The risk of any child developing acute leukaemia is about 1 in 2000 with 400-450 new cases a year in the United Kingdom. Cure rates approaching 75% can be achieved with combination chemotherapy, but this figure disguises success rates that vary from 10% to 90% with the different biological subtypes of the disease. In this review I discuss how new insights into the underlying molecular biology of leukaemia have changed our understanding of the disease. Not only is there the prospect of better treatment and the introduction of new biologically based therapies, but, as the causes of disease are being unravelled, the possibility of prevention may not just be wishful thinking.
Summary points
Different chromosomal and gene abnormalities in leukaemia define biological subsets of disease with prognostic importance
Chromosome translocations generate chimeric fusion genes, which provide stable sensitive markers that are unique for each patient's leukaemic clone and can be used to track its origins and response to treatment
The common chromosome translocations in childhood leukaemia seem to initiate disease and often arise prenatally
One or more postnatal genetic alterations are also needed for leukaemia development, and in childhood acute lymphoblastic leukaemia these may be caused by abnormal immune responses to infection
Proteins coded by fusion genes operate principally by blocking cell differentiation in leukaemic cells and provide potential targets for new treatments
Methods
This article is based on information and views published from my laboratory plus comprehensive, prospective screening of leading journals for leukaemia and cancer research.
A diverse disease with variable clinical outcome
It has long been recognised that childhood leukaemia is not one homogeneous disease. The major morphological division into acute lymphoblastic leukaemia and acute myeloblastic leukaemia is supplemented by the identification of a range of subsets based on gene expression, antigens that delineate cell type or differentiation status, and chromosomal and molecular abnormalities. These include chromosome translocations—exchanges of large tracks of DNA between chromosomes, resulting (at the point of exchange) in the generation of chimeric or fusion genes1—and changes in chromosome number (hyperdiploidy or hypodiploidy). At a more subtle level, there may also be gene deletions or single nucleotide base changes in genes.
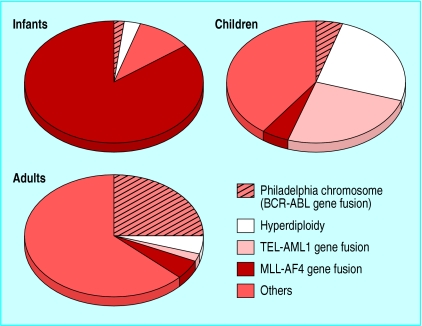
This molecular archaeology has uncovered what has long been suspected—that the acute leukaemias are biologically diverse diseases.2 Moreover, in acute lymphoblastic leukaemia these subgroups segregate with age (fig 1), which may help explain the considerable differences in outcome between infants aged <1 year, children (2-10 years old), and adults.3 A similar spectrum of molecular diversity exists for acute myeloblastic leukaemia. Several of these molecular abnormalities have independent prognostic importance in the context of particular treatment regimens (see table).2
Figure 1.
Major molecular subsets of acute lymphoblastic leukaemia in infants (<1 year old), children (2-10 years old), and adults
What most descriptions of leukaemic cells obscure, however, is the dynamic, evolving nature of the disease, a feature it shares with all other cancers.4 Leukaemia is a clonal disease (originating in a single cell) and evolves by the accrual of mutations within a clone. This results in progressive genetic diversification followed by a “natural selection” of dominant mutant subclones. Clinical outcome depends on not only the nature of the leukaemic clone but how far it has evolved by the time pathological symptoms are recognised, a correct diagnosis is made, and treatment started. Diagnostic delay increases the probability that the clone will have progressed to the point where additional mutations have been acquired, including those endowing drug resistance, rendering eradication more difficult.
