Abstract
Whole genome sequencing (WGS) and data concerning identity and safety for Saccharomyces cerevisiae CBS 493.94 are reported. This strain was isolated from a British brewery in 1958 and deposited at the CBS culture collection Westerdijk Fungal Biodiversity Institute under the accession number CBS 493.94. The long-reads sequencing data, obtained via PacBio Sequel, and short-reads data, via Illumina NovaSeq 6000, were deposited at NCBI under accession number PRJNA1044661. The hybrid assembly was made publicly available via Zenodo and NCBI. For strain identification, data from 18S rRNA, ANI dendrogram and Core Genome single nucleotide polymorphism (SNP) Tree showed that the present isolate belongs to the genus Saccharomyces, species cerevisiae. The potential genes of concern, e.g. antimycotic resestance genes, were not detected. This strain is commonly used as a feed additive for animal health improvement and the present data summarise the unambiguous identity and strain's FKS1 gene does not code for any amino acid variants of concern.
Keywords: Whole genome sequencing, Genes of concern, Microbial safety, Yeast identity, Saccharomyces cerevisiae
Specifications Table
| Subject | Microbiology |
| Specific subject area | Microbial genomics |
| Data format | Raw reads: NCBI BioProject number PRJNA1044661 and Zenodo (https://doi.org/10.5281/zenodo.10083536) Analysed: The genome assembly was deposited in Zenodo (https://doi.org/10.5281/zenodo.10209995) and NCBI (JBCEXF000000000) AGUSTUS coding sequences results were deposited in Zenodo (https://doi.org/10.5281/zenodo.10260328). |
| Type of data | Raw reads and assembled data from the Saccharomyces cerevisiae CBS 493.94 genome sequencing Table Figure |
| Data collection | DNA extraction Whole genome sequencing via Pacific Biosciences (PacBio) Sequel, using SMRT Cell, Illumina NovaSeq 6000 |
| Data source location | Institution: Alltech Inc. City/Town/Region: Nicholasville, Kentucky Country: USA |
| Data accessibility | Repository name: NCBI BioProjects database Data identification number: PRJNA1044661 Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1044661 Repository name: Zenodo Data identification number: 10.5281/zenodo.10209995 Direct URL to data: https://doi.org/10.5281/zenodo.10209995 |
| Related research article | Not applicable |
1. Value of the Data
-
•
Live yeast cells are commonly used as probiotic feed additives in animal nutrition aimed at performance and health improvement.
-
•
Although S. cerevisiae is a Qualified Presumptive Safe (QPS) species in the European Union, the unambiguous strain identity and safety of each strain should be demonstrated via Next Generation Sequencing (NGS) Technologies, being required for feed additives entry in a food supply chain. The present data reports the CBS 493.94 strain identity and safety-related information.
-
•
The dataset reported herein could be valuable information to different research and regulatory sectors.
2. Background
S. cerevisiae is commonly used as a feed additive for animal zootechnical performance improvement [1]. This species is considered a QPS species for humans, animals, and the environment [2]. However, all strains introduced in the food chain should be analysed via NGS for their identity and safety-related traits. For this purpose, we herein report the WGS data, strain taxonomical identity, and a search for genes of concern for CBS94.93 strain, commonly used as a feed additive.
3. Data Description
The WGS of long- and short-reads were obtained via PacBio and Illumina platforms. The raw reads were deposited to the NCBI BioProjects database under identification number PRJNA1044661 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1044661). After assembly, the WGS for CBS 493.94 was composed of 32 contigs. The total length of contigs reached a value of 11,635,572 bp, the mean contig length was 363,611.62, and the N50 value was 676,443 (Table 1).
Table 1.
Assembly results for S. cerevisiae CBS 493.94.
| Assembly Statistics | |||||
|---|---|---|---|---|---|
| Total length (bp) | Number of Contigs (bp) | Mean Contig Length (bp) | Longest Contig (bp) | Shortest Contig (bp) | N50 |
| 11,635,572 | 32 | 363,612 | 1,471,023 | 2511 | 676,443 |
bp=base pairs.
