SUMMARY
The envelope (E) glycoprotein is the primary target of type-specific (TS) neutralizing antibodies (nAbs) after infection with any of the four distinct dengue virus serotypes (DENV1–4). nAbs can be elicited to distinct structural E domains (EDs) I, II, or III. However, the relative contribution of these domain-specific antibodies is unclear. To identify the primary DENV3 nAb targets in sera after natural infection or vaccination, chimeric DENV1 recombinant encoding DENV3 EDI, EDII, or EDIII were generated. DENV3 EDII is the principal target of TS polyclonal nAb responses and encodes two or more neutralizing epitopes. In contrast, some were individuals vaccinated with a DENV3 monovalent vaccine-elicited serum TS nAbs targeting each ED in a subject-dependent fashion, with an emphasis on EDI and EDIII. Vaccine responses were also sensitive to DENV3 genotypic variation. This DENV1/3 panel allows the measurement of serum ED TS nAbs, revealing differences in TS nAb immunity after natural infection or vaccination.
In brief
Munt et al. uses chimeric DENV3 recombinant viruses to map human monoclonal antibody epitopes and demonstrate preferential E-glycoprotein-domain-specific neutralizing antibody responses in polyclonal sera after natural infection or vaccination. The approach provides a metric for evaluating the relationship between antibody neutralization and protective immunity.
Graphical Abstract
INTRODUCTION
Dengue virus (DENV) is a widespread, arthropod-borne RNA virus1,2 that infects an estimated >390 million people worldwide, resulting in 40,000 fatal cases each year.3,4 Approximately 40% of the world’s population live in areas at risk for DENV infection.5–7 Although DENV typically causes flu-like symptoms, 5% of the symptomatic individuals develop dengue shock syndrome (DSS) or dengue hemorrhagic fever (DHF), which can progress to hemorrhage, shock, and death.8–13 The incidence and geographic range of DENV infections are predicted to increase dramatically due to growing urbanization, human travel, and environmental changes, including global warming, heightening the need for effective countermeasures.14
DENV particles encode two surface membrane (M) gly coproteins, the premembrane (prM) and envelope (E) gly coproteins,15 that play essential roles in virion entry, maturation, assembly, and egress. During virus maturation, prM initially presents in immature, rough particles and is subsequently cleaved by host furin proteases into the pr and M proteins. Cleavage promotes dramatic structural changes in the virion, from immature, rough forms that transition into more infectious, smooth, and mature particles that display novel antigenic sites on the E protein.16 The E protein protomer is essential for viral attachment and entry into the cell.17–19 Structurally, E protein monomers contains three distinct domains (EDs), designated EDI, EDII, and ED-III.20,21 On the mature viral surface, E proteins interact to form 90 homodimers that tightly pack to form a protein E with icosahedral symmetry.22 On the viral E, simple and quaternary structure epitopes, spanning two or more E protein monomers, are targeted by both type-specific (TS) and/or cross-reactive (CR) neutralizing antibody (nAb) responses, making E a critical target for evaluating the design and performance of vaccines.23,24 DENV is comprised four distinct serotypes (DENV1–4), which are further subdivided intogenotypeswithineachserotype.25–28Afteraprimaryinfection, individuals typically develop transient serotype CR immunity to other serotypes29,30 and long-lasting, TS protective Ab responses to the homologous serotype.8,31–35 However, recent findings reveal a more nuanced immune protective repertoire, where reinfections by different genotypes within the same serotype are possible.36–41 After secondary infection with a different serotype, more potent and persistent CR Ab responses are elicited, which appear protective against all four serotypes.42,43 Paradoxically, DHF/DSS is also more likely to occur during secondary DENV infection, most often associated with Ab-dependent enhancement (ADE) mechanisms.44–47 After infection with a new serotype, ADE is likely driven by pre-existing CR, non-nAbs that bind the virus and promote interactions with Fc receptors, enhancing virus replication and activation of target immune cells, host cytokine expression, and disease.48,49
Several DENV tetravalent vaccines are in clinical development, including an US Food and Drug Administration (FDA)-licensed chimeric yellow fever-dengue, live-attenuated, tetravalent dengue vaccine (Dengvaxia/CYD-TDV, Sanofi Pasteur) and 2 live-attenuated vaccines in phase III trials including Takeda’s tetravalent dengue vaccine candidate (Tak-003/005—Takeda) and the National Institutes of Health tetravalent admixture of monovalent vaccines (TV003—NIH), the former recently licensed for use under QDENGA in individuals aged 4 and older in Indonesia and the European Union. Dengvaxia generally induced nAbs to all 4 DENV serotypes for 2 years after vaccination in most individuals, although breakthrough infections still occurred in some vaccinees.50–54 Importantly, some naive, vaccinated children in the Philippines were at a higher risk of developing severe dengue upon subsequent infection.54 Currently, Dengvaxia is licensed in the United States and elsewhere for individuals >9 years old with pre-existing DENV immunity (“preimmune”).55,56 Other live-attenuated vaccine candidates stimulate a serotype-specific immunodominant response (Takeda)57 or more balanced, TS Ab responses in up to 76% of the vaccinees (NIH).58 High-titered total (TS + CR) nAbs represent the traditional correlate for vaccine efficacy and protection. Although defined correlates for DENV protective immunity are complex and still understudied, TS nAb titers represent another potential correlate for protective immunity after DENV infection.1,53,59 In DENV3, EDI, and EDII typically encode potent antigenic sites for DENV3 TS nAbs, whereas EDIII contains weaker TS-neutralizing epitopes.60 However, CR prM and EDII fusion-loop epitopes have been linked to responses associated with ADE. Methods that accurately quantify the kinetics, magnitude, durability, and domain-specific targeting of the TS nAb response in serum may shed new insights into the immunologic basis for breakthrough infection and protective immunity. To address these questions, we developed a panel of DENV1 recombinant viruses that encoded different combinations of EDI, EDII, or EDIII from the DENV3 E glycoprotein. These chimeric viruses replicate efficiently and enable the measurements of serum DENV3 ED-specific neutralization responses across the 2-fold, 3-fold, and 5-fold axes on the virion. Herein, we describe a DENV1/3 reagent panel and provide evidence that the ED targeting of serum TS nAbs not only differs after natural DENV3 infection and vaccination but also varies in a genotype-specific fashion.
RESULTS
Design and recovery of DENV1/3 EDII and EDIII recombinant viruses
The DENV1/3 full ED chimeric panel was developed using a quadripartite reverse genetics system41,61 and features the EDI, EDII, or EDIII residues from the DENV3, G-III, Sri Lanka ’89 strain introduced into the DENV1 (Westpac) E glycoprotein (Figures 1A–1C). In addition to the previously described DENV1/3 EDI recombinant virus that encodes the DENV3 3-fold axis in E,60 we designed DENV1/3 EDII and EDIII recombinant virus chimeras that encode the DENV3 E glycoprotein 2-fold and 5-fold symmetry axes, respectively. Initial attempts to recover a recombinant DENV1 virus that encoded the DENV3 EDII domain were not successful. Recovery of viable DENV1/3 EDII chimeras required the inclusion of a serotype-matched chimeric prM protein, designed to maintain favorable DENV3 prM-EDII interface interactions that likely function in maturation and release. Specifically, residues 1–108 of the DENV3 prM were fused with residues 109–166 of the DENV1 M protein, thereby preserving N- and C-terminal interactions with the appropriate homologous DENV1 and DENV3 E protein residues (Figure S1A). Four DENV1/3 EDII constructs (A–D) were designed and recovered, with between 24 and 38 amino acid residues transplanted from DENV3 into DENV1, with DENV1/3 EDIIC and D encoding increasing numbers of DENV3 residues in the EDI-EDII hinge region (Figure S1B). Of the 4 EDII chimeras, DENV1/3 EDIIA (with 33 DENV3 EDII residues), replicated most efficiently in Vero81 non-human primate (NHP) cells (titer = 1 × 107 FFU/mL) (Figure 2A). The DENV1/3 EDIIA virus retained binding to DENV1 TS monoclonal Abs (mAbs) binding to EDI (human mAbs [hmAbs] 14C10 and 1F4) and EDIII, demonstrating the preservation of native DENV1 structure.62,63 With titers approaching 1.3 × 106 FFU, DENV1/3 EDIIC encodes 37 DENV3 EDII residues and presented fully intact 1F4 and partially intact 14C10 epitopes, respectively. DENV1/3 EDIIB and EDIID, encoding 24 and 38 DENV3 residues, were also recovered but replicated less efficiently.
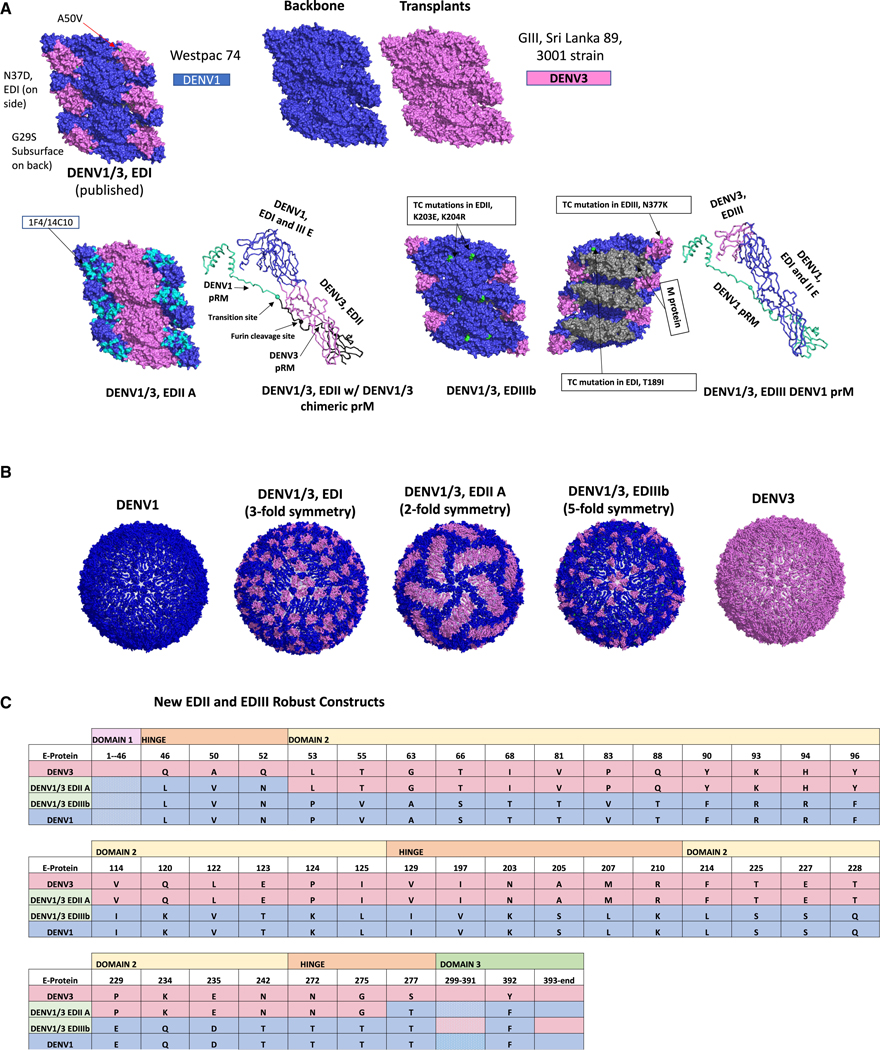
Figure 1. DENV1/3 E glycoprotein recombinant viruses.
(A) Design of DENV1/3 glycoproteins. Structural glycoprotein design. DENV1/3 EDI, EDII, and EDIII chimeras are depicted in a hexameric raft formation with 6 antiparallel E glycoprotein monomers forming 3 parallel dimers (PDB: 3J27, for visualization); DENV1 = dark blue, DENV3 = pink, DENV1 1F4/14C10 epitopes =cyan on the EDII chimera and residue mutations in the EDIII chimera = lime green. The premembrane (prM) protein is pictured to the right of the EDII and EDIII chimeras (PDB: 6IDI, for visualization). For prM, DENV1= black, DENV3 = teal.
(B) DENV1/3 particle symmetry. DENV3 E protein 3-fold, 2-fold, and 5-fold symmetry are represented on the whole virion with DENV1/3 EDI, DENV1/3 EDII, and DENV1/3 EDIII respectively, with DENV3 = pink, DENV1 = dark blue (PDB: 3J6S_*60, for visualization).
(c) Primary sequence variation in chimeric viruses. E glycoprotein DENV3 original encoded changes into DENV1 are represented for DENV1/3 EDII and EDIII chimeras, DENV3 = pink, DENV1 = dark blue.
Figure 2. Phenotypic analysis of DENV1/3 E glycoprotein recombinant viruses.
