Abstract
The Vpr protein of human immunodeficiency virus type 1 (HIV-1) influences the in vivo mutation rate of the virus. Since Vpr interacts with a cellular protein implicated in the DNA repair process, uracil DNA glycosylase (UNG), we have explored the contribution of this interaction to the mutation rate of HIV-1. Single-amino-acid variants of Vpr were characterized for their differential UNG-binding properties and used to trans complement vpr null mutant HIV-1. A striking correlation was established between the abilities of Vpr to interact with UNG and to influence the HIV-1 mutation rate. We demonstrate that Vpr incorporation into virus particles is required to influence the in vivo mutation rate and to mediate virion packaging of the nuclear form of UNG. The recruitment of UNG into virions indicates a mechanism for how Vpr can influence reverse transcription accuracy. Our data suggest that distinct mechanisms evolved in primate and nonprimate lentiviruses to reconcile uracil misincorporation into lentiviral DNA.
Retroviral RNA is copied into DNA by the virus-encoded enzyme reverse transcriptase via a process called reverse transcription (2, 42). The error-prone nature of reverse transcription greatly contributes to the high level of genetic diversity observed within populations of retroviruses (4, 14, 18, 40, 41). Several variables define the diversity of human immunodeficiency virus type 1 (HIV-1) and retrovirus populations: (i) the rate of mutation per replication cycle, (ii) the number of replication cycles, (iii) the fixation rate of mutations, and (iv) the rate of recombination (6, 26). The high rate of HIV-1 replication is an important determinant in driving HIV-1 evolution (5, 13, 47).
A genetically engineered system has been developed for HIV-1 to measure the in vivo rate of forward mutation per replication cycle (27). The mutation rate of HIV-1 in this system was determined to be 3 × 10−5 to 4 × 10−5 mutation per target base pair per cycle (24, 27), where base substitution mutations (G-to-A and C-to-T transitions) and frameshift mutations (−1 frameshifts in runs of T's and A's) were most commonly detected. Replication of HIV-1 in the presence or absence of the auxiliary protein Vpr indicated that the mutation rate was as much as fourfold higher in the absence of Vpr (25, 27). This indicated that Vpr could influence the in vivo mutation rate of HIV-1. The vpr gene encodes a 96-amino-acid (aa) nonstructural protein which is associated with HIV-1 particles at a level comparable to that of the Gag precursor and then accumulates in the nuclei of infected cells (7, 22, 30). Incorporation of Vpr into particles requires a direct interaction with the p6 region of Gag (1, 35). In addition to influencing the mutation rate, Vpr has been implicated in the nuclear translocation of the preintegration complex and in cell arrest in the G2 phase of the cell cycle (11, 12, 17, 33). A recent report has indicated that Vpr alone could decrease the frequency of deletion mutations which occur following introduction of UV-damaged plasmid DNA into cells (16). This phenotype does not appear to be related to Vpr's role in the process and accuracy of reverse transcription (25).
The HIV-1 Vpr protein has been found to interact with several cellular partners (32, 51), in particular with two proteins involved in the DNA repair process, uracil DNA glycosylase (UNG) and the human homologue of the yeast RAD23 protein (HHR23A) (3, 10, 49). UNG is an enzyme involved in the base excision repair pathway which specifically removes the RNA base uracil from DNA (19). Uracil appears in DNA by misincorporation during its synthesis when the dUTP pool level is high or by cytosine deamination of dCMP. When cytosine deamination occurs and is not repaired, the result is a C-to-T transition mutation in that DNA strand (and a G-to-A transition in the opposite strand) in the next round of replication. The human ung gene contains two promoters that are required for generation of the mitochondrial (UNG1) and nuclear (UNG2) forms of the enzyme by alternative splicing (28). UNG1 and UNG2 have 35 and 44 unique N-terminal aa, respectively, while the C-terminal 269 aa are identical and contain the catalytic domain. The Vpr-binding site was mapped within the common C-terminal part of UNG, but the interaction did not perturb in vitro UNG enzymatic activity (3). While recent results suggest that the HHR23A protein is a mediator of Vpr-induced cell cycle arrest (10, 34), a detailed mutational analysis of Vpr revealed that the interaction with UNG is genetically separable from the ability of Vpr to perturb the cell cycle (49). A Trp residue located in position 54 of Vpr was found to be critical for the interaction with UNG, but replacement of this residue did not disrupt the G2 arrest activity. It has been observed that Vpr from simian immunodeficiency virus of sooty mangabeys, but not Vpx, associates with UNG (36).
Based on these observations, we tested the hypothesis that the interaction of Vpr with UNG could influence the in vivo mutation rate of HIV-1. We found that binding of Vpr to UNG correlates with the influence of Vpr on the mutation rate. We demonstrate that Vpr recruits the nuclear form of UNG into HIV-1 virions to influence the in vivo mutation rate. These data indicate a mechanism by which Vpr can influence reverse transcription accuracy and suggest the evolution of distinct mechanisms in primate and nonprimate lentiviruses to reconcile uracil misincorporation into lentiviral DNA.
MATERIALS AND METHODS
HIV-1 vectors and expression plasmids used for mutation rate studies.
The HIV-1 vector used in the in vivo forward mutation rate assay (Fig. 1A) was constructed as previously described (27). A vpr null mutant derivative of this vector was made by a primary-combinatorial two-step PCR protocol (25). Plasmids pSVgagpol-rre-r and pSV-A-MLV-env have been previously described (25). The vectors used for expression of wild-type (wt) Vpr or Vpr variants (pCMVvpr) were constructed by amplifying the wt or mutated vpr gene by PCR and inserting it into plasmid pCR3 (Invitrogen). The Vpr variants (Vpr*W54R and Vpr*R90K) were selected by two-hybrid screening from an HIV-1 Vpr mutant library generated by random error-prone PCR (34).
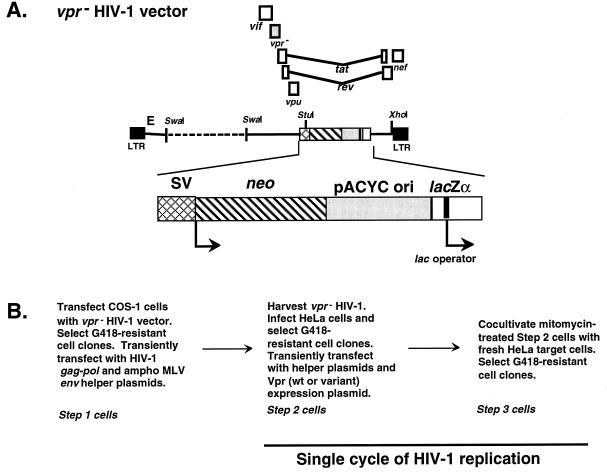
FIG. 1.