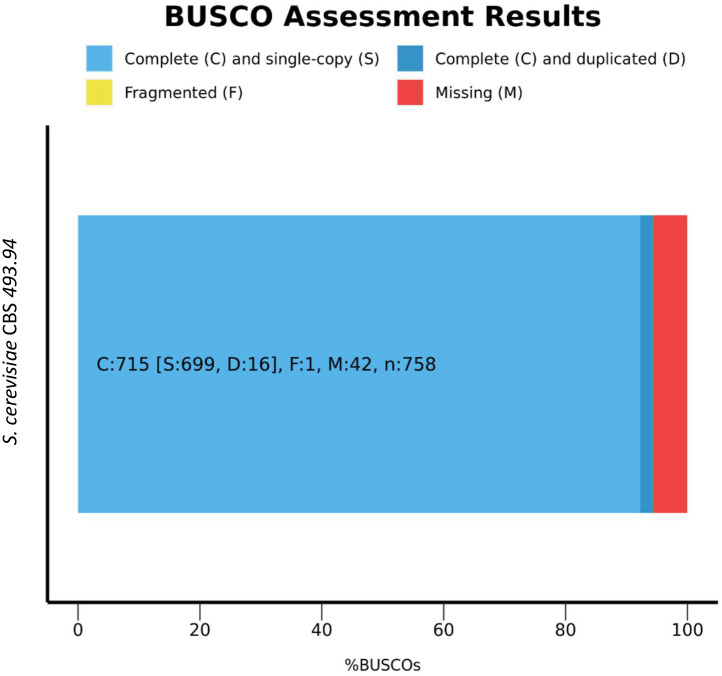
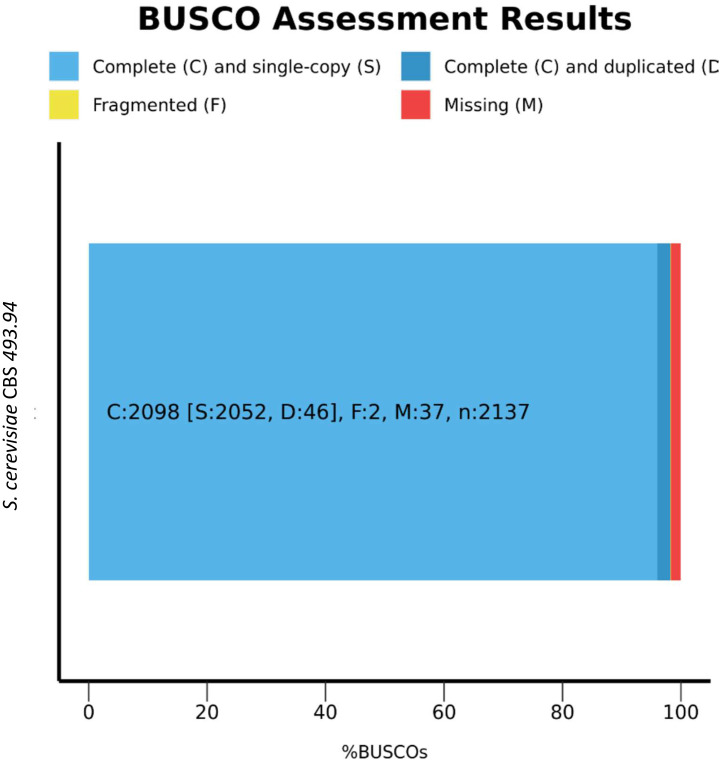
Benchmarking Universal Single-Copy Orthologs (BUSCO) was used to assess the genome assembly completeness and quality of the genome [3]. Results show that more than 90 % completely match to BUSCO gene set from core universal fungal orthologs (Fig. 1) and S. cerevisiae S288C orthologs (Fig. 2).
Fig. 1.
BUSCO analysis results for S. cerevisiae CBS 493.94 against universal fungal orthologs.
Fig. 2.
BUSCO analysis results for S. cerevisiae CBS 493.94 against saccharomycetes_odb10 orthologs.
The expected genome length for the strain S. cerevisiae S288C [4] is 12,157,105, being the total length of the S. cerevisiae CBS 493.94 contigs obtained within +/− 20 % of the expected genome size (11.6 Mb).
Taxonomic strain identification was performed using a comprehensive BLAST analysis. Furthermore, the average nucleotide identity (ANI) and single nucleotide polymorphism (SNP) approaches were used to compare CBS 493.94 strain with sequences of different publicly available strains classified as S. cerevisiae spp.