The fetal origins of childhood leukaemia
There is now compelling evidence that chromosome translocations are often the first or initiating events in leukaemia, occurring prenatally during fetal development. This evidence comes from two sources—identical twin infants or children with concordant acute lymphoblastic leukaemia5,6 and retrospective scrutiny of neonatal blood spots or Guthrie cards.7,8
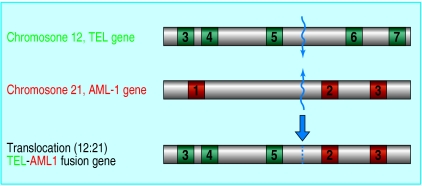
The most common structural genetic abnormality in childhood leukaemia is a fusion of two genes, TEL and AML1. This is generated by a chromosome translocation between chromosomes 12 and 21. Simultaneous breaks in the TEL gene (chromosome 12) and AML1 gene (chromosome 21) are followed by error-prone repair that stitches up the DNA across chromosomes 12 and 21, joining the normally separate TEL and AML1 genes together to form a chimeric or fusion gene (fig 2). As in other chromosomal translocations, the DNA breaks always occur in non-coding regions (introns) of genes. The precise breakpoints in the TEL and AML1 genes can be identified or mapped by the “long distance” polymerase chain reaction (PCR). Breaks always occur, more or less randomly, within a limited region of these genes, but each patient's leukaemic cells have a unique (or clone specific) breakpoint in the DNA sequence.
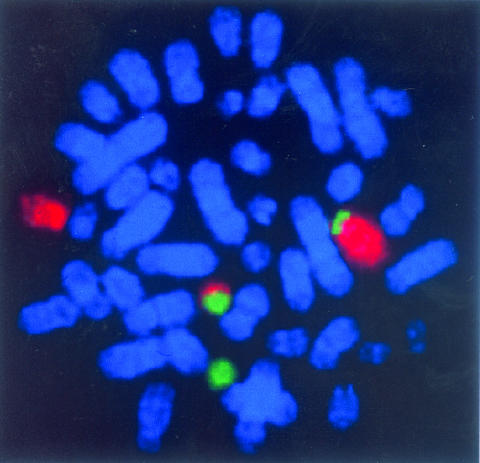
Figure 2.
Chromosomal translocation to form the TEL-AML1 fusion gene in childhood acute lymphoblastic leukaemia. Top: Fluorescence in situ hybridisation labelling of dividing leukaemic cell chromosomes with specific probes for chromosome 12 (red) and chromosome 21 (green) reveal two red and green chromosomes (one large, one small). These are copies of chromosomes 12 and 21 between which there has been a reciprocal exchange of DNA. Bottom: The TEL and AML1 genes lie at the breaks and are brought together by the exchange. The genes break in non-coding (grey) regions between the coding regions (numbered, green or red), and re-joining of the two broken genes forms a novel fusion gene
Analysis of pairs of identical twins with concordant acute lymphoblastic leukaemia shows that leukaemic cells from both twins in a pair share the identical breakpoints in TEL and AML1 genes or, in the case of infant twins with acute lymphoblastic leukaemia, the same breakpoints in the MLL gene.5,6 Monozygotic twins are, of course, themselves monoclonal and genetically identical, but gene breakpoints in leukaemic cells are not inherited—they disappear in remission.
The only plausible explanation for twin leukaemias sharing the same gene breakpoints is that the chromosomal breaks generating the fusion gene must have occurred just once, in one blood stem cell, in one twin in utero. Subsequently, but still in utero, descendent progeny of this transformed cell spread to the other twin, presumably via the anastomoses that exist within shared, single (monochorionic) placentas.5 We assume that at this early stage a clinically silent or covert preleukaemic clone is generated which, after birth, may evolve to full blown leukaemia anything from two months to 14 years later.3,6
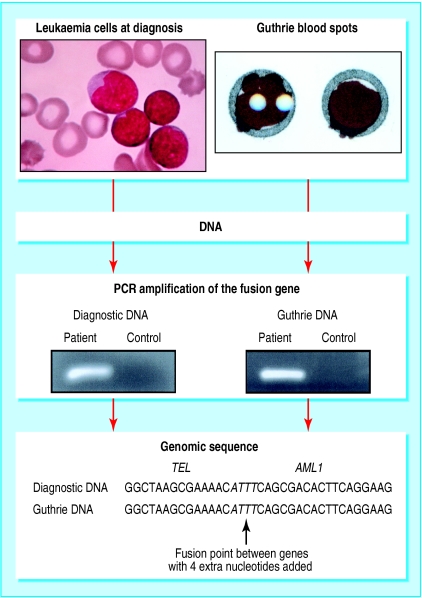
Further evidence that childhood leukaemia can originate before birth comes from scrutiny of neonatal blood spots or Guthrie cards (fig 3). PCR tests for specific fusion genes, designed for each patient, can detect as few as 1-20 leukaemic cells in a blood spot. The presence of the same fusion gene sequence in a neonatal blood spot as is in the patient's leukaemic cells at diagnosis7,8 provides unequivocal evidence that leukaemia has been initiated prenatally, probably by formation of the fusion gene itself. The conclusions from Guthrie card studies is that leukaemia is fetal in origin in all cases of infant leukaemia (with fusions of the MLL gene), in most cases of the common form of childhood acute lymphoblastic leukaemia (with TEL-AML1), and in about half of cases of childhood acute myeloblastic leukaemia (with translocation of chromosomes 8 and 21).