HIV-1 vector used for in vivo forward mutation rate studies. (A) vpr null mutant HIV-1 vector. The vector is shown in the proviral DNA form and has been previously described (25). (B) Protocol for one cycle of HIV-1 vector replication. The steps going from a parental shuttle vector provirus in the step 2 cell to a vector provirus in the step 3 cell constitute a single cycle of replication. LTR, long terminal repeat; SV, simian virus 40; ampho MLV, amphotropic murine leukemia virus.
Cell culture, transfections, infections, and cocultivations.
HeLa and COS-1 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum or 10% fetal bovine serum, respectively. HIV-1 vectors and expression plasmids were transfected into COS-1 or HeLa cells by use of dimethyl sulfoxide-Polybrene (27). HeLa cells were infected in the presence of Polybrene. Infection of HeLa target cells was done by cocultivation of virus-producing cells with target cells (25). The 293T cells used in the virion packaging assay were maintained in DMEM supplemented with 10% fetal calf serum, and 25 mM HEPES was added during virus production. They were transfected with the 22-kDa polyethylenimine (Euromedex) as previously described (35).
Cell culture strategy used to generate a single cycle of HIV-1 replication.
The experimental protocol developed to assay a single cycle of HIV-1 shuttle vector replication is shown in Fig. 1 (25). Step 2 clones were tested by Southern analysis to ensure that only a single vector proviral DNA was present. The lacZα peptide gene in the vector proviral DNA of step 2 clones was sequenced to confirm that no mutation was introduced.
Recovery of HIV-1 vector proviral DNA and sequence analysis of the lacZα peptide region.
Purified genomic DNA from pools of step 3 clones was digested with the restriction enzymes StuI and XhoI to release the neo, pACYC origin of replication, and lacZα peptide gene sequences from the HIV-1 vector. Proviral DNA was purified with the Lac repressor protein as previously described (25). The ratio of the number of white plus light blue bacterial colonies to the total number of colonies observed provided the forward mutation rate for a single retroviral replication cycle. Plasmid DNA was purified and sequenced in the lacZα peptide gene region by an automated DNA sequencer (Applied Biosystems). Mutation rates were calculated as previously described (25).
Yeast two-hybrid assay.
The construction of the HIV-1 Vpr mutant library fused to the DNA-binding domain of the LexA repressor (LexABD) and the two-hybrid screening procedure of the library have already been described (34). Vectors for expression of wt Vpr or the Vpr*W54R and Vpr*R90K variants fused to LexABD were described previously (34), while vectors for expression of UNG1, UNG2, and a truncated form of UNG without the N-terminal part of the protein (UNG57/66) fused to the Gal4 activation domain (Gal4AD) were constructed in the pGAD1318 plasmids (3). The L40 yeast strain was cotransformed with the indicated LexABD and Gal4AD hybrid expression vectors and plated on selective medium. Double transformants were then assayed for β-galactosidase activity and histidine auxotrophy as previously described (34).
Analysis of Vpr and UNG incorporation into HIV-1 virions.
Incorporation of Vpr and UNG was analyzed using a packaging assay in which Vpr and UNG were expressed in trans and incorporated into virions (35). The HIV-1-based packaging vectors pCMVΔR8.9 (lacking the env and auxiliary genes) and pCMVΔR8.2 (lacking only the env gene) and the pMD.G plasmid for expression of the vesicular stomatitis virus G protein were kindly provided by D. Trono (Geneva, Switzerland) (53). Vectors for expression of wt or mutated Vpr and UNG1, UNG2, and UNG57/66 fused to the epitope tag from the influenza virus hemagglutinin (HA) were constructed in pAS1B as previously described (35). For analysis of Vpr-dependent incorporation of UNG, cells were cotransfected with 10 μg of pCMVΔR8.9, 5 μg of pMD.G, 10 μg of pAS1B-UNG57/66, and the indicated amounts of pAS1B-Vpr (wt or mutated). For incorporation analysis of the distinct UNG forms, cells were cotransfected with 10 μg of pCMVΔR8.2, 5 μg of pMD.G, and 10 μg of either pAS1B-UNG1, -UNG2, or -UNG57/66. Cell culture supernatants were collected 48 h after transfection and filtered through 0.45-μm-pore-size filters, and an aliquot was assayed for CAp24 antigen. Virions were collected by ultracentrifugation for 1 h at 100,000 × g and suspended in ice-cold lysis buffer (10 mM Tris [pH 7.6], 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100). For preparation of cell lysates, cells were trypsinized, collected by centrifugation, and suspended in ice-cold lysis buffer. Cell and virion lysates were incubated on ice for 5 min and clarified by centrifugation. The protein concentration of the cell lysates was measured (Bio-Rad). Proteins from cell (50 μg of total protein) and virion (50 ng of CAp24) lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with anti-HA 3F10 (Boehringer) or anti-CAp24 antibodies (39).
RESULTS
Replication of a vpr null mutant HIV-1 vector and genetic trans complementation with Vpr.
The vpr null mutant HIV-1 vector (25) used in these studies was derived from the HIV-1 shuttle vector 3.12 (Fig. 1A). In order to complement this vpr null mutant HIV-1 vector, a plasmid expressing wt Vpr or a Vpr variant was transiently transfected along with gag-pol and env expression plasmids (25). The HIV-1 vector produced from either COS-1 or HeLa cells was used to infect fresh HeLa target cells (Fig. 1B). Cocultivation of mitomycin C-treated step 2 cells with fresh HeLa target cells to produce step 3 cells led to titers that were typically about 8 × 102 to 3 × 103 CFU/2.5 × 105 HeLa target cells. The steps going from a parental shuttle vector provirus in step 2 cells to a vector provirus in step 3 cells constitute a single cycle of replication (Fig. 1B). Southern analysis of total DNA from each step 2 cell clone was done to ensure that each clone contained only one provirus copy (data not shown). Proviral DNA from at least 5 × 105 cells of each step 2 cell clone was purified using the Lac repressor protein and introduced into Escherichia coli to screen for mutations in the lacZα gene region.
Vpr virion incorporation is required to influence the in vivo mutation rate.