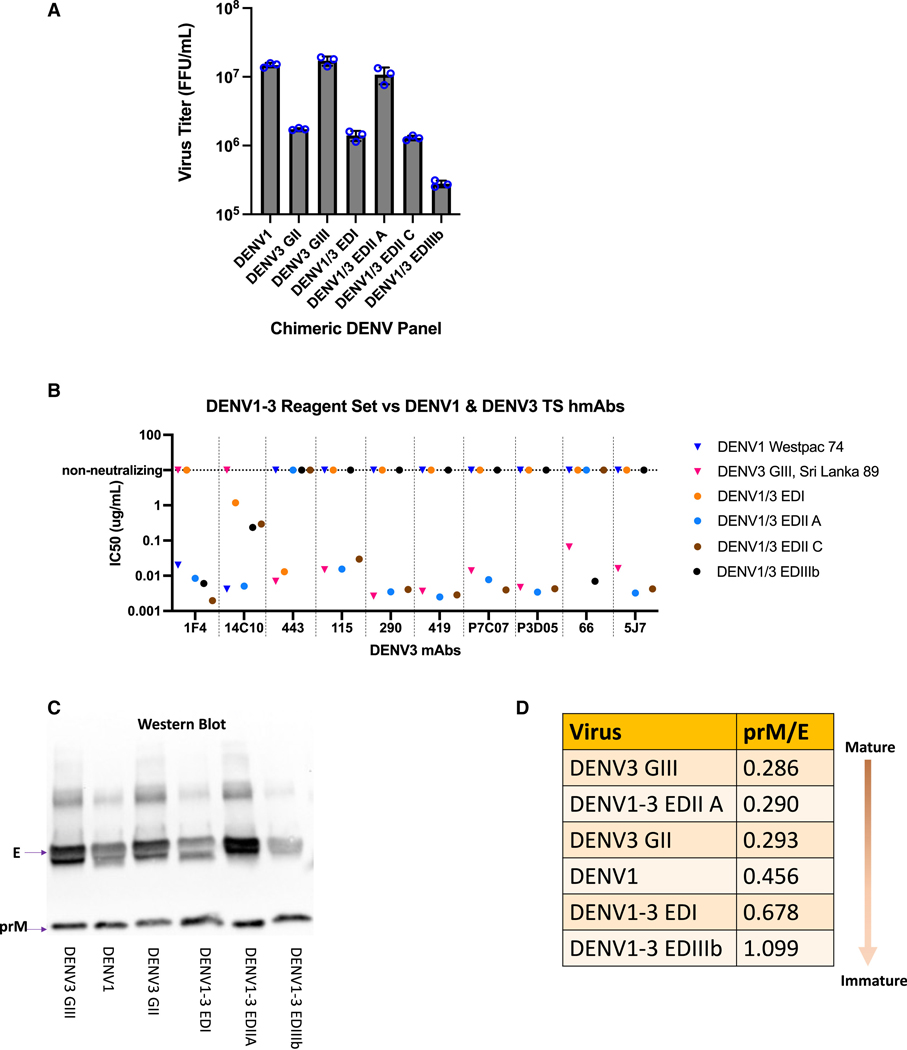
(A) Virus growth. Vero81 NHP cells were inoculated in triplicate and virus titers were measured by fluorescent foci units (FFU) at 48 h after infection, blue circles = triplicate titers.
(B) hmAB neutralization phenotypes. Confirmation of DENV1 and 3 epitopes in the DENV1/3 EDI, EDII, and EDIII panel plus parental strains using DENV1 and 3 mAb arrays.
(C) Western blots. E and prM proteins were visualized through western blot and (D) Virus Maturation Status. prM/E ratios determined, with lower numbers indicating greater maturity.
Four DENV1/3 EDIII mutants were also constructed (A–D), designed to display DENV3 EDIII while preserving DENV1 EDIII residues on the interaction interfaces and select EDIII residues engaging the M and capsid proteins during virion assembly (Figure S1C). All recombinants were recovered on C636 insect cells but had attenuated growth phenotypes in Vero81 NHP cells, especially DENV1/3 EDIII. The most robust mutant, DENV1/3 EDIIIb, retained one DENV1 EDIII residue at position F392 and replicated to titers of 2.8 × 105 fluorescent foci units/ml (FFU/mL) on Vero81 cells (Figure 2A). DENV1/3 EDIIIc was less hardy, and DENV1/3 EDIIIa and EDIIId could not be recovered in Vero81 cells. Virus stocks from the most replication-competent chimeras (DENV1/3 EDIIA, DENV1/3 EDIIC, and DENV1/3 EDIIIb) in Vero81 cells were selected for downstream immunologic and biochemical phenotyping. Deep sequencing data for the E glycoprotein of the recombinant virus stocks revealed mutations that arose in the DENV1–3 EDI and EDIIIb mutants (Figure S2). Deep sequencing revealed three DENV1 reversion mutations in EDI of the DENV1/3 EDI virus at G29S, N37D, and A50V, which likely stabilize the hinge regions and subsurface interactions. DENV1/3 EDIIIb virus stocks contained mutations at position N377K, a subsurface residue in EDIII, T16I, a subsurface residue in EDI, and at surface positions K203E and K204R in EDII. K203E, K204R, and N377K are unique mutations, localized along the interface between two monomers in the dimer. T161I is a reversion to a DENV1 codon and resides on the interface between two monomers in the dimer (Figures 1A and S2).
mAb neutralization phenotypes within the DENV1/3 full panel
Previously, our group mapped 13 DENV3 TS hmAbs to EDI, EDII, and EDIII.60 To evaluate the preservation of DENV3 and DENV1 TS epitopes on the DENV1/3 chimeric and parental viruses, we tested neutralization sensitivity to a panel of DENV3 hmAbs (−443, −115, −290, −419, −66, −5J7, -P3D05, and -P7C07)60,64,65 and DENV1 hmAbs (−1F4 and −14C10) (Figure 2B; Table S2). As designed, DENV1 TS hmAb-1F4 binding to EDI neutralized the DENV1 and DENV1/3 EDIIA, EDIIC, and EDIIIb chimeras, but not DENV3 or the DENV1/3 EDI chimera. DENV1 TS hmAb-14C10, which targets a quaternary epitope centered in EDI that extends into EDIII on an adjacent monomer, potently neutralized DENV1 and DENV1/3 EDIIA but lost ≥56% of its neutralizing activity against the DENV1/3 EDI, EDIIC, and EDIIIb recombinants. DENV3 was not neutralized by hmAb-14C10. Loss of neutralization in DENV1/3 EDIIC was likely due to the introduction of DENV3 residues N52Q and T275G in the EDI/EDII hinge region, which reside within the 14C10 epitope footprint.62 Several natural infection and vaccination-derived hmAbs (e.g., −115, −290, −419, −5J7, -P3D05, and -P7C07) potently neutralized the DENV1/3 EDIIA and EDIIC chimeras and DENV3, but not the other ED chimeras, localizing those Abs to epitopes encoded in DENV3 EDII. Finally, the DENV3 EDIII-targeting TS hmAb-66 efficiently neutralized the DENV1/3 EDIIIb chimera and DENV3, but not the remainder of the chimeric viruses. Importantly, each DENV3 hmAb neutralized only the chimera containing its epitope’s domain, but not the other chimeras, further confirming the overall ED epitope presentation across the DENV1/3 panel. Based on virologic and immunologic phenotyping with hmAbs, DENV1/3 EDIIA was chosen as the best candidate for DENV1/3 EDII domain-specific mapping.
Western blot analysis was performed on the final virus panel to evaluate maturation status after passage on Vero cells. Although all virus stocks were only partially mature, DENV1/3 EDIIIb was more immature than DENV3 WT (prM/E ratios 1.099 and 0.456, respectively), potentially explaining why DENV3 EDIII-targeting hmAb-66 more efficiently neutralized the DENV1/3 EDIII chimera better than wild-type DENV3 (Figures 2C and 2D).
Evidence for multiple unique/overlapping neutralizing epitopes in DENV3 EDII
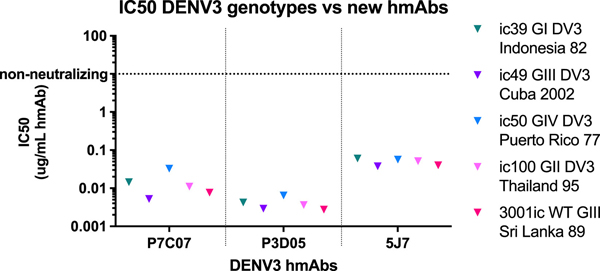
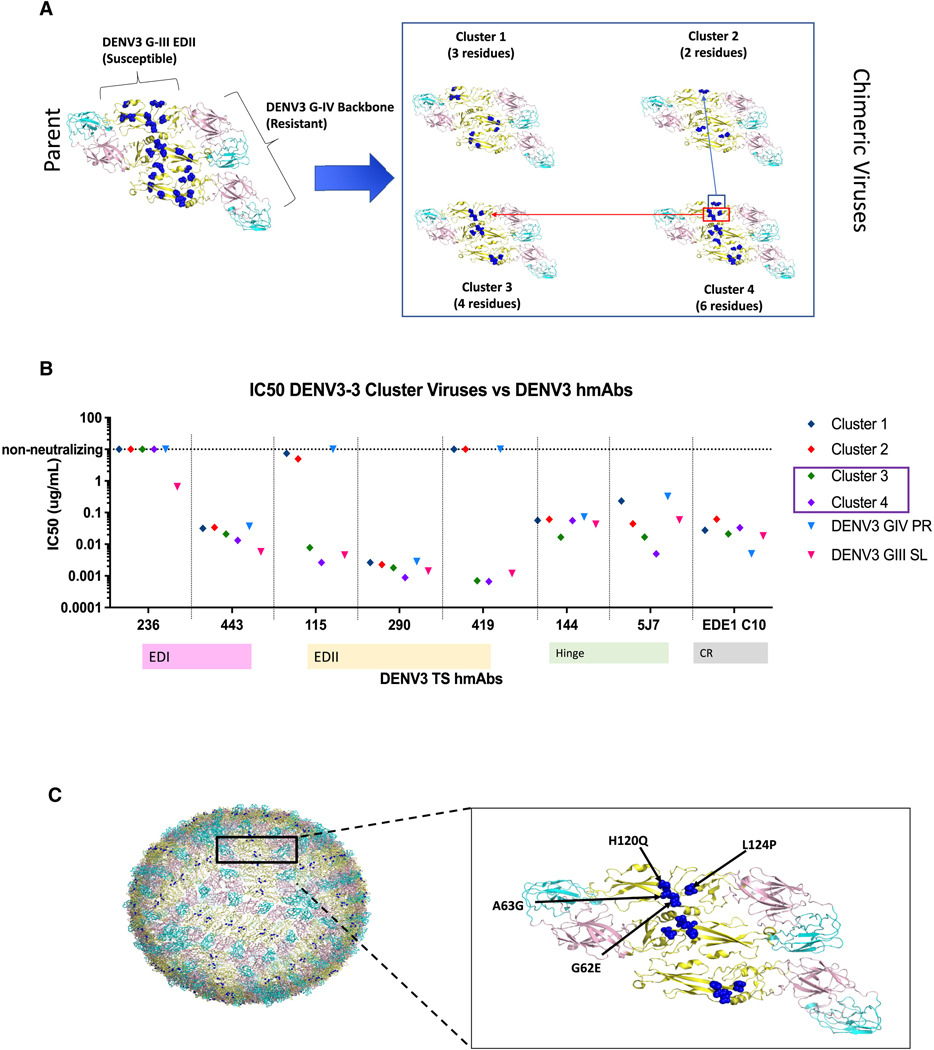
Previous studies demonstrated that natural infection-derived DENV3 TS-neutralizing hmAbs-115 and −419 target an epitope in EDII that is conserved in genotypes I, II, and III, but not IV.60 A third anti-EDII DENV3 TS hmAb, −290, did not exhibit differential neutralization across a panel of DENV3 genotypes, whereas -P3D05 and -P7C07 remained untested. Therefore, hmAbs-P3D05 and -P7CO7, derived after primary DENV3 natural infection followed by tetravalent vaccination, were tested for neutralization of multiple DENV3 genotypes using chimeric DENV3 G-III viruses displaying different genotype E glycoproteins (Figure 3). The hmAbs efficiently neutralized all genotypic variants in the panel, suggesting that hmAbs-P7CO7, -P3D05, and the previously analyzed hmAb-290 either recognized unique EDII epitopes or engaged overlapping epitopes in a unique manner. To functionally map the exact EDII residue clusters that regulated hmAb-115 and −419 genotype-specific neutralization, we created a panel of DENV3 EDII mutants (DENV3/3, clusters 1–4) containing EDII residues from a genotype susceptible to hmAb-115 and −419 neutralization (DENV3 G-III), which were introduced into a genotype backbone resistant to neutralization (DENV3 G-IV) (Figures 4A and S3). As 9 variable residues were spatially clustered on the E glycoprotein dimer (Figure 4A), proximal blocks of 3 or 6 DENV3 G-III, EDII residues were transplanted into the resistant G-IV backbone (e.g., cluster 1: T68I, T81V, and S113L; or cluster 4: G62E, A63G, H120Q, L124P, A224T, and V226T). In parallel, two additional recombinants were designed that further subdivided the six residues in cluster 4 into two smaller clusters (cluster 2: A224T and V226T; cluster 3: G62E, A63G, H120Q, and L124P). All viruses were viable and replicated in cell culture, cluster 1 to 106, cluster 3 to 105, and clusters 2 and 4 to 104 FFU/mL. The susceptible DENV3 G-III and DENV3/3 genotype mutant clusters 3 and 4 were potently neutralized by hmAbs-115 and −419, whereas the resistant DENV3 G-IV, and DENV3/3 cluster 1 and 2 mutants were not neutralized (Figure 4B). These data demonstrate that the cluster 3 mutant contains important functional residues for hmAb-115 and −419 neutralization (e.g., G62E, A63G, H120Q, and L124P) (Figure 4C).
Figure 3. DENV3 vaccine-elicited hmAB neutralize multiple DENV3 genotypes.
Isolated from vaccinee’s, novel EDII-targeting mAbs-P3D05 and -P7C07 were tested via FRNT for neutralization with a panel of DENV3/3 envelope genotype I–IV chimeras, displayed on a DENV3 G-III backbone, and exhibited similar potency between strains.
Figure 4. DENV3 hmAbs-115 and −419 target clustered residues in EDII.
(A) DENV3 EDII epitope mapping strategy. DENV3/3 EDII, G-III into G-IV parent (9 residues) and 4 DENV3/3, G-III into G-IV cluster chimeras (C1–C4) encoding between 2 and 6 DENV3 G-III residues are depicted, G-III changed residues = blue spheres (PDB: 3J27, for visualization). C2 and C3 together make the residues encoded into C4.
(B) hmAB neutralization phenotypes. DENV1/3 EDII-targeting mAbs-115 and −419 neutralize C3 and C4, with the critical residues lying in C3.
(C) Annotated location of nAb epitopes in EDII. The critical mAb-115 and −419 epitope residues are depicted on the whole viron and are located at the EDII-EDII dimer interface (E62, G63, Q120, P124) (PDB: 3J6S_*60, for visualization).
