Abstract
Objective
RNA epigenetic modifications play an important role in regulating immune response of mammals. Bovine mastitis induced by Staphylococcus aureus (S. aureus) is a threat to the health of dairy cattle. There are numerous RNA modifications, and how these modification-associated enzymes systematically coordinate their immunomodulatory effects during bovine mastitis is not well reported. Therefore, the role of common RNA modification-related genes (RMRGs) in bovine S. aureus mastitis was investigated in this study.
Methods
In total, 80 RMRGs were selected for this study. Four public RNA-seq data sets about bovine S. aureus mastitis were collected and one additional RNA-seq data set was generated by this study. Firstly, quantitative trait locus (QTL) database, transcriptome-wide association studies (TWAS) database and differential expression analyses were employed to characterize the potential functions of selected enzyme genes in bovine S. aureus mastitis. Correlation analysis and weighted gene co-expression network analysis (WGCNA) were used to further investigate the relationships of RMRGs from different types at the mRNA expression level. Interference experiments targeting the m6A demethylase FTO and utilizing public MeRIP-seq dataset from bovine Mac-T cells were used to investigate the potential interaction mechanisms among various RNA modifications.
Results
Bovine QTL and TWAS database in cattle revealed associations between RMRGs and immune-related complex traits. S. aureus challenged and control groups were effectively distinguished by principal component analysis based on the expression of selected RMRGs. WGCNA and correlation analysis identified modules grouping different RMRGs, with highly correlated mRNA expression. The m6A modification gene FTO showed significant effects on the expression of m6A and other RMRGs (such as NSUN2, CPSF2, and METTLE), indicating complex co-expression relationships among different RNA modifications in the regulation of bovine S. aureus mastitis.
Conclusion
RNA epigenetic modification genes play important immunoregulatory roles in bovine S. aureus mastitis, and there are extensive interactions of mRNA expression among different RMRGs. It is necessary to investigate the interactions between RNA modification genes regulating complex traits in the future.
Keywords: Mastitis, RNA Modification Gene, RNA-seq, Staphylococcus aureus
INTRODUCTION
Staphylococcus aureus (S. aureus) is a gram-positive opportunistic pathogen [1], and a common cause of mastitis in ruminants, which is a key concern of the dairy industry due to its wide range of species, complex mechanisms of action, and difficulty of cure [2]. Bovine mammary gland tissue and mammary alveolar cells, particularly bovine mammary epithelial cells (Mac-T cells), are popular experimental models in S. aureus mastitis studies [3] and play a critical role in bovine immunological defense [4]. The Mac-T cell line is an immortalized version of bovine mammary epithelial cells, established by introducing the SV-40 large T-antigen into primary bovine mammary alveolar cells [5,6]. The Mac-T cell line retains the typical ‘cobblestone’ structure and specific secretory functions of mammary epithelial cells (MECs), providing a more manageable model of MECs [4,7]. Coupling with its ability to clarify the immunological functions of MECs, the Mac-T cell line is widely used in S. aureus mastitis studies.
RNA epigenetic modification is widespread in mammals and contributes to many cellular biological processes such as immunity, lipid metabolism, biological rhythms, and reproductive development [8,9]. Most studies only investigated the regulatory roles of individual RNA-modifying enzymes or single modifications in complex traits. However, it has been shown that RNA modifications do not work independently, and there are extensive interactions among them [2,10]. RNA epigenetic modifications are mostly reversible, and coordinated with writers (methylation transferases), erasers (demethylases), and readers (proteins that bind specifically to methylation sites) [11].
RNA epigenetic modifications mainly include N6-methyladenosine (m6A), N6, 2’-O-dimethyl adenosine (m6Am), N1-methyladenosine (m1A), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), N4-acetyl cytidine (ac4C), alternative polyadenylation (APA), pseudouridine (Ψ), and N7-methylguanosine (m7G) [12–21]. It has been reported that the RNA epigenetic modifications extensively involved in the regulation of immune processes. For example, the m6A modification crucially regulates immune cell proliferation, differentiation, and function [12]. The m1A modification modulates T-cell immune responses and suppresses inflammation [18]. The hm5C modification potentially impacts immune cell signaling and inflammation, influencing the pathogenesis of immune-related diseases [22]. To understand the effects of RNA modification-related genes (RMRGs) on bovine S. aureus mastitis, we compiled a catalogue of genes linked to RNA modifications and inflammation as well as disease (Supplementary Table S1). Then, 80 RMRGs [12–21] were selected for this study by comparing with the bovine gene database.
In this study, we investigated the roles of RMRGs in bovine mastitis caused by S. aureus using principal component analysis (PCA), weighted gene co-expression network analysis (WGCNA), and correlation analyses. Furthermore, we validated the potential interaction mechanisms of RNA interference. This study on RNA epigenetic changes in the pathogenesis of S. aureus mastitis will be beneficial for reducing the incidence of bovine S. aureus mastitis and promoting the long-term development of the dairy industry.
MATERIALS AND METHODS
Design
To elucidate the roles of RNA epigenetic modification genes in bovine S. aureus mastitis, four types of analyses were performed in this study (Figure 1), including mapping analysis of 80 RMRGs using the animal quantitative trait locus (QTL) and cattle Genotype-Tissue Expression atlas (cGTEx) transcriptome-wide association studies (TWAS) databases, PCA and expression analysis based on five RNA sequencing (RNA-seq) datasets, WGCNA analysis based on dataset four. Additionally, validation of expression regulation was performed by interference experiments (dataset six) and methylated RNA immunoprecipitation sequencing (MeRIP-seq) data (dataset seven). The findings provide evidence for potential interactions among RNA modification genes, highlighting their involvement in the pathogenesis of bovine S. aureus mastitis.
Figure 1.
Technical route of this study.
Data
To obtain public dataset on the transcriptional landscape of bovine S. aureus mastitis and m6A site information in cattle, the searches were performed on the NCBI GEO database platform using the terms “Mac-T cells”, “cattle”, “m6A”, and “Staphylococcus aureus”. In this study, a total of seven datasets were used in analyses, including five public datasets and two datasets generated by the current study. Of which, four raw RNA-seq datasets and one raw MeRIP-seq dataset were downloaded from public database. The underlined two datasets (five and six) were generated by the current study (Table 1).
Table 1.
