Highlights
-
•
The study generated and characterized monoclonal antibodies against E184L ASFV protein.
-
•
Two novel linear epitopes were identified using sequentially truncated overlapping peptides.
-
•
The two epitopes are immunodominant and could be used as potential epitope based antigen markers.
Keywords: African swine fever virus, E184L, Linear B cell epitope, Monoclonal antibody
Abstract
African swine fever virus (ASFV) is a large double-stranded DNA virus with a complex structural architecture and encodes more than 150 proteins, where many are with unknown functions. E184L has been reported as one of the immunogenic ASFV proteins that may contribute to ASFV pathogenesis and immune evasion. However, the antigenic epitopes of E184L are not yet characterized. In this study, recombinant E184L protein was expressed in prokaryotic expression system and four monoclonal antibodies (mAbs), designated as 1A10, 2D2, 3H6, and 4C10 were generated. All four mAbs reacted specifically with ASFV infected cells. To identify the epitopes of the mAbs, a series of overlapped peptides of E184L were designed and expressed as maltose binding fusion proteins. Accordingly, the expressed fusion proteins were probed with each E184L mAb separately by using Western blot. Following a fine mapping, the minimal linear epitope recognized by mAb 1A10 was identified as 119IQRQGFL125, and mAbs 2D2, 3H6, and 4C10 recognized a region located between 153DPTEFF158. Alignment of amino acids of E184L revealed that the two linear epitopes are highly conserved among different ASFV isolates. Furthermore, the potential application of the two epitopes in ASFV diagnosis was assessed through epitope-based ELISA using 24 ASFV positive and 18 negative pig serum and the method were able to distinguish positive and negative samples, indicating the two epitopes are dominant antigenic sites. To our knowledge, this is the first study to characterize the B cell epitopes of the antigenic E184L protein of ASFV, offering valuable tools for future research, as well as laying a foundation for serological diagnosis and epitope-based marker vaccine development.
1. Introduction
African swine fever (ASF) is a highly contagious and lethal hemorrhagic viral disease of pigs. ASF outbreaks are typically associated with huge economic loss, mainly due to the high lethality of the virus to domestic pigs, restrictions of pig movements and the introduction of massive culling campaigns (Sánchez-Cordón et al., 1997). The impact of the disease is even more severe in small holder farmers in developing countries, who depend on pig as additional income and relatively cheap source of protein (Sánchez-Cordón et al., 1997; Chenais et al., 2018).
ASF was first described by Montgomery (2024) in 1921 in Kenya as a disease causing high mortality in domestic pigs, and was limited to eastern and southern Africa for more than thirty years (Penrith, 2020; Penrith and Vosloo, 2009). The first transcontinental transmission of ASF to Europe was recorded in 1957 (Portugal) and 1960 (Spain). Fortunately, it was successfully eradicated from its occurrence outside Africa in the mid-1990s, except from the Italian island of Sardinia where it remains endemic since 1978 (Sánchez-Cordón et al., 1997). However, a second incursion to the Republic of Georgia occurred in 2007 (Rowlands et al., 2008), most likely via contaminated feed of domestic pigs with subsequent fast spread to neighboring countries and further into Eastern Europe (Sanchez-Vizcaino et al., 2015). Prior to August 2018, the disease had never been reported in Asia, but since then, it has entered China possessing half of the world's pig population, and subsequently spread to neighboring countries (Ma et al., 2020; Zhou et al., 2018; Zhao et al., 2019; Wen et al., 2019). Ever since, the disease continues to spread in an unpredictable manner across many European and Asian countries.
The causative agent of the disease is African swine fever virus (ASFV), which is a linear, large double-stranded DNA virus with a genome size varying from 170 to 194 kbp, depending on the virus isolate (Chapman et al., 2011). ASFV encodes approximately 150–167 open reading frames (ORFs), including 68 structural proteins, more than 100 non-structural proteins, and multiple polypeptides (Alejo et al., 2018). The lack of knowledge regarding the structural and functional attributes of the majority of ASFV proteins has impeded the development of vaccines and diagnostics for ASF. Morphologically, ASFV is icosahedral in symmetry and has a complex structural architecture composed of several concentric layers (Dixon et al., 2013; Wang et al., 2019; Yáñez et al., 1995). There are 24 genotypes of ASFV based on the major capsid p72 protein encoding gene B646L, and 8 serotypes based on the viral hemagglutinin CD2-like protein and C-type lectin (Malogolovkin et al., 2015).
The rapid spread of the disease has created an urgent need for research into the structure and function of ASFV proteins, as well as the development of vaccines and diagnostic methods (Wang et al., 2024; Zhao et al., 2023; Chen et al., 2020). It is widely recognized that monoclonal antibodies that are both stable and effective play a significant role in advancing these investigations. The research reports on the E184L ASFV protein are currently limited, but the available studies indicate its immunogenicity and potential significance. Previous study has shown that E184L induced a significant IFNγ response in lymphocytes from at least one pig immunized with a pool of ASFV peptides (Netherton et al., 2019). It has been also reported that E184L gene is a novel virulence-determinant which was confirmed through the deletion of E184L from the highly virulent ASFV Georgia 2010 (ASFV-G) isolate (Ramirez-Medina et al., 2022). Furthermore, recent reports suggest that E184L protein inhibits host innate immune response via targeting the stimulator of IFN genes (STING)-mediated signaling pathway (Zhu et al., 2023). Considering the available data, E184L could be a potential target to be used in serological assays and vaccine formulations. However, it should be noted that there are currently no reports on E184L monoclonal antibodies or identified key epitopes.
In the present study, the antigenicity of E184L protein was preliminarily tested using serum from ASFV-infected pigs and four hybridoma cell lines that stably secrete antibodies against E184L protein were successfully generated and characterized. Accordingly, two linear B cell epitopes were identified, which could be important for the development of epitope-based marker vaccines and diagnostic tools for ASF infection.
2. Materials and methods
2.1. Cells and viruses
The study utilized two ASFV strains HLJ/18 and SD/DY-I/21, which were isolated in our laboratory. The cell lines HEK 293T were cultured in DMEM with a 10 % heat-inactivated fetal bovine serum, while mouse myeloma cells (SP2/0) were cultured in RPMI-1640 medium supplemented with a 20 % heat-inactivated fetal bovine serum. Porcine alveolar macrophage (PAM) cells were isolated from specific pathogen free (SPF) pigs and maintained in RPMI-1640 medium supplemented with 10 % porcine serum. BK2258 cell lines, isolated from a wild boar fetus (Wang et al., 2024), were cultured in DMEM medium supplemented with 10 % FBS. All cells were incubated at 37 °C in a humidified incubator with 5 % CO2. Experiments involving live ASFV were carried out in a biosafety level 3 (P3) facility at the Harbin Veterinary Research Institute (HVRI) and were approved by the Biosafety Committee of State Key Laboratory for Animal Disease Control and Prevention.
