Abstract
In contrast to the vast majority of cellular proteins, rotavirus proteins are translated from capped but nonpolyadenylated mRNAs. The viral nonstructural protein NSP3 specifically binds the 3′-end consensus sequence of viral mRNAs and interacts with the eukaryotic translation initiation factor eIF4G. Here we show that expression of NSP3 in mammalian cells allows the efficient translation of virus-like mRNA. A synergistic effect between the cap structure and the 3′ end of rotavirus mRNA was observed in NSP3-expressing cells. The enhancement of viral mRNA translation by NSP3 was also observed in a rabbit reticulocyte lysate translation system supplemented with recombinant NSP3. The use of NSP3 mutants indicates that its RNA- and eIF4G-binding domains are both required to enhance the translation of viral mRNA. The results reported here show that NSP3 forms a link between viral mRNA and the cellular translation machinery and hence is a functional analogue of cellular poly(A)-binding protein.
The vast majority of cellular mRNAs possess a 3′ terminal poly(A) sequence. This sequence plays a major role in many aspects of cellular mRNA metabolism (11). Together with the 5′ cap structure, poly(A) synergistically enhances the translation of mRNA (8, 12). This effect is mediated by the poly(A)-binding protein (PABP) (20), which interacts with the 3′ poly(A) and the eukaryotic initiation factor eIF4G (14, 23, 33). eIF4G is a scaffold protein that brings together eIF4E (cap-binding protein), eIF4A (a helicase), PABP, and eIF3 (5). As a consequence of these multiple interactions, the 40S subunit of the ribosome, loaded with initiator tRNA and methionine, is brought to the 5′ end of a circularized mRNA and starts scanning the 5′ untranslated region (UTR) for the first initiation codon (21). The circularization of the mRNA via eIF4E-eIF4G-PABP and mRNA interactions (34) is thought to enhance the translation of the mRNA by allowing rapid reinitiation of new rounds of translation. Circularization of the mRNA seems particularly important for efficient and accurate initiation when competition exists between mRNA (27) or when the supply of ribosomes or initiation factors is limited (28).
Rotaviruses are the major cause of diarrhea in young animals and children; they are involved in the death of more than 800,000 children each year worldwide (10). Rotaviruses are members of the Reoviridae family, and their genome is composed of 11 molecules of double-stranded RNA, which encode six structural proteins and five or six nonstructural proteins (6, 17). The virus replication cycle occurs entirely in the cytoplasm. Upon virus entry, the viral transcriptase synthesizes capped but nonpolyadenylated mRNAs (13). The viral mRNAs bear 5′ and 3′ untranslated regions (UTR) of variable length and are flanked by two different sequences common to all genes. In the group A rotaviruses, the 3′-end consensus sequence UGACC is highly conserved among the 11 genes.
We have previously shown that rotavirus NSP3 presents several similarities to PABP; in rotavirus-infected cells, NSP3 can be cross-linked to the 3′ end of rotavirus mRNAs (24, 25) and is coimmunoprecipitated with eIF4G (23). The binding of NSP3A to eIF4G and its specific interaction with the 3′ end of viral mRNA (24, 25) brings the viral mRNA and the translation initiation machinery into contact, thus favoring efficient translation of the viral mRNA. NSP3 interacts with the same region of eIF4G as PABP does (14, 23). As a consequence, during rotavirus infection PABP is evicted from eIF4G, probably impairing the translation of polyadenylated mRNA and leading to the shutoff of cellular mRNA translation observed during rotavirus infection (23).
Here we describe the establishment of a cell line expressing NSP3. We used this cell line to show that NSP3 and the cap structure synergistically enhance rotavirus-like mRNA translation in vivo. Then we used an in vitro assay to study the effect of NSP3 on the translation of reporter mRNAs with poly(A) or rotavirus 3′-end sequences. Using the same in vitro translation assay and NSP3 mutants, we show that the two functional domains of NSP3 are required to enhance rotavirus mRNA translation and that the eIF4G-binding domain of NSP3 inhibits translation when it is separated from the RNA-binding domain.
MATERIALS AND METHODS
Plasmid construction.
Manipulation of nucleic acids was done as described by Sambrook et al. (31) unless otherwise indicated. The cDNA of rotavirus RF gene 7 (RF7) encoding NSP3A has been described previously (25). It was subcloned into pTEJ4 (15), which allows constitutive expression under the control of the ubiquitin promoter. To obtain pT7-RF6-F.Luc-RF6, two complementary synthetic oligonucleotides with the sequence of the T7 promoter fused to the sequence of the 5′ UTR of rotavirus RF6 (with a NcoI site on the ATG) (3) were cloned between the HindIII and XbaI sites of pUC19 to produce pT7-RF6-5′. The 3′ UTR of RF6 bearing a BamHI site at its 5′ end and KspI and EcoRI sites at its 3′ end was amplified by PCR from RF6 cDNA (3) and was cloned between the BamHI and EcoRI sites of pT7-RF6-5′. The firefly luciferase (F. Luc) gene (NcoI-BamHI fragment) was purified from pSP-Luc(+) (Promega) and inserted between the 5′ and 3′ RF6 UTRs to produce pT7-RF6-F.Luc-RF6. The sequences of the RF6 UTRs of pT7-RF6-F.Luc-RF6 were checked by DNA sequencing. pCMVT7-R.Luc-polyA was obtained by cloning a synthetic oligonucleotide bearing 25 adenines, followed by a NsiI restriction site, between the XbaI and BamHI sites of pRL CMV (Promega). pRF6-F.Luc-polyA was obtained by subcloning the HindIII-XbaI fragment from pRF6-F.Luc-RF6 into pCMV-RL-polyA.