The grouping information for all datasets
| Sample | The sample record number of sequence read archive experiment | |||
|---|---|---|---|---|
| Dataset one | SRX1713237 C-1 | SRX1713500 S-1 | ||
| SRX1713308 C-2 | SRX1713526 S-2 | |||
| Dataset two | SRX11650574 C-1 | SRX11650571 S-1 | ||
| SRX11650575 C-2 | SRX11650572 S-2 | |||
| SRX11650576 C-3 | SRX11650573 S-3 | |||
| Dataset three | SRX13089400 C-1 | SRX13089403 S1-1 | SRX13089406 S2-1 | |
| SRX13089401 C-2 | SRX13089404 S1-2 | SRX13089407 S2-2 | ||
| SRX13089402 C-3 | SRX13089405 S1-3 | SRX13089408 S2-3 | ||
| Dataset four | SRX10643265 C-1 | SRX10643269 S1-1 | SRX10643282 S2-1 | SRX10643295 S3-1 |
| SRX10643266 C-2 | SRX10643270 S1-2 | SRX10643283 S2-2 | SRX10643296 S3-2 | |
| SRX10643277 C-3 | SRX10643271 S1-3 | SRX10643284 S2-3 | SRX10643297 S3-3 | |
| SRX10643288 C-4 | SRX10643272 S1-4 | SRX10643285 S2-4 | SRX10643298 S3-4 | |
| SRX10643299 C-5 | SRX10643273 S1-5 | SRX10643286 S2-5 | SRX10643300 S3-5 | |
| SRX10643308 C-6 | SRX10643274 S1-6 | SRX10643287 S2-6 | SRX10643301 S3-6 | |
| Dataset five | SRR24210509 C-1 | SRR24210505 S-1 | ||
| SRR24210508 C-2 | SRR24210504 S-2 | |||
| SRR24210507 C-3 | SRR24210503 S-3 | |||
| SRR24210506 C-4 | SRR24210502 S-4 | |||
| Dataset six | SRR24210509 C-1 | SRR26403327 siFTO-1 | ||
| SRR24210508 C-2 | SRR26403326 siFTO-2 | |||
| SRR24210507 C-3 | SRR26403325 siFTO-3 | |||
| SRR24210506 C-4 | SRR26403324 siFTO-4 | |||
| Dataset seven | SRR13005820 Input-1 | SRR13005811 IP-1 | ||
| SRR13005821 Input-2 | SRR13005812 IP-2 | |||
| SRR13005822 Input-3 | SRR13005813 IP-3 | |||
Note: The underlined datasets were generated by this study. For six RNA-seq datasets, dataset one is at the individual level, whereas dataset two to six are at the cellular level. The MeRIP-seq dataset is dataset seven. “C” represents the control, “S” represents the challenge of S. aureus, and “siFTO” represents the group with FTO gene interference. “Input” represents the total RNA before immunoprecipitation, serving as a control. “IP” represents “Immunoprecipitation,” which is the fraction of RNA enriched for m6A modifications.
Dataset one to five are related to S. aureus infection challenge compared to control. Based on various datasets, the samples from the control group (C) and S. aureus strain challenge group (S) were used in this study. Dataset one included the saline-treated bovine udder quarter (samples C-1 and C-2), and the high-concentration S. aureus-challenge udder quarter (samples S-1 and S-2). Dataset two comprised three groups, including the control group (samples C-1, C-2, and C-3); the Methicillin-Resistant S. aureus (MRSA) group (samples S1–1, S1–2, and S1–3); and the Methicillin-Sensitive S. aureus (MSSA) group (samples S2–1, S2–2, and S2–3) of the S. aureus challenge treatment. Dataset four included the control group (samples C-1, C-2 and C-3) and the MSSA-challenged group (samples S-1, S-2 and S-3). Dataset four generated by our previous study [3], included the control group (samples C-1 to C-6), the S. aureus strains group 1 (samples S1–1 to S1–6), S. aureus strains group 2 (samples S2–1 to S2–6), and S. aureus strains group 3 (samples S3–1 to S3–6). Dataset five generated by the current study, included control group (samples C-1 to C-4) and Newman strain of S. aureus challenged group (sample S-1 to S-4).
Dataset six is related to post-siFTO at the cellular level. In addition, the dataset seven is a MeRIP-seq dataset with three samples.
In addition, QTLs information covering all traits for cattle (ARS UCD1.2), pig (SS11.1), and sheep (OAR rambo1) were downloaded from the Animal QTL database ( www.animalgenome.org/cgi-bin/QTLdb/index ). Trait information predicted by cattle TWAS from the cGTEx database (cgtex.roslin. ed.ac.uk) was used in this study. The mapping analysis was used to integrate the RMRGs with the trait information from the QTLs database and the cattle TWAS data.
RNA-seq data processing
The quality of the raw reads from the RNA-seq data was assessed using FastQC v0.11.9 ( https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Subsequently, the NGS QC Toolkit v2.3.3 [23] was used to remove adaptors and filter out poor-quality reads. The clean reads were aligned to the Bos taurus reference genome (version: ARS-UCD1.2.109) using the Hisat2 v2.2.1 [24]. The aligned SAM files were then converted into BAM files using SAMtools v1.9 ( https://github.com/samtools/samtools/releases/), followed by quantitative analysis with Featurecounts.
Differentially expressed genes analysis
The DESeq2 tool ( https://bioconductor.org/packages/release/bioc/html/DESeq2.html ) in R software was used for data normalization across different samples, and the ComBat package [25] was used to correct batch effects. The DESeq2 tool was employed to obtain differentially expressed genes (DEGs) among different groups, which was determined by implementing the Wald test. Additionally, p-values were adjusted using the Benjamini-Hochberg method. A significance level of p<0.05 was considered to be statistically significant [26].
m6A peak identification from MeRIP-seq data
Conversion of sra files to fasta format was executed with the SRA-Toolkit v.2.9.6 6 ( https://www.ncbi.nlm.nih.gov/books/NBK158900/). Contaminated reads with adapters, those of low quality, and ambiguous reads were removed using the NGS QC Toolkit v2.3.3 [23]. Then, the reads contained rRNA were removed using Bowtie2 v2.4.1 ( https://bowtie-bio.sourceforge.net/bowtie2/index.shtml ). Finally, the clean reads were mapped to the bovine reference genome using Hisat2 v2.2.1 [24]. The SAM files were then converted into BAM files using SAMtools v1.9, with m6A peaks called using the exomepeak2 package in R.
Weighted gene co-expression network analysis
The gene co-expression network was constructed using the WGCNA package in R to detect the correlations among RMRGs. Considering the relatively large data size, dataset four was selected to perform WGCNA.
The top 10,000 highly variable genes were initially selected for WGCNA. Based on a soft threshold power for co-expression similarity, the adjacency was calculated to convert the adjacency matrix into a topology overlap matrix (TOM), and the corresponding difference (1-TOM) was calculated by using the TOM to represent the distance of the relationships between genes. The clustering of genes was then carried out using the 1-TOM. Thirdly, gene modules made up of at least 60 genes are created using dynamic cut-tree function detection and hierarchical clustering to group genes with comparable expression patterns. Finally, the threshold merge CutHeight parameter for merging modules was set to 0.25, and the key module was determined based on the DEGs between the S. aureus challenged group and the control group.