2.2. Expression and purification of the E184L protein
In order to obtain recombinant E184L fusion protein, E184L full length sequences were amplified from the strain ASFV HLJ/18 (GenBank accession number MK333180.1) using two pairs of primers (Supplementary Table 1). Restriction enzymes NdeI and EcoRI were included to the forward and reverse primers, to allow the amplified sequence to be cloned into the pMAL-C5X expression vector (New England Biolabs, NEB #E8200S). Successful cloning was confirmed by sequencing and the recombinant plasmids were named pMAL-C5X-E184L. Accordingly, pMAL-C5X-E184L was transformed into E. coli ER2523, and induced by 0.5 mM isopropy β-d-1-thiogalactopyranoside (IPTG) for 5 h at 37 °C. Thereafter, the expression of E184L fusion protein were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot. To purify E184L protein, bacterial cells were harvested by centrifugation, resuspended in pre-cold PBS (25 mL/liter of bacterial culture), and lysed by sonication. After centrifugation at 12,000 rpm for 30 min, supernatants were collected and filtered through 0.22 μm and purified using pre-packed MBP Trap column, operated by using AKTA AVANT liquid chromatography system. Buffers for purification were prepared according to the manufacturer's instructions (binding buffer = 20 mM Tris–HCl and elution buffer 10 mM maltose in binding buffer).
2.3. Generation of monoclonal antibodies against E184L
To generate monoclonal antibodies, the purified MBP tagged E184L protein was used as an immunogen to inoculate mice and the standard protocol for hybridoma technology was followed as described previously (Tesfagaber et al., 2021). In brief, six-week-old female BALB/c mice were immunized subcutaneously with purified MBP tagged E184L protein emulsified with an equal volume of Freund's complete adjuvant, followed with two booster injections at two-week intervals with the same immunogen and equal volume of Freund's incomplete adjuvant. Mouse with the highest antibody titer detected by indirect ELISA, was euthanized humanly, and spleen cells were harvested and fused with SP2/0 myeloma cells by using polyethylene glycol (PEG). Antibody secreting hybridoma cells were screened by using ELISA plate coated with the recombinant E184L protein.
2.4. Western blot
Purified E184L and bacterial-expressed peptide fusion proteins were separated by 12 % SDS–PAGE, transferred onto PVDF membranes, and blocked with 5 % skimmed milk for 1 h at room temperature. Subsequently, the membranes were probed with E184L mAbs and ASFV positive pig serum for 1 h at 37 °C, washed three times with 0.01 % Tween-20 (PBST), and incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG for 1 h at 37 °C. Following extensive washing, protein bands were visualized using a digital imaging system.
Also, a dot blot assay was performed to confirm the linear epitopes that can be recognized by E184L mAbs. In brief, short peptides (E184L-R3 and E184L-R5 fragments) were expressed as MBP tagged proteins and spotted on nitrocellulose membrane (2 μL each). The whole E184L and MBP protein were used as a positive and negative control, respectively. The membranes were subsequently blocked with 5 % skimmed milk powder for 1 h, followed by a 2h incubation with each E184L mAb at room temperature. After washing the membranes five times with PBST for 5 min each, HRP-labeled goat anti-mouse antibody was added, and the membranes were washed three times with PBST. Finally, enhanced chemiluminescence reagent was used to detect the binding ability of each mAb to the target peptides.
2.5. Enzyme-linked immunosorbent assay (ELISA)
Indirect ELISA was used to screen E184L mAbs secreting hybridoma cell lines. Ninety-six-well plates were coated with MBP-tagged E184L protein (0.2 μg/well) in PBS at 4 °C overnight, followed by blocking with 5 % skimmed milk in PBS for 1 h at 37 °C. Next, fifty microliters (50 μL) of hybridoma supernatant were added to each well and incubated for 2 h at 37 °C. Thereafter, HRP conjugated goat anti-mouse IgG diluted 1:10,000 was added and incubated for 1 h at 37 °C. Of note, after each step, the microtiter plates were washed three times with PBST. Finally, reaction was developed by adding chromogenic substrate solution (TMB) for 10 min, stopped with 2 M sulphuric acid and result was read at OD450 absorbance.
2.6. Immunofluorescence assay (IFA)
To evaluate the specificity and ability of E184L mAbs to detect ASFV infected cells, IFA was performed in HEK 293T cells transfected with E184L recombinant gene (pCGGAS-E184L) and ASFV-infected BK2258 cells. HEK293T cells were cultured in 24-well plate at a density of 1.25×105/well in a complete DMEM medium. After 24 h, cells were transfected pCGGAS-E184L plasmid DNA and incubated at 37 °C in 5 % CO2. At 24 h post transfection, HEK293T cells were washed with PBS and fixed with 4 % formaldehyde for 10 min at room temperature. Similarly, BK2258 cells were seeded in 96-well plates and infected with ASFV SD/DY-I/21 at an MOI of 0.01. At 24 h post-infection (hpi), the cells were washed with PBS and fixed with 4 % formaldehyde. Following washing with PBS, 0.25 % triton was added to each well in order to increase the permeability of the cells and was left at room temperature for 10 min. The cells were then incubated with E184L monoclonal antibodies for 1 h at 37 °C., followed with FITC conjugated goat ant-mouse IgG. After each incubation step, the wells were washed four times with PBS. Finally, results were observed using a fluorescent microscope.
2.7. Construction and expression of short E184L peptide fusion proteins
To map the mAbs, preliminary E184L was divided into two truncated segments, designated as E184L-L1 (N-terminus end) and E184L-R1(C-terminus end) with both E184L-L1 and E184L-R1 having 10 amino acids (aa) in common. The two truncated segments were amplified using the primers listed in supplementary Table 1, cloned to pMAL-C5X, and expressed in E. coli ER2523 as MBP-tagged proteins. Subsequently, expressed truncated segments were probed with each E184L mAb separately. Based on the result found, E184L-R1 was further truncated into five overlapping short peptides spanning the whole E184L-R1 segment, designated as R2, R3, R4, R5, and R6, and the peptides were expressed as MBP fusion proteins in pMAL-C5X. Each of the truncated proteins R2, R3, R4, R5, and R6 contain 30 aa, and overlapped by 15 aa from the succeeding peptide. To precisely map the core sequence of the epitope of each mAb, short peptides that show a reaction to any of the E184L mAbs were further sequentially truncated from the N terminus and the C terminus by deleting one amino acid at a time, respectively. The primers used to amplify each peptide are listed in supplementary Table 1.
2.8. Prediction of the continuous B cell epitope regions of E184L
The continuous B cell epitopes and antigenic regions of E184L were predicted by in silico analysis of the online B cell epitope prediction tools BepiPred (http://tools.iedb.org/bcell/) and ABCPred (https://webs.iiitd.edu.in/raghava/abcpred/index.html). The predicted B cell epitope regions in E184L protein are shown in supplementary figures (Figs. S1 and S2).
3. Result
3.1. Expression, purification, and antigenicity test of E184L recombinant protein
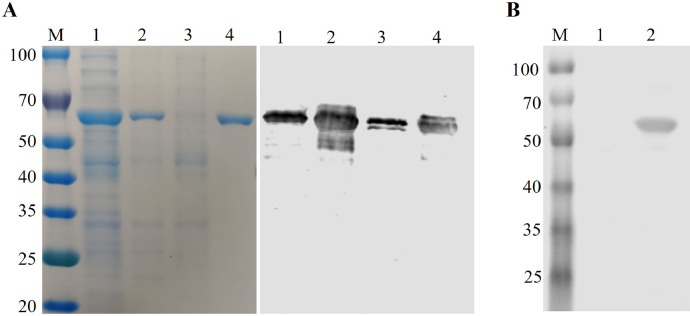
The recombinant pMAL-C5X-E184L plasmid was constructed, transformed into E. coli ER2523, and expressed as MBP-tagged protein. The recombinant E184L protein was successfully expressed, and a strong protein band was observed at the expected size (∼65 kDa) by using SDS-PAGE and Western blot (Fig. 1). E184L protein was purified with a pre-packed MBP-Trap column by using AKTA Avant liquid chemotagraphy purification machine. The purity of E184L protein was confirmed by using SDS-PAGE and Western blot (Fig. 1, lane 4). A nanodrop spectrophotometer was used to quantify the purified E184L protein, which was subsequently frozen at −80 °C for future use.
