Abstract
Human cytomegalovirus (HCMV) infection of permissive cells has been reported to induce a cell cycle halt. One or more viral proteins may be involved in halting progression at different stages of the cell cycle. We investigated how HCMV infection, and specifically IE86 protein expression, affects the cell cycles of permissive and nonpermissive cells. We used a recombinant virus that expresses the green fluorescent protein (GFP) to determine the effects of HCMV on the cell cycle of permissive cells. Fluorescence by GFP allowed us to select for only productively infected cells. Replication-defective adenovirus vectors expressing the IE72 or IE86 protein were also used to efficiently transduce 95% or more of the cells. The adenovirus-expressed IE86 protein was determined to be functional by demonstrating negative autoregulation of the major immediate-early promoter and activation of an early viral promoter in the context of the viral genome. To eliminate adenovirus protein effects, plasmids expressing GFP for fluorescent selection of only transfected cells and wild-type IE86 protein or a mutant IE86 protein were tested in permissive and nonpermissive cells. HCMV infection induced the entry of U373 cells into the S phase. All permissive cells infected with HCMV were blocked in cell cycle progression and could not divide. After either transduction or transfection and IE86 protein expression, the number of all permissive or nonpermissive cell types in the S phase increased significantly, but the cells could no longer divide. The IE72 protein did not have a significant effect on the S phase. Since IE86 protein inhibits cell cycle progression, the IE2 gene in a human fibroblast IE86 protein-expressing cell line was sequenced. The IE86 protein in these retrovirus-transduced cells has mutations in a critical region of the viral protein. The locations of the mutations and the function of the IE86 protein in controlling cell cycle progression are discussed.
Primary infection by human cytomegalovirus (HCMV), a member of the betaherpesvirus family, is usually asymptomatic in immunologically healthy people. In individuals with compromised cellular immunity, HCMV can cause pneumonitis, encephalitis, retinitis, hepatitis, and gastroenteritis. In utero infection can lead to congenital neurological complications, including mental retardation, sensorineural hearing loss, or even death (4, 24). There is also an epidemiological link between HCMV-seropositive individuals and accelerated coronary artery restenosis following angioplasty (50). HCMV replicates in various terminally differentiated cell types, including smooth muscle, endothelial, epithelial, neuronal, and microglial cells, fibroblasts, and differentiated cells of the monocyte/macrophage lineage (12–14, 16, 25, 29, 46).
HCMV gene expression occurs in three temporal phases designated immediate early (IE), early, and late. Transcription of the IE genes occurs at five loci and is independent of any de novo viral protein synthesis. The IE proteins affect transcriptional regulation from both cellular and viral promoters (reviewed in references 48 and 51) and are involved in escape from immune surveillance (3, 27). IE gene expression is required for transcription of the early genes, which encode viral proteins that are required for viral DNA replication. Replication of viral DNA is required for transcription of the late viral genes, which encode structural proteins involved in the synthesis of the virion.
The greatest amount of transcription at IE times after infection occurs from the major IE (MIE) locus (51, 53, 54). Spliced mRNAs transcribed from this locus alternatively encode two well-studied viral proteins, IE72 and IE86, plus several isomers (51) that regulate genes expressed during the S phase of the cell cycle. The IE72 protein activates DNA polymerase α, c-fos, c-myc, and dihydrofolate reductase promoters (9, 21, 58). In addition, it can interact with cellular p107 and consequently activate E2F-responsive promoters (42). The IE86 protein is a strong, promiscuous transcriptional activator that interacts with factors of the basal-transcription machinery (8, 20, 28, 30, 33, 34). In vivo, it can interact with itself and with the early viral protein encoded by the UL84 gene (10, 15, 44, 49). Truncated forms of the IE86 protein are also reported to interact in vitro with Rb, p53, CREB, PML, TBP, TEF-1, Sp-1, TFIIB, c-Jun, and TAFII-130 (2, 8, 19, 20, 30, 45, 47, 50). The IE86 protein has been reported to transactivate the cyclin E promoter (6). Activation of cyclin E is essential for progression of the cell cycle into the S phase.