The virion incorporation of Vpr into HIV-1 particles requires direct interaction with the p6 region of the Gag precursor (1, 35). In order to extend the observation that Vpr virion incorporation is required to influence the HIV-1 mutation rate, we used the assay described in Fig. 1B to compare the effects of Vpr trans complementation on the mutation frequencies of vpr null mutant HIV-1 (expressing a wt gag gene) and p6− vpr null mutant HIV-1 (expressing a mutant gag gene lacking the p6-encoding region). As indicated in Table 1, complementation with Vpr had no influence on the mutation rate of p6− vpr null mutant HIV-1 and resulted in an average mutation frequency comparable to that of noncomplemented vpr null mutant HIV-1 (chi square, 0.009; P >0.95) but significantly higher (chi square, 20; P <0.01) than that of vpr null mutant HIV-1 complemented with Vpr. Levels of Vpr expression were comparable in cells expressing the wt and p6-truncated forms of the Gag precursor (data not shown). These data support the requirement of Vpr incorporation into virions in order for Vpr to influence the HIV-1 mutation rate.
TABLE 1.
Incorporation of Vpr into virions is required to influence the mutation frequency of HIV-1 vectorsa
| Virus variant | No. of mutants/total no. of bacterial colonies | Mutation frequency (no. of mutations/cycle) |
|---|---|---|
| vpr null mutant HIV-1 | 24/1,107 | 0.022 |
| 19/1,183 | 0.016 | |
| vpr null mutant HIV-1 trans complemented with wt Vpr in producer cells | 10/1,520 | 0.007 |
| 7/1,264 | 0.006 | |
| p6−vpr null mutant HIV-1 | 27/1,329 | 0.020 |
| 14/943 | 0.015 | |
| p6−vpr null mutant HIV-1 trans complemented with wt Vpr in producer cells | 33/1,775 | 0.019 |
| 28/1,181 | 0.024 | |
| 19/1,227 | 0.015 | |
| vpr null mutant HIV-1 trans complemented with wt Vpr in target cells | 27/1,329 | 0.020 |
| 14/943 | 0.015 | |
| 30/1,673 | 0.018 |
The average mutation frequency of vpr null mutant HIV-1 in the absence of trans complementation was 0.019 mutation/cycle and was 0.006 mutation/cycle in the presence of Vpr trans complementation. The average mutation frequency of p6− vpr null mutant HIV-1 was 0.018 mutation/cycle. Complementation with wt Vpr of p6− vpr null mutant HIV-1 resulted in an average mutation frequency of 0.019 mutation/cycle (standard deviation of 0.005 mutation/cycle). Complementation with wt Vpr in target cells resulted in an average mutation frequency of 0.018 mutation/cycle (standard deviation of 0.003 mutation/cycle).
To test whether trans complementation of Vpr could be provided in the infected target cells rather than in the virus-producing cells, vpr null mutant HIV-1 was replicated with trans complementation of Vpr in step 3 target cells. Complementation of target cells with Vpr resulted in a mutation frequency significantly higher (chi square, 15; P <0.01) than that observed by complementation in virus-producing cells and equivalent (chi square, 0.033; P >0.5) to the mutation frequency of noncomplemented vpr null mutant HIV-1 (Table 1). Expression of Vpr in target cells was comparable to that in the virus-producing cells (not shown). These data indicate that trans complementation with Vpr in the target cells does not complement vpr null mutant HIV-1 in the mutation rate assay. This further supports the conclusion that Vpr virion incorporation is required to influence the HIV-1 mutation rate.
Vpr binding to UNG correlates with the influence on the HIV-1 in vivo mutation rate.
In order to analyze the potential correlation between the Vpr influence on the HIV-1 mutation frequency and the interaction with UNG, the effect of Vpr variants was analyzed in the mutation rate assay. We had previously reported that a single substitution of the Trp residue in position 54 (Vpr*W54R variant) was sufficient to abolish binding to UNG but did not disrupt the Vpr-induced G2 arrest activity. These data demonstrated the critical role of this residue in the maintenance of Vpr binding to UNG and indicated that this interaction is not involved in the perturbations of the cell cycle (34). The Vpr*R90K variant, containing a conservative substitution of Arg90 located in the C-terminal basic domain of the protein, was included in our analysis to study the relationship between the G2 arrest activity and the influence on the HIV-1 mutation rate. This mutant interacted with UNG as efficiently as wt Vpr but was unable to induce G2 arrest in HeLa cells (34).
Vpr*W54R and Vpr*R90K were analyzed in parallel for their influence on the HIV-1 mutation rate. The vpr null mutant HIV-1 vector was replicated in the absence of Vpr or trans complemented with a wt Vpr, Vpr*W54R, or Vpr*R90K expression plasmid. The proviral DNA from pooled step 3 cells, representing over 50,000 different clones for each experiment, was purified and introduced into E. coli to screen for mutations in the lacZα gene.
Three thousand seven hundred thirty-four bacterial colonies were screened in three replicates where vpr null mutant HIV-1 was trans complemented with Vpr*W54R. Fifty-nine of these colonies had a white or light blue colony color phenotype (Table 2). The average mutation frequency in these experiments was 59 to 3,734 or 0.016 mutation per cycle. The mutation frequency of vpr null mutant HIV-1 complemented with Vpr*W54R was significantly different from that found when it was complemented with wt Vpr (chi square, 17; P <0.01) but not from the mutation frequency of noncomplemented vpr null mutant HIV-1 (chi-square, 0.4; P > 0.5). This indicates that expression of Vpr*W54R leads to a mutation frequency phenotype comparable to that of vpr null mutant HIV-1 alone and therefore does not influence the in vivo mutation rate. Vpr*W54R interacted with the Gag precursor (data not shown) and was incorporated into HIV-1 particles as efficiently as wt Vpr (see Fig. 3C and reference 35), indicating that the mutation frequency phenotype of Vpr*W54R was not related to inefficient virion incorporation.
TABLE 2.