Fig. 1.
A) Expression and purification of E184L protein with MBP-tag. Bacterial lysates from E. coli. ER2523 with recombinant plasmids pMAL-C5x-E184L and purified E184L protein were subjected to SDS-PAGE (right) and Western blot analysis with anti-MBP mAb (left). Sample order in both SDS-PAGE and Western blot were as follows: lane 1, the supernatant of ER2523 with pMAL-C5x- E184L induced by IPTG after sonication; lane 2, the precipitation of ER2523 with pMAL-C5x- E184L; Lane 3, pMAL-C5x- E184L without IPTG addition; and lane 4, the purified E184L protein. B) Reactivity of E184L to ASFV-positive pig serum, lane 1 was loaded with MBP (negative control) and lane 2 was loaded with E184L protein and stained with ASFV-positive serum at a dilution of 1:1000.
Identifying immunogenic and antigenic viral proteins play a fundamental role in developing a diagnostic assay and vaccine design. Herein, we conducted preliminary tests with serum from ASFV-infected pigs to assess the antigenicity of E184L protein by using Western blot. In brief, purified E184L protein was separated by SDS-PAGE, transferred to PVDF membrane, stained with ASFV positive serum, and a clear protein band was observed at the expected size (Fig. 1B). These results indicate that E184L is an antigenic ASFV protein and has the potential to be a target antigen to develop a diagnostic assay.
3.2. Production and characterization of monoclonal antibodies against E184L
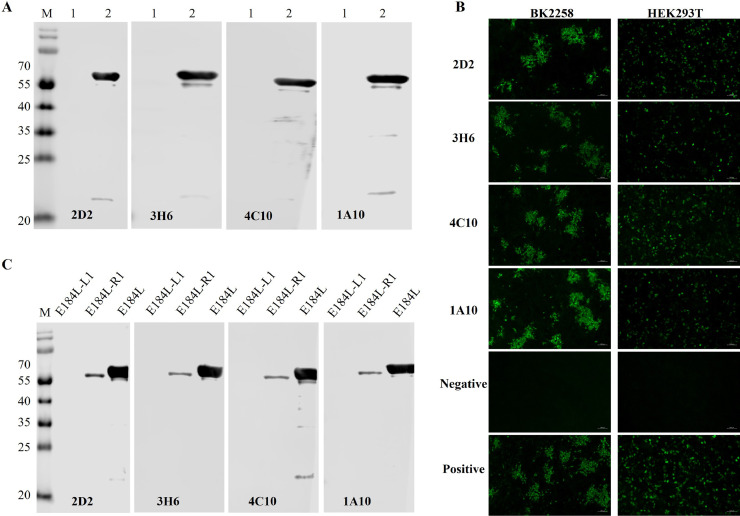
To produce monoclonal antibodies (mAbs), female BALB/c mice were immunized with purified E184L protein. Spleen cells from mice with high titers of E184L antibody were then fused with SP2/0 mouse myeloma cells. The resulting hybridoma cells, which secreted E184L antibodies, were screened by using an indirect ELISA with E184L protein as the coating antigen. Ultimately, four monoclonal hybridoma cell lines (1A10, 2D2, 3H6, and 4C10) that strongly and specifically reacted with the E184L protein were obtained. To further determine the specificity of the mAbs, Western blot and IFA were performed. All four mAbs specifically recognized the entire E184L protein but did not react with the MBP protein (Fig. 2A). Similarly, IFA results from both HEK 293T cells transfected with the pCAGGS-E184L plasmid and BK2258 cells infected with ASFV showed that all mAbs (1A10, 2D2, 3H6, and 4C10) strongly reacted with the recombinant E184L protein and ASFV-infected cells (Fig. 2B). Furthermore, the isotype of the mAbs were characterized by using Mouse Monoclonal Antibody Isotype ELISA Kit. The results showed that 2D2, 3H6, and 4C10 were the IgG1 subclass, 1A10 was the IgG2a subclass, and the light chain type of all four mAbs were Kappa (Table 1).
Fig. 2.
Characterization of E184L mAbs. A) Characterization of E184L mAbs using Western blot; lane 1 was loaded with MBP protein and lane 2 was loaded with purified MBP tagged E184L protein. 3H6, 1A10, 2D2, and 4C10 mAbs were used as primary antibody and all specifically reacted to E184L protein. B) Characterization of E184L mAbs using IFA. All four mAbs recognized ASFV-infected BK2258 cells and HEK 293T transfected with pCAGGS-E184L recombinant plasmid. C) Preliminary antigenic epitope identification (Round 1). The whole E84L protein was truncated into two fragments (E184L-L1 and E184L-R1) and were probed with each mAb separately after expression. The four E184L mAbs recognized the C terminus end of the protein (E184L-R1).
Table 1.
Identification of subclasses of E184L monoclonal antibodies.
| Monoclonal antibodies |
||||
|---|---|---|---|---|
| 1A10 | 2D2 | 3H6 | 4C10 | |
| Ig subclass | IgG2a | IgG1 | IgG1 | IgG1 |
| Light chain type | κ | κ | κ | κ |
3.3. Identification of linear B cell epitopes
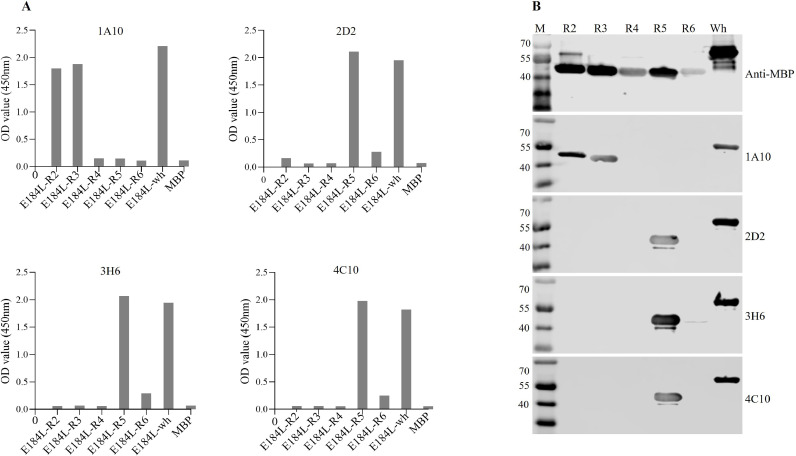
The successful expression of the preliminary truncated and secondary truncated E184L proteins was evaluated using anti-MBP antibody, and the amino acid sequences for the preliminary truncated and secondary truncated E184L proteins are listed in Table 2, Table 3, respectively. After initially truncating the full length E184L into two fragments (E184L-L1 and E184L-R1), the general recognition regions of the four mAbs were determined by using Western blot and all four mAbs reacted to E184L-R1 (95–184 aa) (Fig. 2C). Based on the preliminary result, peptide E184L-R1 was further truncated into five short overlapped peptides (R2, R3, R4, R5, and R6) spanning the whole length of the E184L-R1(Fig. 3). After successful expression of the five E184L-R1 fragments, their reactivity to the four mAbs were tested by indirect ELISA (Fig. 4A) and Western blotting (Fig. 4B). One of the mAbs (1A10) reacted with E184L-R2 and E184L-R3 (95–140 aa) (Fig. 4). As the two peptides (R2 and R3) are overlapped by 16 aa, the recognition region to mAb 1A10 was deducted to 110RLCKVFSIMIQRQGFL125. Whereas the three mAbs (2D2, 3H6, and 4C10) reacted to peptide E184L-R5 (Fig. 4), suggesting the linear epitope for the three mAbs lay between the amino acid region 141ILMVADNLYGEQDPTEFFSLIIEQTKTIKK170.