Strategy for mapping DENV3 E-glycoprotein-domain-specific neutralizing phenotypes in polyclonal sera
To define E-glycoprotein-domain-specific structural targets of the DENV3 TS human polyclonal Ab response, DENV3 primary convalescent or vaccinated human serum samples were first depleted all DENV3-binding Abs or just DENV serotype CR Abs (retaining any DENV3 TS Abs) using beads coated with a DENV3 G-II reference strain (homotypic depletion) or with a mixture of DENV1, 2, and 4 virions (heterotypic depletion)43 (Figure S4A). To demonstrate the efficiency of depletion studies, DENV3 EDI-, EDII-, and EDIII-directed TS hmAbs were introduced into heat-inactivated normal human sera (NHS), depleted using the three conditions above, and then assayed by focus reduction neutralization test (FRNT). Sample A was comprised hmAbs-290 (EDII) and −443 (EDI), whereas sample B contained hmAbs-236 (EDI), −115 (EDII), and −66 (EDIII). Under these conditions, little, if any, reductions in neutralization titers were noted after applying the control and DENV1, 2, 4 depletions. Under identical conditions, all DENV3 hmAb neutralization titers were lost after DENV3 homotypic depletion (Figure S4B).
DENV3 ED-specific and genotype-nAb responses after primary natural infection
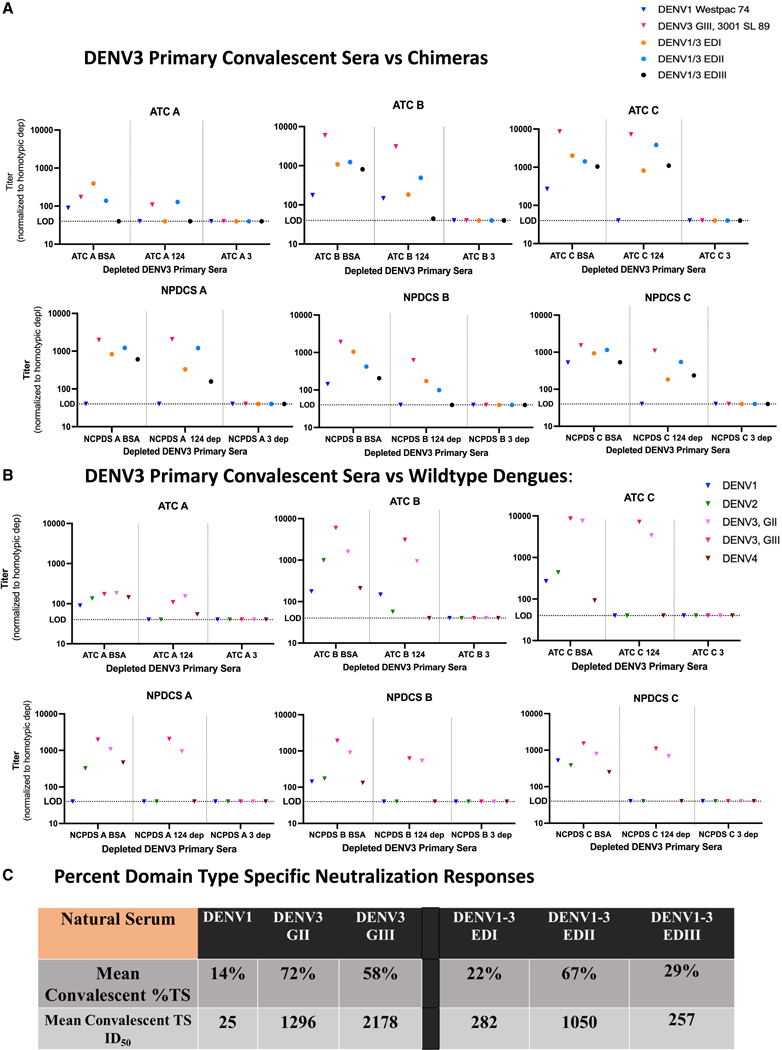
Six primary human DENV3 (G-III) convalescent natural infection serum samples were obtained from the Nicaraguan Pediatric Dengue Cohort Study (NPDCS) (n = 3)66 and the adult Arbovirus traveler cohort (ATC) (n = 3)67 (Table S1). The depleted natural infection serum samples (n = 6) were tested against the DENV1/3 EDI, II, and III chimeric virus set. After heterotypic depletion with DENV1, 2, and 4, the primary DENV3 TS neutralization response neutralized DENV1/3 EDIIA either the highest (5/6) or the second highest (1/6), suggesting that EDII is the dominant target of DENV3 TS sera in these natural infection samples (Figure 5A). However, variations in the response were also noted across the panel, as 3/6 produced nAbs to all three EDs, 2/6 targeted two different EDs, and 1/6 only recognized a single ED. For example, neutralization titers from one subject from Nicaragua targeted DENV1/3 EDI over DENV1/3 EDIIA, although at similar levels (ID50 = 173 vs. 99) (Table S3A). Most of the convalescent subjects also had lower levels of DENV3 TS nAbs that were tracked with EDI (5/6) and/or EDIII (3/6), demonstrating subdominant responses.
Figure 5. Primary DENV3 TS serum neutralization responses primarily target EDII after infection.
(A) ED-targeting of late convalescent primary infection sera. Six DENV3 primary natural serum samples, 3 from the adult Arbovirus Travelers Cohort (ATC A–C) and three from the Nicaraguan Pediatric Dengue Cohort Study (NPDCS A–C) were depleted with bovine serum albumin (BSA), DENV1, 2, and 4, or DENV3 virus on a magnetic bead, and the remaining sera tested for neutralization with the DENV1/3 EDI-EDIII panel.
(B) Wild-type and genotype outcomes. Late convalescent sera were used to neutralize wild-type viruses (DENV1, 2, 4, and DENV3 genotypes G-II and G-III). The BSA control represents total neutralization, the middle column (DENV1, 2, 4 depletion) represents the DENV3 TS nAb response, and the DENV3 depleted column represents any background neutralization which was then subtracted from the other columns.
(C) ED specific neutralization phenotypes. The mean DENV3 %TS antibody responses and mean DENV3 TS ID50 titers were calculated from the BSA and 1, 2, 4 depleted fractions to each virus across the late convalescent sera and represent the mean percentage or ID50 titer to each virus that is DENV3 TS compared with the control depletions, which contain both TS and CR antibody fractions.
The depleted late convalescent sera from the six individuals exposed to DENV3 infections were also tested for neutralization of DENV3 G-III and G-II strains to determine the breadth of TS neutralization (Figure 5B). The DENV3 TS serum Abs similarly neutralized both genotypes in most individuals (mean fold-change from DENV3 G-III to G-II = 0.03) demonstrating neutralization breadth across these two DENV3 genotypes (Figures 5B and S6).
On average after natural primary DENV3 infection, 22%, 67%, and 29% of the DENV3 TS response targeted the DENV3 EDI, EDII, and EDIII viruses, respectively, further supporting that EDII is the primary antigenic target of naturally derived DENV3-specific Abs (Figure 5C).
DENV3 genotype-specific neutralization responses following monovalent vaccination
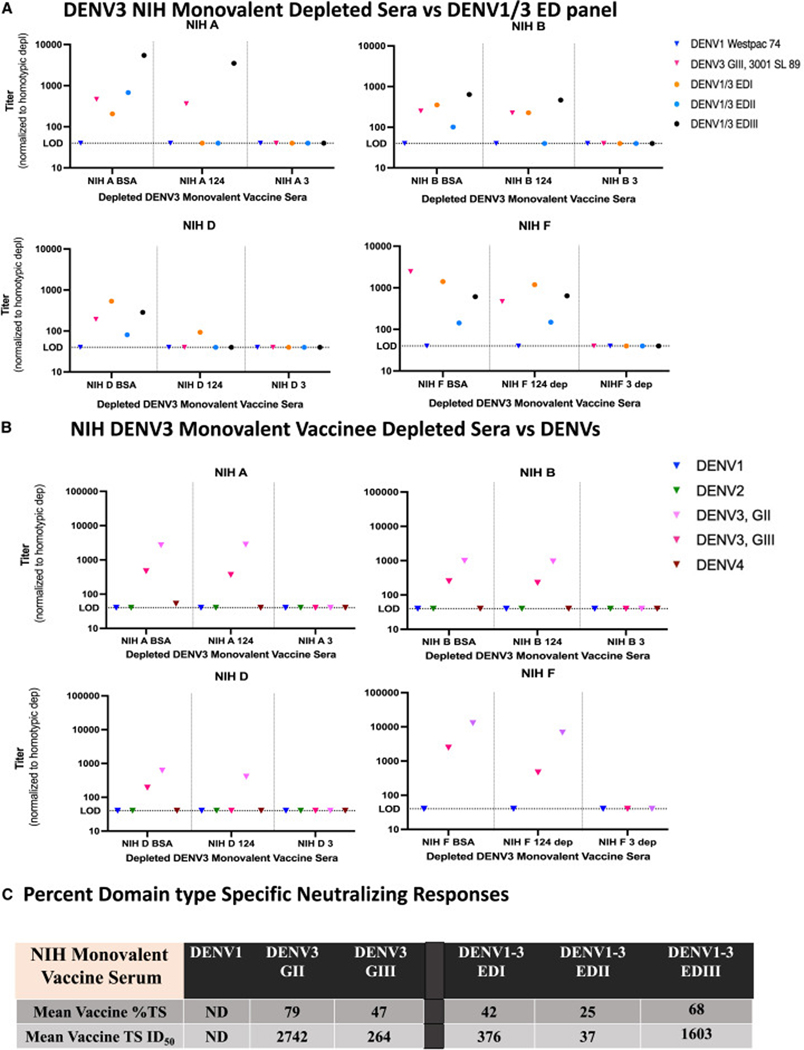
Next, we analyzed six DENV3 (G-I) NIH monovalent vaccine serum samples collected 180 days after prime/boost68 (Table S1). Using undepleted serum samples, variable neutralization titers were noted across the DENV1/3 panel (Figure S5), with the highest responses targeting EDIII (3/6), EDI (2/6), and EDII (1/6). As high cross-domain responses were noted in some undepleted samples, four samples (NIH A, B, D, and F; 3 high titers, 1 low titer) were depleted to further confirm the diminished TS response recognition pattern of DENV1/3 EDII. The depleted NIH monovalent serum samples (n = 4) were tested with the DENV1/3 chimeric panel, with 50% of the TS sera recognizing only one ED, 25% recognizing two different EDs, and 25% recognizing all three ED regions. Depletion clarified the responses in the samples, showing preferential targeting of EDIII (NIHA), EDIII > EDI (NIH B), or EDI (NIH D). Of the 3, none had measurable TS nAbs to DENV3 EDII. In sample NIH F, TS nAbs mostly tracked with EDI > EDIII domains, with a smaller portion of the TS nAbs targeting EDII, but at a much lower level than EDI (8-fold) and EDIII (4-fold). As clear differences were noted in some undepleted and depleted DENV3 serum samples such as NIH B and NIH D, the data support a requirement for depletions when mapping ED responses in the DENV1/3 chimeric panel. However, as one of the undepleted samples, NIH E, did not have any response to EDII, together, these data still support the hypothesis that monovalent responses preferentially targeted EDIII and EDI over EDII.
When the depleted NIH serum samples were tested against two different DENV3 genotypes, the DENV3 TS nAbs neutralized DENV3 G-II more efficiently than DENV3 G-III in all cases (mean fold-change = 10.68) (Figures 6B and S6). Comparing mean fold-change, the DENV3 TS monovalent vaccine sera were significantly more likely (p = 0.0095/356-fold) to neutralize DENV3 G-II over G-III than the natural infection sera (Figure S6). On average, after monovalent DENV3 vaccination, 42%, 25%, and 68% of the DENV3 TS serum Ab responses targeted the DENV3 EDI, EDII, and EDIII viruses, respectively (Figure 6C). In contrast to natural infection sera, analyses of these four vaccine samples suggest that NIH DENV3 monovalent vaccination preferentially induces DENV3 TS Abs that target EDI and EDIII and are more effective against DENV3 G-II than G-III. Together, these data suggest significant differences between ED targeting frequency and focus, which culminates in unique collections of genotype distinct Ab populations derived after natural infection and vaccination.
Figure 6. Monovalent DENV3 vaccine serum preferentially targets EDIII and EDI over EDII.
(A) ED-targeting of DENV3 monovalent vaccine serum. Four DENV3 NIH monovalent vaccinee serum samples (NIH A, B, D, and F) were depleted with BSA, DENV1, 2, and 4 or DENV3 on a magnetic bead and the remaining sera tested for neutralization with the DENV1/3 EDI-EDIII panel.
(B) Wild-type and genotype outcomes. DENV3 vaccine sera were used to neutralize wild-type viruses (DENV1, 2, 4, and DENV3 genotypes G-II and G-III). The BSA-depleted denotes the control where total antibodies are present. The DENV1, 2, 4 column denotes DENV3 TS antibodies and the DENV3 column denotes any background neutralization, which was then subtracted from the BSA and DENV1, 2, and 4 columns.
(C) ED specific neutralization phenotypes. The mean DENV3 %TS antibody responses and mean DENV3 TS ID50 titers were calculated from the BSA and 1, 2, 4 depleted fractions to each virus across NIH monovalent and represent the percentage or ID50 titer to each virus that is DENV3 TS compared with the control depletions, which contain both TS and CR antibody fractions.