Mutation frequency in recovered proviruses of vpr null mutant HIV-1 complemented in trans with Vpr variants with differential binding to UNGa
| trans-Vpr variant | No. of mutants/total no. of bacterial colonies | Mutation frequency (no. of mutations/cycle) |
|---|---|---|
| No trans complementation | 13/928 | 0.014 |
| 18/773 | 0.023 | |
| wt Vpr | 10/2,029 | 0.005 |
| 12/1,726 | 0.007 | |
| Vpr*W54R | 23/1,639 | 0.014 |
| 19/1,285 | 0.015 | |
| 17/810 | 0.021 | |
| Vpr*R90K | 11/1,340 | 0.008 |
| 3/709 | 0.004 | |
| 8/1,233 | 0.007 |
The average mutation frequency of vpr null mutant HIV-1 in the absence of trans complementation was 0.018 mutation/cycle. Complementation with wt Vpr resulted in an average mutation frequency of 0.006 mutation/cycle. Complementation with Vpr variants Vpr*W54R and Vpr*R90K resulted in average mutation frequencies of 0.016 mutation/cycle (standard deviation of 0.004 mutation/cycle) and 0.007 mutation/cycle (standard deviation of 0.002 mutation/cycle), respectively.
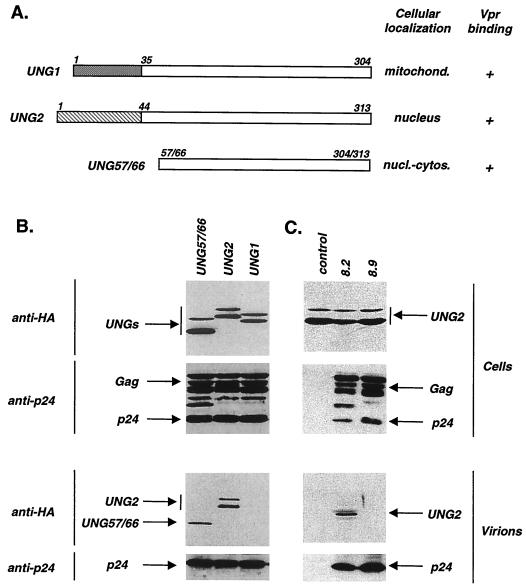
FIG. 3.
Vpr-dependent recruitment of UNG into HIV-1 particles. (A) Virion incorporation of UNG57/66. 293T cells were cotransfected with pCMVΔR8.2 (containing an intact vpr gene), pMD.G, and either pAS1B-UNG57/66 (+) or pAS1B without an insert (−). (B) Vpr dose-dependent incorporation of UNG. Cells were cotransfected with pCMVΔR8.9 (lacking vpr) and pMD.G in combination with 10 μg of pAS1B-UNG57/66 and the indicated increasing amounts of pAS1B-Vprwt. NT, nontransfected cells. (C) Failure of Vpr*W54R to recruit UNG into virions. Cells were cotransfected with pCMVΔR8.9 and pMD.G with or without 10 μg of pAS1B-UNG57/66 in combination with 20 μg of either pAS1B-Vprwt, pAS1B-Vpr*W54R, or pAS1B without an insert (−). Proteins from cell and virion lysates were separated by SDS-PAGE and analyzed by Western blotting with anti-HA (cells and virions, upper panels) or anti-CAp24 (cells and virions, lower panels) antibody.
Three thousand two hundred eighty-two bacterial colonies were screened in three replicates where vpr null mutant HIV-1 was complemented with Vpr*R90K. Twenty-two of these colonies had a white or light blue colony color phenotype (Table 2). The average mutation frequency was thus 22 to 3,282 or 0.007 mutation per cycle. The mutation frequency of vpr null mutant HIV-1 complemented with Vpr*R90K was not significantly different from that of vpr null mutant HIV-1 complemented with wt Vpr (chi square, 0.02; P >0.5) but was significantly different from the mutation frequency of noncom- plemented vpr null mutant HIV-1 (chi square, 15; P < 0.01). We have previously shown that Vpr*R90K bound to the Gag precursor and was efficiently incorporated into HIV-1 particles (35). These results indicate that the expression of Vpr*R90K influences the in vivo mutation rate in a manner comparable to that of wt Vpr. Since Vpr*R90K completely fails to induce a G2 arrest (34), they indicate that the effects of Vpr on the cell cycle are genetically separable from those on the reverse transcription process.
Complementation of vpr mutant HIV-1 with a Vpr*W54R UNG binding-deficient mutant protein leads to a mutation phenotype similar to that observed with vpr null mutant HIV-1 alone.
In order to compare the mutation phenotype of vpr mutant HIV-1 complemented with Vpr*W54R to that of noncomplemented vpr null mutant HIV-1, the types of mutations that led to the white or light blue colony color phenotype were determined by DNA sequencing of the lacZα gene (Fig. 2). Twenty (34%) of the 59 mutants sequenced from vpr null mutant HIV-1 complemented with Vpr*W54R had a single G-to-A base pair transition mutation, which was the predominant substitution observed. This percentage is comparable to what was observed for single G-to-A substitution mutations in vpr null mutant HIV-1 alone (11 [37%] of 30). Six hypermutants that each contained multiple mutations were observed for vpr null mutant HIV-1 complemented with Vpr*W54R (Fig. 2), and five of them contained at least one G-to-A transition. Therefore, 25 (44%) of the 59 mutants sequenced for vpr null mutant HIV-1 complemented with Vpr*W54R had G-to-A mutations. In comparison, two hypermutants (Fig. 2) were observed for vpr null mutant HIV-1 alone and both had at least one G-to-A mutation, indicating that 13 (43%) of the 30 mutants had G-to-A mutations. These data indicate that the rates of G-to-A mutation in both vpr null mutant HIV-1 complemented with Vpr*W54R and vpr null mutant HIV-1 alone are comparable and three- to fourfold higher than that of vpr null mutant HIV-1 complemented with wt Vpr (data not shown).
FIG. 2.
Plus strand nucleotide sequence of the lacZα gene region in a vpr null mutant HIV-1 vector complemented with Vpr*W54R. The start for nucleotide numbering is the beginning of the 5′ long terminal repeat of the HIV-1 vector. The locations of base substitution, frameshift, and deletion mutations in vpr null mutant HIV-1 vectors from parallel experiments that were complemented with Vpr*W54R or noncomplemented are shown above and below the nucleotide sequence, respectively. The start and stop codons of the lacZα open reading frame and the lac operator sequence are boxed. Nucleotide positions of base pair substitutions (letters above or below the sequence), +1 frameshifts (letters with ▾), −1 frameshifts (▿) and deletions (solid black lines with arrows and adjacent to the deletion names) are indicated. Mutations from the same mutant are designated with a Greek letter adjacent to each mutation; identical Greek letters indicate mutations from the same mutant.