Table 2.
Sequences of the initial overlapped truncated E184L fusion peptides.
| Peptide Name | Amino acid sequence |
|---|---|
| R2(95–125) | 95KFQKMLNAITEQLMSRLCKVFSIMIQRQGFL125 |
| R3(110–140) | 110RLCKVFSIMIQRQGFLKTQTLMYSHLFTILS140 |
| R4(126–155) | 126KTQTLMYSHLFTILSILMVADNLYGEQDPT155 |
| R5(141–170) | 141ILMVADNLYGEQDPTEFFSLIIEQTKTIKK170 |
| R6(156–184) | 156EFFSLIIEQTKTIKKKKKSGSEEEESHEE184 |
Table 3.
Sequences of the secondary truncated E184L fusion peptides (fine mapping).
| Peptide Name | Amino acid sequence | Peptide Name | Amino acid sequence |
|---|---|---|---|
| R5–1(142–170) | LMVADNLYGEQDPTEFFSLIIEQTKTIKK | R3–1(111–125) | LCKVFSIMIQRQGFL |
| R5–2(143–170) | MVADNLYGEQDPTEFFSLIIEQTKTIKK | R3–2(112–125) | CKVFSIMIQRQGFL |
| R5–3(144–170) | VADNLYGEQDPTEFFSLIIEQTKTIKK | R3–3(113–125) | KVFSIMIQRQGFL |
| R5–4(145–170) | ADNLYGEQDPTEFFSLIIEQTKTIKK | R3–4(114–125) | VFSIMIQRQGFL |
| R5–5(146–170) | DNLYGEQDPTEFFSLIIEQTKTIKK | R3–5(115–125) | FSIMIQRQGFL |
| R5–6(147–170) | NLYGEQDPTEFFSLIIEQTKTIKK | R3–6(116–125) | SIMIQRQGFL |
| R5–7(148–170) | LYGEQDPTEFFSLIIEQTKTIKK | R3–7(117–125) | IMIQRQGFL |
| R5–8(149–170) | YGEQDPTEFFSLIIEQTKTIKK | R3–8(118–125) | MIQRQGFL |
| R5–9(150–170) | GEQDPTEFFSLIIEQTKTIKK | R3–9 (119–125) | IQRQGFL |
| R5–10(151–170) | EQDPTEFFSLIIEQTKTIKK | R3–10(120–125) | QRQGFL |
| R5–11(152–170) | DPTEFFSLIIEQTKTIKK | R3–11(110–124) | RLCKVFSIMIQRQGF |
| R5–12(153–170) | PTEFFSLIIEQTKTIKK | R3–12(110–123) | RLCKVFSIMIQRQG |
| R5–13(154–170) | TEFFSLIIEQTKTIKK | R3–13(110–122) | RLCKVFSIMIQRQ |
| R5–14(155–170) | EFFSLIIEQTKTIKK | R3–14(110–121) | RLCKVFSIMIQR |
| R5–15(141–168) | ILMVADNLYGEQDPTEFFSLIIEQTKTI | ||
| R5–16(141–166) | ILMVADNLYGEQDPTEFFSLIIEQTK | ||
| R5–17(141–164) | ILMVADNLYGEQDPTEFFSLIIEQ | ||
| R5–18(141–162) | ILMVADNLYGEQDPTEFFSLII | ||
| R5–19(141–160) | ILMVADNLYGEQDPTEFFSL | ||
| R5–20(141–158) | ILMVADNLYGEQDPTEFF | ||
| R5–20b(141–158 | ILMVADNLYGEQDPTEF | ||
| R5–21(141–156) | ILMVADNLYGEQDPTE | ||
| R5–22(141–154) | ILMVADNLYGEQDP | ||
| R5–23(141–152) | ILMVADNLYGEQ | ||
| R5–24(141–150) | ILMVADNLYG |
Fig. 3.
Schematic diagram showing the design of truncated overlapping short peptides spanning the full length of E184L protein and the sequential deletion of amino acid for fine mapping.
Fig. 4.
Identification of antigenic linear epitopes on the E184L protein (round 2) using Enzyme-linked immunosorbent assay (ELISA) (A) and Western blot (B). The results showed that mAb 1A10 recognized peptide R2 and R3, and mAbs 2D2, 3H6, and 4C10 recognized peptide R5. The full length E184L protein was used as a positive control and is indicated as “wh” in lane 6.
3.4. Fine mapping of the target epitope for 1A10 mAb
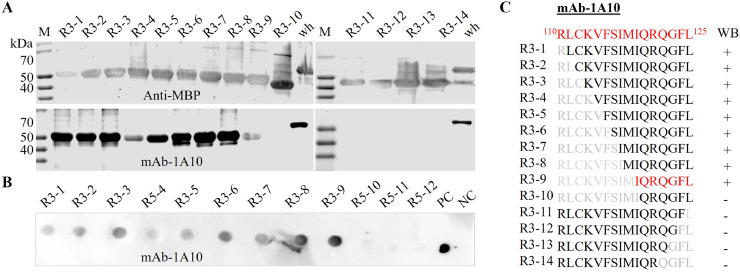
To precisely locate the site of mAb 1A10, the peptide 110RLCKVFSIMIQRQGFL125 was sequentially truncated from the N-terminus side, designated as R3–1, R3–2, R3–3, R3–4, R3–5, R3–6, R3–7, R3–8, R3–9, and R3–10. The truncated peptides were expressed as MBP fusion products and probed with mAb 1A10 by using Western blot. The results showed that when nine amino acid residues (R110 to M118) were serially removed from the N terminus, it showed no marked decrease in Western blot analysis (Fig. 5A). However, the removal of “I119” amino acid (R3–10) had completely decreased the binding ability of the peptide R3–10 to mAb 1A10 (Fig. 5A). To further confirm the core amino acid, the peptide 110RLCKVFSIMIQRQGFL125 was sequentially truncated from the C-terminus side, designated as R3–11, R3–12, R3–13, and R3–14 (Fig. 5B). After each peptide was expressed and detected with mAb 1A10, none of the peptides (R3–11, R3–12, R3–13, and R3–14) reacted to mAb 1A10 (Fig. 5A). These results indicate that the amino acid Isoleucine at position 119 (I119) and Leucine at position 125 (L125) are the key amino acid sites for mAb 1A10 binding and the core peptide sequence is 119IQRQGFL125. To validate the result, a dot blot assay was also performed on twelve E184L-R3 peptides (R3–1 to R12). Similar to the Western blot result, mAb 1A10 reacted to peptide R3–1 to R3–9, but not to Peptide R3–10 to R3–12 (Fig. 5C), which further confirms the core amino acid sequence for mAb 1A10 is 119IQRQGFL125.
Fig. 5.