Figure 3.
Identification of fusion genes in neonatal blood spots of patients with leukaemia. At diagnosis of childhood acute lymphoblastic leukaemia, a TEL-AML1 fusion gene can be identified in the leukaemic cells. The TEL-AML1 sequence is first determined by long range PCR, then oligonucleotide primers are designed for that unique sequence and for use in short range (conventional) PCR. DNA is extracted from a diagnostic sample for PCR and, in parallel, a segment from a neonatal blood spot is subjected to PCR. If successful, both samples from the patient amplify to produce a nucleotide sequence visualised as a band in a gel. Sequencing of these bands shows them to be identical
The “two hit” model for childhood leukaemia
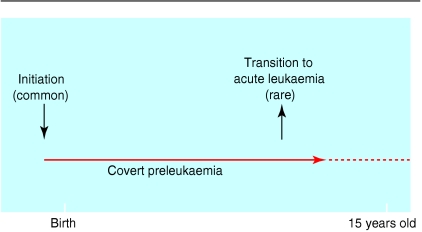
Although there are no accurate data for concordance rates of leukaemia in infant twins, anecdotally it seems to be exceptionally high, perhaps approaching 100% (that is, if one twin has it, so will the other). If correct, this suggests that MLL gene fusion in utero has a dramatic impact, ensuring subsequent leukaemia. But for children aged 2-6 years (with acute lymphoblastic leukaemia) the concordance rate is considerably lower, around 5%. This still represents a 100-fold extra risk of leukaemia for the twin of a patient with acute lymphoblastic leukaemia but also indicates the need for some additional postnatal event(s) for which there is a 1 in 20 chance, or 95% discordance. This suggests, at a minimum, a “two hit” model for the natural course of childhood leukaemia (fig 4).3
Figure 4.
Natural course of childhood leukaemia
If this model of leukaemia development is correct, then, for every child with acute lymphoblastic leukaemia diagnosed, there should be at least 20 healthy children who have had a chromosome translocation, a functional leukaemia fusion gene, and a covert preleukaemic clone generated in utero. This possibility has been investigated by screening unselected samples of newborn cord blood for fusion genes. About 600 samples have been screened, and around 1% have a leukaemic TEL-AML1 fusion gene (H Mori, et al, personal communication). This 1% represents 100 times the cumulative rate or risk of acute lymphoblastic leukaemia (with a TEL-AML1 gene), indicating that the frequency of conversion of the preleukaemic clone to overt disease is low. The real bottleneck in development of acute lymphoblastic leukaemia therefore seems to be a stringent requirement for a second “hit” after birth—that is, exposure and additional chromosomal or molecular abnormality.
Causal mechanisms
A key issue to resolve is what exposures or events might precipitate the chromosome breaks whose improper repair initiates or promotes childhood leukaemia. Given the biological diversity of leukaemia, it is highly unlikely that there is a single cause. Even for a defined biological subtype of the disease, there probably isn't one cause as such but a causal mechanism. As with other cancers, this is likely to involve an interaction of exposure (exogenous or endogenous) with inherent genetic susceptibility, and chance.4
Epidemiological evidence suggests that ionising radiation, certain chemicals (such as benzene), viruses (human T cell leukaemia/lymphoma virus type I, Epstein-Barr virus), and bacteria (Helicobacter pylori) may play a part in the development of some subtypes of leukaemia and lymphoma in adults and children. Whether any of these exposures have a major role in childhood leukaemia is uncertain, but large scale case-control molecular epidemiological studies in Britain and the United States may provide answers. The UK children's cancer study (UKCCS) seeks to address several hypotheses on different exposures, combined with definition of biological subtypes of disease and genetic studies.9 It and a parallel US study have already ruled out electromagnetic fields as a major factor in leukaemia aetiology.10
A critical role for infection?