Expression profile and correlation analysis
By employing DESeq2 and Combat [25] in R, read counts were normalization correction standardized. PCA results were shown with the ggplot2 ( https://ggplot2.tidyverse.org/) program, which was colored by distinct groupings. PCA was used to analyze specific changes in the expression of RMRGs in individual level of cattle and Mac-T cells after treatment.
Correlation heatmaps of mRNA expressions among various RNA epigenetic modifying enzymes were generated using Corrplot program in R. The Pearson correlations of RMRGs expressions among the control group in dataset four were calculated using the cor.test package in R. The hierarchical clustering (hclust) order technique was used to rank the correlation coefficients for Corplot plotting.
Functional and pathway enrichment analysis
Kyoto encyclopedia of genes and genomes (KEGG) and gene ontology (GO) enrichment analysis were conducted on the gene modules of WGCNA and the DEGs following FTO interference by using KOBAS 3.0 ( https://bioinfo.org/kobas/genelist/). The R package ImageGP and ggplot2 were used to visualize 20 GO terms with a significance level of p<0.05.
Cell culture, S. aureus challenged of Mac-T Cells, siRNA transfection
All the cells were grown at 37°C and 5% CO2. Mac-T cells were grown in the complete medium (Dulbecco’s modified eagle medium [DMEM]) supplemented with 10% fetal bovine serum and a penicillin/streptomycin mixture of 100 U/mL). Following three consecutive generations of cell culture (48 hours per generation), stable Mac-T cells were resuspended in a complete culture medium and seeded in a six-well plate with an inoculation density of 1×106 cells/well. The culture medium was discarded upon reaching the logarithmic growth phase. Subsequently, the cells were challenged with a solution of Newman strain of S. aureus for a duration of 6 hours, with a bacteria-to-cell ratio (multiplicity of infection) of 10:1. Following the S. aureus challenge, the six-well plate was washed three times with sterile phosphate-buffered saline (PBS) Subsequently, 1 mL of Trizol was added to lyse the cells in the plate, and the RNA was extracted from the lysate for library preparation and sequencing. After library preparation, RNA-seq sequencing was performed using the Illumina HiSeq2500 platform at Novogene Corporation, generating Data two in our study.
The siRNAs employed in this study included siFTO, and a non-targeting control siRNA (siNC). All siRNA transfections were carried out according to the manufacturer’s instructions using the Lipofectamine 2000 transfection reagent (ThermoFisher, Waltham, MA, USA). The interfering fragments employed in this study were specifically designed and provided by Jintuosi Company (Beijing, China). The siRNA sequence for the sense strand (5’-3’) is GCGAGUUCAGA UGGGACAUTT, and the antisense strand (5’–3’) sequence is AUGUCCCAUCUGAACUCGCTT. A cell seeding of 5×105 cells per well was conducted in a six-well plate. When the cell confluence reached 60% to 80%, the complete cell culture medium was removed, and the cells were subjected to two washes with sterile PBS buffer. Subsequently, the fresh complete cell culture medium (DMEM supplemented with 10% fetal bovine serum) was added, supplemented with 100 pmol of siRNA and 5 μL of transfection reagent (lipofectamine 2000) per well. The cells were then incubated at 37°C in a cell culture incubator for a duration of 36 hours. After the incubation period, cells were lysed using TRIzol for RNA extraction. The extracted RNA was then used for library preparation and sequenced on the Illumina HiSeq2500 platform provided by Novogene Company, which constituted dataset six.
Quantitative real-time polymerase chain reaction
The quantitative real-time polymerase chain reaction (qRT-PCR) procedure for target genes was performed as follows. Total RNA was extracted using the Trizol reagent. The concentration and quality of RNA was assessed using a Nanodrop 2000 spectrophotometer. After confirming the primer specificity through conventional PCR, qRT-PCR was performed with two technical replicates for each sample. The average Ct value of the technical replicates for each sample was used for subsequent analysis. The 2−ΔΔCt method [26] was employed to calculate the relative expression levels of the target genes among the samples.
Western blot
Cells were collected from a single well of a six-well plate and lysed with a mixture of radio immunoprecipitation assay (Beyotime, Shanghai, China; P0013B) buffer and phenylmethylsulfonyl fluoride (Solarbio, Beijing, China; P0100) (100:1, 200 μL). After centrifugation at 12,000 g, the protein concentration in the supernatants was determined using the Beyotime Enhanced BCA Protein Assay Kit (P0010). The samples were appropriately diluted and mixed with 5× sodium dodecyl sulfate, followed by boiling at 95°C for 10 minutes to denature the proteins. Solarbio Precast gels (PG01215-S) were employed for protein separation based on molecular weight, and the proteins were transferred onto a 0.45 μm polyvinylidene fluoride membrane using a wet transfer apparatus. The membrane was then blocked with 5% non-fat dry milk in 1× TBST (Tris 10 mM, NaCl 150 mM, 0.05% (v/v) Tween-20, pH 7.5) at room temperature for 2 hours. Subsequently, the membrane was incubated overnight at 4°C with primary antibodies, including Anti-GAPDH antibody (1:3,000; Solarbio, China; K200057M) and Anti-FTO antibody (1:1,000; Abcam, Cambridge, UK; EPR24440-12). After washing five times with 1× TBST for 5 minutes each, the membrane was incubated with a goat anti-rabbit immunoglobulin G (IgG) secondary antibody (1:3,000; Beyotime, China; A0208) for 2 hours at room temperature. The membrane was washed five times with TBST for 5 minutes each. Finally, the membrane was visualized using the BeyoECL Plus chemiluminescent substrate (P0018S).
Statistical analysis
Complementary statistical evaluations were conducted using GraphPad Prism version 9.3.1. RMRGs exhibiting consistent expression across over five challenge groups were further selected for their trends. By calculating the average value for each group, a singular paired t-test was carried out, wherein a p-value less than 0.05 was taken as indicative of statistical significance.
RESULTS
Immune-related QTLs neighbor the mammalian RMRGs
To explore the roles of RMRGs in cattle complex traits, 80 genes selected in this study were evaluated by mapping QTLs in public databases (Supplementary Table S1). As shown in Figure 2, a total of 186 immune-related QTLs were identified within 1 kb upstream and downstream of the RNA modification-related enzyme genes, including QTLs associated with IgG level, somatic cell score (SCS), and clinical mastitis. The QTLs of SCC neighbor 22 RMRGs, including CPSF1, TET3, EIF3G, CFI, TET2, METTL16, NUDT21, LRPPRC, YTHDF3, ALKBH5, METTL4, IGF2BP1, EIF3A, PUS7, CBLL1, SRSF2, CSTF1, NSUN7, DNMT3B, VIRMA, DNMT3A, and YTHDC1. The RMRGs that identified the QTLs for clinical mastitis included LRPPRC, YTHDF3, EIF3A, EIF3H, METTL1, NSUN7, VIRMA, WTAP, NSUN4, and YBX1. Furthermore, a large number of immune-related QTLs also neighbor RMRGs in pig and sheep (Supplementary Figure S1). Collectively, these RMRGs may play an important role in animal immunity traits.
