→A) in both cell types suggested that these substitutions may promote replication of the virus in lymphoblastoid cells. Here, we present data from a completely different line of investigation that provides further support for this hypothesis. Our results indicate that these 5′NTR substitutions result in moderate but highly reproducible and statistically significant increases in the activity of the HCV IRES in some, but not all, lymphoblastoid cells. We studied a total of nine different cell lines and found that these substitutions enhanced translation in three of five lymphoblastoid cell lines but not in any of the four nonlymphoblastoid cell lines tested or in cell-free translation assays carried out with reticulocyte lysates in vitro (Fig. 4). The fact that we observed significant increases in the activity of the A-A-A IRES in three of five lymphoblastoid cell lines suggests that this cell type-specific enhancement of HCV translation may be a feature that is broadly shared by many cells of lymphoid origin.
It was surprising to find no increase in the activity of the A-A-A IRES in HPBMa10-2 cells, since this was one of the two cell lines for which Nakajima et al. (23) described the selection of the A-A-A quasispecies during the replication of the H77 virus. Nonetheless, there are at least two possible explanations for this finding. First, the translational advantage conferred by the A-A-A substitutions within the HCV IRES may be dependent upon the abundance of the viral RNA. The reporter transcripts that were generated under the control of the CMV promoter in transfected cells were certainly more abundant than the viral RNA in HCV-infected HPBMa10-2 cells (23, 33), and at a higher RNA abundance other factors could become limiting for IRES-directed translation. This situation could potentially mask a translational advantage of one IRES sequence over another that would be evident at a lower RNA abundance. Thus, the in vivo transfection approach used in the experiments described in this communication may have been insufficiently sensitive to detect a difference in the translational activity of the A-A-A IRES in HPBMa10-2 cells. Alternatively, previous studies have demonstrated significant clonal variation in the permissiveness of HPBMa cells, the progenitor of the clonal HPBMa10-2 cell line, for HCV replication (33). The basis for this variation is unknown, but it could relate to differences in the ability of HPBMa cell clones to support HCV translation. Although the HPBMa10-2 clone was selected originally for its ability to support HCV infection (33), the stability of this phenotype is not well established. Moreover, the HPBMa10-2 cells used for the translation studies described in this communication were not evaluated directly for their ability to support HCV replication. Unfortunately, we could not achieve a sufficient level of transfection efficiency to determine the relative translational activity of the A-A-A IRES in Daudi cells, the other cell line for which Nakajima et al. (23) noted the selection of these nucleotide substitutions. Molt4 cells, for which we found the greatest increase in the translational activity of the A-A-A IRES (Fig. 4), are permissive for HCV replication (33), while nothing is known about the ability of Jurkat cells to be infected with HCV.
Cell type-specific differences in IRES activity have been demonstrated previously for picornaviruses, including, in particular, poliovirus and HAV (7, 10, 15, 29). Mutations within the 5′NTR of HAV that specifically promote viral translation in African green monkey kidney cells are selected for during the adaptation of this virus to growth in these cells (5, 6, 29). Unlike that of HCV, the replication of HAV in cultured cells is sufficiently robust to allow the demonstration of quantitative increases in replication associated with mutations in the 5′NTR that facilitate IRES-directed translation (5–7). Interestingly, in the case of HAV, substantial increases in viral replication can result from only limited enhancement of the efficiency of translation, on the order of that observed with the A-A-A IRES in Bjab or Molt4 cells. With HAV, the cell type-specific differences in IRES activity are likely to reflect at least in part the relative abundance of two cellular proteins that compete for binding to the viral RNA, glyceraldehyde-3-phosphate dehydrogenase and polypyrimidine tract binding protein, and that have opposing effects on the efficiency of IRES-directed translation (8, 28, 36). The HCV IRES differs from picornavirus IRES elements not only in its secondary RNA structure but also with respect to its ability to bind to the 40S ribosome subunit in the absence of either canonical or noncanonical translation initiation factors (26). Nonetheless, several cellular proteins, including polypyrimidine tract binding protein (1, 8), the La autoantigen (2), and heterogeneous nuclear ribonucleoprotein L (9), have been suggested to bind to the HCV 5′NTR and specifically enhance HCV translation. It is likely that the nucleotide differences that distinguish the G-C-G from the A-A-A variants of H77 influence the binding of one or more such proteins to the IRES in a way that optimizes these interactions in cells of lymphoid origin.