Two hypotheses have suggested that an abnormal response to common infections plays a decisive role in the development of childhood acute lymphoblastic leukaemia. One proposes that transiently increased rates of leukaemia (of any subtype), sometimes in clear geographical clusters, are due to population mobility and mixing resulting in infection occurring in previously unexposed or susceptible individuals.11 The “delayed infection” hypothesis suggests that acute lymphoblastic leukaemia in children is caused by a lack of exposure to infection and a failure of immune system modulation in infancy.12 Later, an abnormal immune response occurs to one or more common bacterial or viral infections incurred after (delayed) mixing with infectious carriers, such as other children in playgroups or schools.
This second hypothesis is similar to the “hygiene hypothesis” put forward to explain allergies and asthma and type 1 diabetes.13 It suggests that it is the aberrant response to infection that promotes the crucial second, postnatal event.12 Epidemiological support for the delayed infection hypothesis come from studies of children with acute lymphoblastic leukaemia. These show that such children are less likely to have had some common infections in infancy, have had fewer social contacts in infancy, are more likely to have been first born, and are less likely to have received certain vaccinations, particularly for Haemophilus influenzae.14,15
Over the next year or two, it should become clear whether childhood leukaemia involves infectious exposures. If it does, this raises the possibility of prevention, but it cannot apply to all types of childhood leukaemia. For acute lymphoblastic leukaemia in infants (with MLL gene fusions), aetiological hypotheses and the available epidemiological data are distinct. A recent international epidemiological study of infant leukaemia has implicated transplacental chemical exposures to pesticides (Baygon) and a drug (dipyrone) during pregnancy.16
Inherited susceptibility
Inherited genetic variation is likely to be important in determining differential susceptibility to leukaemia, as in other cancers and diseases. Risk of infant leukaemia has been associated with polymorphic variants of the NQ01 gene, which codes for an enzyme that detoxifies benzene metabolites and quinone-containing flavonoids and other substances.17 For typical childhood acute lymphoblastic leukaemia, there is preliminary evidence that HLA class II alleles influence risk,18 and inherited variations in other immune genes that influence responses to infection probably play a role in susceptibility. Finally, risk of acute lymphoblastic leukaemia in infants, older children, and adults has been linked to inheritance of alleles of MTHFR, a key gene in the folate metabolism pathway.19 The reason for this association may lie in the way that folate metabolism affects the fidelity of DNA replication and, possibly, vulnerability to chromosomal breaks. Recent data suggest that folate intake in pregnant women may be an important dietary modifier of risk for paediatric acute lymphoblastic leukaemia.20
New therapeutic targets and better indicators of prognosis
Genetic alterations in leukaemia (and other cancers) affect the complex signalling networks that control cell proliferation, cell differentiation, and cell death by apoptosis.21 These effects help to explain why certain molecular abnormalities result in adverse clinical outcomes. They also present new opportunities for targeting treatments. For example, the BCR-ABL fusion gene (found in the Philadelphia chromosome associated with acute lymphoblastic leukaemia and chronic myeloid leukaemia) results in the production of an active kinase enzyme (from the ABL part of the gene) that drives cell proliferation independently of normal requirements for growth factor and blocks apoptosis and therefore drug responsiveness pathways. Normal p53 protein in cells is required to induce cell death after anoxia or DNA damage from exposure to drugs or irradiation. Mutations or deletions in the p53 gene are rare at presentation of leukaemia but are more common at relapse, helping to explain the therapeutic “resistance” of more advanced disease.
Additional education resources
Leukaemia Research Fund (www.lrf.org.uk). Information on booklets available on leukaemia for parents and patients as well as both reference works and books for medical students and doctors
Seminars in Haematology. 2000;37(4). Review articles on chromosome changes in leukaemia and related diseases
Henderson ES, Lister TA, Greaves MF, eds. Leukemia. 6th ed. Philadelphia: WB Saunders, 1996. Standard textbook on leukaemia covering biology and treatment in adults and children
The fusion genes generated by chromosome translocation (TEL-AML1 in acute lymphoblastic leukaemia, AML1-ETO in acute myeloblastic leukaemia, and PML-RARA in acute promyelocytic leukaemia) primarily block cell differentiation. The aberrant proteins produced by these genes inhibit gene activity and differentiation by recruiting repressor molecules.22 These repressors include histone deacetylase enzymes. These enzymes can, however, be counteracted by selective inhibitors, and one promising line of treatment—at present targeted at acute promyeloblastic leukaemia with PML-RARA fusions—is to use such drugs to reverse the block to normal cell development.23 The success in using a derivative of retinoic acid to induce remission in acute promyeloblastic leukaemia24 is also encouraging in this respect.