C-to-T transition mutations were the second most common type of mutation detected for both vpr null mutant HIV-1 complemented with Vpr*W54R and vpr null mutant HIV-1 alone (Fig. 2). For vpr null mutant HIV-1 complemented with Vpr*W54R, 10 (17%) of 59 mutants contained a single C-to-T mutation. Two of the six hypermutants had one C-to-T transition, for a total of 12 (20%) of 59 mutants having C-to-T transitions. For vpr null mutant HIV-1 alone, 6 (20%) of 30 mutants had single C-to-T transitions, both hypermutants had C-to-T transitions, and thus a total of 8 (27%) of the 30 mutants had C-to-T transitions. This indicates that the rates of C-to-T transitions are similar for both vpr null mutant HIV-1 complemented with Vpr*W54R and vpr null mutant HIV-1 alone and are over twofold higher than that of vpr null mutant HIV-1 complemented with wt Vpr (data not shown).
Single frameshift mutations as an entire group of mutations (but primarily −1 or +1 frameshifts in runs of T's and A's) were identified in 14 (24%) of 59 mutants for vpr null mutant HIV-1 complemented with Vpr*W54R, and 8 (27%) of 30 for vpr null mutant HIV-1 alone. One hypermutant from vpr null mutant HIV-1 complemented with Vpr*W54R had two −1 frameshifts in runs of A's. The rates of frameshift mutations observed for vpr null mutant HIV-1 complemented with Vpr*W54R and for vpr null mutant HIV-1 alone are comparable to what was observed in vpr null mutant HIV-1 complemented with wt Vpr (not shown). Similarly, the rates of deletion mutations detected for both vpr null mutant HIV-1 complemented with Vpr*W54R (1 [2%] of 59) and for vpr null mutant HIV-1 alone (1 [3%] of 30) are comparable to that observed in vpr null mutant HIV-1 complemented with wt Vpr (not shown). These data confirm that these two types of mutations are not influenced by expression of Vpr (25).
Based upon the characterization of the types of mutations that occurred, the calculated in vivo mutation rate for vpr null mutant HIV-1 complemented with Vpr*W54R is 13 × 10−5 mutation per target base pair per cycle, which is fourfold higher than that of vpr null mutant HIV-1 complemented with wt Vpr.
UNG is recruited into HIV-1 particles through Vpr incorporation.
Since virion incorporation of Vpr is required to influence the in vivo mutation rate, we have explored whether UNG could be recruited into virus particles. Incorporation into virions was analyzed using a packaging assay in which UNG fused to the HA epitope (HA-tagged UNG) was expressed in trans in virus-producing cells (35). We first analyzed the incorporation of a truncated form of UNG containing a deletion of the N-terminal part of the protein (UNGΔ57/66) because it corresponded to the UNG clone initially isolated in the two-hybrid screening performed to identify Vpr-interacting proteins (3). 293T cells were transfected with the HA-tagged UNG expression vector in combination with a HIV-1-based packaging vector (pCMVΔR8.2) containing an intact vpr gene (53), and the virion- and cell-associated UNG was then assessed by Western blot analysis using an anti-HA monoclonal antibody (MAb) (Fig. 3A). The HA-tagged version of UNG57/66 was detected in the supernatant of transfected cells (lower panel), indicating that it is incorporated into virions. To determine if the recruitment of UNG into virions is dependent on Vpr incorporation, we used the same virion packaging assay described above but HA-tagged Vpr and UNG57/66 were both expressed in trans in virus-producing cells transfected with a HIV-1 packaging vector lacking the auxiliary genes (pCMVΔR8.9). Parallel experiments were performed in which 293T cells were cotransfected with pCMVΔR8.9 and a constant amount of the HA-tagged UNG57/66 expression vector, in combination with increasing amounts of the HA-tagged Vpr vector. The virion- and cell-associated UNG and Vpr were then assessed by Western blot analysis with the anti-HA MAb (Fig. 3B). No HA-UNG57/66 was detected in virions when Vpr was not expressed in virus-producing cells, indicating that UNG is incorporated in a Vpr-dependent manner. In contrast, UNG57/66 was found in virions when the amount of Vpr expressed in cells was increased. Both HA-Vpr and UNG were simultaneously detected in a range of 10 to 20 μg of Vpr plasmid that was transfected into cells. The same blots were probed with an anti-CAp24 MAb to verify that similar amounts of virions were produced in all of the transfections. These results demonstrate that Vpr incorporation is required to recruit UNG into HIV-1 particles.
We then analyzed whether the mutation frequency phenotype of the Vpr*W54R UNG binding-deficient variant was related to a defect in UNG incorporation into virions. Cells were thus transfected with pCMVΔR8.9 and the HA-tagged UNG57/66 expression vectors, in combination with the HA-tagged wt Vpr or Vpr*W54R expression plasmid (Fig. 3C). As previously, UNG57/66 was detectable in virions only when wt Vpr was coexpressed in virus-producing cells, and both UNG and wt Vpr were thus incorporated. In contrast, no UNG was detected in virions produced from cells expressing high levels of Vpr*W54R, even though this variant was incorporated as efficiently as wt Vpr. These data demonstrate that a UNG binding-deficient Vpr variant does not recruit UNG into virions, suggesting that the mutation frequency phenotype of Vpr*W54R results from its inability to allow the recruitment of UNG into HIV-1 particles.
The nuclear form of UNG is preferentially incorporated into HIV-1 particles.
Since UNG exists as mitochondrial (UNG1) and nuclear (UNG2) isoforms whose N-terminal sequences differ (see Fig. 4A), we have explored whether both forms of UNG could be recruited into virions. Each of these UNG forms fused to Gal4AD bound to LexABD-Vpr as efficiently as did UNG57/66 in a yeast two-hybrid assay (Fig. 4A), confirming that the N-terminal portions of UNG1 and UNG2 do not influence Vpr binding. The incorporation of UNG1 and UNG2 into virions was analyzed by transfection of 293T cells with the HIV-1 pCMVΔR8.2 vector and the HA-tagged UNG1, UNG2, or UNG57/66 expression vector. Virion- and cell-associated UNGs were then assessed by Western blot analysis using the anti-HA MAb (Fig. 4B). UNG2 and UNG57/66 were efficiently incorporated into virions, since the HA-tagged version of each form was detected in the supernatants of transfected cells (lower panel). In contrast, mitochondrial UNG1 was not incorporated into virions despite detectable level of the protein in transfected cell lysate (upper panel). UNG2 was incorporated in a Vpr-dependent manner, since it was detected in virions when 293T cells were transfected with pCMVΔR8.2 but not when cells were transfected with pCMVΔR8.9 lacking the vpr gene (Fig. 4C). These results indicate that nuclear UNG2 is the preferential form incorporated into HIV-1 particles.