Fine mapping of the target epitope for E184L mAb 1A10 by sequential amino acid deletion from the N and C-terminus of the peptide 110RLCKVFSIMIQRQGFL125. A) Analysis of the reactivity of the truncated peptides (R3 peptides) to mAb 1A10 using Western blot. The MBP fused peptides were probed with 1A10 mAb. The mAb-1A10 reacts to peptide R3–1 to R3–9, but not to peptide R3–10 to R3- 14 which indicates the core amino acid for these antibodies lay between 119IQRQGFL125. B) Dot blot assay to further confirm the linear core amino acid sequence of mAb 1A10. After succseful expresion of E184L-R3 peptides (R3–1 to R3–12), 2 μL form each was spoted into nitrocellulose membrane and tested against mAb 1A10. The full length E184L and MBP pprotein were used as postive and negative control and are indicated as “PC” and “NC”, respectively. C) Schematic design showing the sequential deletion of amino acid and its respective result of Western blot after staining with mAb 1A10. MBP antibody was used to test the successful expression of each peptide.
3.5. Fine mapping of the target epitope for 2D2, 3H6, and 4C10 mAbs
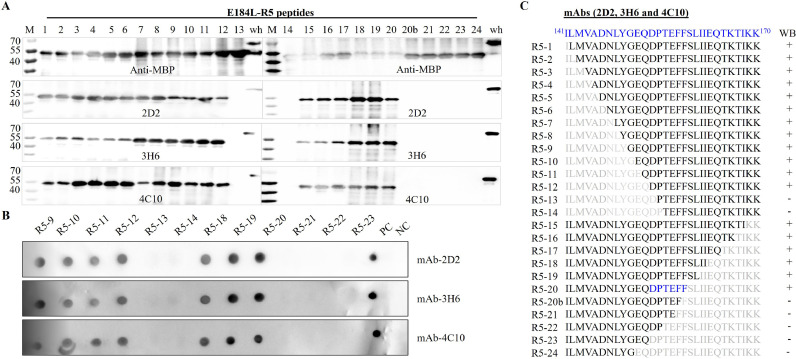
To precisely map the core sequence of the epitope for 2D2, 3H6, and 4C10 mAbs, the peptide 141ILMVADNLYGEQDPTEFFSLIIEQTKTIKK170 was sequentially truncated from the N and the C-terminus, respectively. The series of truncated peptides were expressed as MBP fusion products and probed with 2D2, 3H6, and 4C10 mAbs by using Western blot. The results demonstrated that the deletion of twelve amino acid residues from N terminus (R5–1 to R5–12) did not affect the reactivity of 2D2, 3H6, and 4C10. However, when the amino acid “D153” (peptide R5–13) was further removed, the reactivity of the three mAbs to peptide-13 (R5–13) was completely lost (Fig. 6A). On the other hand, the peptide fusion protein maintained its reactivity with 2D2, 3H6, and 4C10 mAbs when 12 amino acids were sequentially removed from the C terminus. However, when the amino acid phenylalanine at position 158 (F158) was removed from the C terminus, the resulting fusion protein (R5–20b) completely lost its reactivity to 2D2, 3H6, and 4C10 (Fig. 6A). These findings indicate that the amino acid Aspartic acid at position 153 (D153) and Phenylalanine at position 158 (F158) are the key amino acid sites for mAbs 2D2, 3H6, and 4C10 binding, and 153DPTEFF158 is the core sequence. To further confirm the core amino acid sequence, a dot blot assay was also performed. Based on the Western blot results, twelve E184L-R5 peptides (R5–9 to R5–14 and R5–18 to R5–23) were selected, spotted on to nitrocellulose membrane, and probed with mAbs 2D2, 3H6, and 4C10. As expected, the result was consistent with the results of Western blot, where mAbs 2D2, 3H6, and 4C10 reacted with R5–9 to R5–12 (N terminus deletion) and R5–18-R5–20 (C-terminus deletion) (Fig. 6B). However, all three mAbs failed to react with peptide R5–13 (D153 deleted) and peptide R5–21 (F158 deleted) (Fig. 6B), which further confirms the core amino acid sequence for mAbs 2D2, 3H6, and 4C10 is 153DPTEFF158.
Fig. 6.
Fine mapping of the target epitope for E184L mAbs (2D2, 3H6, and 4C10) by sequential amino acid deletion from the N and C-terminus end of the peptide 141ILMVADNLYGEQDPTEFFSLIIEQTKTIKK170. A) Analysis of the reactivity of the truncated peptides (R5 peptides) to 2D2, 3H6, and 4C10 mAbs by using Western blot. The MBP fused peptides were probed with each mAb separately. The three mAbs reacted to peptide R5–1 to R5–12 until the Aspartic acid at position 153 (D153) was removed from the N terminus end, and to peptide R5–15 to R5–20 until the phenylalanine at position 158 (F158) was removed from the C terminus end. These results indicate that the core amino acid for 2D2,3H6 and 4C10 mAbs lay between 153DPTEFF158. C) Dot blot assay to further confirm the linear core amino acid sequence of mAbs 2D2, 3H6 and 4C10. E184L-R5 peptides (R5–9 to R5–14 and R5–18 to R5–23) were expressed, spoted into nitrocellulose membrane and probed with 2D2,3H6 and 4C10 mAbs. The full length E184L and MBP pprotein were used as postive and negative control and are indicated as “PC” and “NC”, respectively. C) schematic design showing the sequential deletion of amino acid and its respective result of Western blot after staining with 2D2,3H6, and 4C10 mAbs. MBP antibody was used to test the successful expression of each peptide.
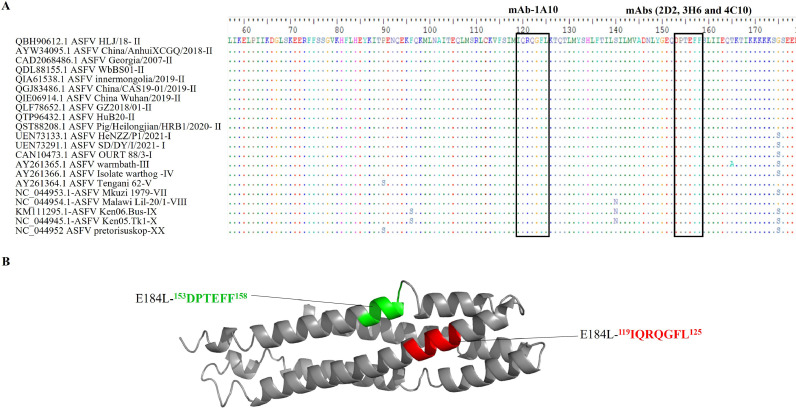
3.6. Homology analysis of the identified E184L epitopes
To evaluate whether the epitope region is conserved among different ASFV genotypes, we analysed E184L protein sequence from 21 ASFV isolates, including 10 genotypes. Sequence alignment of these 21 ASFV isolates revealed that the two novel epitopes (119IQRQGFL125 and 153DPTEFF158) identified in this study are highly conserved (Fig. 7A). Moreover, the 3D structure of E184L protein was predicted using an online protein structure prediction tool I-TASSER (https://zhanggroup.org/I-TASSER/) and PyMOL software was employed for the visualization of the spatial structure of the epitopes (Fig. 7B).
Fig. 7.