Visualizing the antigenic relationships between viruses and polyclonal sera
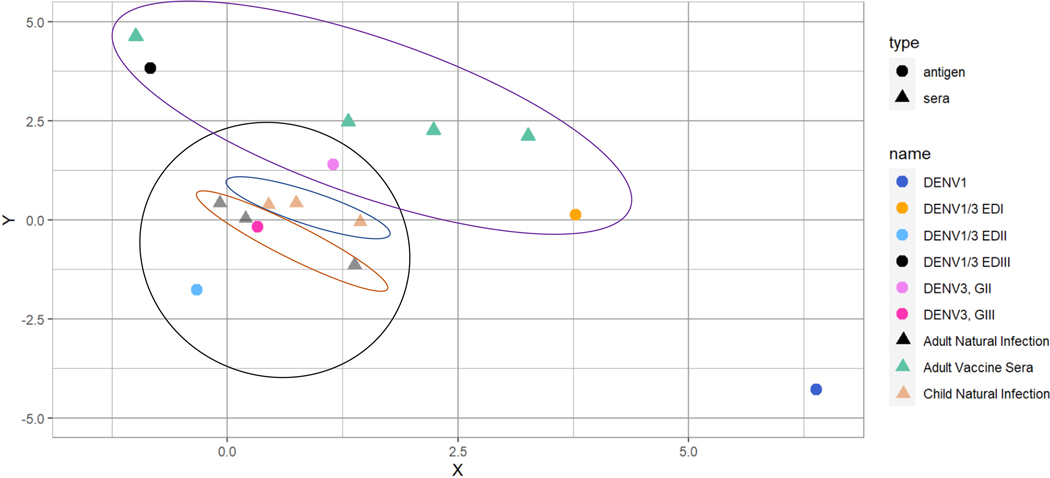
Antigenic cartography was employed to better visualize the ED-specific antigenic relationships between the chimeric viruses, based on the TS serum ID50 responses (Figure 7).69,70 After natural infection, TS nAb responses mapped closest to DENV3 G-III, rather than to DENV3 G-II strains, and were also in close proximity to DENV1/3 EDIIA, indicating that the antigenic juxtaposition of these sample sets was heavily regulated by nAbs preferentially targeting DENV3 EDII. There were two clear phenotypes exhibited in the convalescent serum Ab population (n = 6). The NPDCS children’s sera (n = 3) are positioned near equidistant between DENV3 G-II and G-III, whereas the ATC adult natural sera (n = 3) grouped closer to DENV3 G-III than to G-II. DENV1/3 EDI and EDIIIb were more antigenically distant than DENV1/3 EDIIA from both DENV3 genotypes. In relation to DENV1, DENV1/3 EDI was antigenically positioned closest, followed by DENV1/3 EDIIA, and then DENV1/3 EDIIIb. In contrast to natural infection, DENV3 TS polyclonal Ab responses in NIH monovalent vaccine sera mapped more closely to DENV3 G-II than to G-III and chimeras DENV1/3 EDI and EDIIIb, but not to DENV1/3 EDIIA. These data clearly support the existence of large antigenic shifts in ED epitope targeting between Abs in natural infection and NIH vaccine-recipient sera.
Figure 7. Cartography reveals distinct clusters of natural infection and vaccine serum responses.
Antigenic cartography of the DENV wild-type and recombinant viruses and serum ID50s are depicted using the Smith et al. method. Viruses are shown as circles and serum samples are shown as triangles, corresponding to the colored key on the right. Convalescent sera are grouped together in the black circle, and further separated into pediatric sera (blue oval) and adult sera (rust colored oval). Vaccine sera are grouped together in the purple oval. Each grid box represents a 2-fold change in titer.
DISCUSSION
DENV remains a major global health threat due to the geographic prevalence of all four serotypes, the potential for disease enhancement following secondary infection and vaccine obstacles, which are further complicated by the lack of clearly defined correlates of protection. Large, phase III clinical studies have demonstrated that total neutralization titers are important, yet complex correlates of protection, since breakthrough infections have been frequently observed, even in some participants with high-titer serum nAb responses.50–54 Recent studies also support the hypothesis that total TS nAb responses may represent correlates of protective immunity as well.37,54,59,71 TS nAbs have been shown to target epitopes in EDI, EDII, and EDIII in DENV3,60 and similar findings are reported across all DENVs.34,62,63,65,72–74 After infection or vaccination, one potential strategy to measure protective Ab-mediated immunity is to develop methods that measure the E-glycoprotein ED-specific antigenic contributions to the overall nAb responses in polyclonal sera.75 By manipulating the prM-E structural interactions in DENV virions, a panel of well-characterized DENV1/3 chimeric ED recombinant viruses was isolated and used to empirically support the hypothesis that EDII is the primary target for DENV3 TS nAbs responses after natural infection. The DENV3 EDIIA antigenic site appears to contain at least three unique and/or overlapping nAb epitopes (e.g., those recognized by hmAb-5J7, −115/−419, or −290/-P3D05/-P7C07), supporting a need for future high-resolution structures. One antigenic site, recognized by hmAbs-115/−419, was highly sensitive to natural genetic variation encoded by a currently circulating DENV3 genotype (IV), supporting potential Ab-driven evolution over time.
Although little is known regarding the prM content of most DENV serotypes produced in vivo, DENV1 particles were shown to be mostly mature.76 Maturation status is known to influence the magnitude of the DENV neutralization response for some monoclonal and polyclonal sera.77 However, DENV prM cleavage is inefficient and can subtly alter virion maturation, epitope presentation, and neutralization across natural isolates, leading to altered disease severity.78 As the maturation status of DENV3 genotypes in vivo remains uncertain, we evaluated ED-specific primary nAb responses following natural infection and vaccination, using partially mature viruses throughout. Future studies may reveal subtle differences in ED-specific neutralizing titers using mature and immature DENV1/3 ED recombinant virus panels after natural infection and vaccination.
Previously, the findings of Messer et al. displayed evidence that most people who experienced a natural DENV3 infection exhibited effective neutralization potency against multiple DENV3 genotypes but that the level of neutralization varied by individual.41 Our data also support the hypothesis that serum nAb ED-specificity oftentimes neutralizes across genotypes and can vary across individuals while dictating the need for larger collections of well-characterized primary infection and vaccine cohorts and the use of depletion studies to accurately measure TS ED responses in polyclonal sera. Although the sample size and volume were small, and future studies are needed across vaccine cohorts from different ages and geographic regions, DENV3 NIH monovalent vaccine-induced serum nAbs not only appear to preferentially target EDIII and EDI, over EDII, but were also less potent against some DENV3 genotypes. In addition to the genetic variation in the EDII region of DENV3 G-IV, there is genetic variation in the EDI-EDII and EDI-EDIII hinge regions between DENV3 G-III and DENV3 G-I/G-II, which could also alter neutralization potency after vaccination with NIH’s DENV3 G-I monovalent vaccine strain (Figure S7). These data support the need to develop matched DENV3 ED genotype-serum matched reagents designed to tease out subtle differences associated with natural variation and neutralization within genotypes of each serotype. As similar results have been reported following DENV4 vaccination,37,52 naive populations may be less protected against DENV genotypic variants as well. Our data support the concept that both the total number of and preferential specificity of ED-specific TS nAb epitopes may provide correlates for understanding the mechanisms of DENV3 protective Ab-mediated immunity.
The development of the chimeric DENV1/3 ED recombinant virus panel required creating DENV1 virions that encoded DENV3 residues at the 2-fold or 5-fold axes, designated DENV1/3 EDIIA or DENV1/3 EDIIIb, respectively. Although recovery of the DENV1/3 EDIIIb recombinant virus was based on previously developed chimeras,75,79 recovery of DENV1/3 EDIIA and DENV1/3 EDIIC required more extensive structure-guided designs that incorporated prM and E protein-protein interaction networks, as initial studies revealed that a DENV1/3 EDII chimera with a DENV1 prM was not viable. Structural studies suggested that the DENV3 prM N-terminal domain residues through residue 108 should provide a more compatible interaction interface with portions of DENV3 EDII. In parallel, DENV1 prM residues from position 109 to the C-terminal end should interact more favorably with DENV1 E residues. The recovery of replication-competent DENV1/3 EDII recombinant viruses supported these hypotheses. Importantly, experimental evolution in Vero81 cells identified several second-site reversions in the DENV1/3 chimeras at interfaces between E dimers and E-prM. The DENV3 EDIIIb Vero81-adapted virus also contained two tissue culture-adapted mutations at the EDII-EDII intradomain interface and two subsurface mutations along the EDI-EDIII interface across monomers, which are close in proximity to M in mature particles and could be important for the E-M interaction matrix. Notably, Abs like hmAb-5J7, which spans multiple monomers and different domains, retained potent neutralizing activity to both DENV1/3 EDII chimeras. These data provide further evidence that residues in the intradomain and interprotein interfaces, and in the interdomain hinge regions, are likely critical for maintaining virus stability and fusion capacity.80–82 Chimeric E-glycoproteins provide a platform to map critical interaction residues in E and prM that regulate virion function. Future structural studies on early and evolved recombinants may reveal patterns of variation that are critical for virion stability and infectivity.
In individual recipients, tetravalent DENV vaccines induce heterogeneous neutralizing titers, potentially reflecting the different replication efficiencies of each serotype in heterogeneous human populations.53,55,57,58,83 The DENV1/3 EDIIA virus preserved both major DENV1 epitopes for neutralizing hmAbs-1F4 and −14C10, and multiple potent DENV3 TS EDII-directed neutralizing epitopes. This bivalent live virus immunogen, coupled with efficient replication at greater than 107 FFU/mL in culture, highlights its potential as a single dose, bivalent vaccine for DENV1 and DENV3. The larger DENV1/3 EDIIC transplant virus also preserved all DENV3 EDII epitopes tested, including the DENV1 hmAb epitope-1F4 and −14C10 epitope, although the latter Ab displayed a ~56-fold reduction in neutralization performance. Alternatively, the DENV1/3 EDIIC virus replicated slightly less efficiently in vitro but preserved the hmAb-1F4 and −14C10 epitopes, providing a second potential candidate vaccine for testing in primates as described by our group.60,84 As DENV1 and DENV3 are phylogenetically close, our data support the need for depletion studies when evaluating ED-specific responses in polyclonal primary or secondary serum with these recombinant viruses.
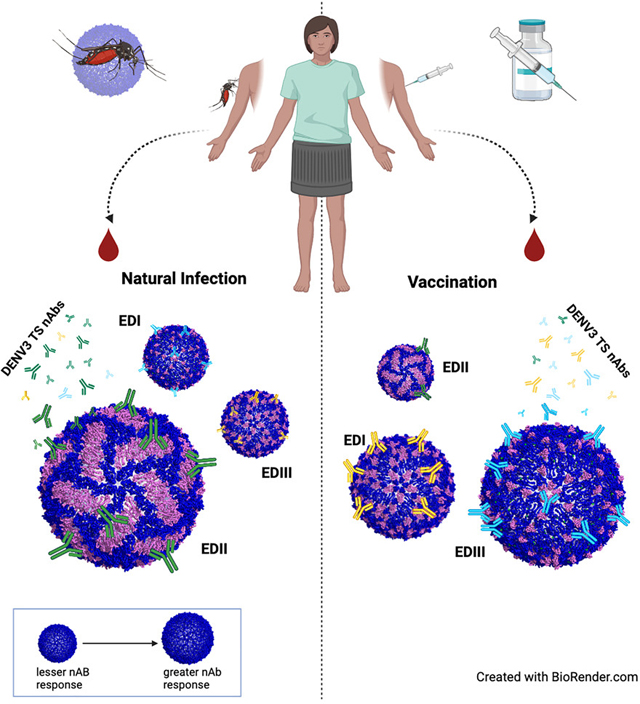
Serum TS nAbs may target one or more EDs in a hierarchical fashion, which may differentially influence protective immunity and the frequency of breakthrough infections after primary infection or vaccination. Our data suggest that EDII is the principal target of TS nAbs after DENV3 primary infection, with lower-level nAb responses targeting EDI and/or EDIII in a patient-specific manner. In contrast, monovalent NIH DENV3 vaccination primarily elicited TS nAbs to EDI and EDIII, whereas EDII was only weakly targeted in about 25% of the samples tested, potentially providing opportunities for genotype-driven breakthrough infections, like those reported for DENV4.37,52 The DENV3 component in the TAK tetravalent vaccines replicated less efficiently than wild-type DENV3 in animal models and some humans, potentially altering ED-specific breadth.57,83,85–90 These data make a compelling argument for new research campaigns that fully characterize hmAb neutralizing and binding repertoires to DENV1–4 TS and CR nAb epitopes. Differences in genotype and ED targeting after infection and vaccination are likely influenced by significant differences in serotype replication efficiency, preexposure histories, vaccine strain, and natural variation across domains.
Cartography provides a visualization platform that reveals antigenic relationships between patient serum and ED-specific polyclonal responses after infection and vaccination.91 Not surprisingly, DENV3 TS primary serum Ab responses clustered much nearer to DENV3 G-III, G-II, and DENV1/3 EDII, than Ab responses targeting DENV1/3 EDI and DENV1/3 EDIII. Furthermore, late convalescent serum from the naturally infected children grouped closely and were relatively equidistant from DENV3 G-II and G-III, compared with the adult convalescent sera, which clustered nearest to DENV3 G-III. Although speculative, given the small sample size, these unique antigenic groupings between adults and children may suggest structural targeting differences in an age-dependent manner. In contrast, adult monovalent vaccine-recipient serum Abs mapped nearest to DENV3 G-II, a strain more closely related to the DENV3 G-I Sleman ‘78 strain present in the vaccine formulation, supporting the hypothesis that monovalent vaccine responses could be more genotype-dependent than responses seen after natural infection.
In summary, we identified genetic features in prM and E that likely co-evolve and interact to regulate DENV virion assembly, function, and viability, supporting earlier studies that implicated intimate prM/E interactions in virus viability and fitness. The recombinant DENV1/3 chimeric and DENV3/3 genotype panels and parental strains identified three or more hmAb epitopes in EDII and dictate a need for Ab-epitope high-resolution structures. One of the EDII epitopes appears to be under strong immune selection, leading to evolutionary changes that escape select hmAb neutralization, further supporting a need for additional structures. The DENV1/3 chimeric panel identifies differences in the targeting of TS responses across EDI, EDII, and EDIII after DENV3 infection and vaccination. A strength of our mutant/depletion neutralization assay model is the presence of clear differences in DENV3 TS targeting of the 3 domains that emerge following infection and vaccination, which could explain vaccine-induced Ab breadth and durability. Future studies using ED-specific neutralization metrics may reveal new mechanistic correlates for understanding breakthrough infections. Our data also suggest that new reagents will be needed to dissect the complex relationships between correlates of protective immunity across DENV serotypes and genotypes and the relationships with ADE. One limitation of the study was that sample sets were small and of low volume, hampering depletion studies against a large panel of virus strains. Additionally, due to sourcing constraints, it happened that all our depleted serum samples were female in origin. Larger cohorts of well-characterized natural infection and vaccine-recipient serum samples, including representative genders and those that experienced breakthrough infections, will be critical to confirm any potential associations between ED nAb responses and protective immunity. Another interesting limitation is that EDIII-exchanged viruses occasionally show increased sensitivity to neutralization in comparison with the parental strain, suggesting that maturation status or the potential emergence of cryptic epitopes may exist, requiring structure-function investigations. We have solved many of the structural constraints that restrict ED exchange between serotypes, isolated highly replication-competent recombinant viruses encoding chimeric DENV1 and DENV3 immunogens and measured ED-specific nAbs in sera. These recombinant viruses may also have utility as a future bivalent vaccine candidate for in vivo testing in primates and then perhaps in humans.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Please direct requests for further information, resources, and/or reagents to Ralph Baric (rbaric@email.unc.edu).