Figure 2.
The top 20 immunity-associated quantitative trait loci (QTLs) identified by RNA modification-related genes.
Mapping analysis of TWAS database was further performed on the bovine RMRGs. As shown in Table 2, the gene expression of RNA modifying-related enzymes is associated with immune system activation and plays a role in regulating responses to disease. For example, HAKAI was associated with mastitis, METTL3 with metritis, and HNRNPA2B1 and NOP2 with somatic cell score. Those results further showed the functions of RMRGs in regulating the bovine immune response.
Table 2.
Trait prediction of RMRGs in TWAS data
| RNA modification | Gene | Tissue | Trait | Zscore1) | p-value | Pred_Perf_Pval2) |
|---|---|---|---|---|---|---|
| m6A | METTL3 | Liver | Metritis | −2.3909 | 0.0168 | 0.0028 |
| VIRMA | Intramuscular_fat | Foot angle | −2.4427 | 0.0146 | 0.0133 | |
| RBM15B | Blood | Rear teat placement | −2.6011 | 0.0093 | 0.0070 | |
| HAKAI | Blood | Mastitis | 2.4212 | 0.0155 | 0.0060 | |
| ALKBH5 | Jejunum | Heifer conception rate | −2.4593 | 0.0139 | 0.0137 | |
| YTHDF1 | Intramuscular_fat | Daughter pregnacy rate | 4.1217 | <0.0001 | 0.0038 | |
| IGF2BP3 | Mammary | Displaced abomasum | −2.5360 | 0.0112 | 0.0370 | |
| PRRC2A | Liver | Age at first calving | −3.6954 | 0.0002 | <0.0001 | |
| HNRNPA2B1 | Lymph_node | Somatic cell score | −3.2516 | 0.0011 | 0.0175 | |
| HUR | Blood | Feet and legs composite | 2.6296 | 0.0085 | <0.0001 | |
| EIF3A | Intramuscular_fat | Front teat placement | 3.4039 | 0.0007 | <0.0001 | |
| EIF3B | Liver | Rump width | −2.3750 | 0.0175 | 0.0009 | |
| EIF3D | Hypothalamus | Protein percentage | 2.4948 | 0.0126 | 0.0020 | |
| EIF3G | Adipose | Fore udder attachment | −2.4327 | 0.0150 | 0.0207 | |
| EIF3H | Intramuscular_fat | Body depth | 2.8686 | 0.0041 | 0.0119 | |
| A-To-I | ADAR | Lung | Protein percentage | −2.7649 | 0.0057 | 0.0018 |
| ADARB2 | Lymph_node | Protein percentage in milk | −2.6487 | 0.0081 | <0.0001 | |
| m1A | TRMT61A | Blood | Dairy form | −1.9728 | 0.0485 | 0.0045 |
| YTHDF1 | Intramuscular_fat | Daughter pregnacy rate | 4.1217 | <0.0001 | 0.0038 | |
| m5C | NSUN1 | Blood | Somatic cell score | 2.7819 | 0.0054 | <0.0001 |
| NSUN3 | Liver | Hypocalcemia | −2.7430 | 0.0061 | 0.0017 | |
| NSUN6 | Adipose | Ketosis | 1.9764 | 0.0481 | 0.0015 | |
| TET1 | Macrophage | Body depth | −1.9606 | 0.0499 | 0.0002 | |
| TET2 | Muscle | Daughter still birth | −2.3387 | 0.0193 | 0.0062 | |
| ALYREF | Blood | Stature | 2.7530 | 0.0059 | <0.0001 | |
| ψ | PUS3 | Blood | Sire still birth | −2.1355 | 0.0327 | 0.0016 |
| hm5C | TET2 | Muscle | Daughter still birth | −2.3387 | 0.0193 | 0.0062 |
| m7G | METTL1 | Liver | Net merit | 3.5597 | 0.0004 | 0.0021 |
| APA | CPSF1 | Macrophage | Fat percentage in milk | −16.3369 | <0.0001 | <0.0001 |
| CPSF3 | Mammary | Stature | 2.4495 | 0.0143 | 0.0319 | |
| CPSF4 | Macrophage | Rump width | 3.1984 | 0.0014 | 0.0011 | |
| CSTF3 | Lymph_node | Rump angle | −2.4693 | 0.0135 | 0.0037 |
RMRGs, RNA modification-related genes; TWAS, transcriptome-wide association studies; m6A, N6-methyladenosine; m6Am, N6, 2’-O-dimethyl adenosine; m1A, N1-methyladenosine; m5C, 5-methylcytosine; hm5C, 5-hydroxymethylcytosine; ac4C, N4-acetyl cytidine; APA, alternative polyadenylation; Ψ, pseudouridine; m7G, N7-methylguanosine.
Zscore: S-PrediXcan’s association result for the gene, typically HUGO for a gene.
Pred_Perf_Pval: p-value of tissue model’s correlation to gene’s measured transcriptome (prediction performance).
Expression of RMRGs can well characterize bovine S. aureus mastitis
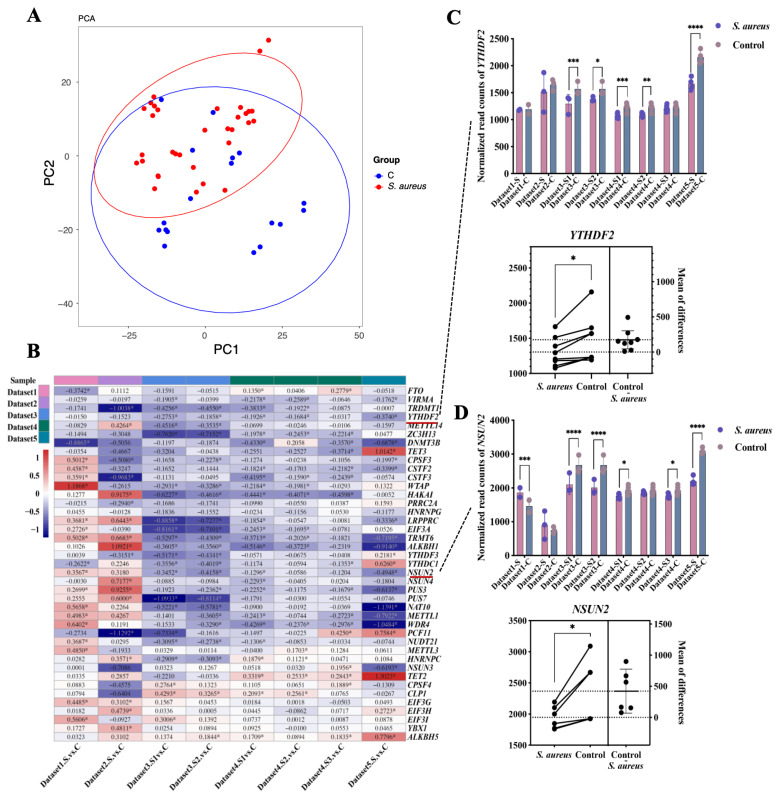
To investigate the functions of RMRGs in the immune response of bovine S. aureus mastitis, the downloaded RNA-seq datasets related to bovine S. aureus mastitis from public databases were subjected to unified standardized data processing. Genes associated with RNA modification enzymes were then selected for subsequent analysis. PCA of all the data shows that the expression of RMRGs could well distinguish the control and S. aureus challenged groups (Figure 3A; Supplementary Figure S2). In dataset one, the S. aureus challenge caused physiological and morphological changes, such as increased body temperature, SCC at the individual-level. These individual-level changes are consistent with the clinical feature of mastitis in dairy cows, and the differential expression of RMRGs is a significant response to the S. aureus challenge in bovine mastitis.