Characterization of the E184L linear epitopes. A) Sequence alignment of the identified epitopes and the amino acid sequences of all epitopes among different ASFV strains were analyzed by using BioEdit software. B) Localization of the epitopes. The identifiied Epitopes were located in simulated 3D model of E184L protein by PyMol software. The 3D structure of E184L protein was predicted using an online protein structure prediction tool I-TASSER (https://zhanggroup.org/I-TASSER/).
3.7. The two novel E184L epitopes are antigenic and have the potential to be applied for diagnostic assay development
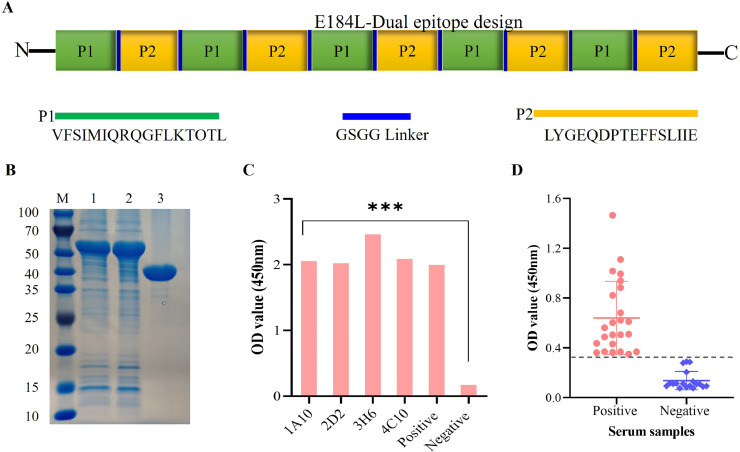
To determine whether the two linear epitopes identified in our study are antigenic, the reactivity of these two epitopes to ASFV-positive serum were tested. In brief, E184L dual epitope containing VFSIMIQRQGFLKTOTL and LYGEQDPTEFFSLIIE was designed, and the pMAL-C5X-E184L-epitope plasmid (named as E184L-dual epitope) was synthesized by GenScript Biotech Co., Ltd. (Nanjing, China). The schematic presentation of the E184L dual epitope design is shown in Fig. 8A. After successful expression and purification (Fig. 8B), the reaction of the four E184L mAbs against the synthesized protein were tested, and as expected all the four E184L mAbs reacted strongly (Fig. 8C), which further confirms their core amino acid sequence is within the synthesized protein. Subsequently, a dual epitope-based ELISA using pig serum from ASFV-infected pigs was performed. In brief, ninety-six-well ELISA plate was coated with the E184L-dual epitope at a concentration of 0.4 mg/mL and was used to detect ASFV antibodies. After testing 24 ASFV positive and 18 negative serum samples, the E184L epitope-based ELISA was able to distinguish positive and negative samples, where the OD value of ASFV positive samples were ranging from 0.36 to 1.46 and OD value of negative samples were between 0.08 to 0.28 (Fig. 8D). These results suggest that the two E184L epitopes are dominant and antigenic and could be used as target antigens to develop a diagnostic test.
Fig. 8.
Assessing the potential application of the two nove E184L epitopes. A) Schematic presentation of the E184L-dual epitope design. B) Purification of the E184L-dual epitope protein, lane 1 and lane 2 were loaded with 20 μL purified protein and lane 3 was loaded with 20 μL purified MBP protein (control). C) Reactivity test of the synthesized E184L-dual epitope protein to the four mAbs prepared in the study. Anti-E184L polyclonal mouse serum and SP2/0 cell supernantnts were used as positive and negative control, respectively. D) Dual epitope-based ELISA to confirm the antigenecity and potential application of the two novel E184L epitopes.
4. Discussion
ASF is a significant swine viral disease that leads to substantial economic losses in the global pig industry. Currently, there are no effective vaccines or treatments available for ASF. Therefore, the control and prevention of the disease primarily relies on accurate diagnosis, herd isolation, and the culling of infected animals. In order to develop safe and effective subunit vaccines or precise diagnostic assays, it is vital to identify immunogenic and antigenic protein targets of ASFV. However, the understanding of the structure, function, and antigenic determinants of most ASFV proteins remains limited. Some immunological determinants of ASFV proteins have been identified and reported, including p72, p30, p54, pp62, CD2v, B602L, and p22 (Cubillos et al., 2013; Gallardo et al., 2019; Peng-fei et al., 2022; Tsegay et al., 2022). Additionally, the antigenic epitopes of several ASFV proteins, such as p72 (Heimerman et al., 2018; Miao et al., 2023), p30 (Zhou et al., 2022; Si-hui et al., 2023), p54 (Zheng et al., 2023), CD2v (Jia et al., 2022; Ren et al., 2022), K205R (Zhang et al., 2023), CP312R (Hagoss et al., 2023), and pC129R (Wang et al., 2023), have been reported.
ASFV E184L protein has been reported as a significant immune determinant of ASFV (Ramirez-Medina et al., 2022), however, little is known about the antigenic structure of the E184L protein. In the present study, E184L protein was expressed in E. coli, purified, and confirmed by using anti-MBP antibody. In agreement with previous studies (Ramirez-Medina et al., 2022), we found that the purified E184L protein strongly reacts with anti-ASFV antibody serum which suggests that the E184L could be a good antigenic target for developing a subunit vaccine and diagnostic assays. Monoclonal antibodies have the advantages of high purity, high specificity for binding to the antigen, and unlimited supply as compared to polyclonal antibodies. Using the purified E184L protein as an immunogen, we generated and characterized four mAbs against E184L protein. Analysis of those mAbs with ELISA, Western blot, and IFA showed that all four mAbs (1A10, 2D2, 3H6, and 4C10) were able to recognize the immunizing E184L antigen.
E184L protein has been reported to inhibit host innate immune response via targeting the stimulator of IFN genes (STING)-mediated signaling pathway (Zhu et al., 2023), indicating a role of E184L in ASFV immune escape. Identifying key antigenic epitopes of E184L could help to further explore the protein function. Moreover, identification of B cell epitopes in viral structural proteins plays a crucial role in understanding virus-host interactions and developing effective diagnostic tools and vaccines (Gershoni et al., 2007; Wang and Yu, 2004). To identify the linear epitope of these mAbs, a series of truncated overlapping peptides covering the whole length of E184L gene were expressed as MBP fusion proteins for peptide scanning. Preliminary, E184L was divided into two truncated fragments (E184L-L1 and E184L-R1), expressed, and probed with the four E184L mAbs separately. Notably, all four mAbs recognized the short truncated E184L-R1 fusion protein located in a region between amino acid residues 95 and 184 of the E184L protein. Based on the strategy of expressing truncated peptides as fusion proteins, we further identified the shortest possible core sequences that can be recognized by three mAbs (2D2, 3H6, and 4C10) located in a narrow range between 141ILMVADNLYGEQDPTEFFSLIIEQTKTIKK170, and mAb 1A10 recognized a region between 110RLCKVFSIMIQRQGFL125. By sequential amino acid deletion, the key epitopes were further shortened and two novel linear B cell epitopes 153DPTEFF158 for 2D2, 3H6, and 4C10 mAbs and 119IQRQGFL125 for 1A10 mAb were identified. The B cell epitope regions in E184L were also predicted by in silico analysis of online Linear Epitope Prediction tools. The prediction results revealed that the peptides 153DPTEFF158 and 119IQRQGFL125 had a high prediction score, suggesting that these two epitopes identified in the present study are among the antigenic regions in E184L protein (Figs. S1 and S2).