FIG. 4.
Preferential incorporation of nuclear UNG2 into HIV-1 virions. (A) Cellular distribution and Vpr binding of UNG1, UNG2, and UNG57/66. UNG1 and UNG2 have 35 and 44 unique N-terminal aa, respectively, while the C-terminal 269 aa are identical. Cellular distribution was analyzed by indirect immunofluorescence assay of 293T cells transfected with plasmid pAS1B-UNG1, -UNG2, or -UNG57/66. Vpr binding was determined in a two-hybrid assay of L40 yeast cells expressing the LexABD-Vprwt hybrid and either UNG1, UNG2, or the UNG57/66 Gal4AD hybrid. mitochon., mitochondria; nucl., nucleus; cytos., cytoplasm. (B) Virion incorporation analysis of the two UNG forms. 293T cells were cotransfected with pCMVΔR8.2, pMD.G, and either plasmid pAS1B-UNG1, -UNG2, or -UNG57/66. (C) Vpr-dependent incorporation of UNG2 into virions. 293T cells were cotransfected with pMD.G, pAS1B-UNG2, and either plasmid pCMVΔR8.2 or pCMVΔR8.9. In panels B and C, proteins from cell and virion lysates were separated by SDS-PAGE and analyzed by Western blotting with anti-HA (cells and virions, upper panels) or anti-CAp24 (cells and virions, lower panels) antibody.
DISCUSSION
This work focused on the functional characterization of the interaction of HIV-1 Vpr with UNG, a cellular protein that is implicated in the DNA repair process. The results indicate the contribution of the binding of Vpr to UNG to the in vivo mutation rate of HIV-1. Further data are presented which show that the Vpr recruitment of the nuclear form of UNG into HIV-1 particles is required for Vpr to influence the in vivo mutation rate. The correlation between the capacity of Vpr to interact with UNG and its ability to both influence the HIV-1 mutation rate and mediate virion packaging of UNG supports this conclusion. In contrast, Vpr binding to HHR23A, the other Vpr-interacting DNA repair protein, does not correlate with the influence of Vpr on the mutation rate (L.M.M. and S.B., unpublished results).
Two Vpr variants were tested for their influence on the in vivo mutation rate of HIV-1 in order to assess whether the interaction of Vpr with the DNA repair protein UNG could be correlated with the influence of Vpr on the mutation rate. The Vpr*W54R variant does not bind UNG, does interact with the Gag precursor, is efficiently incorporated into virus particles, and causes cell cycle arrest. This indicates that Vpr*W54R displays a wt phenotype, with the exception of its inability to bind UNG. When Vpr*W54R was used for trans complementation of vpr null mutant HIV-1 in a single cycle of replication, it was found that the rate of mutation was comparable to that of noncomplemented vpr null mutant HIV-1 alone. In addition, vpr null mutant HIV-1 complemented with Vpr*W54R had a spectrum of mutations similar to that of noncomplemented vpr null mutant HIV-1. This provides genetic evidence in support of the conclusion that the inability of Vpr*W54R to interact with UNG influences the in vivo mutation rate of HIV-1. In contrast, Vpr*R90K influences the in vivo mutation rate in a manner comparable to that of wt Vpr but completely fails to induce a G2 arrest (34). Therefore, the Vpr effects on the cell cycle are genetically separable from those on the HIV-1 mutation rate since they are related to distinct regions of Vpr. Alpha-helical region II of the protein contributes to reverse transcription accuracy, while the C-terminal basic domain is crucial for the G2 arrest activity (8, 52).
The absence of UNG in HIV-1 particles that have efficiently incorporated Vpr*W54R indicates that the failure of Vpr*W54R to interact with UNG not only prevents virion incorporation of UNG but also affects the influence of Vpr on the HIV-1 mutation rate. The Vpr dose dependence for incorporation of UNG into HIV-1 particles also provides evidence for a Vpr-specific mechanism of UNG incorporation. These observations indicate that for Vpr to influence the in vivo mutation rate of HIV-1, both Vpr and UNG must be efficiently incorporated into HIV-1 particles. Nuclear UNG2 is the predominant form of UNG that is incorporated into virions, whereas the mitochondrial UNG1 form is not efficiently incorporated. Like UNG2 (28), HIV-1 Vpr displays evident karyophilic properties (15, 31, 46), suggesting that the Vpr-UNG2 complex takes place in the nuclei of infected cells, migrates to the cytoplasm, and then is incorporated into virions through the interaction of Vpr with the Gag precursor protein. The inability of Vpr*W54R to mediate UNG incorporation is not related to a defect of nuclear import of this Vpr variant, since it localizes to the nucleus as efficiently as the wt protein (not shown). Alternatively, the Vpr-UNG2 complex could be formed in the cytoplasm and targeted to the plasma membrane before nuclear import of both Vpr and UNG2. Since the Trp54 residue located in C-terminal alpha-helical region II of Vpr is crucial for the maintenance of UNG binding (34), it appears that Vpr may also simultaneously interact with the Gag precursor through N-terminal alpha-helical region I (8, 23, 35, 50). In contrast, the UNG1 form sequestered into mitochondria fails to access the core of HIV-1 virions although it displays the ability to physically interact with Vpr. It was recently reported that UNG was detected in HIV-1 virions in the absence of Vpr, requiring the presence of the viral integrase protein when Vpr is absent for UNG incorporation (48). Determination of UNG incorporation with a Vpr mutant that was deficient in UNG binding but was efficiently incorporated into HIV-1 particles was not analyzed in this study. Our data indicate that when Vpr is not present in HIV-1 particles, there is no detectable UNG incorporation. While we cannot formally exclude the possibility that integrase also contributes to UNG incorporation in the virion packaging assay used in the present study, our results suggest that the interaction of UNG with Vpr is the major pathway for UNG incorporation into HIV-1 particles.