Materials availability
Plasmids and chimeric viruses generated in this study are available upon request.
There are restrictions with regard to availability of serum reagents due to low volume of the samples.
Data and code availability
Deep sequencing data have been deposited in the NCBI database and are publicly available upon this manuscript’s publication date. Accession numbers are listed in the key resources table (SRR23674710, SRR23675053, SRR23675073).
No original code was developed for this manuscript.
Any additional information required to reanalyze data reported in this manuscript is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| DENV-443 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943662 |
| DENV-115 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943663 |
| DENV-290 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943664 |
| DENV-419 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943665 |
| DENV-P7C07 | Magnani et al.64/JVI.00867–17 | RRID:AB_2943666 |
| DENV-P3D05 | Magnani et al.64/JVI.00867–17 | RRID:AB_2943667 |
| DENV-66 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943668 |
| DENV-5J7 (plasmid produced Ig) | Beltramello et al.92; https://doi.org/10.1016/j.chom.2010.08.007 | RRID:AB_2943669 |
| DENV-1F4 (hybridoma produced) | Fibriansah et al.63 | RRID:AB_2943670 |
| DENV-14C10 (plasmid produced Ig) | Teoh et al.62 | RRID:AB_2943671 |
| Goat anti-human IgG (Fc)-AP | Meridian Life Science, Inc | Cat#W99008A; RRID: AB_151913 |
| DENV-4G2 (hybridoma produced Ig) | ATCC HB-112 | D1–4G2–4-15; RRID: AB-2722738 |
| DENV-2H2 (hybridoma produced Ig) | ATCC HB-114 | D3–2H2–9-21; RRID: AB-95338 |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| DENV1ic West Pac 74 | Gallichotte et al., 201793; https://doi.org/10.1128/mSphere.00380–16 | N/A |
| DENV2ic 16803 | Gallichotte et al., 201594; https://doi.org/10.1128/mBio.01461–15 | N/A |
| DENV3 Thailand/CH53489/1973 | Wahala et al.95; https://doi.org/10.1371/journal.ppat.1000821 | N/A |
| DENV3ic Sri Lanka 89 G-III | Messer et al.61 | N/A |
| DENV3 Indonesia 1982, G-I | Messer et al.41 | N/A |
| DENV3 Thailand 1995, G-II | Messer et al.41 | N/A |
| DENV3 Cuba 2002, G-III | Messer et al.41 | N/A |
| DENV3 Puerto Rico 1977, G-IV | Messer et al.41 | N/A |
| DENV4ic Sri Lanka 92 | Widman et al.96; https://10.1586/14760584.2015.961431 | N/A |
| DENV1/3 EDI B | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | NCBI accession #:SRR23674710 |
| DENV1/3 EDII A | This Manuscript | NCBI accession #: SRR23675053 |
| DENV1/3 EDII B | This Manuscript | N/A |
| DENV1/3 EDII C | This Manuscript | N/A |
| DENV1/3 EDII D | This Manuscript | N/A |
| DENV1/3 EDIIIa | This Manuscript | N/A |
| DENV1/3 EDIIIb | This Manuscript | NCBI accession #: SRR23675073 |
| DENV1/3 EDIIIc | This Manuscript | N/A |
| DENV1/3 EDIIId | This Manuscript | N/A |
| DENV3/3 C1 | This Manuscript | N/A |
| DENV3/3 C2 | This Manuscript | N/A |
| DENV3/3 C3 | This Manuscript | N/A |
| DENV3/3 C4 | This Manuscript | N/A |
|
| ||
| Biological Samples | ||
|
| ||
| Serum Samples from DENV3 Primary Infection Survivors | This Manuscript | Adult Arbovirus Travelers Cohort, UNC; DT103/ATC A, DT133/ATC B, DT226/ATC C |
| Serum Samples from DENV3 Primary Infection Survivors | This Manuscript | Nicaraguan Pediatric Dengue Cohort Study, Donor IDs: 4082.7/NPDCS A, 4862.7/NPDCS B, 5862.7/NPDCS C |
| Serum Samples from DENV3 NIH monovalent vaccine trial | Swanstrom et al.68; PMCID: PMC6581895 | Depeleted IDs: 257.01.02/NIH A, 257.01.16/NIH B, 257.03.47/NIH D, 257.03.50/NIH F; Undepleted IDs: 257.03.45/NIH C, 257.03.49/NIH E |
| Normal Human Serum (NHS) Sample | Henein et al.53; PMCID: PMC8245170 | DT236 |
|
| ||
| Chemicals, Peptides, & Recombinant Proteins | ||
|
| ||
| DMEM, high glucose, GlutaMAX™ Supplement | Thermo Fisher Scientific | Cat# 10566–024 |
| Dulbecco’s modified Eagle’s/Ham’s F-12 50/50 Mix | Gibco | 10–092-CV |
| Opti-MEM I | Gibco | 31985–070 |
| TrueBlue substrate | KPL | 0510–0050 |
| TRIzol LS | Thermo Fisher Scientific | 10296028 |
| Non-fat dry milk | Bio Rad | Cat# 1706404 |
|
| ||
| Cell Lines | ||
|
| ||
| C6/36 cells | ATCC | CRL-1660; RRID: CV_CL_Z230 |
| Vero-81 cells | ATCC | CCL-81; RRID: CV-CL_0059 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| Illumina Stranded Total RNA Prep | Illumina | Ribozero Plus Protocol; 20040525 |
| MiSeq Reagent Kit v3 | Illumina | MS-102–3001 |
|
| ||
| Deposited Data | ||
|
| ||
| DENV1/3 EDI B | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | NCBI accession #:SRR23674710 |
| DENV1/3 EDII A | This Manuscript | NCBI accession #: SRR23675053 |
| DENV1/3 EDIIIb | This Manuscript | NCBI accession #: SRR23675073 |
|
| ||
| Recombinant DNA | ||
|
| ||
| Seq & PCR Primer: DENV3 (G-IV), 916F: GCC CAT TAC ATA GGC ACT TC | This Manuscript | Cluster 916F |
| Seq & PCR Primer: DENV3 (G-IV), 2143R: CGA GCT TCC TTT CTT GTA CC | This Manuscript | Cluster 2143R |
| Seq Primer, Cluster 935R: GAA GTG CCT ATG TAA TGG GC | This Manuscript | Cluster 935R |
| Seq Primer, Cluster 1420F: GTT CAC ACA GGA GAT CAA CAC C | This Manuscript | Cluster 1420F |
| Seq Primer, Cluster 1441R: GGT GTT GAT CTC CTG TGT GAA C | This Manuscript | Cluster 1441R |
| Seq Primer, Cluster 1735F: GTG GTT GTT CTT GGA TCG CAA G | This Manuscript | Cluster 1735F |
| Seq Primer, Cluster 1756R: CTT GCG ATC CAA GAA CAA CCA C | This Manuscript | Cluster 1756 R |
| PCR Primer: DENV1/3 Panel; A1F: CAT GCT CCT CAT GCT GCT GC | This Manuscript | A1F |
| PCR Primer: DENV1/3 Panel; U24R: GCC TTT CCA GTT GAT TAC AC | This Manuscript | U24R |
| Seq Primer: DENV1/3 Panel; 1215F: CGA TTC GAG ATG TCC AAC ACA AGG | This Manuscript | 1215 F |
| Seq Primer: DENV1/3 Panel; 1238R: CCT TGT GTT GGA CAT CTC GAA TCG | This Manuscript | 1238 R |
| Seq Primer: DENV1/3 Panel; 1765R: CTA GTA CGA CTA CTT CCT GC | This Manuscript | 1765 R |
| Seq Primer: DENV1/3 Panel; 1650F: CTT ACC ACT GCC TTG GAC CTC | This Manuscript | 1650 F |
| Seq Primer: DENV1/3 Panel; 1670R: GAG GTC CAA GGC AGT GGT AAG | This Manuscript | 1670R |
|
| ||
| Software and Algorithms | ||
|
| ||
| GraphPad Prism, v9.0 | GraphPad Software, Inc. | https://www.graphpad.com |
| PyMOL | Schrödinger, LLC | https://www.pymol.org/ |
|
| ||
| Other | ||
|
| ||
| MiSeq | Illumina | N/A |
| EL406 washer dispenser | BioTek | N/A |
| Biostack microplate stacker | BioTek | N/A |
| Immunospot | CTL | N/A |
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Human Serum Samples
Natural Infection Sera
Sera were obtained from three participants from the Nicaraguan Pediatric Dengue Cohort Study (NPDCS), all were derived from females aged 4–7 years.47,66 The samples were obtained during the annual samples collection 5.5–8.5 months after symptomatic primary DENV3 infections (see Table S1). All NPDCS samples were obtained with full informed consent from the parents or participant assent for children aged 5 years and above. The study was approved by the IRB at the University of California, Berkeley, the International Vaccine Initiative, and the Nicaraguan Ministry of Health (Centro Naticional de Diagnostico y Referencia and HIMJR). Sera were also obtained from three female participants, aged 28–42 years, in the Adult Arbovirus Traveler Cohort at the University of North Carolina, Chapel Hill, 1.5–18 years after natural symptomatic DENV3 infection.67 The Traveler cohort samples were obtained with full informed consent from travelers with self-reported Dengue symptoms (IRB no. 08–0895).
Vaccine Sera
6 adult volunteers from the NCT00831012 clinical trial, using vaccine: rDENV3Δ30/31 and study protocol: CIF-257. Sera were obtained 6–7.5 months post vaccination.68 NIH A, B, D, and F were all from female participants, aged 28–46 years. Monovalent vaccine sera were obtained through recruited volunteers at John Hopkins Bloomberg School of Public Health Center for Immunization Research (CIR). The study protocol was approved by the Joint Committee for Clinical Medicine and each volunteer was fully informed and consented.
Normal Human Serum (NHS)
NHS was obtained from a member of the Adult Arbovirus Traveler Cohort who was negative for all anti-dengue antibodies by ELISA and neutralization assays.
Cell Culture
Vero 81 cells were maintained in medium (DMEM 50:50 F12 medium containing 5% FBS and 1% PenStrep, NEAA, Glutamax and bicarbonate) and incubated at 37°C as previously described.60 Vero81 cells were split using 0.05%trypsin/EDTA. C636 cells were maintained in 1XMEM media with 5% FBS, 1% Pen-Strep, and 1% NEAA at 32°C, as previously described.79
Escherichia coli, for cloning purposes, were cultured at 37C in LB using selective antibodies.
METHOD DETAILS
DENV Wildtype Strains
Several DENV isolates were used during this study including DENV1 (Nauru/1974/Westpac), DENV2 (Thailand/1974/S16803), DENV3 G-II (CH53489/1973/Thailand), DENV3 G-III (Sri Lanka/1989), DENV4 (Sri Lanka/1992). Virus stocks were propagated in Vero81 cells for polyclonal studies, as previously described by our group.79
Tissue Culture
Vero 81 cells were maintained in medium (DMEM 50:50 F12 medium containing 5% FBS and 1% PenStrep, NEAA, Glutamax and bicarbonate) and incubated at 37°C as previously described.60 Vero81 cells were split using 0.05%trypsin/EDTA. C636 cells were maintained in 1XMEM media with 5% FBS and 1% Pen-Strep at 32°C, as previously described.79
Design and Recovery of Recombinant Viruses
Four chimeric DENV1/3 EDII, 4 EDIII and 4 DENV3/3 recombinant viruses were designed using our previously described tetrapartite DENV1 and DENV3 reverse genetics platforms.41,60,61 Briefly, 4 DNA fragments, which are flanked by class IIS restriction endonucleases, are excised from agarose gels, and ligated together to produce a full length DENV 1 wildtype and derivative molecular clone encoding each chimeric E glycoprotein construct. To generate chimeric E glycoproteins, DENV3 EDI, II and III sequences were synthesized in the DENV1 E glycoprotein backbone encoded in the A clone by BioBasic and assembled using Golden Gate cloning and molecular techniques. The DENV3/3 genotype panel was engineered to encode DENV3 G-III natural variation within the DENV3 G-IV A plasmid backbone using BioBasic and a full-length clone was assembled as described above. Full-length viral RNA was then transcribed and electroporated into Vero81 cells, a non-human primate cell line, or in C636 mosquito cells and incubated for 4 to 7 days at 37 or 32°C, respectively. The supernatant was harvested, and cells were removed by centrifugation before freezing at −80°C. Virus stocks were titered by fluorescent foci assay and Sanger sequenced as previously described by our group.60 The final set of chimeras used for polyclonal analysis were subject to full-genome sequencing (DENV1/3 EDI, EDIIA, and EDIIIb chimeras) as previously described.97 Wildtype DENV3 G-III and DENV1 Westpac were passage 3, the EDI virus was passage 4, EDIIA was passage 1, and due to Vero adaptation and low replication efficiency, EDIIIb was passaged 7 times before deriving individual stocks for sequencing.