In vitro synthesis of mRNAs.
F.Luc-rota and F.Luc-nonrota mRNAs were synthesized from pT7-RF6-F.Luc-RF6 templates linearized with KspI or EcoRI, respectively. The pT7-RF6-F.Luc-RF6 linearized with KspI was treated with T4 polynucleotide kinase in the absence of dCTP to ensure the production of 3′ recessed ends. Renilla luciferase (R.Luc) poly(A) and F.Luc poly(A) mRNA were obtained respectively from pCMV-T7-R.Luc-poly(A) and pRF6-F.Luc-poly(A), linearized with NsiI and treated with T4 DNA polymerase in the absence of dATP to obtain 3′ recessed ends.
T7 polymerase transcription systems supplied or not supplied with cap analogue (Message Machine [Ambion] and Ribomax [Promega], respectively) were used to synthesize capped and uncapped mRNA. In all cases, tritiated UTP (800 mCi/mmol [Amersham]) was added in trace amount (40 μCi) to determine the quantity of mRNA synthesized and to monitor the purification of the mRNAs. After treatment with RNase-free DNase I for 10 min at 37°C, RNAs were purified from nucleotides and cap analogue by phenol extraction and repeated ethanol precipitation in the presence of ammonium acetate or on Sephadex G-10 push columns (Stratagene). In both cases, measurement of trichloroacetic acid-precipitable radioactivity by scintillation counting indicated that more than 90% of the unincorporated nucleoside triphosphates were removed. The purified RNA was finally analyzed by agarose gel electrophoresis.
DNA and RNA transfections.
ST (swine testis) cells were cultivated in Earle's minimal essential medium supplemented with 10% fetal calf serum. They were transfected with pTEJ4 RF7 (10 μg) by lipofection (Lipofectin; Life Technologies) together with pX343 (1 μg). Stable antibiotic-resistant clones were selected by the presence of 100 μg of hygromycin per ml for 10 days. Clones were isolated using cloning cylinders and screened for NSP3 expression by indirect immunofluorescence with monoclonal antibody ID3 as previously described (26).
ST or ST-NSP3 cells grown to 70% confluence in 12-well plates were transfected with cationic liposomes (DRMIE-C; Life Technologies) with both 400 ng of F.Luc mRNA and 4 ng of R.Luc mRNA. After 5 h of lipofection, the RNA mixture was removed and fresh medium was added. Cells were lysed 20 h later for the measurement of luciferase activities by the dual-luciferase method (Promega).
For electroporation of RNA, ST-NSP3 cells grown to 70% confluence were trypsinized and adjusted to 107 cells/ml of Earle's minimal essential medium. The cells were transferred to a sterile electroporation cuvette, mixed with 200 ng of F.Luc mRNA and 10 ng of capped-R.Luc-poly(A) mRNA, and electroporated (Easyject+) at 260 V and 1,050 μF. Immediately after electroporation, MEM with 10% serum was added and the cells were incubated at 37°C for 5 to 24 h.
Immunoprecipitation and growth curves.
ST and ST-NSP3 cells in 100-mm-diameter dishes were labeled for 2 h with 20 μCi of Tran35S-label (38 TBq/mol; ICN) as previously described (24). Immunoprecipitation with an anti-NSP3 monoclonal antibody was carried out as described previously (24), except that cell lysates were precleared by a 1-h incubation with protein A-Sepharose before being subjected to immunoprecipitation.
Plates (35 mm) were seeded with 6 × 105 ST or ST-NSP3 cells, and living cells were counted at different times by trypan blue exclusion after trypsination.
Mutagenesis and expression of recombinant NSP3.
Expression of NSP3 fused to a track of 6 histidine residues in Escherichia coli using the T7 expression system (32) has been described previously (22). Two mutations that improve the expression and renaturation of the protein were introduced into NSP3 cDNA by using the Quick-Change (Stratagene) protocol; cysteine 227 was changed to asparagine to impair the formation of disulfide bonds during purification of the protein, and methionine 206 was changed to isoleucine to impair internal translation initiation and copurification of a truncated C-terminal fragment of NSP3. All the NSP3 mutants used in this study were obtained by site-directed mutagenesis (Quick-Change) of this modified cDNA. The resulting plasmids were transfected into E. coli BL21/DE3, and the expression of NSP3 was induced by the addition of IPTG (isopropyl-β-d-thiogalactoside; final concentration, 1 mM). Induction and purification of the recombinant protein were conducted as described previously (22). Renaturation of recombinant proteins eluted from nickel-chelating Sepharose columns (Pharmacia) was modified to improve the solubility and stability of the proteins. Wild-type NSP3 and RNA-binding mutants were renatured by stepwise dialysis with decreasing concentrations of urea in 10 mM Tris (pH 7.4)–10% glycerol–1 mM EDTA–10 mM dithiothreitol. NSP3 mutants on the eIF4G-binding domain were renatured by stepwise dialysis against 10 mM Tris (pH 8)–10% glycerol–1 mM EDTA–10 mM dithiothreitol.
Analysis of the RNA-binding and eIF4G-binding properties of recombinant NSP3.
Purified recombinant proteins were tested for their RNA-binding activity by UV cross-linking and gel retardation as previously described (22, 25). The eIF4G-binding capacity of NSP34–290 was tested by the two-hybrid assay after transfer of the NSP34–290 gene into the yeast vector pGBT9 NS (22, 23). Yeast strain HF7C was transfected with pGAD424-eIF4GI/+37:174 and pGBT9-NSP34–290 or pGBT9-NSP34–313 and spread onto medium without tryptophan and leucine to monitor the cotransfection efficiency and onto medium without tryptophan, leucine, or histidine to assess the interaction between the two fusion proteins.