The observation that Vpr binding to UNG correlates with the in vivo mutation rate of HIV-1 implies a role for UNG in the accuracy of the reverse transcription process. UNG functions in cells as a DNA repair enzyme that specifically removes from DNA the RNA base uracil, which appears by misincorporation during DNA synthesis when the dUTP pool is high or by cytosine deamination of dCMP. When cytosine deamination occurs, the result is a C-to-T transition mutation in that DNA strand and a G-to-A transition in the opposite strand in the next round of replication. The data presented in Fig. 2 indicate that the predominant types of mutations detected in vpr null mutant HIV-1 both alone and trans complemented with Vpr*W54R were G-to-A and C-to-T transition mutations. Based upon what is known of UNG function, a G-to-A transition mutation in the HIV-1 plus-strand DNA, in the absence of functional UNG activity, could be an indication of cytosine deamination in the minus strand DNA made during reverse transcription. In the presence of UNG activity, the uracil created by cytosine deamination would be in the DNA strand of a DNA-RNA hybrid, assuming that the cytosine deamination occurred during the minus strand DNA synthesis step of reverse transcription. Little is known regarding the function of UNG in removing uracil from DNA that is in a DNA-RNA hybrid. The presence in HIV-1 particles of other repair enzymes which participate in the uracil excision repair pathway could help to support the specific role in virion packaging of UNG and the HIV-1 mutation rate. However, we failed to detect in HIV-1 virions the apurinic/apyrimidinic (AP) endonuclease (known as HAP, APEX, or Ref-1), the second enzyme involved in this pathway (19), suggesting that the other enzymes are recruited after viral entry into the target cells. Although the data presented here indicate that UNG enzymatic activity directly influences the HIV-1 mutation rate, UNG may also influence the mutation rate by other mechanisms, such as modulation of the access of deoxynucleoside triphosphates to reverse transcriptase or interaction of the Vpr-UNG complex with the reverse transcriptase to influence its enzymatic fidelity.
Most nonprimate lentiviruses are known to encode and package into virus particles a dUTPase, an enzyme that regulates the levels of dUTP in cells and therefore influences the potential misincorporation of uracil into viral DNA (9, 20, 21, 38, 43–45). Replication of nonprimate lentiviruses that lack functional dUTPase activity leads to misincorporation of uracil into viral DNA, a reduced level of replication in macrophages (i.e., nondividing cells), and an increased level of G-to-A transition mutations. The inhibition of dUTPase activity leading to an increased level of G-to-A transitions appears to have a phenotype similar to that of vpr null mutant HIV-1 or vpr null mutant HIV-1 trans complemented with Vpr*W54R in the mutation rate assay. Both primate and nonprimate lentiviruses have the ability to replicate in nondividing cells, which are presumed to have low levels of S-phase cellular enzymes involved in DNA synthesis and repair, such as dUTPase and UNG (19, 29, 37). The encoding of dUTPase by nonprimate lentiviruses and the incorporation of UNG by primate lentiviruses such as HIV-1 by its interaction with Vpr support the hypothesis that these different mechanisms evolved in order for these viruses to remove uracil from their DNA when replicating in nondividing cells.
ACKNOWLEDGMENTS
We thank L. Bernard and R. Casseron for outstanding technical assistance and D. Trono and the National Institutes of Health AIDS Reagent Program for the kind gift of various reagents. We also thank M. Emerman for stimulating conversations and M. Williams for comments on the manuscript.
This work was supported by grant GM 56615 from the Public Health Service (to L.M.M.), by the French National Agency for AIDS Research (R.B.), and by SIDACTION (S.B.).
REFERENCES
- 1.Bachand F, Yao X-J, Hrimech M, Rougeau N, Cohen E A. Incorporation of Vpr into human immunodeficiency virus type 1 requires a direct interaction with the p6 domain of the p55 gag precursor. J Biol Chem. 1999;274:9083–9091. doi: 10.1074/jbc.274.13.9083. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumor viruses. Nature (London) 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 3.Bouhamdan M, Benichou S, Rey F, Navarro J-M, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, Sire J. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffin J. Genetic diversity and evolution of retroviruses. Curr Top Microbiol Immunol. 1992;176:143–164. doi: 10.1007/978-3-642-77011-1_10. [DOI] [PubMed] [Google Scholar]
- 5.Coffin J. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 6.Coffin J M. Retroviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1767–1847. [Google Scholar]
- 7.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. Mutational analysis of cell cycle arrest, nuclear localization, and virion packaging of human immunodeficiency virus type 1 Vpr. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elder J H, Lerner D L, Hasselkus-Light C S, Fontenot D J, Hunter E, Luciw P A, Montelaro R C, Phillips T R. Distinct subsets of retroviruses encode dUTPase. J Virol. 1992;66:1791–1794. doi: 10.1128/jvi.66.3.1791-1794.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gragerov A, Kino T, Ilyina-Gragerova G, Chrousos G P, Pavlakis G N. HHR23A, the human homologue of the yeast repair protein RAD23, interacts specifically with Vpr protein and prevents cell cycle arrest but not the transcriptional effects of Vpr. Virology. 1998;245:323–330. doi: 10.1006/viro.1998.9138. [DOI] [PubMed] [Google Scholar]
- 11.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M-A, Gendelman H E, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 14.Holland J J, De La Torre J C, Steinhauer D A. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins Y, McEntee M, Weis K, Greene W C. Characterization of HIV-1 vpr nuclear import: analysis of signals and pathways. J Cell Biol. 1998;143:875–885. doi: 10.1083/jcb.143.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jowett J B, Xie Y-M, Chen I S Y. The presence of human immunodeficiency virus type 1 Vpr correlates with a decrease in the frequency of mutations in a plasmid shuttle vector. J Virol. 1999;73:7132–7137. doi: 10.1128/jvi.73.9.7132-7137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M-L, Chen I S Y. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz R A, Skalka A M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- 19.Krokan H E, Standal R, Slupphaug G. DNA glycosylases in the base excision repair of DNA. Biochem J. 1997;325:1–16. doi: 10.1042/bj3250001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lerner D L, Wagaman P C, Phillips T R, Prospero-Garcia O, Henriksen S J, Fox H S, Bloom F E, Elder J H. Increased mutation frequency of feline immunodeficency virus lacking functional deoxyuridine-triphosphatase. Proc Natl Acad Sci USA. 1995;92:7480–7484. doi: 10.1073/pnas.92.16.