Further evidence for the hypothesis that the A-A-A substitutions confer a replication advantage in cells of lymphoid origin comes from the fact that the dominant viral quasispecies identified in PBMC from three chimpanzees infected with the H77 inoculum contained these substitutions (30). In contrast, the dominant viral quasispecies in the serum and liver of these animals contained the G-A-A variant. More recently, we had the opportunity to examine serum and PBMC collected in 1990 and 1995 from the same patient (patient H) who had donated the H77 inoculum in 1977. Strikingly, we found by direct sequencing of amplified cDNA that the dominant quasispecies recovered from PBMC in 1990 and 1995 contained the A-A-A variant (Y. K. Shimizu, unpublished data). The dominant quasispecies present in serum collected in 1995 was G-A-A, but in 1990 it was A-A-A, the putative lymphotropic variant. This latter finding is of particular interest, as it suggests that the dominant quasispecies present in serum in 1990 was virus that was replicated in the PBMC compartment. An alternative interpretation would be that the A-A-A IRES is fully functional in the liver and that there is no adverse selective pressure against this sequence in infected hepatocytes in situ. Such an interpretation would be consistent with the lack of a difference in the activities of the G-C-G IRES and the A-A-A IRES in Huh-7 cells (Fig. 4) but would fail to explain the apparent strong preference for G107 in virus recovered from the liver of chimpanzees (30). Unfortunately, no liver tissue is available from patient H in 1990 to help resolve this issue.
Our results add to previous studies suggesting that the replication of HCV in lymphoid cells may play a role in the pathogenesis of hepatitis C. Further studies are necessary to determine whether steps in the viral life cycle other than translation, such as viral entry and RNA synthesis, also contribute to the ability of certain viral variants to replicate preferentially in lymphoid cells. Unanswered questions concern the relative magnitude of replication in the lymphoid compartment compared with that in the liver, the extent to which viruses traffic between these compartments, and the impact of this on disease pathogenesis.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (U19-AI40035 and RO1-AI32599) and the Texas Advanced Technology Program. H.L. was supported in part by a fellowship from the Association pour la Recherche sur le Cancer.
REFERENCES
- 1.Ali N, Siddiqui A. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J Virol. 1995;69:6367–6375. doi: 10.1128/jvi.69.10.6367-6375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard M R, Abell G, Honda M, Carroll A, Gartland M, Clarke B, Suzuki K, Lanford R, Sangar D V, Lemon S M. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology. 1999;30:316–324. doi: 10.1002/hep.510300137. [DOI] [PubMed] [Google Scholar]
- 4.Chang T T, Young K C, Yang Y J, Lei H Y, Wu H L. Hepatitis C virus RNA in peripheral blood mononuclear cells: comparing acute and chronic hepatitis C virus infection. Hepatology. 1996;23:977–981. doi: 10.1002/hep.510230506. [DOI] [PubMed] [Google Scholar]
- 5.Day S P, Lemon S M. A single base mutation in the 5′ noncoding region of HAV enhances replication of virus in vitro. In: Brown F, Chanock R M, Ginsberg H S, Lerner R A, editors. Vaccines 90: modern approaches to new vaccines including prevention of AIDS. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. pp. 175–178. [Google Scholar]
- 6.Day S P, Murphy P, Brown E A, Lemon S M. Mutations within the 5′ nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J Virol. 1992;66:6533–6540. doi: 10.1128/jvi.66.11.6533-6540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funkhouser A W, Schultz D E, Lemon S M, Purcell R H, Emerson S U. Hepatitis A virus translation is rate-limiting for virus replication in MRC-5 cells. Virology. 1999;254:268–278. doi: 10.1006/viro.1998.9548. [DOI] [PubMed] [Google Scholar]
- 8.Gosert R, Chang K H, Rijnbrand R, Yi M K, Sangar D V, Lemon S M. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol Cell Biol. 2000;20:1583–1595. doi: 10.1128/mcb.20.5.1583-1595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahm B, Kim Y K, Kim J H, Kim T Y, Jang S K. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J Virol. 1998;72:8782–8788. doi: 10.1128/jvi.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haller A A, Stewart S R, Semler B L. Attenuation stem-loop lesions in the 5′ noncoding region of poliovirus RNA: neuronal cell-specific translation defects. J Virol. 1996;70:1467–1474. doi: 10.1128/jvi.70.3.1467-1474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda M, Beard M R, Ping L H, Lemon S M. A phylogenetically conserved stem-loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J Virol. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda M, Kaneko S, Matsushita E, Kobayashi K, Abell G, Lemon S M. Cell cycle regulation of hepatitis C virus IRES-directed translation. Gastroenterology. 2000;118:152–162. doi: 10.1016/s0016-5085(00)70424-0. [DOI] [PubMed] [Google Scholar]