In order to investigate the conservation of epitope sequences across different strains and genotypes of ASFV, a total of 21 ASFV E184L protein sequences were aligned. The alignment revealed a high degree of conservation for the two epitopes 153DPTEFF158 and 119IQRQGFL125 among various ASFV strains. These newly identified epitopes hold great potential for advancing the understanding of the E184L protein's structure and function through further research. Additionally, our findings offer valuable insights for the development of subunit vaccines and the establishment of serological diagnostic methods. To the best of our knowledge, this is the first study to characterize the epitopes of the antigenic E184L protein of ASFV.
CRediT authorship contribution statement
Weldu Tesfagaber: Writing – review & editing, Writing – original draft, Methodology, Formal analysis. Desong Lan: Methodology, Formal analysis, Conceptualization. Wan Wang: Visualization, Validation, Investigation, Formal analysis, Data curation. Rui Zhao: Visualization, Validation, Investigation, Formal analysis, Data curation. Li Yin: Visualization, Validation, Investigation, Formal analysis, Data curation. Mingyang Yang: Visualization, Validation, Investigation, Formal analysis, Data curation. Yuanmao Zhu: Visualization, Validation, Investigation, Formal analysis, Data curation. Encheng Sun: Visualization, Validation, Investigation, Formal analysis, Data curation. Renqiang Liu: Visualization, Validation, Investigation, Formal analysis, Data curation. Wenjun Lin: Visualization, Validation, Investigation, Formal analysis, Data curation. Zhigao Bu: Supervision, Conceptualization. Fang Li: Supervision, Data curation, Conceptualization. Dongming Zhao: Writing – review & editing, Writing – original draft, Supervision, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding statement
This work was supported by the National Key R&D Program of China (2022YFD1800600, 2021YFD1800101), the Heilongjiang Provincial Natural Science Foundation of China (JQ2023C005), Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-CSLPDCP-202301), and Central public-interest Scientific Institution Basal Research Fund (No. 1610302022003).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.virusres.2024.199412.
Contributor Information
Fang Li, Email: lifang01@caas.cn.
Dongming Zhao, Email: zhaodongming@caas.cn.
Appendix. Supplementary materials
Data availability
No data was used for the research described in the article.
References
- Sánchez-Cordón P.J., Montoya M., Reis A.L., Dixon L.K. African swine fever: a re-emerging viral disease threatening the global pig industry. Veter. J. 1997;233(2018):41–48. doi: 10.1016/j.tvjl.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenais E., Ståhl K., Guberti V., Depner K. Identification of wild boar-habitat epidemiologic cycle in African swine fever epizootic. Emerg. Infect. Dis. 2018;24(4):810–812. doi: 10.3201/eid2404.172127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery R.E. On a form of swine fever occurring in British East Africa (Kenya Colony) J. Comp. Pathol. 2024;34:159–191. [Google Scholar]
- Penrith M.L. Current status of African swine fever. CABI Agricult. Biosci. 2020;1(1):11. [Google Scholar]
- Penrith M.L., Vosloo W. Review of African swine fever: transmission, spread and control. J. S. Afr. Vet. Assoc. 2009;80(2):58–62. doi: 10.4102/jsava.v80i2.172. [DOI] [PubMed] [Google Scholar]
- Rowlands R.J., Michaud V., Heath L., Hutchings G., Oura C., Vosloo W., Dwarka R., Onashvili T., Albina E., Dixon L.K. African swine fever virus isolate, Georgia, 2007. Emerg. Infect. Dis. 2008;14(12):1870–1874. doi: 10.3201/eid1412.080591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vizcaino J.M., Mur L., Gomez-Villamandos J.C., Carrasco L. An update on the epidemiology and pathology of African swine fever. J. Comp. Pathol. 2015;152(1):9–21. doi: 10.1016/j.jcpa.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Ma J., Chen H., Gao X., Xiao J., Wang H. African swine fever emerging in China: distribution characteristics and high-risk areas. Prev. Vet. Med. 2020;175 doi: 10.1016/j.prevetmed.2019.104861. [DOI] [PubMed] [Google Scholar]
- Zhou X., Li N., Luo Y., Liu Y., Miao F., Chen T., Zhang S., Cao P., Li X., Tian K., Qiu H.J., Hu R. Emergence of African Swine Fever in China, 2018. Transbound. Emerg. Dis. 2018;65(6):1482–1484. doi: 10.1111/tbed.12989. [DOI] [PubMed] [Google Scholar]
- Zhao D., Liu R., Zhang X., Li F., Wang J., Zhang J., Liu X., Wang L., Zhang J., Wu X., Guan Y., Chen W., Wang X., He X., Bu Z. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg. Microbes. Infect. 2019;8(1):438–447. doi: 10.1080/22221751.2019.1590128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., He X., Zhang X., Zhang X., Liu L., Guan Y., Zhang Y., Bu Z. Genome sequences derived from pig and dried blood pig feed samples provide important insights into the transmission of African swine fever virus in China in 2018. Emerg. Microbes. Infect. 2019;8(1):303–306. doi: 10.1080/22221751.2019.1565915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman D.A., Darby A.C., Da Silva M., Upton C., Radford A.D., Dixon L.K. Genomic analysis of highly virulent Georgia 2007/1 isolate of African swine fever virus. Emerg. Infect. Dis. 2011;17(4):599–605. doi: 10.3201/eid1704.101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejo A., Matamoros T., Guerra M., Andrés G., Shisler J.L. A proteomic atlas of the African swine fever virus particle. J. Virol. 2018;92(23):e01293. doi: 10.1128/JVI.01293-18. -18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon L.K., Chapman D.A., Netherton C.L., Upton C. African swine fever virus replication and genomics. Virus Res. 2013;173(1):3–14. doi: 10.1016/j.virusres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- Wang N., Zhao D., Wang J., Zhang Y., Wang M., Gao Y., Li F., Wang J., Bu Z., Rao Z., Wang X. Architecture of African swine fever virus and implications for viral assembly. Science. 2019;366(6465):640–644. doi: 10.1126/science.aaz1439. [DOI] [PubMed] [Google Scholar]