Measurement of luciferase activities and synergy.
After lipofection or electroporation of ST or ST-NSP3 cells with firefly and Renilla reporter mRNAs, cells were lysed and luciferase activities were measured using the Dual Luciferase kit as specified by the supplier (Promega) and an automatic luminometer (Berthold). The luciferase activities of cells transfected with the DNA used as matrices for in vitro RNA transcription were measured and found to be equal to the luciferase activities of mock-transfected cells. Luciferase activities were standardized to the protein concentration of the samples.
The ratio of the capped-Luc-poly(A) mRNA translation to the sum of capped-Luc-nonrota mRNA and uncapped-Luc poly(A) mRNA gives the amount of synergy between cap and poly(A) (21). The ratio of the capped-Luc-rota mRNA translation to the sum of capped-Luc-nonrota mRNA and uncapped-Luc rotavirus mRNA gives the amount of synergy between cap and rotavirus 3′ end.
In vitro translation.
Nuclease-treated rabbit reticulocyte lysate (FlexiRRL; Promega) adjusted to 110 mM KCl was used to translate reporter mRNAs in the presence of recombinant NSP3. Each translation mixture consisted of 70% of RRL (Promega), 10 U of RNasin RNase inhibitor per μl, 20 μM amino acid mixture, 5 ng of mRNA (1 nM), and 100 ng of purified NSP3 (230 nM) or the same volume of the same batch of NSP3 final dialysis buffer. After a 1-h incubation at 30°C, the luciferase activity of samples was measured using the F Luciferase kit as recommended by the supplier (Promega).
RESULTS
NSP3 expression enhances the expression of rotavirus-like mRNA in vivo.
To investigate the role of NSP3 in rotavirus translation, we first established a stable cell line expressing NSP3. The cell line generally used to propagate rotaviruses is MA104. In our hands, MA104 cells age poorly, as seen by reduced growth capacity and susceptibility to rotavirus infection with increased passage number. This cell line was therefore not suited for our purpose; instead, we chose to use the ST cell line, whose growth is not restricted by passage number and which is fully susceptible to rotavirus infection (19).
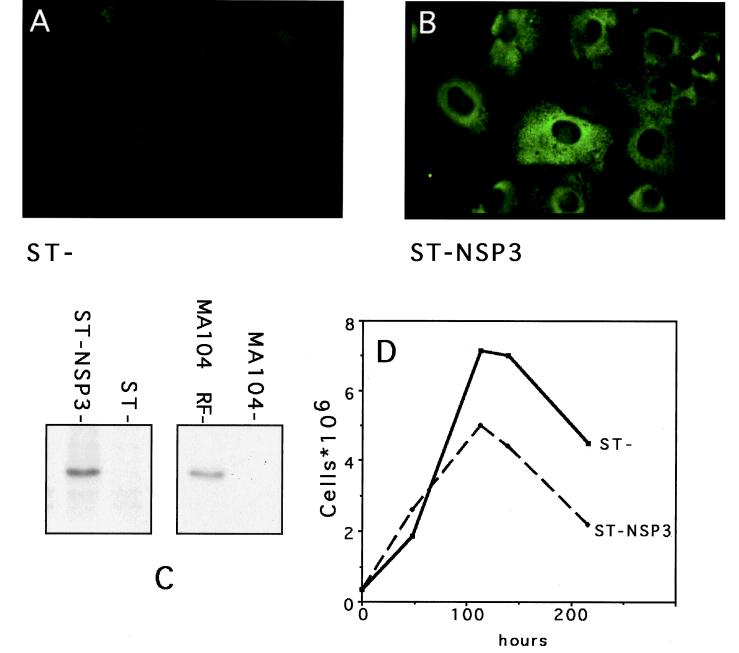
ST cells constitutively expressing NSP3 (under the control of the ubiquitin promoter [15]) were screened by indirect immunofluorescence (Fig. 1A and B) with a monoclonal antibody directed against NSP3 (2). Only a few cell clones were positive for NSP3 expression, and one (called ST-NSP3) that synthesizes a detectable amount of NSP3 by immunoprecipitation (Fig. 1C) was selected for this study. The cells were affected by expression of NSP3; ST-NSP3 cells seemed slightly larger than the parental ST cell, they were more robust when electroporated (see below), and their growth (doubling time, 48 h) was slower than that of the parental cell line (Fig. 1D; doubling time, 35 h).
FIG. 1.
Expression of NSP3 in ST-NSP3 cells. (A and B) Expression of NSP3 in ST (A) and ST-NSP3 (B) cells was detected by indirect immunofluorescence using monoclonal antibody ID3 directed against NSP3. (C) Radiolabeled proteins from 3 × 106 ST-NSP3 cells and ST cell lysates were immunoprecipitated with monoclonal antibody ID3 and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). An autoradiogram of the gel is shown. Controls are provided by immunoprecipitation of radiolabeled proteins from 3 × 105 MA104 cells infected (MA104-RF) or mock infected (MA104) with rotavirus RF strain. (D) Growth curves of ST and ST-NSP3 cells.
NSP3 produced by ST-NSP3 cells had the same molecular weight as NSP3 produced during virus infection (Fig. 1C), was recognized by the monoclonal antibody against NSP3, and had an intracytoplasmic distribution (Fig. 1D) similar to that of NSP3 produced during rotavirus infection (data not shown).