Wildtype and Recombinant Virus Quantification
Cells were plated on tissue-culture-treated 96-well plates at 2 × 104 cells/well the day before the assay. Wildtype and derivative virus were serially diluted, plated onto the cells in duplicate at 50 μL per well and then incubated at 37°C for 1 hour. Overlay medium (2% FBS DMEM, 1% P/S/NEAA, 5 g methylcellulose) was placed over the top at 125 μL per well after 1 hour and the titer plate was incubated for 48 hours at 37 °C. After incubation, cells were fixed in 10% formaldehyde for 20 to 30 minutes and washed in permeabilization buffer (Invitrogen) three times. 5% powdered milk (Bio-Rad)/permeabilization buffer was used to block nonspecific binding for 10 mins, and then incubated with primary DENV antibody (2H2/4G2) for ~1 hr. After washing, a secondary antibody (HRP goat-anti-mouse/Sigma) was incubated for 45 minutes, washed extensively, and then developed with True Blue solution (KPL/Sera Care). Foci were visualized, counted and quality controlled with an Elispot analyzer (C.T.L.).
Strategy for Mapping DENV3 E Glycoprotein Domain Specific Neutralizing Phenotypes in Polyclonal Sera
Beads were coated with either a bovine serum albumin (BSA) control, a combination DENV1 (Westpac), 2 (S16803) and 4 (Sri Lanka/1992) (heterotypic depletion), or with a WHO DENV3 G-II reference strain (homotypic depletion). Depleted sera were tested via FRNT, and the resulting dilution at which 50% maximal inhibition occurred (Inhibitory Dilutions 50 or ID50s) were normalized to the DENV3 homotypic depletion for each subject. For proof of concept, DENV3 EDI, EDII and EDIII-directed TS hmAbs were introduced into heat-inactivated normal human sera (NHS) at a value 103 their ID50 dilution, depleted using the three conditions above, and then assayed by FRNT. Sample A was comprised of hmAbs-290 (EDII) and −443 (EDI), while Sample B contained hmAbs-236 (EDI), −115 (EDII) and −66 (EDIII). Percent TS responses were determined for each virus for each serum sample using the following calculations: %TS response = (TS-depleted value/BSA-depleted value) *100. These percentages are not additive, but rather represent the fraction of the total polyclonal antibody population directed at a specific ED that is DENV3 TS. Mean DENV3 TS ID50 Titer was calculated and values <40 were listed as not detected (ND). Mean fold-change of the DENV3 TS ID50 titers were determined using the following formula: Mean fold change = (DENV3 G-II TS value - DENV3 G-III TS value)/DENV3 G-III TS value and values of <40 were designated with a value of “20” for purposes of determining a fold-change.
Wildtype and Recombinant Virus Focus Reduction Neutralization Test (FRNT)
Using 3,200 FFU/mL of each virus, 33 μmg/mL of each monoclonal antibody or polyclonal sera was diluted and then incubated with virus at 37°C for 1 hour. The virus/antibody mixture was then placed onto cells (plated at 2 × 104 cells/well 24 hours in advance) at 50 μL per well in duplicate and incubated for 1 hour at 37°C. After, overlay (OptiMEM I, 5% carboxymethylcellulose, 2% FBS, 1% PenStrep) was then placed over the top of the cells at 125 μL/well, the plates were incubated at 37°C for 48 hours, stained and virus foci enumerated using a CTL Elispot analyzer. As a note, some serum samples were tested with DENV2 and DENV4 to look for background. Little to no background was present in the natural sera and no background existed in any monovalent sample tested.
Western Blot
Purified supernatant from the virus stocks were diluted with 4× Laemmli Sample buffer (Bio-Rad) and boiled at 70°C for 15 minutes. Following SDS-PAGE electrophoresis, proteins were transferred to PVDF membrane and blocked in blocking buffer (5% non-fat milk in PBS + 0.05% Tween-20, PBS-T) overnight. The membrane was incubated with polyclonal rabbit anti-prM (1:1,000) (Invitrogen, Cat. #PA5–34966) and purified human anti-Env (fusion loop), 1M7 (2 μg/ml) in 2% BSA + PBS-T solution for 1 h at 37°C. The primary antigen-antibody complex was detected by incubating the blot with goat anti-rabbit IgG HRP (1:10,000, Jackson-ImmunoLab) and sheep anti-human IgG HRP (1:5,000, GE Healthcare) in 5% milk, for 1 h at room temperature. Membranes were developed by Super-signal West Pico PLUS Chemiluminescent Substrate (ThermoFisher). The pixel intensity of individual bands was measured using ImageJ, and maturation was calculated by using the following equation: (prM/Env).
QUANTIFICATION AND STATISTICAL ANALYSIS
50% inhibitory concentration (IC50) and 50% inhibitory dose (ID50) graphical analysis was completed using GraphPad Prism, v.9, variable slope sigmoidal dose-response curve (Figures 2, 3, 4, 5, and 6). A non-parametric Mann-Whitney test was used to compare mean-fold change between 2 groups. P-values are indicated by symbols: **=p=0.0095 (Figure S6).
Supplementary Material
Highlights.
EDII is the main target of TS polyclonal Abs after DENV3 infection
TS polyclonal Abs post-DENV3 monovalent vaccination mainly target DENV3 EDI and EDIII
DENV3 G-I monovalent vaccination makes more genotype-sensitive Abs than G-III infection
There are 3 or more functional monoclonal antibody epitopes in DENV3 EDII
ACKNOWLEDGMENTS
We thank members of the study teams at the Centro de Salud Socrates Flores Vivas, the National Virology Laboratory in the Centro Nacional de Diagnostico y Referencia, the Sustainable Sciences Institute in Nicaragua, and Alena Markman at University of North Carolina, Chapel Hill for their dedication and high-quality work in conducting the studies and preparing the convalescent samples. We also thank Anna Durbin and Steve Whitehead for DENV3 NIH monovalent samples. Additionally, we are grateful to the study participants and their families. This project received support from NIH grants P01 AI106695 to E.H. and AI107731 to A.M.D. This work was also supported by the National Center for Research Resources, grant UL1 RR024975-01, and the National Center for Advancing Translational Sciences, grant 2 UL1 TR000445-06. The Nicaraguan Pediatric Cohort Study was also supported by NIH grant R01AI099631 to A.B. and grant VE-1 to E.H. from the Pediatric Dengue Vaccine Initiative. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
DECLARATION OF INTERESTS
R.S.B. and E.H. have served on the Scientific Advisory Boards for Takeda vaccines, VaxArt, and Invivyd Therapeutics and has collaborations with Gilead, Janssen Pharmaceuticals, Pardas Biosciences, and Chimerix. A.M.D. has served as an unpaid consultant for Moderna and Takeda vaccines and is an unpaid member of Merck’s dengue vaccine Scientific Advisory Board, and R.S.B. and A.M.D. are inventors on pending and approved flavivirus vaccine and diagnostic patents filed by the University of North Carolina at Chapel Hill. A.M.D. is co-directing a partnership program between UNC and Moderna to develop flavivirus vaccines. UNC has applied for a patent related to the chimeric viruses. J.E.C. has served as a consultant for Takeda vaccines, Sanofi Pasteur, Pfizer, and Novavax; is on the Scientific Advisory Boards of CompuVax and Meissa Vaccines; and is a Founder of IDBiologics, Inc.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.chom.2023.10.004.
REFERENCES
- 1.de Silva AM, and Harris E. (2018). Which dengue vaccine approach is the most promising, and should we be concerned about enhanced disease after vaccination? The path to a dengue vaccine: learning from human natural dengue infection studies and vaccine trials. Cold Spring Harb. Perspect. Biol. 10, a029371. 10.1101/cshperspect.a029371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierson TC, and Diamond MS (2015). A game of numbers: the stoichiometry of antibody-mediated neutralization of Flavivirus infection. Prog. Mol. Biol. Transl. Sci. 129, 141–166. 10.1016/bs.pmbts.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, et al. (2013). The global distribution and burden of dengue. Nature 496, 504–507. 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC (2021). About dengue. https://www.cdc.gov/dengue/about/index.html.
- 5.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Moyes CL, Farlow AW, Scott TW, and Hay SI (2012). Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl. Trop. Dis. 6, e1760. 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duong V, Lambrechts L, Paul RE, Ly S, Lay RS, Long KC, Huy R, Tarantola A, Scott TW, Sakuntabhai A, and Buchy P. (2015). Asymptomatic humans transmit dengue virus to mosquitoes. Proc. Natl. Acad. Sci. USA 112, 14688–14693. 10.1073/pnas.1508114112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grange L, Simon-Loriere E, Sakuntabhai A, Gresh L, Paul R, andHarris E. (2014). Epidemiological risk factors associated with high global frequency of inapparent dengue virus infections. Front. Immunol. 5, 280. 10.3389/fimmu.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halstead SB (2015). Pathogenesis of dengue: dawn of a New Era. F1000Res 4. F1000 Faculty Rev-1353. 10.12688/f1000research.7024.1. [DOI] [Google Scholar]
- 9.Halstead SB, Scanlon JE, Umpaivit P, and Udomsakdi S. (1969). Dengue and Chikungunya virus infection in man in Thailand, 1962–1964. IV. Epidemiologic studies in the Bangkok metropolitan area. Am. J. Trop. Med. Hyg. 18, 997–1021. 10.4269/ajtmh.1969.18.997. [DOI] [PubMed] [Google Scholar]
- 10.Russell PK, Yuill TM, Nisalak A, Udomsakdi S, Gould DJ, and Winter PE (1968). An insular outbreak of dengue hemorrhagic fever. II. Virologic and serologic studies. Am. J. Trop. Med. Hyg. 17, 600–608. 10.4269/ajtmh.1968.17.600. [DOI] [PubMed] [Google Scholar]
- 11.Winter PE, Nantapanich S, Nisalak A, Udomsakdi S, Dewey RW, and Russell PK (1969). Recurrence of epidemic dengue hemorrhagic fever in an insular setting. Am. J. Trop. Med. Hyg. 18, 573–579. 10.4269/ajtmh.1969.18.573. [DOI] [PubMed] [Google Scholar]
- 12.Winter PE, Yuill TM, Udomsakdi S, Gould D, Nantapanich S, and Russell PK (1968). An insular outbreak of dengue hemorrhagic fever. I. Epidemiologic observations. Am. J. Trop. Med. Hyg. 17, 590–599. 10.4269/ajtmh.1968.17.590. [DOI] [PubMed] [Google Scholar]
- 13.Katzelnick LC, Montoya M, Gresh L, Balmaseda A, and Harris E. (2016). Neutralizing antibody titers against dengue virus correlate with protection from symptomatic infection in a longitudinal cohort. Proc. Natl. Acad. Sci. USA 113, 728–733. 10.1073/pnas.1522136113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell LP, Luther C, Moo-Llanes D, Ramsey JM, Danis-Lozano R, and Peterson AT (2015). Climate change influences on global distributions of dengue and Chikungunya virus vectors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370. 10.1098/rstb.2014.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mackenzie JM, and Westaway EG (2001). Assembly and maturation of the Flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75, 10787–10799. 10.1128/JVI.75.22.10787-10799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadler K, Allison SL, Schalich J, and Heinz FX (1997). Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71, 8475–8481. 10.1128/JVI.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modis Y, Ogata S, Clements D, and Harrison SC (2004). Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319. 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 18.Anderson R, King AD, and Innis BL (1992). Correlation of E protein binding with cell susceptibility to dengue 4 virus infection. J. Gen. Virol. 73, 2155–2159. 10.1099/0022-1317-73-8-2155. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Maguire T, and Marks RM (1996). Demonstration of binding of dengue virus envelope protein to target cells. J. Virol. 70, 8765–8772. 10.1128/JVI.70.12.8765-8772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandl CW, Guirakhoo F, Holzmann H, Heinz FX, and Kunz C. (1989). Antigenic structure of the Flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J. Virol. 63, 564–571. 10.1128/JVI.63.2.564-571.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinz FX (1986). Epitope mapping of Flavivirus glycoproteins. Adv. Virus Res. 31, 103–168. 10.1016/s0065-3527(08)60263-8. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, and Rossmann MG (2003). Structures of immature Flavivirus particles. EMBO J. 22, 2604–2613. 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn RJ, Dowd KA, Beth Post C, and Pierson TC (2015). Shake,rattle, and roll: impact of the dynamics of Flavivirus particles on their interactions with the host. Virology 479–480, 508–517. 10.1016/j.virol.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson TC, and Diamond MS (2008). Molecular mechanisms of antibody-mediated neutralisation of flavivirus infection. Expert Rev. Mol. Med. 10, e12. 10.1017/S1462399408000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen R, and Vasilakis N. (2011). Dengue–quo tu et quo vadis? Viruses 3, 1562–1608. 10.3390/v3091562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons CP, Farrar JJ, Nguyen v V., and Wills B. (2012). Dengue. N. Engl. J. Med. 366, 1423–1432. 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 27.Weaver SC, and Vasilakis N. (2009). Molecular evolution of dengue viruses: contributions of phylogenetics to understanding the history and epidemiology of the preeminent arboviral disease. Infect. Genet. Evol. 9, 523–540. 10.1016/j.meegid.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rico-Hesse R. (2003). Microevolution and virulence of dengue viruses. Adv. Virus Res. 59, 315–341. 10.1016/s0065-3527(03)59009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wahala WM, and Silva AM (2011). The human antibody response todengue virus infection. Viruses 3, 2374–2395. 10.3390/v3122374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katzelnick LC, and Harris E. (2018). The use of longitudinal cohorts forstudies of dengue viral pathogenesis and protection. Curr. Opin. Virol. 29, 51–61. 10.1016/j.coviro.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halstead SB (1988). Pathogenesis of dengue: challenges to molecular biology. Science 239, 476–481. 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 32.Kouri GP, Guzmán MG, and Bravo JR (1987). Why dengue haemorrhagic fever in Cuba? 2. An integral analysis. Trans. R. Soc. Trop. Med. Hyg. 81, 821–823. 10.1016/0035-9203(87)90042-3. [DOI] [PubMed] [Google Scholar]