Another promising approach is the use of STI-571, a selective inhibitor of Abl protein and related kinases, which is has been shown to have a major impact on chronic myeloid leukaemia.25,26 Hopefully, other small compounds can be designed that will block the signalling pathways that are overactive in paediatric and adult leukaemic cells, preferably in a non-toxic fashion as with STI-571.
Another hope is that the ability to obtain more comprehensive or complete genetic profiles of leukaemic cells may allow prognosis to be defined extremely accurately.27 Prognosis in paediatric acute lymphoblastic leukaemia can already be assessed with some accuracy using highly sensitive polymerase chain reaction methods to detect molecular markers of leukaemic cells, such as clonal rearrangements of immunoglobulin genes or T cell receptor genes or the unique leukaemia fusion genes. With such methods, quantitative assessment of levels of residual leukaemic cells during treatment is predictive of later outcome28 and may therefore provide a good guide to individual patient management.
Table.
Prognostic correlates of molecular genetic abnormalities in childhood leukaemia
| Molecular marker | Associated prognosis |
|---|---|
| Infant acute lymphoblastic leukaemia | |
| MLL-AF4 fusion gene | Very poor |
| Acute lymphoblastic leukaemia | |
| Hyperdiploidy* | Very good |
| TEL-AML1 fusion gene | Good to very good† |
| Hypodiploidy‡ | Very poor |
| BCR-ABL fusion gene§ | Very poor |
| CDK4I/p16del¶ | Very poor |
| Acute myeloblastic leukaemia | |
| AML1-ETO fusion gene | Intermediate |
| MLL-AF10 fusion gene | Intermediate |
All fusion genes are generated by chromosome exchanges (translocations).
Extra copies (such as 3 instead of 2) of certain chromosomes.
Patients with TEL-AML1 fusion gene seem less likely to relapse if treated with regimens that include high dose L-asparaginase.
Loss of some whole chromosomes.
This alteration (the Philadelphia chromosome) is much more common in adult acute lymphoblastic leukaemia.
CDK4I codes for cyclin D kinase inhibitor, which restrains the cell cycle.
Acknowledgments
I thank Dr C Harrison for supplying the figure of fluorescence in situ hybridisation labelling of leukaemic cell chromosomes.
Molecular genetics provide exciting new insights into the pathogenesis of childhood leukaemia
Footnotes
Funding: Work in my laboratory is supported by the Leukaemia Research Fund and the Kay Kendall Leukaemia Fund.
Competing interests: None declared.