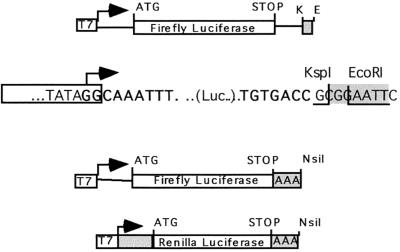
Rotavirus-like reporter mRNAs (F.Luc-rota) were obtained by subcloning the firefly luciferase gene (F.Luc) between the 5′ UTR and 3′ UTR of RF6 (3) (Fig. 2). The addition of a KspI restriction site overlapping the rotavirus 3′ consensus sequence UGACC allowed mRNAs to be synthesized, in vitro, with a UGACC sequence precisely at their 3′ end. In contrast, if the same plasmid is linearized with EcoRI prior to transcription, the UGACC sequence lies 4 nucleotides upstream of the AAUU sequence at the 3′ end of the mRNA. Such mRNAs serve as control reporter (F.Luc-nonrota), since we have previously shown that NSP3 recognizes the UGACC sequence only if it is located at the 3′ end of the RNA (24, 25). Thus, F.Luc-rota and F.Luc-nonrota mRNAs differ only in their very 3′ ends. To study the effect of NSP3 on the expression of poly(A) mRNA, a 25-base poly(A) sequence was inserted in place of the 3′ RF6 UTR; this mRNA is referred to as F.Luc-poly(A). To compare the effect of the 3′ ends on the expression of the various reporter capped mRNAs, capped F.Luc-rota, F.Luc-nonrota, and F.Luc-poly(A) mRNAs were introduced by lipofection into ST and ST-NSP3 cells. Figure 3 illustrates the results of such experiments. Poly(A) mRNAs were efficiently translated in ST and ST-NSP3 cells. The F.Luc-pA translation in ST-NSP3 cells was slightly diminished, probably reflecting a slight inhibition of poly(A)-dependent translation in this cell line. Non-poly(A) mRNA were barely translated in ST cells irrespective of their (rota or nonrota) 3′ ends. However, F.Luc-rota mRNA was efficiently translated in ST-NSP3 cells (Fig. 3). The major difference in ST-NSP3 cells is the sevenfold increase in F.Luc-rota translation efficiency, showing that the expression of NSP3 in mammalian cells promotes efficient translation of rotavirus-like mRNAs.
FIG. 2.
Rotavirus-like and poly(A) reporter mRNAs. The salient features of the DNA matrices used for in vitro synthesis of the different mRNAs used are shown (not to scale). (Top) The firefly luciferase gene was cloned between the 5′ and 3′ UTRs of RF6 downstream of the T7 promoter (T7). F.Luc-rota mRNA was obtained from KspI (K)-linearized matrix, and F.Luc-nonrota was obtained from EcoRI (E)-linearized matrix. The DNA sequences at the junction between the T7 promoter and the 5′ UTR (bold type) and between the 3′ UTR (bold type) and the vector are indicated. The site of cleavage on the top strand of the DNA template and the restriction enzymes used are indicated. (Middle) The 3′ RF6 UTR was replaced by a poly(A)25 sequence, and F.Luc poly(A) mRNA was obtained from the NsiI-linearized matrix. (Bottom) A poly(A)25 sequence was cloned downstream of the R.Luc gene and RLuc poly(A) mRNA was obtained with NsiI-linearized matrix. The box labeled T7 indicates the T7 promoter;arrows indicate the T7 transcription start site; AAA indicates poly(A)25; K indicates the KspI site; E indicates the EcoRI site; ATG indicates the initiation codon; STOP indicates the termination codon. Boxes indicate nonrotavirus sequences, and shaded areas indicate transcribed vector sequences.
FIG. 3.
Translation of capped rotavirus-like and poly(A) mRNA in ST and ST-NSP3 cells. F.Luc activity was measured 24 h after lipofection of capped F.Luc-rota (black columns), capped F.Luc-nonrota (hatched columns), and capped-F.Luc-poly(A) (white columns) mRNAs into ST (left) or ST-NSP3 (right) cells. The level of translation is expressed in arbitrary firefly translation units. These units were standardized with respect to concomitantly transfected R.Luc-poly(A) mRNA so that variation due to different transfection efficiencies could be discounted.
Synergy between the 5′ cap and 3′ rotavirus ends.
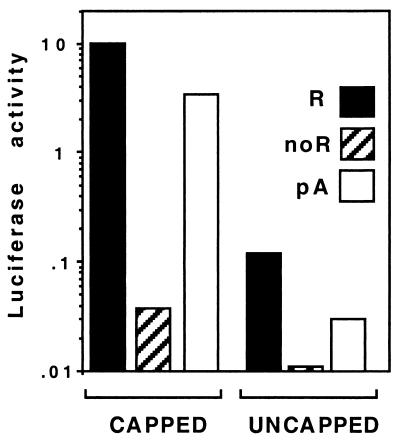
The 5′ cap structures and the 3′-end poly(A) sequences act synergistically to enhance mRNA translation. Synergy can be measured by the ratio between the F.Luc activity obtained with capped F.Luc-poly(A) mRNA and the sum of F.Luc activities obtained with uncapped F.Luc-poly(A) and capped F.Luc-nonrota mRNA (20). Measuring synergy in vivo requires measuring the activity of uncapped mRNA. When introduced into ST-NSP3 cells by lipofection, uncapped mRNAs were very poorly translated (data not shown), precluding the use of this method to study synergy. Alternatively, RNA can be introduced into cells by electroporation (8). Electroporation has the advantages over lipofection of being much quicker and synchronous for all cells and allowing the rapid recovery of cells after introduction of mRNA.