Figure 3.
Principal component analysis (PCA) and Log2fold change (FC) for gene expression of RNA modification-related genes in control and Staphylococcus aureus challenged groups. (A) presents the PCA results of control and S. aureus challenged groups from all datasets. (B) presents Log2FC between the S. aureus challenged group and control group in each data. * Means the p-value of less than 0.05. (C)-(D) Presents the normalized expression levels of the NSUN2 and YTHDF2 genes across eight distinct groups. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. Paired t-test representing the significantly consistent trend group of gene changes.
There were eight strains of S. aureus involved in these five datasets. For RMRGs, only that with the same change direction of differential expression in more than five strains challenged groups were considered to be important S. aureus mastitis-related genes. In total, 41 important genes were selected from the 80 RMRGs (Table 3; Figure 3B). Large differences were found in the expression patterns between individual-level and cell-level, while the change trend of RNA-changed gene expression at the cellular level was almost consistent across the different datasets. Among 41 selected RMRGs, the expression levels of 29 genes were decreased after the S. aureus challenged treatment. In this study, some significant differences of expression level between the S. aureus challenged and control groups were observed by employing paired t-test, such as YTHDF2 and NSUN2 presented in Figure 3C–3D. These findings indicate that RMRGs may collectively contribute to the regulation of inflammatory responses in bovine S. aureus mastitis.
Table 3.
RMRGs chosen in this study1)
| RNA modification | S. aureus challenged | Writer | Eraser | Reader |
|---|---|---|---|---|
| m6A (18/34) | Up-regulated | ALKBH5 | HNRNPC, FMR1, EIF3G, EIF3H, EIF3I | |
| Down-regulated | METTL14, WTAP, VIRMA, ZC3H13, HAKAI | YTHDC1, YTHDC2, YTHDF2, YTHDF3, HNRNPG, LRPPRC, EIF3A | ||
| Non-significant | METTL3, METTL16, RBM15, RBM15B | FTO | YTHDF1, IGF2BP1, IGF2BP2, IGF2BP3, PRRC2A, HNRNPA2B1, SRSF2, HUR, EIF3B, EIF3C, EIF3D | |
| m5C (9/17) | Up-regulated | NSUN3 | TET2 | YBX1 |
| Down-regulated | NSUN2, NSUN6, DNMT2, DNMT3B | TET3 | YTHDF2 | |
| Non-significant | NSUN1, NSUN4, NSUN5, NSUN7, DNMT1, DNMT3A | TET1 | ALYREF | |
| m1A (6/13) | Down-regulated | TRMT6 | ALKBH1 | YTHDF2, YTHDF3, YTHDC1, YTHDC2 |
| Non-significant | TRMT61A, TRMT61B, TRMT10C, NML | ALKBH3, FTO | YTHDF1 | |
| m6Am (1/3) | Down-regulated | METTL4 | ||
| Non-significant | PCIF1 | FTO | ||
| Adenosine-to-inosine editing (A-to-I) | Non-significant | ADAR, ADARB1, ADARB2 | ||
| Pseudouridine (ψ) (2/7) | Down-regulated | PUS3, PUS7 | ||
| Non-significant | PUS1, PUS2, PUS4, PUS6, PUS9 | |||
| ac4C (1/1) | Down-regulated | NAT10 | ||
| m7G (2/2) | Down-regulated | METTL1, WDR4 | ||
| APA (Alternative polyadenylation)(8/12) | Up-regulated | CPSF1, CPSF4, CLP1 | ||
| Down-regulated | CPSF3, CSTF2, CSTF3, PCF11, NUDT21 | |||
| Non-significant | CPSF2, CSTF1, CFI, PABPN1 | |||
| hm5C (1/1) | Up-regulated | TET2 |
RMRGs, RNA modification-related genes.
The percentage of RMRGs showing consistent modifications, as classified by our analysis, is presented in brackets alongside the respective modification genes.
Significant correlation of expression among RMRGs
Based on dataset four, WGCNA were constructed to further investigate the interactions of RMRGs. The top 10,000 genes with highly variable expression were clustered into 15 modules, including 14 co-expression modules and one module consisting of other genes.
The relationship between the expression of the module gene and the treatment groups was analyzed. In module 8, the changes in the challenged S1 group were more pronounced than in the S2 and S3 groups, compared to the control group (Figure 4A). In module 14, the control group had the highest positive correlation (p<0.05), while negative correlations were found with the challenged groups S1, S2, and S3, respectively. Thus, modules 8 and 14 were selected for further investigation. Within modules 8 and 14, there are many types of enzyme genes correlated with RNA modifications. There was the largest number of RNA modified gene enriched in module 8 (Table 4), including m6A modified writer (ZC3HB, RBM15B, and HAKAI), eraser (FTO), and reader (EIF3D); m5C modified writer (NSUN2), and reader (ALKBH1); m1A modified writer (TRMT6), the reader (ALKBH1), the eraser (FTO); writers of A-to-I (CPSF1, CPSF2, CPSF3, CPSF4, and CSTF2); writers of ψ (PUS3); writer of m7G (METTL1). Due to similar expression patterns, enzyme genes correlated with RNA modifications from various types were clustered into a same module, which suggested potential interactions among RMRGs.
Figure 4.
RMRGs exhibit interactions and participate in immune regulation. (A) Weighted correlation network analysis for different Staphylococcus aureus challenged groups. (B) Correlation of gene expression among RMRGs within Module 8. (C)–(D) Gene ontology analysis of the genes for modules 8 and 14. RMRGs, RNA modification-related genes. * p<0.05, ** p<0.01, and *** p<0.001, respectively.
Table 4.