Concerning the BLAST analysis, identity percentages, referred to 18S rRNA, reached 100 % in the top 5 results, shown in Table 2.
Table 2.
Identification of S. cerevisiae CBS 493.94 by 18S rRNA gene.
| Description | Max score | Total score | Query cover | E value | % identity | Accession length (bp) | Accession |
|---|---|---|---|---|---|---|---|
| S. cerevisiae strain NCIM3186 chromosome XII sequence | 3321 | 6642 | 1 | 0 | 100 | 1,078,087 | CP011821.1 |
| S. cerevisiae YJM693 chromosome XII sequence | 3321 | 534,700 | 1 | 0 | 100 | 2,458,844 | CP006458.1 |
| S. cerevisiae YJM1433 chromosome XII sequence | 3321 | 342,100 | 1 | 0 | 100 | 1,874,567 | CP006416.1 |
| S. cerevisiae YJM1383 chromosome XII sequence | 3321 | 318,900 | 1 | 0 | 100 | 1,869,821 | CP006402.1 |
| S. cerevisiae YJM1355 chromosome XII sequence | 3321 | 425,100 | 1 | 0 | 100 | 2,249,646 | CP006399.1 |
The publicly available S. cerevisiae spp. data used for the ANI and SNP comparison is shown in Table 3.
Table 3.
Reference genomes used for identification of S. cerevisiae CBS 493.94.
| Reference Genome Sequences |
|---|
| GCA_000146045_2_Saccharomyces_cerevisiae_S288C_strain |
| GCA_001051215_1_Saccharomyces_cerevisiae_strain_ySR127 |
| GCA_003086655_1_Saccharomyces_cerevisiae_strain_BY4742 |
| GCA_003709285_1_Saccharomyces_cerevisiae_strain_KSD |
| GCA_004014915_1_Saccharomyces_cerevisiae_strain_Makgeolli |
| GCA_004328465_1_Saccharomyces_cerevisiae_strain_ySR128 |
| GCA_018219195_1_Saccharomyces_cerevisiae_strain_IMF17 |
| GCA_021172205_1_Saccharomyces_cerevisiae_strain_S288C |
| GCA_022695735_1_Saccharomyces_cerevisiae_strain_UWOPS83 |
| GCA_023508825_1_Saccharomyces_cerevisiae_strain_CICC |
| GCA_024732265_1_Saccharomyces_cerevisiae_strain_PY0001 |
| GCA_024972935_1_Saccharomyces_cerevisiae_strain_L261col5 |
| GCA_024972955_1_Saccharomyces_cerevisiae_strain_L261 |
| GCA_030607045_1_Saccharomyces_cerevisiae_strain_2 |
| GCA_903819125_2_Saccharomyces_cerevisiae_strain_HN1 |
| GCA_903819135_2_Saccharomyces_cerevisiae_strain_Y55 |
| GCA_903819145_2_Saccharomyces_cerevisiae_strain_BJ4 |
| GCA_903819155_2_Saccharomyces_cerevisiae_strain_HLJ1 |
| GCA_903819175_2_Saccharomyces_cerevisiae_strain_SX2 |
| GCA_903819185_2_Saccharomyces_cerevisiae_strain_JXXY16 |
| GCA_903819195_2_Saccharomyces_cerevisiae_strain_XXYS1 |
| GCA_903819205_2_Saccharomyces_cerevisiae_strain_EM14S01 |
| GCF_000146045_2_Saccharomyces_cerevisiae_S288C.fasta |
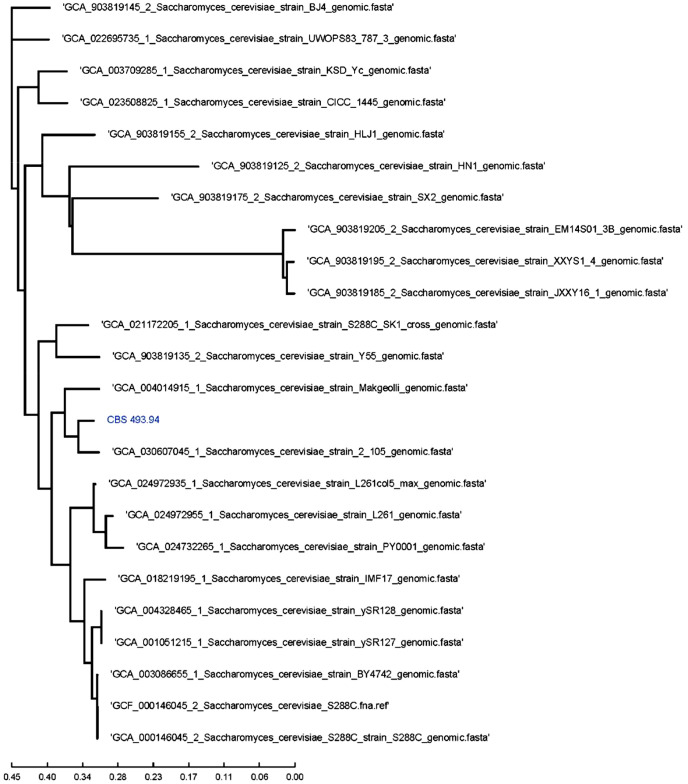
The achieved identity percentage when the sample isolate was compared with these sequences was above 98 % (Fig. 3). As a phylogenetic tree is recommended, a SNP tree was generated. A core genome alignment percentage of 82.2 % between the Saccharomyces genome sequences was achieved, with S. cerevisiae 2-105 the taxonomically closest strain (Fig. 4). Furthermore, SNP distance matrix results are shown in Annex 1 (Zenodo repository: https://zenodo.org/records/10262057).
Fig. 3.