We set up electroporation conditions for ST-NSP3 cells, but for unknown reasons, ST cells did not sustain the conditions used to electroporate ST-NSP3 cells and could not be used in the following experiments. mRNAs with different 3′ ends were electroporated into ST-NSP3 cells and the F.Luc activity was measured 5 h after electroporation (Fig. 4) (Note that the F.Luc translation is different from that in Fig. 2 due to the different proportions of R.Luc and F.Luc mRNAs introduced into the cells.) The F.Luc-rota and F.Luc-poly(A) mRNAs were translated more efficiently than the F.Luc-nonrota mRNA was, irrespective of their 5′-end structure. This result showed that F.Luc-rota is efficiently translated in ST-NSP3 cells when introduced by electroporation and confirmed the results obtained with lipofection.
FIG. 4.
Translation of capped and uncapped rotavirus-like and poly(A) mRNA in ST-NSP3 cells. F.Luc activity was measured 5 h after electroporation of capped (left) or uncapped (right) F.Luc-rota (black columns), F.Luc-nonrota (hatched columns), and F.Luc-poly(A) (white columns) mRNAs into ST-NSP3 cells. The level of translation is expressed by the ratio of F.Luc to R.Luc activities. The level of translation is expressed in arbitrary firefly translation units. These units were standardized with respect to concomitantly transfected R.Luc-poly(A) mRNA so that variation due to different transfection efficiencies could be discounted.
Synergy between mRNA 3′ and 5′ ends can be calculated from the results in Fig. 4. Synergy values of 21 (standard deviation = 1, n = 2) between cap and poly(A) were calculated when the luciferase activity was measured 5 h after the introduction of F.Luc-poly(A) mRNA into ST-NSP3 cells. Under the same conditions, a synergy value of 35 (standard deviation = 5, n = 2) was calculated for rotavirus-like mRNA, showing that the UGACC sequence can also cooperate with the NSP3 5′ cap structure to enhance the translation of rotavirus mRNA.
NSP3 stimulates the translation of virus-like mRNA in vitro when bound to eIF4G.
To examine directly the enhancement of translation of virus-like mRNA by NSP3, we turned to in vitro translation systems. Purified recombinant NSP3 expressed in E. coli (NSP34–313) was added to RRL programmed with uncapped mRNAs with different 3′ ends (Fig. 5). The effect of NSP3 (added at a final concentration of 20 or 200 mM) on translation efficiency was expressed by the ratio of F.Luc activity obtained in the presence of NSP3 to the F.Luc activity measured in the presence of NSP3 renaturation buffer (Fig. 5). Adding NSP3 to uncapped-F.Luc-rota mRNAs increased their translation nearly 100-fold (Fig. 5, 200 mM). At the same concentration, NSP3 increased the translation of uncapped-F.Luc-nonrota or uncapped-F.Luc-poly(A) mRNA only 15- and 10-fold, respectively. With both concentrations of NSP3, the ratio of specific to nonspecific enhancement of translation was nearly 10 (Fig. 5).
FIG. 5.
Stimulation of translation by recombinant wild-type NSP3 of rotavirus-like, non-poly(A), and poly(A) mRNAs in vitro. Wild-type NSP3 (NSP34–313) was added to RRL programmed with different mRNAs (same abbreviations as in Fig. 3) at a 200- or 20-fold excess over the RNA 3′ end. Fold stimulation is expressed by the ratio of F.Luc activity obtained in the presence of recombinant protein to the activity obtained in the absence of recombinant protein. Bars indicate standard deviation calculated from three independent experiments.
Addition of a cap to the 5′ end of F.Luc mRNAs enhances the translation (in the absence of NSP3) of any F.Luc mRNA 50-fold (data not shown). Thus, when the same experiments were conducted with capped-F.Luc-rota, only a modest additional twofold increase in translation was obtained on addition of NSP3 (data not shown). However, a clear effect of NSP3 on translation of rotavirus-like mRNA could be observed in vitro with uncapped mRNAs which were used for the in vitro experiments described below.
Next we wanted to study the relative importance of the RNA- and eIF4G-binding activities of NSP3 in the enhancement of translation of rotavirus mRNA. The in vitro assay described above was used to test NSP3 mutants. A deletion of the last 24 amino acids of NSP3 abolishes the interaction between NSP3 and eIF4G when tested by the two-hybrid assay (23) but has no effect on the RNA-binding activity (22) (see Fig. 7B and C). The same mutant was produced in E. coli, and the purified recombinant protein (NSP33–290) was used in the in vitro translation assay. NSP33–290 does not enhance the translation of the uncapped-F.Luc-rota mRNA (Fig. 6). Even the nonspecific stimulation of F.Luc-nonrota and F.Luc-poly(A) translation observed with NSP3 was abolished. We concluded from this experiment that an interaction between NSP3 and eIF4G is required to enhance the translation of rotavirus-like mRNA.
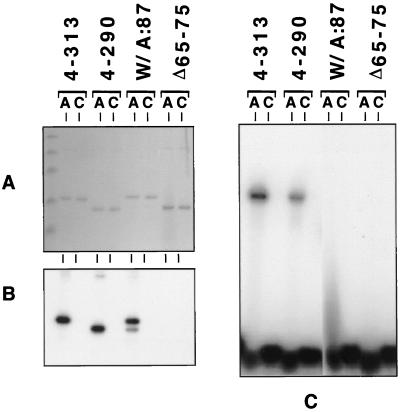
FIG. 7.
Characterization of the RNA-binding properties of wild-type NSP3 and NSP3 mutants. Wild-type NSP3 (NSP34–313), and mutants with mutations of the eIF4G-binding domain (NSP34–290) or the RNA-binding domains (NSP3W/A:87 and NSP3Δ65–75) were expressed in E. coli and purified. Recombinant proteins were incubated with 5′-end radiolabeled RNA bearing the group A rotavirus 3′-end sequence (AUAUGACC) (lanes A) or the group C rotavirus 3′-end sequence (AUAUGGCU) (lanes C). Samples were analyzed, after UV cross-linking, by SDS-PAGE and Coomassie blue staining (A) followed by autoradiography (B) (autoradiography of the gel shown in panel A). Alternatively, samples were directly analyzed in a gel retardation assay by nondenaturing PAGE and autoradiography (C).