The distribution of different RMRGs in different modules
| Item | In-module gene counts | RMRGs count | RMRGs |
|---|---|---|---|
| module8 | 1,887 | 15 | ZC3H13, RBM15B, HAKAI, FTO, EIF3D, TRMT6, ALKBH1, NSUN2, PUS3, METTL1, CPSF1, CPSF2, CPSF3, CPSF4, CSTF2 |
| module13 | 1,328 | 12 | HNRNPA2B1, HNRNPC, HUR, LRPPRC, EIF3A, TRMT10C, ALKBH3, YBX1, PUS1, NAT10, WDR4, CSTF3 |
| module11 | 760 | 9 | ALKBH5, YTHDC1, EIF3H, EIF3I, PCIF1, NSUN6, DNMT3B, ALYREF, PUS7 |
| module14 | 685 | 7 | METTL16, WTAP, VIRMA, YTHDF2, NSUN1, CLP1, NUDT21 |
| module5 | 1,311 | 5 | YTHDF1, SRSF2, NSUN3, DNMT1, DNMT3A |
| module2 | 636 | 5 | YTHDC2, EIF3C, EIF3G, NML, TET3 |
| module3 | 942 | 3 | RBM15, IGF2BP2, TET2 |
| module6 | 599 | 3 | PRRC2A, HNRNPG, ADARB1 |
| unknown | 295 | 3 | METTL14, YTHDF3, CSTF1 |
| module12 | 114 | 3 | NSUN4, DNMT2, TRDMT1 |
| module10 | 415 | 2 | FMR1, ADAR |
| module1 | 259 | 2 | EIF3B, NSUN5 |
| module4 | 181 | 2 | METTL3, PABPN1 |
| module9 | 490 | 1 | PCF11 |
| module7 | 98 | 1 | TRMT61A |
RMRGs, RNA modification-related genes.
To further investigate the relationships among RMRGs, expression correlations of selected genes from module 8 were calculated based on dataset four (Figure 4B). Significant positive correlations were observed in 39 gene pairs, while significant negative correlations were observed in 8 gene pairs. Interactions were found among genes from a same modification, such as a significant negative correlation between the m6A writer HAKAI and the m6A eraser FTO. Interactions between different RNA modifications were also identified, including a significant negative correlation between the m6A eraser FTO and the m5C writer NSUN2, as well as a significant negative correlation between FTO and the APA writer CPSF2. Similar results were found for other modules (Supplementary Figure S3). These findings suggest potentially synergistic or antagonistic effects of RNA modifications in bovine S. aureus mastitis.
The GO enrichment analysis demonstrated that genes within modules 8 and 14 are closely associated with the S. aureus challenge, and some significant (p<0.05) terms associated with immunity and inflammation were found. For example, GO terms macroautophagy, apoptotic process, and mRNA methylation were enriched in module 8 (Figure 4C), and terms RNA splicing, negative regulation of epithelial cell migration, apoptosis process, and cell adhesion molecule binding were enriched in module 14 (Figure 4D). In this study, different RNA epigenetic modifications can be clustered in the same co-expression module and are associated with immune responses.
Interactions between RNA modifications are partly explained by the existence of m6A modification within RMRGs
In this study, a significant correlation between RMRGs, however, the mechanism behind this has to be investigated further. The genes within module 8 showed the strongest positive correlation and the largest number of clustered genes in the control group based on the WGCNA enrichment analysis. Importantly, this module also exhibited the highest abundance of RMRGs among all the identified modules. m6A modification is extensively studied in RNA modification research, with FTO playing a significant role as an m6A demethylase in its intricate regulation. This study supports the association of FTO with S. aureus mastitis at the individual level within dataset one. Thus, we chose FTO, a gene in module 8 for validation. To confirm the connection of gene expression between RMRGs, an RNA interference experiment was performed for FTO.
According to the interference experiments, siRNA significantly (p<0.0001) knocked down the expression and protein level of FTO (Figure 5A–5C). Interference of gene FTO significantly increased the expression level of CPSF2 (Figure 5D), which confirmed the negative correlation between FTO and CPSF2 found in this study (Figure 4B). Furthermore, through analysis of downloaded MeRIP-seq data, we identified the presence of m6A modification on the CPSF2 gene, and the m6A peak decreased after S. aureus challenged. (Figure 5F). Therefore, FTO likely regulates the expression of CPSF2 by modulating its m6A modification. Based on our findings, the FTO gene affects the expression of RMRGs that harbor m6A modification sites, which is likely attributed to the presence of m6A modifications within the genes involved in RNA modification. The initial findings on the function of RMRGs in S. aureus mastitis and the role of FTO in this biological process have the potential to partially elucidate the molecular mechanisms responsible for S. aureus mastitis in bovine.
Figure 5.
FTO influence the expression of RNA-modified genes containing m6A modification sites. (A) Interference result for FTO. **** p<0.001. (B)–(C) Total cell extracts were harvested from siFTO and NC Mac-T cells and subjected to Western blot. (D) Expression changes of CPSF2 gene after cellular-level interference with FTO. * p<0.05. (E) CPSF2 gene expression significantly correlated with FTO. (F) Variations in the m6A modification peak within the CPSF2 gene between the control group and Staphylococcus aureus challenged group. (G) Kyoto encyclopedia of genes and genomes analysis of differentially expressed genes (DEGs) with m6A peaks post-FTO interference. Circle attributes indicate p-value significance and gene count. (H) Gene ontology enrichment of DEGs with m6A peaks post-FTO interference. Circle attributes indicate p-value significance and gene count. m6A, N6-methyladenosine.
In integration of MeRIP-seq and RNA-seq following FTO gene interference, it found a set of 3,224 DEGs, of which 1,164 DEGs exhibited m6A modification peaks. The KEGG pathway enrichment (Figure 5G) revealed a significant enrichment of these m6A-marked DEGs in inflammation-associated pathways, such as the Wnt signaling pathway, the Measles pathway, and the Hepatitis B pathway. GO enrichment analysis for 1,164 m6A-marked DEGs (Figure 5H) revealed a strong enrichment of m6A marked genes in biological processes and molecular functions related to transcription factor binding, RNA splicing and RNA binding. The potential impact of FTO on processes related to transcriptional regulation and post-transcriptional modification was emphasized by the prominence of categories related to transcriptional regulation and post-transcriptional modification. Interestingly, the term associated with “m6A-methyladenosine-containing RNA binding” was also enriched. These findings show how RNA modifications can affect bovine S. aureus mastitis, by illustrating the potential underlying molecular mechanisms resulting from FTO interference.
DISCUSSION
This study revealed the association between RMRGs and the immune system, specifically in bovine S. aureus mastitis. Analysis of RNA-seq data from five datasets showed consistency within challenged and control groups at individual and cellular levels, as well as heterogeneity between groups. We identified 41 key genes with consistent differential expression patterns across datasets, and correlation analysis demonstrated interactions between homologous and different RNA modifications. Furthermore, we validated the regulation of RMRGs at the cell level using MeRIP-seq data and the interference experiment, providing additional evidence for potential interactions between different RNA modifications.