References
- 1.Rowley JD. The critical role of chromosome translocations in human leukemias. Annu Rev Genet. 1998;32:495–519. doi: 10.1146/annurev.genet.32.1.495. [DOI] [PubMed] [Google Scholar]
- 2.Kersey JH. Fifty years of studies of the biology and therapy of childhood leukemia. Blood. 1997;90:4243–4251. [PubMed] [Google Scholar]
- 3.Greaves M. Molecular genetics, natural history and the demise of childhood leukaemia. Eur J Cancer. 1999;35:173–185. doi: 10.1016/s0959-8049(98)00433-x. [DOI] [PubMed] [Google Scholar]
- 4.Greaves M. Cancer. The evolutionary legacy. Oxford: Oxford University Press; 2000. [Google Scholar]
- 5.Ford AM, Ridge SA, Cabrera ME, Mahmoud H, Steel CM, Chan LC, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 6.Wiemels JL, Ford AM, Van Wering ER, Postma A, Greaves M. Protracted and variable latency of acute lymphoblastic leukemia after TEL-AML1 gene fusion in utero. Blood. 1999;94:1057–1062. [PubMed] [Google Scholar]
- 7.Gale KB, Ford AM, Repp R, Borkhardt A, Keller C, Eden OB, et al. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc Natl Acad Sci USA. 1997;94:13950–13954. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–1503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 9.UK Childhood Cancer Study Investigators. The United Kingdom childhood cancer study: objectives, materials and methods. Br J Cancer. 2000;82:1073–1102. doi: 10.1054/bjoc.1999.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.UK Childhood Cancer Study Investigators. Childhood cancer and residential proximity to power lines. Br J Cancer. 2000;83:1573–1580. doi: 10.1054/bjoc.2000.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kinlen LJ. Epidemiological evidence for an infective basis in childhood leukaemia. Br J Cancer. 1995;71:1–5. doi: 10.1038/bjc.1995.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greaves MF. Aetiology of acute leukaemia. Lancet. 1997;349:344–349. doi: 10.1016/s0140-6736(96)09412-3. [DOI] [PubMed] [Google Scholar]
- 13.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev. 2001;1:69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 14.Dockerty JD, Draper G, Vincent T, Rowan SD, Bunch KJ. Case-control study of parental age, parity and socioeconomic level in relation to childhood cancers. Int J Epidemiol (in press). [DOI] [PubMed]
- 15.Auvinen A, Hakulinen T, Groves F. Haemophilus influenzae type B vaccination and risk of childhood leukaemia in a vaccine trial in Finland. Br J Cancer. 2000;83:956–958. doi: 10.1054/bjoc.2000.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexander FE, Patheal SL, Biondi A, Brandalise S, Cabrera M-E, Chan LC, et al. Transplacental chemical exposure and risk of infant leukemia with MLL gene fusion. Cancer Res. 2001;61:2542–2546. [PubMed] [Google Scholar]
- 17.Wiemels JL, Pagnamenta A, Taylor GM, Eden OB, Alexander FE, Greaves MF, et al. A lack of a functional NAD(P)H:quinone oxidoreductase allele is selectively associated with pediatric leukemias that have MLL fusions. Cancer Res. 1999;59:4095–4099. [PubMed] [Google Scholar]
- 18.Taylor GM, Dearden S, Payne N, Ayres M, Gokhale DA, Birch JM, et al. Evidence that an HLA-DQA1-DQB1 haplotype influences susceptibility to childhood common acute lymphoblastic leukaemia in males provides further support for an infection-related aetiology. Br J Cancer. 1998;78:561–565. doi: 10.1038/bjc.1998.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiemels JL, Smith RN, Taylor GM, Eden OB, Alexander FE, Greaves MF, et al. Methylenetetrahydrofolate reductase (MTHFR) polymorphisms and risk of molecularly defined subtypes of childhood acute leukemia. Proc Natl Acad Sci USA. 2001;98:4004–4009. doi: 10.1073/pnas.061408298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson JR, FitzGerald P, Willoughby MLN, Armstrong BK. Maternal folate supplementation in pregnancy and protection against common acute lymphoblastic leukaemia in childhood: a case-control study. Lancet. 2001;358:1935–1940. doi: 10.1016/S0140-6736(01)06959-8. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 22.Guidez F, Zelent A. Role of nuclear receptor co-repressors in leukemogenesis. Curr Top Microbiol Immunol. 2001;254:165–185. doi: 10.1007/978-3-662-10595-5_9. [DOI] [PubMed] [Google Scholar]
- 23.Redner RL, Wang J, Liu J. Chromatin remodelling and leukemia: new therapeutic paradigms. Blood. 1999;94:417–428. [PubMed] [Google Scholar]
- 24.Fenaux P, Chomienne C, Degos L. Treatment of acute promyelocytic leukaemia. Best Pract Res Clin Haematol. 2001;14:153–174. doi: 10.1053/beha.2000.0121. [DOI] [PubMed] [Google Scholar]
- 25.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 26.Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Fernandes Reese S, Ford JM, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 27.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 28.Van Dongen JJM, Seriu T, Panzer-Grümayer ER, Biondi A, Pongers-Willemse MJ, Corral L, et al. Prognostic value of minimal residual disease in childhood acute lymphoblastic leukemia. Lancet. 1998;352:1731–1738. doi: 10.1016/S0140-6736(98)04058-6. [DOI] [PubMed] [Google Scholar]