FIG. 6.
Effect of recombinant wild-type NSP3 and the eIF4G-binding mutant of NSP3 on the translation of rotavirus-like, non-poly(A), and poly(A) mRNAs in vitro. Wild-type NSP3 (NSP34–313) (left) or NSP3 mutated in its eIF4G-binding domain (NSP34–290) (right) was added to RRL programmed with different mRNAs (same abbreviations as in Fig. 3). Fold stimulation is expressed by the ratio of F.Luc activity obtained in the presence of recombinant protein to the activity obtained in the absence of recombinant protein.
Enhanced translation correlates with the RNA-binding capacity of NSP3.
The RNA-binding domain of NSP3 has been mapped to the N-terminal half of the protein (22). Two mutations were introduced into this domain: a conserved tryptophan (29) at position 87 was changed to alanine to give NSP3W/A:87, and a deletion of 11 amino acids was introduced at positions 65 to 75 to give NSP3Δ65–75. Figure 7A depicts an example of the purified NSP3 used in the experiments described below. The RNA-binding activity of these two mutants was tested in a UV cross-linking assay with synthetic RNA (Fig. 7A and B) and in a gel retardation assay (Fig. 7C) as described previously (25). The two assays are not exactly equivalent. Gel retardation is dynamic, so that protein and RNA can dissociate during the running of the gel, whereas UV cross-linking creates a covalent link between molecules and even brief interactions occurring during the UV irradiation can be observed. NSP3W/A:87 could be UV cross-linked to the rotavirus group A 3′-end sequence (Fig. 7B) but not to the rotavirus group C 3′-end sequence. However, NSP3W/A:87 was able to make only an unstable complex with the rotavirus group A 3′-end RNA sequence, as evidenced by the absence of a neat band in a gel retardation assay (Fig. 7C). Instead, the presence of a smear on the autoradiogram reflected the instability of the RNA-NSP3W/A:87 complexes that dissociated during electrophoresis (16). The deletion of amino acids 65 to 75 totally abolished the capacity of NSP3 to bind RNA when tested either by UV cross-linking (Fig. 7B) or by gel retardation (Fig. 7C).
When NSP3W/A:87 was used in the in vitro translation assay, it stimulated rotavirus-like mRNA translation better than it stimulated nonrotavirus mRNA translation but to a much lower extent than did wild-type NSP34–313 (Fig. 8). With the deletion mutant NSP3Δ65–75, a twofold nonspecific stimulation of translation was observed with the three kinds of RNA tested. Thus, the strength of the interaction of NSP3 with RNA correlates with its translation stimulation capacities and specificity.
FIG. 8.
Effect of RNA-binding mutants of NSP3 on the translation of rotavirus-like non-poly(A) and poly(A) mRNA in vitro. Wild-type NSP3 (NSP34–313) (left) or NSP3 mutated in its RNA-binding domain (NSP3WA:87 [middle] and NSP3Δ65–75 [right]) was added to RRL programmed with different mRNAs (same abbreviations as in Fig. 3). Fold stimulation is expressed as the ratio of F.Luc activity obtained in the presence of recombinant protein to the activity in the absence of recombinant protein.
Inhibition of translation by NSP3.
The ability of NSP3 to bind to eIF4G and to evict PABP from eIF4F is thought to be the cause of the shutoff of cellular mRNA translation observed during rotavirus infection (23). If this is the case, addition of NSP3 to the in vitro translation assay might be expected to inhibit the translation of polyadenylated mRNA. However, the addition of wild-type NSP3 or the RNA-binding mutant of NSP3 to the in vitro translation assay mixture programmed with uncapped poly(A) mRNA slightly stimulated the translation (Fig. 5, 6, and 8). Two possible explanations for this observation can be provided. First, the recombinant wild-type and mutant NSP3 have a nonspecific RNA-binding activity not detected by the RNA-binding assays used here. This nonspecific RNA-binding activity could be sufficient to enhance the translation of any mRNA added to the RRL. Second, this effect could be similar to the trans-activation properties described recently for yeast PABP (20). Otero et al. have shown that addition of PABP, disabled for poly(A) or eIF4G binding, to yeast cell extract stimulates the translation of non-poly(A)+ mRNA (20). This property of PABP has been called trans-activation.
To attempt to discriminate between these two possibilities, two new NSP3 mutants were designed; NSP3163–313 bears an entire deletion of the RNA-binding domain but is still able to bind eIF4G (22), and NSP3163–290 contains an additional deletion of the last C-terminal 24 amino acids of NSP3, which completely disables its eIF4G-binding domain (22). The complete deletion of the RNA-binding domain totally abolished the translation stimulation properties of NSP3 (Fig. 9) and even induced a 50% reduction in the translation of any mRNA. This inhibition of translation was not the result of the toxicity of NSP3163–313 but, rather, was the result of its interaction with eIF4G, since the addition of NSP3163–290 prepared under the same conditions had no inhibitory effect on translation (Fig. 9).
FIG. 9.
Effect of the eIF4G-binding domain of NSP3 on the translation of rotavirus-like and poly(A) mRNAs in vitro. Recombinant NSP3 from which the entire RNA-binding domain NSP3163–313) (left) or both the RNA- and eIF4G-binding domains (NSP3163–290) (right) were deleted, was added to RRL programmed with different mRNAs (same abbreviations as in Fig. 3). Fold stimulation is expressed by the ratio of F.Luc activity in the presence of recombinant protein to the activity in the absence of recombinant protein.