- Yáñez R.J., Rodríguez J.M., Nogal M.L., Yuste L., Enríquez C., Rodriguez J.F., Viñuela E. Analysis of the complete nucleotide sequence of African swine fever virus. Virology. 1995;208(1):249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- Malogolovkin A., Burmakina G., Titov I., Sereda A., Gogin A., Baryshnikova E., Kolbasov D. Comparative analysis of African swine fever virus genotypes and serogroups. Emerg. Infect. Dis. 2015;21(2):312–315. doi: 10.3201/eid2102.140649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhang J., Li F., Zhang Z., Chen W., Zhang X., Sun E., Zhu Y., Liu R., He X., Bu Z., Zhao D. The attenuated African swine fever vaccine HLJ/18-7GD provides protection against emerging prevalent genotype II variants in China. Emerg. Microbes. Infect. 2024;13(1) doi: 10.1080/22221751.2023.2300464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Sun E., Huang L., Ding L., Zhu Y., Zhang J., Shen D., Zhang X., Zhang Z., Ren T., Wang W., Li F., He X., Bu Z. Highly lethal genotype I and II recombinant African swine fever viruses detected in pigs. Nat. Commun. 2023;14(1):3096. doi: 10.1038/s41467-023-38868-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhao D., He X., Liu R., Wang Z., Zhang X., Li F., Shan D., Chen H., Zhang J., Wang L., Wen Z., Wang X., Guan Y., Liu J., Bu Z. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020;63(5):623–634. doi: 10.1007/s11427-020-1657-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netherton C.L., Goatley L.C., Reis A.L., Portugal R., Nash R.H., Morgan S.B., Gault L., Nieto R., Norlin V., Gallardo C., Ho C.S., Sanchez-Cordon P.J., Taylor G., Dixon L.K. Identification and immunogenicity of African swine fever virus antigens. Front. Immunol. 2019;10:1318. doi: 10.3389/fimmu.2019.01318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Medina E., Vuono E., Rai A., Pruitt S., Espinoza N., Velazquez-Salinas L., Pina-Pedrero S., Zhu J., Rodriguez F., Borca M.V., Gladue D.P. Deletion of E184L, a putative DIVA target from the pandemic strain of African swine fever virus, produces a reduction in virulence and protection against virulent challenge. J. Virol. 2022;96(1) doi: 10.1128/JVI.01419-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Li S., Ma C., Yang F., Cao W., Liu H., Chen X., Feng T., Shi Z., Tian H. African swine fever virus E184L protein interacts with innate immune adaptor STING to block IFN production for viral replication and pathogenesis. J. Immunol. 2023;210(4):442–458. doi: 10.4049/jimmunol.2200357. [DOI] [PubMed] [Google Scholar]
- Wang W., Zhang Z.J., Tesfagaber W., Zhang J.W., Li F., Sun E.C., Tang L.J., Bu Z.G., Zhu Y.M., Zhao D.M. Establishment of an indirect immunofluorescence assay for the detection of African swine fever virus antibodies. J. Integr. Agric. 2024;23(1):228–238. [Google Scholar]
- Tesfagaber W., Wang L., Tsegay G., Hagoss Y.T., Zhang Z., Zhang J., Huangfu H., Xi F., Li F., Sun E., Bu Z., Zhao D. Characterization of Anti-p54 monoclonal antibodies and their potential use for African swine fever virus diagnosis. Pathogens. 2021;10(2):178. doi: 10.3390/pathogens10020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos C., Gómez-Sebastian S., Moreno N., Nuñez M.C., Mulumba-Mfumu L.K., Quembo C.J., Heath L., Etter E.M.C., Jori F., Escribano J.M., Blanco E. African swine fever virus serodiagnosis: a general review with a focus on the analyses of African serum samples. Virus Res. 2013;173(1):159–167. doi: 10.1016/j.virusres.2012.10.021. [DOI] [PubMed] [Google Scholar]
- Gallardo C., Fernández-Pinero J., Arias M. African swine fever (ASF) diagnosis, an essential tool in the epidemiological investigation. Virus. Res. 2019;271 doi: 10.1016/j.virusres.2019.197676. [DOI] [PubMed] [Google Scholar]
- Peng-fei W.A.N.G., Ming W.A.N.G., Zhi-bin S.H.I., Zhen-zhao S.U.N., Li-li W.E.I., Zai-si L.I.U., Shi-da W.A.N.G., Xi-jun H.E., WANG J.-f. Development of a recombinant pB602L-based indirect ELISA assay for detecting antibodies against African swine fever virus in pigs. J. Integr. Agric. 2022;21(3):819–825. [Google Scholar]
- Tsegay G., Tesfagaber W., Zhu Y., He X., Wang W., Zhang Z., Sun E., Zhang J., Guan Y., Li F., Liu R., Bu Z., Zhao D. Novel P22-monoclonal antibody based blocking ELISA for the detection of African swine fever virus antibodies in serum. Biosafety Health. 2022;4(4):234–243. [Google Scholar]
- Heimerman M.E., Murgia M.V., Wu P., Lowe A.D., Jia W., Rowland R.R. Linear epitopes in African swine fever virus p72 recognized by monoclonal antibodies prepared against baculovirus-expressed antigen. J. Veter. Diagn. Invest. 2018;30(3):406–412. doi: 10.1177/1040638717753966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C., Yang S., Shao J., Zhou G., Ma Y., Wen S., Hou Z., Peng D., Guo H., Liu W., Chang H. Identification of p72 epitopes of African swine fever virus and preliminary application. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1126794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Shi Z., Luo J., Cao L., Yang B., Wan Y., Wang L., Song R., Ma Y., Tian H., Zheng H. Preparation and epitope mapping of monoclonal antibodies against African swine fever virus P30 protein. Appl. Microbiol. Biotechnol. 2022;106(3):1199–1210. doi: 10.1007/s00253-022-11784-7. [DOI] [PubMed] [Google Scholar]
- Si-hui S.Z, Jing-jing Y., Yi-ning L., Chang-de W.U., Zhen-jiang Z., Yuan-mao Z.H.U., Bo M., Jia-xing Z., Xue-xia W., Ying Z. Identification of two novel linear epitopes on the p30 protein of African swine fever virus. J. Integr. Agric. 2023;22(6):1945–1949. [Google Scholar]
- Zheng N., Li C., Hou H., Chen Y., Zhang A., Han S., Wan B., Wu Y., He H., Wang N., Du Y. A novel linear B-cell epitope on the p54 protein of African swine fever virus identified using monoclonal antibodies. Viruses. 2023;15(4):867. doi: 10.3390/v15040867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R., Zhang G., Bai Y., Liu H., Chen Y., Ding P., Zhou J., Feng H., Li M., Tian Y., Wang A. Identification of linear B cell epitopes on CD2V protein of African Swine fever virus by monoclonal antibodies. Microbiol. Spectr. 2022;10(2):e01052. doi: 10.1128/spectrum.01052-21. -21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Ding P., Liu S., Zhang N., Chen Y., Li Q., Fan L., Chang Z., Zhang G. Development and characterization of recombinant ASFV CD2v protein nanoparticle-induced monoclonal antibody. Int. J. Biol. Macromol. 2022;209:533–541. doi: 10.1016/j.ijbiomac.2022.03.069. [DOI] [PubMed] [Google Scholar]
- Zhang S.-J., Liu J., Niu B., Zhu Y.-M., Zhao D.-M., Chen W.-Y., Liu R.-Q., Bu Z.-G., Hua R.-H. Comprehensive mapping of antigenic linear B-cell epitopes on K205R protein of African swine fever virus with monoclonal antibodies. Virus Res. 2023;328 doi: 10.1016/j.virusres.2023.199085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagoss Y.T., Shen D., Zhang Z., Li F., Bu Z., Zhao D. Novel epitopes mapping of African swine fever virus CP312R protein using monoclonal antibodies. Viruses. 2023;15(2):557. doi: 10.3390/v15020557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Bai J., Zhang L., Xia T., Yang X., Zhang K., Gao Y., Jiang P. A new B cell epitope of pC129R protein of African swine fever virus identified by monoclonal antibodies. Vet. Microbiol. 2023;282 doi: 10.1016/j.vetmic.2023.109744. [DOI] [PubMed] [Google Scholar]
- Gershoni J.M., Roitburd-Berman A., Siman-Tov D.D., Freund N.T., Weiss Y. Epitope mapping: the first step in developing epitope-based vaccines. BioDrugs. 2007;21:145–156. doi: 10.2165/00063030-200721030-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-F., Yu M. Epitope identification and discovery using phage display libraries: applications in vaccine development and diagnostics. Curr. Drug. Targets. 2004;5(1):1–15. doi: 10.2174/1389450043490668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.