Average Nucleotide Identity (ANI) dendrogram of S. cerevisiae CBS 493.94.
Fig. 4.
SNP Tree for S. cerevisiae CBS 493.94 based on Core Genome Phylogeny using Harvest.
The potential presence of antimycotic resistance was assessed via the Mycotic Antifungal Resistance Database (MARDy) [5]. The former database reports that antimycotic resistance is conferred by amino acid substitutions in FKS1 gene that are present in isolates, demonstrating the corresponding MIC breakpoint for arborcandin C and caspofungin (according to the CLSI Subcommittee for Antifungal Testing). These amino acid substitutions were not found in the genome assembly. AGUSTUS coding sequences results are available in Annex 2 and Annex 3, deposited at Zenodo repository (https://doi.org/10.5281/zenodo.10260328).
4. Experimental Design, Materials and Methods
4.1. DNA extraction
A freeze-dried pure culture was obtained from the CBS culture collection Westerdijk Fungal Biodiversity Institute (Utrecht,The Netherlands), grown in liquid nutrient media for 48 h at 30 °C, and the DNA was extracted. Methodology details relating to the strain growth and DNA extraction are covered by the intellectual property rights of Baseclear B.V (Leiden, The Netherlands). In brief, a fresh CBS 493.94 cell suspension was used for the DNA extraction using the Quick-DNA Fungal/Bacterial Miniprep kit (Zymo Research, USA) according to manufacturer's instructions; and the lysis was performed in combination with zymolyase (Zymo Research, USA; 5 units/μl final concentration).
4.2. Sequencing strategy, quality control and assembly
DNA from S. cerevisiae CBS 493.94 was used to create a 10-kb single-plex sequencing library sequenced on a SMRT Cell on the PacBio Sequel instrument that generated 7 Gb sequenced bases and 860,922 number of reads. For shot-reads Nextera XT sequencing library (Illumina) was used on the NovaSeq 6000 instrument that generated 4.96 Gb sequenced bases and 16,822,463 number of reads of paired-end 150 nucleotides.
Raw paired end reads were trimmed and processed using BBDuk, version 39.01 (BBMap – Bushnell B. – sourceforge.net/projects/bbmap/) with a read quality trimming parameter of 22. The parameters used were minlen=36, qtrim=rl, and trimq=22. The parameter “qtrim=rl” allows trimming on both ends of the reads; “minlen” establishes reads shorter than after trimmings were discarded; and regions with average quality below “trimq” value were trimmed. 94.37 % of reads were retained after trimming. PacBio CLR reads do not have a PHRED score associated. Illumina reads were used for post assembly polishing of PacBio long reads assembly to correct for errors in long reads assembly.
De novo assembly was performed with Flye 2.9.1-b1780, and the parameters used were “flye –pacbio-raw $fastq -o flye_out –threads 32″. Pilon [6], version 1.24 with default parameters, was the tool used to correct errors from PacBio assembly. Assembly statistics were checked using assemblystats.