DISCUSSION
NSP3 specifically enhances the translation of rotavirus mRNA.
The properties of NSP3 described previously (23, 24) strongly suggest that it is involved in the translation of rotavirus mRNAs. To definitively establish the role of NSP3 in translation, we generated a mammalian cell line expressing NSP3, into which we introduced mRNA by lipofection or electroporation. To our knowledge, this is the first time that a cell line expressing a rotavirus protein has been described. Unlike poly(A) mRNAs, rotavirus-like mRNAs are not translated efficiently when introduced into ordinary cells. However, expression of NSP3, even at a low level, allowed efficient translation of rotavirus-like mRNA. As expected from the RNA-binding specificity of NSP3, the enhancement of translation was specific for mRNAs ending with the rotavirus 3′-end UGACC sequence whereas mRNAs ending with AAUU were poorly translated in either normal or NSP3-expressing cells.
It has been shown that the 3′ poly(A) and 5′ cap structure act synergistically to enhance the translation of cellular mRNA (8) in vivo. Synergy between poly(A) and cap has also been observed in yeast cells and in yeast cell extracts (12, 20), with which a synergy value of 7 has been measured (20). For mammalian CHO cells, a synergy value of 27 (5 h postelectroporation) can be calculated from Fig. 3 of reference 8 and synergy value of 16 and up to 15 has been found with carrot protoplasts and Drosophila embryo cells extract, respectively (9, 18). The value of 20 for synergy between poly(A) and cap measured in ST-NSP3 cells (from Fig. 4) is comparable to the value obtained with CHO cells. A value of 35 for synergy between the cap and the rotavirus 3′ ends could be measured in vivo in the presence of NSP3. This is similar to the synergy between cap and poly(A) and suggests that NSP3 and PABP use the same mechanisms to enhance translation.
An effect of NSP3 on rotavirus mRNA stability, similar to the effect of PABP (4) on poly(A) mRNA, could not be entirely excluded. However, the role of NSP3 as a viral translation factor was confirmed by using recombinant proteins and an in vitro translation assay in which little mRNA degradation occurs (7, 8). The use of NSP3 mutants in this system allowed the roles of the different domains identified on NSP3 to be determined precisely. NSP3 mutants disabled in their ability to bind either eIF4G or RNA were not functional. These results strongly suggest that NSP3 acts as a physical link between viral mRNA and the cellular translation initiation complex. NSP3 brings the viral mRNA in contact with the translation initiation complex eIF4F by interacting simultaneously with eIF4G and the viral mRNA 3′ end. In vitro, these interactions enhance viral mRNA translation independently of the 5′ end of the viral mRNA, but in vivo, a strong synergy occurs between the 5′ cap structure and the 3′ end. For poly(A) mRNA, this role is fulfilled by PABP, and the results described here demonstrate that NSP3 is a functional mimic of PABP.
Effect of NSP3 on the translation of other mRNAs.
NSP3 has been suspected to be involved in the shutoff of cellular mRNA translation (22). Nevertheless, we managed to isolate a cell line expressing a low level of NSP3. ST-NSP3 cells present some differences from the parental ST cell line; notably, its growth is slightly slower (Fig. 1D). It is not known if this results from a general decrease in the translation of poly(A) mRNAs induced by expression of NSP3 or from the specific decrease in the translation of few cellular mRNAs. An inducible expression system for NSP3 is needed to precisely answer this question.
A slight decrease of the F.Luc-poly(A) translation in ST-NSP3 cells compared to ST cells was noted (Fig. 3). This observation might reflect an increased competition for poly(A) mRNA translation in ST-NSP3 cells, induced by the interaction of NSP3 with eIF4G. Eviction of PABP from eIF4F (23) impairs the use of eIF4G by PABP, and consequently less eIF4G is available for the translation of poly(A) mRNAs. However, similar results were obtained when F.Luc poly(A) was normalized to protein concentration instead of R.Luc-poly(A) translation. This suggests that R.Luc poly(A) mRNAs perform better than F.Luc-poly(A) mRNAs in ST-NSP3 cells.
Using a deletion mutant of the whole RNA-binding domain, we showed that NSP3 has the capacity to inhibit translation in vitro. This inhibition is due to its interaction with eIF4G, as demonstrated by the lack of effect of NSP34–290 and NSP3163–290 on translation. The exact concentration of eIF4G in RRL is not known and can only be roughly estimated. The upper estimate of the eIF4E concentration in RRL is 400 nM (30), and the concentration of eIF4G is estimated to be 1/10 the concentration of eIF4E (1). Therefore, the concentration of eIF4G in RRL could be around 40 nM. The amount of NSP3 used in our assay (200 nM) is thus sufficient to recruit all the eIF4G and inhibit translation. This observation indicates that non-poly(A)- and poly(A)-dependent translations use the same translation initiation pathway, which can be blocked by NSP3. However, we observed that a fraction of the mRNA was still translated in the presence of a vast excess of NSP3. This residual translation could be due to a fraction of PABP that could not be removed from eIF4G or to an alternative translation initiation pathway not sensitive to NSP3.
ACKNOWLEDGMENTS
We are indebted to Katherine Kean (Pasteur Institute) for critical and constructive reading of the manuscript. We acknowledge stimulating discussions with J. Cohen throughout this work. We acknowledge the skillful technical assistance of C. Jaegger.
M. Piron was supported by a fellowship from the French Ministère de l'Education Nationale de la Recherche et de la Technologie. This work was funded in part by “Programme de Recherches Fondamentales en Microbiologie, Maladies Infectieuses et Parasitologie” of MENRT.