- 33.Coloma J, and Harris E. (2015). Broad and strong: the ultimate antibody to dengue virus. Nat. Immunol. 16, 135–137. 10.1038/ni.3081. [DOI] [PubMed] [Google Scholar]
- 34.de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE Jr., and de Silva AM (2012). Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl. Acad. Sci. USA 109, 7439–7444. 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy BR, and Whitehead SS (2011). Immune response to dengue virus and prospects for a vaccine. Annu. Rev. Immunol. 29, 587–619. 10.1146/annurev-immunol-031210-101315. [DOI] [PubMed] [Google Scholar]
- 36.Martinez DR, Yount B, Nivarthi U, Munt JE, Delacruz MJ, Whitehead SS, Durbin AP, de Silva AM, and Baric RS (2020). Antigenic variation of the dengue virus 2 genotypes impacts the neutralization activity of human antibodies in vaccinees. Cell Rep. 33, 108226. 10.1016/j.celrep.2020.108226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallichotte EN, Baric TJ, Nivarthi U, Delacruz MJ, Graham R, Widman DG, Yount BL, Durbin AP, Whitehead SS, de Silva AM, and Baric RS (2018). Genetic variation between dengue virus Type 4 strains impacts human antibody binding and neutralization. Cell Rep. 25, 1214–1224. 10.1016/j.celrep.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zulueta A, Martín J, Hermida L, Alvarez M, Valdés I, Prado I, Chinea G, Rosario D, Guillén G, and Guzmán MG (2006). Amino acid changes in the recombinant Dengue 3 Envelope domain III determine its antigenicity and immunogenicity in mice. Virus Res. 121, 65–73. 10.1016/j.virusres.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Waggoner JJ, Balmaseda A, Gresh L, Sahoo MK, Montoya M, Wang C, Abeynayake J, Kuan G, Pinsky BA, and Harris E. (2016). Homotypic dengue virus reinfections in Nicaraguan children. J. Infect. Dis. 214, 986–993. 10.1093/infdis/jiw099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Forshey BM, Stoddard ST, and Morrison AC (2016). Dengue viruses and lifelong immunity: reevaluating the conventional wisdom. J. Infect. Dis. 214, 979–981. 10.1093/infdis/jiw102. [DOI] [PubMed] [Google Scholar]
- 41.Messer WB, Yount B, Hacker KE, Donaldson EF, Huynh JP, de Silva AM, and Baric RS (2012). Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization. PLoS Negl. Trop. Dis. 6, e1486. 10.1371/journal.pntd.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinsky A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T, et al. (2015). Erratum: Corrigendum: A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 785. 10.1038/ni0715-785a. [DOI] [PubMed] [Google Scholar]
- 43.Patel B, Longo P, Miley MJ, Montoya M, Harris E, and de Silva AM (2017). Dissecting the human serum antibody response to secondary dengue virus infections. PLoS Negl. Trop. Dis. 11, e0005554. 10.1371/journal.pntd.0005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halstead SB (1982). Immune enhancement of viral infection. Prog. Allergy 31, 301–364. [PubMed] [Google Scholar]
- 45.de Alwis R, Williams KL, Schmid MA, Lai CY, Patel B, Smith SA, Crowe JE, Wang WK, Harris E, and de Silva AM (2014). Dengue viruses are enhanced by distinct populations of serotype cross-reactive antibodies in human immune sera. PLoS Pathog. 10, e1004386. 10.1371/journal.ppat.1004386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salje H, Cummings DAT, Rodriguez-Barraquer I, Katzelnick LC, Lessler J, Klungthong C, Thaisomboonsuk B, Nisalak A, Weg A, Ellison D, et al. (2018). Reconstruction of antibody dynamics and infection histories to evaluate dengue risk. Nature 557, 719–723. 10.1038/s41586-018-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, and Harris E. (2017). Antibody-dependent enhancement of severe dengue disease in humans. Science 358, 929–932. 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lidbury BA, and Mahalingam S. (2000). Specific ablation of antiviral gene expression in macrophages by antibody-dependent enhancement of Ross River virus infection. J. Virol. 74, 8376–8381. 10.1128/jvi.74.18.8376-8381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chareonsirisuthigul T, Kalayanarooj S, and Ubol S. (2007). Dengue virus (DENV) antibody-dependent enhancement of infection upregulates the production of anti-inflammatory cytokines, but suppresses anti-DENV free radical and pro-inflammatory cytokine production, in THP-1 cells. J. Gen. Virol. 88, 365–375. 10.1099/vir.0.82537-0. [DOI] [PubMed] [Google Scholar]
- 50.Dayan GH, Langevin E, Forrat R, Zambrano B, Noriega F, Frago C, Bouckenooghe A, Machabert T, Savarino S, and DiazGranados CA (2020). Efficacy after 1 and 2 doses of CYD-TDV in dengue endemic areas by dengue serostatus. Vaccine 38, 6472–6477. 10.1016/j.vaccine.2020.07.056. [DOI] [PubMed] [Google Scholar]
- 51.Dayan GH, Langevin E, Gilbert PB, Wu Y, Moodie Z, Forrat R, Price B, Frago C, Bouckenooghe A, Cortes M, et al. (2020). Assessment of the long-term efficacy of a dengue vaccine against symptomatic, virologically-confirmed dengue disease by baseline dengue serostatus. Vaccine 38, 3531–3536. 10.1016/j.vaccine.2020.03.029. [DOI] [PubMed] [Google Scholar]
- 52.Gallichotte EN, Henein S, Nivarthi U, Delacruz M, Scobey T, Bonaparte M, Moser J, Munteanu A, Baric R, and de Silva AM (2022). Vaccine-induced antibodies to contemporary strains of dengue virus type 4 show a mechanistic correlate of protective immunity. Cell Rep. 39, 110930. 10.1016/j.celrep.2022.110930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henein S, Adams C, Bonaparte M, Moser JM, Munteanu A, Baric R, and de Silva AM (2021). Dengue vaccine breakthrough infections reveal properties of neutralizing antibodies linked to protection. J. Clin. Invest. 131, e147066. 10.1172/JCI147066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sridhar S, Luedtke A, Langevin E, Zhu M, Bonaparte M, Machabert T, Savarino S, Zambrano B, Moureau A, Khromava A, et al. (2018). Effect of dengue serostatus on dengue vaccine safety and efficacy. N. Engl. J. Med. 379, 327–340. 10.1056/NEJMoa1800820. [DOI] [PubMed] [Google Scholar]
- 55.Capeding MR, Tran NH, Hadinegoro SR, Ismail HI, Chotpitayasunondh T, Chua MN, Luong CQ, Rusmil K, Wirawan DN, Nallusamy R, et al. (2014). Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 384, 1358–1365. 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- 56.Deng SQ, Yang X, Wei Y, Chen JT, Wang XJ, and Peng HJ (2020). A review on dengue vaccine development. Vaccines 8, 63. 10.3390/vaccines8010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biswal S, Reynales H, Saez-Llorens X, Lopez P, Borja-Tabora C, Kosalaraksa P, Sirivichayakul C, Watanaveeradej V, Rivera L, Espinoza F, et al. (2019). Efficacy of a tetravalent dengue vaccine in healthy children and adolescents. N. Engl. J. Med. 381, 2009–2019. 10.1056/NEJMoa1903869. [DOI] [PubMed] [Google Scholar]
- 58.Nivarthi UK, Swanstrom J, Delacruz MJ, Patel B, Durbin AP, Whitehead SS, Kirkpatrick BD, Pierce KK, Diehl SA, Katzelnick L, et al. (2021). A tetravalent live attenuated dengue virus vaccine stimulates balanced immunity to multiple serotypes in humans. Nat. Commun. 12, 1102. 10.1038/s41467-021-21384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henein S, Swanstrom J, Byers AM, Moser JM, Shaik SF, Bonaparte M, Jackson N, Guy B, Baric R, and de Silva AM (2017). Dissecting antibodies induced by a chimeric yellow fever-dengue, live-attenuated, tetravalent dengue vaccine (CYD-TDV) in naive and dengue-exposed individuals. J. Infect. Dis. 215, 351–358. 10.1093/infdis/jiw576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young E, Carnahan RH, Andrade DV, Kose N, Nargi RS, Fritch EJ, Munt JE, Doyle MP, White L, Baric TJ, et al. (2020). Identification of dengue virus Serotype 3 specific antigenic sites targeted by neutralizing human antibodies. Cell Host Microbe 27, 710–724.e7. 10.1016/j.chom.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Messer WB, Yount BL, Royal SR, de Alwis R, Widman DG, Smith SA, Crowe JE Jr., Pfaff JM, Kahle KM, Doranz BJ, et al. (2016). Functional transplant of a dengue virus Serotype 3 (DENV3)-specific human monoclonal antibody epitope into DENV1. J. Virol. 90, 5090–5097. 10.1128/JVI.00155-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, et al. (2012). The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci. Transl. Med. 4, 139ra83. 10.1126/scitranslmed.3003888. [DOI] [PubMed] [Google Scholar]
- 63.Fibriansah G, Tan JL, Smith SA, de Alwis AR, Ng TS, Kostyuchenko VA, Ibarra KD, Wang J, Harris E, de Silva A, et al. (2014). A potent anti-dengue human antibody preferentially recognizes the conformation of E protein monomers assembled on the virus surface. EMBO Mol. Med. 6, 358–371. 10.1002/emmm.201303404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magnani DM, Silveira CGT, Ricciardi MJ, Gonzalez-Nieto L, Pedreño-Lopez N, Bailey VK, Gutman MJ, Maxwell HS, Domingues A, Costa PR, et al. (2017). Potent plasmablast-derived antibodies elicited by the National Institutes of Health dengue vaccine. J. Virol. 91. e00867–e00817. 10.1128/JVI.00867-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fibriansah G, Tan JL, Smith SA, de Alwis R, Ng TS, Kostyuchenko VA, Jadi RS, Kukkaro P, de Silva AM, Crowe JE, and Lok SM (2015). A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat. Commun. 6, 6341. 10.1038/ncomms7341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kuan G, Gordon A, Avilés W, Ortega O, Hammond SN, Elizondo D, Nuñez A, Coloma J, Balmaseda A, and Harris E. (2009). The Nicaraguan pediatric dengue cohort study: study design, methods, use of information technology, and extension to other infectious diseases. Am. J. Epidemiol. 170, 120–129. 10.1093/aje/kwp092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith SA, de Alwis AR, Kose N, Jadi RS, de Silva AM, and Crowe JE Jr. (2014). Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J. Virol. 88, 12233–12241. 10.1128/JVI.00247-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swanstrom JA, Nivarthi UK, Patel B, Delacruz MJ, Yount B, Widman DG, Durbin AP, Whitehead SS, De Silva AM, and Baric RS (2019). Beyond neutralizing antibody levels: the epitope specificity of antibodies induced by National Institutes of Health monovalent dengue virus vaccines. J. Infect. Dis. 220, 219–227. 10.1093/infdis/jiz109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, and Fouchier RA (2004). Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376. 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 70.Lindesmith LC, Boshier FAT, Brewer-Jensen PD, Roy S, Costantini V, Mallory ML, Zweigart M, May SR, Conrad H, O’Reilly KM, et al. (2022). Immune imprinting drives human Norovirus potential for global spread. mBio 13, e0186122. 10.1128/mbio.01861-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moodie Z, Juraska M, Huang Y, Zhuang Y, Fong Y, Carpp LN, Self SG, Chambonneau L, Small R, Jackson N, et al. (2018). Neutralizing antibody correlates analysis of tetravalent dengue vaccine efficacy trials in Asia and Latin America. J. Infect. Dis. 217, 742–753. 10.1093/infdis/jix609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fibriansah G, Ibarra KD, Ng TS, Smith SA, Tan JL, Lim XN, Ooi JS, Kostyuchenko VA, Wang J, de Silva AM, et al. (2015). Dengue virus. Cryo-EM structure of an antibody that neutralizes dengue virus type 2 by locking E protein dimers. Science 349, 88–91. 10.1126/science.aaa8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dejnirattisai W, Wongwiwat W, Supasa S, Zhang X, Dai X, Rouvinski A, Jumnainsong A, Edwards C, Quyen NTH, Duangchinda T, et al. (2015). A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat. Immunol. 16, 170–177. 10.1038/ni.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA,Chipman PR, Kuhn RJ, Diamond MS, and Rossmann MG (2010). Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc. Natl. Acad. Sci. USA 107, 18950–18955. 10.1073/pnas.1011036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gallichotte EN, Young EF, Baric TJ, Yount BL, Metz SW, Begley MC, de Silva AM, and Baric RS (2019). Role of Zika virus envelope protein domain III as a target of human neutralizing antibodies. mBio 10. e01485–e01419. 