BUSCO v5.4.4 was run with default parameters by running the genome assembly against: (1) the core universal fungal orthologs, and (2) against saccharomycetes_odb10 ortholog sets.
4.3. Taxonomical identification
Barrnap version 0.9 And Web Blastn for 18S rRNA were used for gene extraction and sequence identification, respectively. To ensure the accuracy of this prediction and verify its identity, the extracted sequence was subjected to a comprehensive BLAST analysis. BLAST nucleotide database with default parameters was used to perform the analysis against known S. cerevisiae 18S rRNA sequences.
Furthermore, an average nucleotide identity (ANI) approach using MUMmer 3.0 and dRep version 3.4.2 with default parameters was performed [7]. Comparative measurements between two genome sequences, called Overall Genome Relatedness Indices (OGRI), were developed, and proposed to provide a cut-off or define boundaries between species [8]. Among them, average nucleotide identity (ANI) is the most widely used, with a proposed species boundary cut-off of 95–96 % [9].
The isolate was assessed and compared with different whole sequences of strains classified as S. cerevisiae spp. publicly available sequences in NCBI [2]. Data from comparison sequences is shown in Table 3. The achieved identity percentage when the sample isolate was compared with these sequences was all above 98 % (Fig. 3).
The phylogenetic tree is recommended, particularly for taxa with a high identity level between related species [10]. An SNP tree was also made based on core genome phylogeny using Harvest (parsnp-1.7.4) with default parameters [11].
4.4. Identification of genes of potential concern
MARDy VERSION 1.1DB:1.3WS (BETA) was used to identify antimycotic resistance genes. Genes prediction of the identified coding sequences was performed using AUGUSTUS [12] (Model: S. cerevisiae), version 3.3.3, with default parameters.
Limitations
Not applicable.
Ethics Statement
The authors have read and follow the ethical requirements for publication in Data in Brief and confirming that the current work does not involve human subjects, animal experiments, or any data collected from social media platforms.
CRediT Author Statement
Ivana Nikodinoska: Conceptualisation, Writing - Original Draft, Data Curation Colm Moran: Conceptualisation, Writing - Review & Editing, Funding, Project administration.
Acknowledgments
The whole genome sequencing was performed at Baseclear and the bioinformatic analysis at CosmosID and Sandwalk Bioventures.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: The authors I.N and C.A.M. are employees of Alltech, which produces S. cerevisiae CBS 493.94 described in this study.
Data Availability
References
- 1.Desnoyers M., Giger-Reverdin S., Bertin G., Duvaux-Ponter C., Sauvant D. Meta-analysis of the influence of Saccharomyces cerevisiae supplementation on ruminal parameters and milk production of ruminants. J. Dairy Sci. 2009;92(4):1620–1632. doi: 10.3168/jds.2008-1414. [DOI] [PubMed] [Google Scholar]
- 2.Panel EFSA BIOHAZ. Statement on the update of the list of qualified presumption of safety (QPS) recommended microbiological agents intentionally added to food or feed as notified to EFSA 17: suitability of taxonomic units notified to EFSA until September 2022. EFSA J. 2023:7746–7782. doi: 10.2903/j.efsa.2023.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 4.NCBI. https://www.ncbi.nlm.nih.gov/.
- 5.Nash A., Sewell T., Farrer R.A., Abdolrasouli A., Shelton J.M.G., Fisher M.C., Rhodes J. MARDy: mycology antifungal resistance database. Bioinformatics. 2018;34:3233–3234. doi: 10.1093/bioinformatics/bty321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., et al. Pilon: an Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., Salzberg S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun J., Rainey F.A. Integrating genomics into the taxonomy and systematics of Bacteria and Archaea. Int. J. Syst. Evol. Microbiol. 2014;64:316–324. doi: 10.1099/ijs.0.054171-0. [DOI] [PubMed] [Google Scholar]
- 9.Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 10.European Food Safety Authority (EFSA) EFSA statement on the requirements for whole genome sequence analysis of microorganisms intentionally used in the food chain. EFSA J. 2021;19:6506–6520. doi: 10.2903/j.efsa.2021.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:435–439. doi: 10.1093/nar/gkl200. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.