REFERENCES
- 1.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J. 1997;16:1114–1121. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aponte C, Mattion N M, Estes M K, Charpilienne A, Cohen J. Expression of two bovine rotavirus non-structural proteins (NSP2, NSP3) in the baculovirus system and production of monoclonal antibodies directed against the expressed proteins. Arch Virol. 1993;133:85–95. doi: 10.1007/BF01309746. [DOI] [PubMed] [Google Scholar]
- 3.Cohen J, Lefevre F, Estes M K, Bremont M. Cloning of bovine rotavirus (RF strain): nucleotide sequence of the gene coding for the major capsid protein. Virology. 1984;138:1780–1782. doi: 10.1016/0042-6822(84)90159-4. [DOI] [PubMed] [Google Scholar]
- 4.Coller J M, Gray N K, Wickens M P. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dever T E. Translation initiation: adept at adapting. Trends Biochem Sci. 1999;24:398–403. doi: 10.1016/s0968-0004(99)01457-7. [DOI] [PubMed] [Google Scholar]
- 6.Estes M K, Cohen J. Rotavirus gene structure and function. Microbiol Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuichi Y, LaFiandra A, Shatkin A J. 5′-Terminal structure and mRNA stability. Nature. 1977;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 8.Gallie D R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 9.Gebauer F, Corona D F, Preiss T, Becker P B, Hentze M W. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 1999;18:6146–6154. doi: 10.1093/emboj/18.21.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glass R I, Bresee J S, Parashar U, Miller M, Gentsch J R. Rotavirus vaccines at the threshold. Nat Med. 1997;3:1324–1325. doi: 10.1038/nm1297-1324. [DOI] [PubMed] [Google Scholar]
- 11.Gray N K, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 12.Iizuka N, Najita L, Franzusoff A, Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imai M, Akatani K, Ikegami N, Furuichi Y. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J Virol. 1983;47:125–136. doi: 10.1128/jvi.47.1.125-136.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A) binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansen T E, Scholler M S, Tolstoy S, Schwartz T W. Biosynthesis of peptide precursors and protease inhibitors using new constitutive and inducible eukaryotic expression vectors. FEBS Lett. 1990;267:289–294. doi: 10.1016/0014-5793(90)80947-h. [DOI] [PubMed] [Google Scholar]
- 16.Lane D, Prentki P, Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992;56:509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattion N M, Cohen J, Estes M K. Rotavirus proteins. In: Kapikian A, editor. Viral infections of the gastrointestinal tract. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 169–249. [Google Scholar]
- 18.Niepel M, Ling J, Gallie D R. Secondary structure in the 5′-leader or 3′-untranslated region reduces protein yield but does not affect the functional interaction between the 5′-cap and the poly(A) tail. FEBS Lett. 1999;462:79–84. doi: 10.1016/s0014-5793(99)01514-8. [DOI] [PubMed] [Google Scholar]
- 19.Nilsson M, von Bonsdorff C H, Svensson L. Biosynthesis and morphogenesis of group C rotavirus in swine testicular cells. Arch Virol. 1993;133:21–37. doi: 10.1007/BF01309741. [DOI] [PubMed] [Google Scholar]
- 20.Otero L J, Ashe M P, Sachs A B. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 1999;18:3153–3163. doi: 10.1093/emboj/18.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pestova T V, Hellen C U. Ribosome recruitment and scanning: what's new? Trends Biochem Sci. 1999;24:85–87. doi: 10.1016/s0968-0004(99)01356-0. [DOI] [PubMed] [Google Scholar]
- 22.Piron M, Delaunay T, Grosclaude J, Poncet D. Identification of the RNA-binding, dimerization and eIF4GI-binding domains of rotavirus NSP3. J Virol. 1999;73:5411–5421. doi: 10.1128/jvi.73.7.5411-5421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piron M, Vende P, Cohen J, Poncet D. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 1998;17:5811–5821. doi: 10.1093/emboj/17.19.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poncet D, Aponte C, Cohen J. Rotavirus protein NSP3 (NS34) is bound to the 3′ end consensus sequence of viral mRNAs in infected cells. J Virol. 1993;67:3159–3165. doi: 10.1128/jvi.67.6.3159-3165.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poncet D, Laurent S, Cohen J. Four nucleotides are the minimal requirement for RNA recognition by rotavirus non-structural protein NSP3. EMBO J. 1994;13:4165–4173. doi: 10.1002/j.1460-2075.1994.tb06734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poncet D, Lindenbaum P, L'Haridon R, Cohen J. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J Virol. 1997;71:34–41. doi: 10.1128/jvi.71.1.34-41.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preiss T, Hentze M W. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 28.Proweller A, Butler S. Ribosome concentration contributes to discrimination against poly(A)− mRNA during translation initiation in Saccharomyces cerevisiae. J Biol Chem. 1997;272:6004–6010. doi: 10.1074/jbc.272.9.6004. [DOI] [PubMed] [Google Scholar]
- 29.Rao C D, Das M, Ilango P, Lalwani R, Rao B S, Gowda K. Comparative nucleotide and amino acid sequence analysis of the sequence-specific RNA-binding rotavirus nonstructural protein NSP3. Virology. 1995;207:327–333. doi: 10.1006/viro.1995.1087. [DOI] [PubMed] [Google Scholar]
- 30.Rau M, Ohlmann T, Morley S J, Pain V M. A reevaluation of the cap-binding protein, eIF4E, as a rate-limiting factor for initiation of translation in reticulocyte lysate. J Biol Chem. 1996;271:8983–8990. doi: 10.1074/jbc.271.15.8983. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 32.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 33.Tarun S Z, Jr, Sachs A B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 34.Wells S E, Hillner P E, Vale R D, Sachs A B. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]