10.1128/mBio.01485-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raut R, Corbett KS, Tennekoon RN, Premawansa S, Wijewickrama A, Premawansa G, Mieczkowski P, Rückert C, Ebel GD, De Silva AD, and de Silva AM (2019). Dengue type 1 viruses circulating in humans are highly infectious and poorly neutralized by human antibodies. Proc. Natl. Acad. Sci. USA 116, 227–232. 10.1073/pnas.1812055115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tse LV, Meganck RM, Dong S, Adams LE, White LJ, Mallory ML, Jadi R, de Silva AM, and Baric RS (2022). Generation of mature DENVs via genetic modification and directed evolution. mBio 13, e0038622. 10.1128/mbio.00386-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.A Dowd K, Sirohi D, D Speer S, VanBlargan LA, Chen RE, Mukherjee S, Whitener BM, Govero J, Aleshnick M, Larman B, et al. (2023). prM-reactive antibodies reveal a role for partially mature virions in dengue virus pathogenesis. Proc. Natl. Acad. Sci. USA 120, e2218899120. 10.1073/pnas.2218899120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallichotte EN, Baric TJ, Yount BL Jr., Widman DG, Durbin A, Whitehead S, Baric RS, and de Silva AM (2018). Human dengue virus serotype 2 neutralizing antibodies target two distinct quaternary epitopes. PLoS Pathog. 14, e1006934. 10.1371/journal.ppat.1006934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dias RS, Teixeira MD, Xisto MF, Prates JWO, Silva JDD, Mello IO, Silva CCD, and De Paula SO (2021). DENV-3 precursor membrane (prM) glycoprotein enhances E protein immunogenicity and confers protection against DENV-2 infections in a murine model. Hum. Vaccin. Immunother. 17, 1271–1277. 10.1080/21645515.2020.1826798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Allison SL, Schalich J, Stiasny K, Mandl CW, and Heinz FX (2001). Mutational evidence for an internal fusion peptide in Flavivirus envelope protein E. J. Virol. 75, 4268–4275. 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dubey KD, Chaubey AK, and Ojha RP (2013). Stability of trimeric DENV envelope protein at low and neutral pH: an insight from MD study. Biochim. Biophys. Acta 1834, 53–64. 10.1016/j.bbapap.2012.08.014. [DOI] [PubMed] [Google Scholar]
- 83.Biswal S, Borja-Tabora C, Martinez Vargas L, Velásquez H, Theresa Alera M, Sierra V, Johana Rodriguez-Arenales E, Yu D, Wickramasinghe VP, Duarte Moreira E Jr., et al. (2020). Efficacy of a tetravalent dengue vaccine in healthy children aged 4–16 years: a randomised, placebo-controlled, phase 3 trial. Lancet 395, 1423–1433. 10.1016/S0140-6736(20)30414-1. [DOI] [PubMed] [Google Scholar]
- 84.Young E, Yount B, Pantoja P, Henein S, Meganck RM, McBride J, Munt JE, Baric TJ, Zhu D, Scobey T, et al. (2023). A live dengue virus vaccine carrying a chimeric envelope glycoprotein elicits dual DENV2-DENV4 serotype-specific immunity. Nat. Commun. 14, 1371. 10.1038/s41467-023-36702-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rupp R, Luckasen GJ, Kirstein JL, Osorio JE, Santangelo JD, Raanan M, Smith MK, Wallace D, Gordon GS, and Stinchcomb DT (2015). Safety and immunogenicity of different doses and schedules of a live attenuated tetravalent dengue vaccine (TDV) in healthy adults: A Phase 1b randomized study. Vaccine 33, 6351–6359. 10.1016/j.vaccine.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 86.Tricou V, Low JG, Oh HM, Leo YS, Kalimuddin S, Wijaya L, Pang J, Ling LM, Lee TH, Brose M, et al. (2020). Safety and immunogenicity of a single dose of a tetravalent dengue vaccine with two different serotype-2 potencies in adults in Singapore: A phase 2, double-blind, randomised, controlled trial. Vaccine 38, 1513–1519. 10.1016/j.vaccine.2019.11.061. [DOI] [PubMed] [Google Scholar]
- 87.White LJ, Young EF, Stoops MJ, Henein SR, Adams EC, Baric RS, and de Silva AM (2021). Defining levels of dengue virus serotype-specific neutralizing antibodies induced by a live attenuated tetravalent dengue vaccine (TAK-003). PLoS Negl. Trop. Dis. 15, e0009258. 10.1371/journal.pntd.0009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Osorio JE, Brewoo JN, Silengo SJ, Arguello J, Moldovan IR, Tary-Lehmann M, Powell TD, Livengood JA, Kinney RM, Huang CY, and Stinchcomb DT (2011). Efficacy of a tetravalent chimeric dengue vaccine (DENVax) in Cynomolgus macaques. Am. J. Trop. Med. Hyg. 84, 978–987. 10.4269/ajtmh.2011.10-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Osorio JE, Huang CY, Kinney RM, and Stinchcomb DT (2011). Development of DENVax: a chimeric dengue-2 PDK-53-based tetravalent vaccine for protection against dengue fever. Vaccine 29, 7251–7260. 10.1016/j.vaccine.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang CY, Butrapet S, Tsuchiya KR, Bhamarapravati N, Gubler DJ, and Kinney RM (2003). Dengue 2 PDK-53 virus as a chimeric carrier for tetravalent dengue vaccine development. J. Virol. 77, 11436–11447. 10.1128/jvi.77.21.11436-11447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andrade DV, Warnes C, Young E, Katzelnick LC, Balmaseda A, de Silva AM, Baric RS, and Harris E. (2019). Tracking the polyclonal neutralizing antibody response to a dengue virus serotype 1 type-specific epitope across two populations in Asia and the Americas. Sci. Rep. 9, 16258. 10.1038/s41598-019-52511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beltramello M, Williams KL, Simmons CP, Macagno A, Simonelli L, Quyen NT, Sukupolvi-Petty S, Navarro-Sanchez E, Young PR, de Silva AM, et al. (2010). The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 8, 271–283. 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gallichotte EN, Widman DG, Yount BL, Wahala WM, Durbin A, Whitehead S, Sariol CA, Crowe JE Jr., de Silva AM, and Baric RS (2015). A new quaternary structure epitope on dengue virus serotype 2 is the target of durable type-specific neutralizing antibodies. mBio 6. e01461–15. 10.1128/mBio.01461-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gallichotte EN, Menachery VD, Yount BL Jr., Widman DG, Dinnon KH 3rd, Hartman S, de Silva AM, and Baric RS (2017). Epitope Addition and Ablation via Manipulation of a Dengue Virus Serotype 1 Infectious Clone. mSphere 2. e00380–16. 10.1128/mSphere.00380-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wahala WMPB, Donaldson EF, de Alwis R, Accavitti-Loper MA, Baric RS, et al. (2010). Natural Strain Variation and Antibody Neutralization of Dengue Serotype 3 Viruses. PLOS Pathogens 6, e1000821. 10.1371/journal.ppat.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Widman DG, and Baric RS (2015). Dengue virus envelope protein domain I/II hinge: a key target for dengue virus vaccine design? Expert Rev Vaccines 14, 5–8. 10.1586/14760584.2015.961431. [DOI] [PubMed] [Google Scholar]
- 97.Tse LV, Meganck RM, Araba KC, Yount BL, Shaffer KM, Hou YJ, Munt JE, Adams LE, Wykoff JA, Morowitz JM, et al. (2022). Genomewide CRISPR knockout screen identified PLAC8 as an essential factor for SADS-CoVs infection. Proc. Natl. Acad. Sci. USA 119, e2118126119. 10.1073/pnas.2118126119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep sequencing data have been deposited in the NCBI database and are publicly available upon this manuscript’s publication date. Accession numbers are listed in the key resources table (SRR23674710, SRR23675053, SRR23675073).
No original code was developed for this manuscript.
Any additional information required to reanalyze data reported in this manuscript is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| DENV-443 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943662 |
| DENV-115 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943663 |
| DENV-290 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943664 |
| DENV-419 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943665 |
| DENV-P7C07 | Magnani et al.64/JVI.00867–17 | RRID:AB_2943666 |
| DENV-P3D05 | Magnani et al.64/JVI.00867–17 | RRID:AB_2943667 |
| DENV-66 (hybridoma produced) | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | RRID:AB_2943668 |
| DENV-5J7 (plasmid produced Ig) | Beltramello et al.92; https://doi.org/10.1016/j.chom.2010.08.007 | RRID:AB_2943669 |
| DENV-1F4 (hybridoma produced) | Fibriansah et al.63 | RRID:AB_2943670 |
| DENV-14C10 (plasmid produced Ig) | Teoh et al.62 | RRID:AB_2943671 |
| Goat anti-human IgG (Fc)-AP | Meridian Life Science, Inc | Cat#W99008A; RRID: AB_151913 |
| DENV-4G2 (hybridoma produced Ig) | ATCC HB-112 | D1–4G2–4-15; RRID: AB-2722738 |
| DENV-2H2 (hybridoma produced Ig) | ATCC HB-114 | D3–2H2–9-21; RRID: AB-95338 |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| DENV1ic West Pac 74 | Gallichotte et al., 201793; https://doi.org/10.1128/mSphere.00380–16 | N/A |
| DENV2ic 16803 | Gallichotte et al., 201594; https://doi.org/10.1128/mBio.01461–15 | N/A |
| DENV3 Thailand/CH53489/1973 | Wahala et al.95; https://doi.org/10.1371/journal.ppat.1000821 | N/A |
| DENV3ic Sri Lanka 89 G-III | Messer et al.61 | N/A |
| DENV3 Indonesia 1982, G-I | Messer et al.41 | N/A |
| DENV3 Thailand 1995, G-II | Messer et al.41 | N/A |
| DENV3 Cuba 2002, G-III | Messer et al.41 | N/A |
| DENV3 Puerto Rico 1977, G-IV | Messer et al.41 | N/A |
| DENV4ic Sri Lanka 92 | Widman et al.96; https://10.1586/14760584.2015.961431 | N/A |
| DENV1/3 EDI B | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | NCBI accession #:SRR23674710 |
| DENV1/3 EDII A | This Manuscript | NCBI accession #: SRR23675053 |
| DENV1/3 EDII B | This Manuscript | N/A |
| DENV1/3 EDII C | This Manuscript | N/A |
| DENV1/3 EDII D | This Manuscript | N/A |
| DENV1/3 EDIIIa | This Manuscript | N/A |
| DENV1/3 EDIIIb | This Manuscript | NCBI accession #: SRR23675073 |
| DENV1/3 EDIIIc | This Manuscript | N/A |
| DENV1/3 EDIIId | This Manuscript | N/A |
| DENV3/3 C1 | This Manuscript | N/A |
| DENV3/3 C2 | This Manuscript | N/A |
| DENV3/3 C3 | This Manuscript | N/A |
| DENV3/3 C4 | This Manuscript | N/A |
|
| ||
| Biological Samples | ||
|
| ||
| Serum Samples from DENV3 Primary Infection Survivors | This Manuscript | Adult Arbovirus Travelers Cohort, UNC; DT103/ATC A, DT133/ATC B, DT226/ATC C |
| Serum Samples from DENV3 Primary Infection Survivors | This Manuscript | Nicaraguan Pediatric Dengue Cohort Study, Donor IDs: 4082.7/NPDCS A, 4862.7/NPDCS B, 5862.7/NPDCS C |
| Serum Samples from DENV3 NIH monovalent vaccine trial | Swanstrom et al.68; PMCID: PMC6581895 | Depeleted IDs: 257.01.02/NIH A, 257.01.16/NIH B, 257.03.47/NIH D, 257.03.50/NIH F; Undepleted IDs: 257.03.45/NIH C, 257.03.49/NIH E |
| Normal Human Serum (NHS) Sample | Henein et al.53; PMCID: PMC8245170 | DT236 |
|
| ||
| Chemicals, Peptides, & Recombinant Proteins | ||
|
| ||
| DMEM, high glucose, GlutaMAX™ Supplement | Thermo Fisher Scientific | Cat# 10566–024 |
| Dulbecco’s modified Eagle’s/Ham’s F-12 50/50 Mix | Gibco | 10–092-CV |
| Opti-MEM I | Gibco | 31985–070 |
| TrueBlue substrate | KPL | 0510–0050 |
| TRIzol LS | Thermo Fisher Scientific | 10296028 |
| Non-fat dry milk | Bio Rad | Cat# 1706404 |
|
| ||
| Cell Lines | ||
|
| ||
| C6/36 cells | ATCC | CRL-1660; RRID: CV_CL_Z230 |
| Vero-81 cells | ATCC | CCL-81; RRID: CV-CL_0059 |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| Illumina Stranded Total RNA Prep | Illumina | Ribozero Plus Protocol; 20040525 |
| MiSeq Reagent Kit v3 | Illumina | MS-102–3001 |
|
| ||
| Deposited Data | ||
|
| ||
| DENV1/3 EDI B | Young et al.60; https://doi.org/10.1016/j.chom.2020.04.007 | NCBI accession #:SRR23674710 |
| DENV1/3 EDII A | This Manuscript | NCBI accession #: SRR23675053 |
| DENV1/3 EDIIIb | This Manuscript | NCBI accession #: SRR23675073 |
|
| ||
| Recombinant DNA | ||
|
| ||
| Seq & PCR Primer: DENV3 (G-IV), 916F: GCC CAT TAC ATA GGC ACT TC | This Manuscript | Cluster 916F |
| Seq & PCR Primer: DENV3 (G-IV), 2143R: CGA GCT TCC TTT CTT GTA CC | This Manuscript | Cluster 2143R |
| Seq Primer, Cluster 935R: GAA GTG CCT ATG TAA TGG GC | This Manuscript | Cluster 935R |
| Seq Primer, Cluster 1420F: GTT CAC ACA GGA GAT CAA CAC C | This Manuscript | Cluster 1420F |
| Seq Primer, Cluster 1441R: GGT GTT GAT CTC CTG TGT GAA C | This Manuscript | Cluster 1441R |
| Seq Primer, Cluster 1735F: GTG GTT GTT CTT GGA TCG CAA G | This Manuscript | Cluster 1735F |
| Seq Primer, Cluster 1756R: CTT GCG ATC CAA GAA CAA CCA C | This Manuscript | Cluster 1756 R |
| PCR Primer: DENV1/3 Panel; A1F: CAT GCT CCT CAT GCT GCT GC | This Manuscript | A1F |
| PCR Primer: DENV1/3 Panel; U24R: GCC TTT CCA GTT GAT TAC AC | This Manuscript | U24R |
| Seq Primer: DENV1/3 Panel; 1215F: CGA TTC GAG ATG TCC AAC ACA AGG | This Manuscript | 1215 F |
| Seq Primer: DENV1/3 Panel; 1238R: CCT TGT GTT GGA CAT CTC GAA TCG | This Manuscript | 1238 R |
| Seq Primer: DENV1/3 Panel; 1765R: CTA GTA CGA CTA CTT CCT GC | This Manuscript | 1765 R |
| Seq Primer: DENV1/3 Panel; 1650F: CTT ACC ACT GCC TTG GAC CTC | This Manuscript | 1650 F |
| Seq Primer: DENV1/3 Panel; 1670R: GAG GTC CAA GGC AGT GGT AAG | This Manuscript | 1670R |
|
| ||
| Software and Algorithms | ||
|
| ||
| GraphPad Prism, v9.0 | GraphPad Software, Inc. | https://www.graphpad.com |
| PyMOL | Schrödinger, LLC | https://www.pymol.org/ |
|
| ||
| Other | ||
|
| ||
| MiSeq | Illumina | N/A |
| EL406 washer dispenser | BioTek | N/A |
| Biostack microplate stacker | BioTek | N/A |
| Immunospot | CTL | N/A |