For bovine S. aureus mastitis, we conducted an analysis of the ten well-characterized RNA modifications. The key genes identified by the current study accounted for more than 50% of the m1A, m5C, and m6A modified genes reported in previous studies. Therefore, the m1A, m5C, and m6A modifications should be given special attention in studies of bovine S. aureus mastitis. Previous studies have indicated that m5C modification is associated with the development of systemic lupus erythematosus through its influence on CD4+ T cells involved in immune responses [27]. A-to-I and m6A modifications have regulatory functions in human cardiovascular disease [28,29]. Furthermore, RNA modification plays a crucial role in maintaining immune cell homeostasis and function [30,31]. Notably, m6A modification, regulated by the m6A reading protein YTHDF2, suppresses circRNA-mediated immune responses in humans [32]. Additionally, genes modified by m6A are enriched in several immune signaling pathways in the bovine Escherichia coli mastitis [33]. In this study, the regulatory role of RNA modifications in immune complex traits were investigated by bovine QTL and TWAS databases. This study identified a strong association between RMRGs and immune complex traits in cattle, such as clinical mastitis. PCA revealed distinct clustering patterns of RMRGs between the S. aureus challenged group and the control group, indicating their essential role in bovine S. aureus mastitis. Consistent changes in the expression of RMRGs were also observed across different cellular-level RNA-seq datasets. It is necessary to further investigate the mechanisms underlying RNA modification genes in S. aureus bovine mastitis.
Recent studies have indicated that m6A modification exerts an influence on immune regulation by regulating processes, such as RNA alternative splicing and APA [34]; thereby impacting the expression and function of immune-related genes [35]. FTO plays a crucial role in controlling APA and subsequently determining the length of 3’ UTRs, as evidenced by the up-regulated expression of the last exon in cells lacking FTO [36]. CPSF2 is a member of the protein complex associated with RNA 3’ end processing and is involved in the APA process [37,38]. In the present study, interference with the FTO gene significantly increased the expression level of CPSF2 containing m6A modification locus; and the m6A level in CPSF2 was reduced after S. aureus challenge. Consequently, we hypothesize that FTO, through its m6A modification, may regulate the expression of CPSF2 and be involved in the APA process in response to immune regulation. In the future, more thorough work to understand the underlying mechanisms of these interactions would be essential, aiming to gain a comprehensive understanding of how m6A modification collaboratively contributes to immune regulation through APA.
This study revealed potential interactions between enzymes involved in RNA modifications within the same category and across different categories. We propose a mechanism involving RNA modification genes that regulate these interactions. The importance of RNA modification interactions in immune regulation was supported by many studies in various species [10]. For example, the regulation of RNA methylation (m1A, m5C, m6A, and m7G) in brain tissue damage was explored using zebrafish, revealing specific changes in these modifications under hypoxic conditions [39]. Crosstalk mechanisms were also investigated in colon cancer treatment, and the regulatory role of different RNA modifications and their impact on the immune microenvironment were reported [40,41]. In hepatocellular carcinoma, it was shown the regulation of immune-related genes through protein-protein interactions [41,42]. Interaction studies in these species provide support for findings of this study, suggesting the need for in-depth exploration of the mechanisms governing the interplay of RNA modifications in bovine immune regulation.
Compared to the cellular level, the expression changes of RMRGs for bovine S. aureus mastitis showed slight differences at the individual level. These inconsistencies may be due to the different environments of cell lines and individual tissues. Based on our findings, we cautiously speculate that there are three possible causes: on the first hand, the results at the individual level are less consistent with the phenomenon at the cellular level due to the influence of only two individual cows; on the next hand, individual-level mammary tissue contains various classes of cell types and the mechanisms regulating phenotypes in individuals are complex multifactorial influences [3]; on the other hand, the sampling at the individual-level has multiple influences and is variable in time-space compared to the cellular level. Therefore, genes with strong consistency between the individual and cellular levels can be selected as stable genetic markers for disease-resistance breeding in bovines.
In conclusion, RNA modifications were found to play an important role in bovine S. aureus mastitis. The effect of different S. aureus strains on the expression of RMRGs varied after challenge with mammary epithelial cells. RMRGs can be used as potential indicators to assess the status of bovine mastitis. Focusing on the interactions between RMRGs is essential to reveal the genetic architecture of complex traits. The findings from this study are beneficial for understanding the regulatory mechanism of RMRGs on complex traits in cattle.
ACKNOWLEDGMENTS
The authors gratefully acknowledge the support of High-performance Computing Platform of China Agricultural University.
Footnotes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
FUNDING
This research was financially supported by the National Key R&D Program of China (2023YFF1000902, 2021YFD1200903, 2021YFD1200900), National Natural Science Funds of China (32302706), Science and Technology Reward Project in Hebei Province, China (23227602Z), and the Earmarked Fund for China Agriculture Research System (CARS-36).
DATA AVAILAVILITY
All raw and processed sequencing data generated in this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA957225 ( https://www.ncbi.nlm.nih.gov/bioproject/PRJNA957225).
SUPPLEMENTARY MATERIAL
Supplementary file is available from: https://doi.org/10.5713/ab.23.0323
Supplementary Figure S1. The immunity-associated QTLs identified by RMRGs in pig and sheep
Supplementary Figure S2. (A) presents the PCA results of control and S. aureus challenged groups. (B)–(F) presents the PCA results of dataset one to five, respectively.
Supplementary Figure S3. Correlation of gene expression among RMRGs within Module 14.