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichtenstein D L, Rushlow K E, Cook R F, Raabe M L, Swardson C J, Kociba G J, Issel C J, Montelaro R C. Replication in vitro and in vivo of an equine infectious anemia virus mutant deficient in dUTPase activity. J Virol. 1995;69:2881–2888. doi: 10.1128/jvi.69.5.2881-2888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu Y-L, Spearman P, Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mahalingam S, Khan S A, Murali R, Jabbar M A, Monken C E, Collman R G, Srinivasan A. Mutagenesis of the putative alpha-helical domain of the Vpr protein of human immunodeficiency virus type 1: effect on stability and virion incorporation. Proc Natl Acad Sci USA. 1995;92:3794–3798. doi: 10.1073/pnas.92.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansky L M. Forward mutation rate of human immunodeficiency virus type 1 in a T-lymphoid cell line. AIDS Res Hum Retroviruses. 1996;12:307–314. doi: 10.1089/aid.1996.12.307. [DOI] [PubMed] [Google Scholar]
- 25.Mansky L M. The mutation rate of human immunodeficiency virus type 1 is influenced by the vpr gene. Virology. 1996;222:391–400. doi: 10.1006/viro.1996.0436. [DOI] [PubMed] [Google Scholar]
- 26.Mansky L M. Retrovirus mutation rates and their role in genetic variation. J Gen Virol. 1998;79:1337–1345. doi: 10.1099/0022-1317-79-6-1337. [DOI] [PubMed] [Google Scholar]
- 27.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus T A, Skorpen F, Krokan H E. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pardo E G, Gutierrez C. Cell cycle- and differentiation stage-dependent variation of dUTPase activity in higher plant cells. Exp Cell Res. 1990;186:90–98. doi: 10.1016/0014-4827(90)90214-u. [DOI] [PubMed] [Google Scholar]
- 30.Paxton W, Connor R I, Landau N R. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popov S, Rexach M, Zybarth G, Reiling N, Lee M A, Ratner L, Lane C M, Moore M S, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Refaeli Y, Levy D N, Weiner D B. The glucocorticoid receptor type II complex is a target of the HIV-1 vpr gene product. Proc Natl Acad Sci USA. 1995;92:3621–3625. doi: 10.1073/pnas.92.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogel M E, Wu L I, Emerman M. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selig L, Benichou S, Rogel M E, Wu L I, Vodicka M A, Sire J, Benarous R, Emerman M. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J Virol. 1997;71:4842–4846. doi: 10.1128/jvi.71.6.4842-4846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selig L, Pages J-C, Tanchou V, Prévéral S, Berlioz-Torrent C, Liu L X, Erdtmann L, Darlix J-L, Benarous R, Benichou S. Interaction with the p6 domain of the Gag precursor mediates incorporation into virions of Vpr and Vpx proteins from primate lentiviruses. J Virol. 1999;73:592–600. doi: 10.1128/jvi.73.1.592-600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sleigh R, Sharkey M, Newman M A, Hahn B, Stevenson M. Differential association of uracil DNA glycosylase with SIVsm Vpr and Vpx proteins. Virology. 1998;245:338–343. doi: 10.1006/viro.1998.9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spector R, Boose B. Development and regional distribution of deoxyuridine 5′-triphosphate in rabbit brain. J Neurochem. 1983;41:1192–1195. doi: 10.1111/j.1471-4159.1983.tb09073.x. [DOI] [PubMed] [Google Scholar]
- 38.Steagall W K, Robek M D, Perry S T, Fuller F J, Payne S L. Incorporation of uracil into viral DNA correlates with reduced replication of EIAV in macrophages. Virology. 1995;210:302–313. doi: 10.1006/viro.1995.1347. [DOI] [PubMed] [Google Scholar]
- 39.Steimer K S, Puma J P, Power M D, Powers M A, George-Nascimento C, Stephans J C, Levy J A, Sanchez-Pescador R, Luciw P A, Barr P J, et al. Differential antibody responses of individuals infected with AIDS-associated retroviruses surveyed using the viral core antigen p25gag expressed in bacteria. Virology. 1986;150:283–290. doi: 10.1016/0042-6822(86)90289-8. [DOI] [PubMed] [Google Scholar]
- 40.Temin H M. Mixed infection with two types of Rous sarcoma virus. Virology. 1961;13:158–163. doi: 10.1016/0042-6822(61)90049-6. [DOI] [PubMed] [Google Scholar]
- 41.Temin H M. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci USA. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Temin H M, Mitzutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature (London) 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 43.Threadgill D S, Steagall W K, Flaherty M T, Fuller F J, Perry S T, Rushlow K E, LeGrice S F J, Payne S L. Characterization of equine infectious anemia virus dUTPase: growth properties of a dUTPase-deficient mutant. J Virol. 1993;67:2592–2600. doi: 10.1128/jvi.67.5.2592-2600.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turelli P, Guiguen F, Mornex J-F, Vigne R, Querat G. dUTPase-minus caprine arthritis-encephalitis virus is attenuated for pathogenesis and accumulates G-to-A substitutions. J Virol. 1997;71:4522–4530. doi: 10.1128/jvi.71.6.4522-4530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turelli P, Petursson G, Guiguen F, Mornex J-F, Vigne R, Querat G. Replication properties of dUTPase-deficient mutants of caprine and ovine lentiviruses. J Virol. 1996;70:1213–1217. doi: 10.1128/jvi.70.2.1213-1217.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vodicka M A, Koepp D M, Silver P A, Emerman M. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 1998;12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 48.Willetts K E, Rey F, Agostini I, Navarro J-M, Baudat Y, Vigne R, Sire J. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J Virol. 1999;73:1682–1688. doi: 10.1128/jvi.73.2.1682-1688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Withers-Ward E S, Jowett J B, Stewart S A, Xie Y M, Garfinkel A, Shibagaki Y, Chow S A, Shah N, Hanaoka F, Sawitz D G, Armstrong R W, Souza L M, Chen I S. Human immunodeficiency virus type 1 Vpr interacts with HHR23A, a cellular protein implicated in nucleotide excision DNA repair. J Virol. 1997;71:9732–9742. doi: 10.1128/jvi.71.12.9732-9742.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao X-J, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. Mutageneic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao L-J, Mukherjee S, Narayan O. Biochemical mechanism of HIV-1 Vpr function. J Biol Chem. 1994;269:15577–15582. [PubMed] [Google Scholar]
- 52.Zhou Y, Lu Y, Ratner L. Arginine residues in the C-terminus of HIV-1 Vpr are important for nuclear localization and cell cycle arrest. Virology. 1998;242:414–424. doi: 10.1006/viro.1998.9028. [DOI] [PubMed] [Google Scholar]
- 53.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]