- 13.Honda M, Rijnbrand R, Abell G, Kim D, Lemon S M. Natural variation in translational activities of the 5′ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: evidence for a long-range RNA-RNA interaction outside of the internal ribosomal entry site. J Virol. 1999;73:4941–4951. doi: 10.1128/jvi.73.6.4941-4951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato N, Nakazawa T, Mizutani T, Shimotohno K. Susceptibility of human T-lymphotropic virus type I infected cell line MT-2 to hepatitis C virus infection. Biochem Biophys Res Commun. 1995;206:863–869. doi: 10.1006/bbrc.1995.1123. [DOI] [PubMed] [Google Scholar]
- 15.La Monica N, Racaniello V R. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J Virol. 1989;63:2357–2360. doi: 10.1128/jvi.63.5.2357-2360.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanford R E, Chavez D, Chisari F V, Sureau C. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J Virol. 1995;69:8079–8083. doi: 10.1128/jvi.69.12.8079-8083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laskus T, Radkowski M, Wang L F, Jang S J, Vargas H, Rakela J. Hepatitis C virus quasispecies in patients infected with HIV-1: correlation with extrahepatic viral replication. Virology. 1998;248:164–171. doi: 10.1006/viro.1998.9269. [DOI] [PubMed] [Google Scholar]
- 18.Lerat H, Berby F, Trabaud M A, Vidalin O, Major M, Trepo C, Inchauspe G. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Investig. 1996;97:845–851. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lerat H, Rumin S, Habersetzer F, Berby F, Trabaud M A, Trepo C, Inchauspe G. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood. 1998;91:3841–3849. [PubMed] [Google Scholar]
- 20.Maggi F, Fornai C, Vatteroni M L, Giorgi M, Morrica A, Pistello M, Cammarota G, Marchi S, Ciccorossi P, Bionda A, Bendinelli M. Differences in hepatitis C virus quasispecies composition between liver, peripheral blood mononuclear cells and plasma. J Gen Virol. 1997;78:1521–1525. doi: 10.1099/0022-1317-78-7-1521. [DOI] [PubMed] [Google Scholar]
- 21.Major M E, Feinstone S M. The molecular virology of hepatitis C. Hepatology. 1997;25:1527–1538. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 22.Mihm S, Hartmann H, Ramadori G. A reevaluation of the association of hepatitis C virus replicative intermediates with peripheral blood cells including granulocytes by a tagged reverse transcription/polymerase chain reaction technique. J Hepatol. 1996;24:491–497. doi: 10.1016/s0168-8278(96)80171-1. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima N, Hijikata M, Yoshikura H, Shimizu Y K. Characterization of long-term cultures of hepatitis C virus. J Virol. 1996;70:3325–3329. doi: 10.1128/jvi.70.5.3325-3329.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navas S, Martin J, Quiroga J A, Castillo I, Carreno V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–1646. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissen E, Höhne M, Schreier E. In vitro replication of hepatitis C virus in a human lymphoid cell line (H9) J Hepatol. 1994;20:437. doi: 10.1016/s0168-8278(94)80023-5. [DOI] [PubMed] [Google Scholar]
- 26.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rijnbrand R C A, Lemon S M. Internal ribosome entry site-mediated translation in hepatitis C virus replication. In: Hagedorn C H, Rice C M, editors. The hepatitis C viruses. Berlin, Germany: Springer-Verlag KG; 1999. pp. 85–116. [DOI] [PubMed] [Google Scholar]
- 28.Schultz D E, Hardin C C, Lemon S M. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′ nontranslated RNA of hepatitis A virus. J Biol Chem. 1996;271:14134–14142. doi: 10.1074/jbc.271.24.14134. [DOI] [PubMed] [Google Scholar]
- 29.Schultz D E, Honda M, Whetter L E, McKnight K L, Lemon S M. Mutations within the 5′ nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J Virol. 1996;70:1041–1049. doi: 10.1128/jvi.70.2.1041-1049.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu Y K, Igarashi H, Kanematu T, Fujiwara K, Wong D C, Purcell R H, Yoshikura H. Sequence analysis of the hepatitis C virus genome recovered from serum, liver, and peripheral blood mononuclear cells of infected chimpanzees. J Virol. 1997;71:5769–5773. doi: 10.1128/jvi.71.8.5769-5773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu Y K, Igarashi H, Kiyohara T, Shapiro M, Wong D C, Purcell R H, Yoshikura H. Infection of a chimpanzee with hepatitis C virus grown in cell culture. J Gen Virol. 1998;79:1383–1386. doi: 10.1099/0022-1317-79-6-1383. [DOI] [PubMed] [Google Scholar]
- 32.Shimizu Y K, Iwamoto A, Hijikata M, Purcell R H, Yoshikura H. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc Natl Acad Sci USA. 1992;89:5477–5481. doi: 10.1073/pnas.89.12.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimizu Y K, Purcell R H, Yoshikura H. Correlation between the infectivity of hepatitis C virus in vivo and its infectivity in vitro. Proc Natl Acad Sci USA. 1993;90:6037–6041. doi: 10.1073/pnas.90.13.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimizu Y K, Yoshikura H. Multicycle infection of hepatitis C virus in cell culture and inhibition by alpha and beta interferons. J Virol. 1994;68:8406–8408. doi: 10.1128/jvi.68.12.8406-8408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yi M, Schultz D E, Lemon S M. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J Virol. 2000;74:6459–6468. doi: 10.1128/jvi.74.14.6459-6468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]