Supplementary Table S1. The RMRGs selected for this research
REFERENCES
- 1.Becker K, Schaumburg F, Kearns A, et al. Implications of identifying the recently defined members of the Staphylococcus aureus complex S. argenteus and S. schweitzeri: a position paper of members of the ESCMID Study Group for Staphylococci and Staphylococcal Diseases (ESGS) Clin Microbiol Infect. 2019;25:1064–70. doi: 10.1016/j.cmi.2019.02.028. [DOI] [PubMed] [Google Scholar]
- 2.Chan CTY, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mi S, Tang Y, Dari G, et al. Transcriptome sequencing analysis for the identification of stable lncRNAs associated with bovine Staphylococcus aureus mastitis. J Anim Sci Biotechnol. 2021;12:120. doi: 10.1186/s40104-021-00639-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Yang J, Huang Z, et al. Exosomal lnc-AFTR as a novel translation regulator of FAS ameliorates Staphylococcus aureus-induced mastitis. Biofactors. 2022;48:148–63. doi: 10.1002/biof.1806. [DOI] [PubMed] [Google Scholar]
- 5.Huynh HT, Robitaille G, Turner JD. Establishment of bovine mammary epithelial cells (MAC-T): an in vitro model for bovine lactation. Exp Cell Res. 1991;197:191–9. doi: 10.1016/0014-4827(91)90422-q. [DOI] [PubMed] [Google Scholar]
- 6.Ogunnaike M, Wang H, Zempleni J. Bovine mammary alveolar MAC-T cells afford a tool for studies of bovine milk exosomes in drug delivery. Int J Pharm. 2021;610:121263. doi: 10.1016/j.ijpharm.2021.121263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Günther J, Koy M, Berthold A, Schuberth HJ, Seyfert HM. Comparison of the pathogen species-specific immune response in udder derived cell types and their models. Vet Res. 2016;47:22. doi: 10.1186/s13567-016-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352:1408–12. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Song J, Yuan W, Zhang W, Sun Z. Roles of RNA methylation on tumor immunity and clinical implications. Front Immunol. 2021;12:641507. doi: 10.3389/fimmu.2021.641507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song P, Tayier S, Cai Z, Jia G. RNA methylation in mammalian development and cancer. Cell Biol Toxicol. 2021;37:811–31. doi: 10.1007/s10565-021-09627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Gu L, Orellana EA, et al. METTL4 is an snRNA m(6)Am methyltransferase that regulates RNA splicing. Cell Res. 2020;30:544–7. doi: 10.1038/s41422-019-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 14.He C, Bozler J, Janssen KA, et al. TET2 chemically modifies tRNAs and regulates tRNA fragment levels. Nat Struct Mol Biol. 2021;28:62–70. doi: 10.1038/s41594-020-00526-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He PC, He C. mRNA acetylation: a new addition to the epitranscriptome. Cell Res. 2019;29:91–2. doi: 10.1038/s41422-018-0135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol. 2016;17:83–96. doi: 10.1038/nrm.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nombela P, Miguel-López B, Blanco S. The role of m(6)A, m(5)C and Ψ RNA modifications in cancer: novel therapeutic opportunities. Mol Cancer. 2021;20:18. doi: 10.1186/s12943-020-01263-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safra M, Sas-Chen A, Nir R, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature. 2017;551:251–5. doi: 10.1038/nature24456. [DOI] [PubMed] [Google Scholar]
- 19.Selmi T, Hussain S, Dietmann S, et al. Sequence- and structure-specific cytosine-5 mRNA methylation by NSUN6. Nucleic Acids Res. 2021;49:1006–22. doi: 10.1093/nar/gkaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, Yang Y, Sun BF, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606–25. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang LS, Liu C, Ma H, et al. Transcriptome-wide mapping of internal N(7)-Methylguanosine methylome in mammalian mRNA. Mol Cell. 2019;74:1304–16e8. doi: 10.1016/j.molcel.2019.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Zhang X, Lu X, et al. 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res. 2017;27:1243–57. doi: 10.1038/cr.2017.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–3. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Zhernakova A, van Diemen CC, Wijmenga C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat Rev Genet. 2009;10:43–55. doi: 10.1038/nrg2489. [DOI] [PubMed] [Google Scholar]
- 28.Delatte B, Wang F, Ngoc LV, et al. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science. 2016;351:282–5. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- 29.Gatsiou A, Stellos K. RNA modifications in cardiovascular health and disease. Nat Rev Cardiol. 2023;20:325–46. doi: 10.1038/s41569-022-00804-8. [DOI] [PubMed] [Google Scholar]
- 30.Esteve-Puig R, Climent F, Piñeyro D, et al. Epigenetic loss of m1A RNA demethylase ALKBH3 in Hodgkin lymphoma targets collagen, conferring poor clinical outcome. Blood. 2021;137:994–9. doi: 10.1182/blood.2020005823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H, Wang Y, Xiang Y, et al. FMRP promotes transcription-coupled homologous recombination via facilitating TET1-mediated m5C RNA modification demethylation. Proc Natl Acad Sci USA. 2022;119:e2116251119. doi: 10.1073/pnas.2116251119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo J, Wang F, Sun F, et al. Targeted inhibition of FTO demethylase protects mice against LPS-induced septic shock by suppressing NLRP3 inflammasome. Front Immunol. 2021;12:663295. doi: 10.3389/fimmu.2021.663295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Lin C, Zhu Y, et al. Transcriptome profiling of m(6)A mRNA modification in bovine mammary epithelial cells treated with Escherichia coli. Int J Mol Sci. 2021;22:6254. doi: 10.3390/ijms22126254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ke S, Pandya-Jones A, Saito Y, et al. m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31:990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai Q, Shi M, Sun X, et al. Comprehensive analysis of the m6A-related molecular patterns and diagnostic biomarkers in osteoporosis. Front Endocrinol (Lausanne) 2022;13:957742. doi: 10.3389/fendo.2022.957742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ke S, Alemu EA, Mertens C, et al. A majority of m6A residues are in the last exons, allowing the potential for 3’ UTR regulation. Genes Dev. 2015;29:2037–53. doi: 10.1101/gad.269415.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinske LC, Galante PA, Limbeck E, et al. Alternative polyadenylation allows differential negative feedback of human miRNA miR-579 on its host gene ZFR. PLoS One. 2015;10:e0121507. doi: 10.1371/journal.pone.0121507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilubol N, Boufraqech M, Zhang L, Kebebew E. Loss of CPSF2 expression is associated with increased thyroid cancer cellular invasion and cancer stem cell population, and more aggressive disease. J Clin Endocrinol Metab. 2014;99:E1173–82. doi: 10.1210/jc.2013-4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Li X, Ma X, Xiao W, Zhang J. Mapping the m1A, m5C, m6A and m7G methylation atlas in zebrafish brain under hypoxic conditions by MeRIP-seq. BMC Genomics. 2022;23:105. doi: 10.1186/s12864-022-08350-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Yao J, Bao R, et al. Cross-talk of four types of RNA modification writers defines tumor microenvironment and pharmacogenomic landscape in colorectal cancer. Mol Cancer. 2021;20:29. doi: 10.1186/s12943-021-01322-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, Li K, Zhang W, et al. The m6A/m5C/m1A regulated gene signature predicts the prognosis and correlates with the immune status of hepatocellular carcinoma. Front Immunol. 2022;13:918140. doi: 10.3389/fimmu.2022.918140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q, Li X, Tang H, et al. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J Cell Biochem. 2017;118:2587–98. doi: 10.1002/jcb.25957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. The immunity-associated QTLs identified by RMRGs in pig and sheep
Supplementary Figure S2. (A) presents the PCA results of control and S. aureus challenged groups. (B)–(F) presents the PCA results of dataset one to five, respectively.
Supplementary Figure S3. Correlation of gene expression among RMRGs within Module 14.
Supplementary Table S1. The RMRGs selected for this research
Data Availability Statement
All raw and processed sequencing data generated in this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number PRJNA957225 ( https://www.ncbi.nlm.nih.gov/bioproject/PRJNA957225).