A halt in the cell cycle of human fibroblasts (HF) infected with HCMV appears to occur at different stages, including the G1/S and the G2/M transition points, depending on the cell cycle stage at the time of infection (31, 43). For example, HCMV infection of serum-starved HF blocked the cells primarily at the G1/S transition point. The virus may also express several different factors that act at different points in the cell cycle. For example, the virion-associated tegument protein UL69 blocks the cell cycle primarily at the G1/S transition point (32). The IE86 protein of HCMV is also reported to block cell cycle progression (59; E. A. Murphy and M. F. Stinski, 23rd Int. Herpesvirus Workshop, abstr. 290, 1998, and 24th Int. Herpesvirus Workshop, abstr. 3.001, 1999), but the location of the block is not yet known. Since the viral IE86 protein affects a variety of cellular proteins that promote the S phase of the cell cycle, we examined the effect of this viral protein on the permissive U373 cells. The various mechanisms used by HCMV to control cell cycle progression are significant for our understanding of the replication and pathogenesis of the virus because the efficiency of viral DNA replication in infected cells may be increased by reducing competition for cellular DNA precursors.
Here we compare how HCMV affects the cell cycle in permissive U373 cells as well as smooth muscle cells (SMCs), human aortic endothelial cells (HAECs), and human papilloma virus-transformed aortic endothelial cells (HPVAECs). For these studies, we used fluorescence-activated cell sorting (FACS) and a recombinant virus that expresses the green fluorescent protein (GFP) soon after infection. In addition, we examined the specific effects of the IE72 and IE86 proteins expressed from replication-defective adenovirus vectors. We also used expression plasmids expressing both GFP and the IE86 protein to determine the isolated effects of the IE86 protein on the cell cycle. A previous report by Weibusch and Hagemeier (59) using HCMV or IE86 protein expression vectors without selecting cells for GFP expression indicated that the virus or the IE86 protein caused a cell cycle block at the G1/S transition point of U373 cells. When only GFP-expressing cells were analyzed, our results indicated that IE86 protein expression increases the number of permissive U373 or nonpermissive 293T cells in the S-phase component of the cell cycle but blocks cell cycle progression.
MATERIALS AND METHODS
Cell culture, virus, and adenovirus vectors.
Primary human foreskin fibroblasts (HFF) were grown in Eagle's minimal essential medium (Life Technologies, Gaithersburg, Md., or Mediatech, Herndon, Va.) supplemented with 10% newborn calf serum (Sigma, St. Louis, Mo.), penicillin (100 U/ml), and streptomycin (100 μg/ml). U373 cells, a neuroblastoma cell line, were grown in Dulbecco's minimal essential medium (Life Technologies) supplemented with penicillin, streptomycin, and either 5% (high) or 0.2% (low) fetal calf serum (Sigma). 293 or 293T cells were grown as described for U373 cells except either 10% (high) or 0.05% (low) fetal calf serum was used. The LXSN-IE86 cells, which contain the HCMV IE2 gene, were a gift from James McDougall (University of Washington) and were grown as described previously (5). Human medial SMCs and HAECs were grown in media obtained from Clonetics (Walkersville, Md.) and supplemented with 10% fetal bovine serum.
Maintenance and propagation of the HCMV Towne strain has been described previously (52). The recombinant HCMV RΔ-582/-1108gfp contains a deletion between −1140 and −583 relative to the MIE promoter transcription start site (39). The UL127 gene and the strong repressor boundary element (35) between the UL127 promoter and the MIE enhancer were replaced by the adenovirus E1b promoter driving expression of the humanized GFP.
Replication-defective E1a− and E1b− recombinant adenovirus vectors expressing either the “Tet-off” transactivator (Ad-Trans), the IE86 protein (Ad-IE86), or the IE72 protein (Ad-IE72) were constructed by one of the authors (D. Streblow) using shuttle vectors as described previously (55). The cDNA of the IE1 or IE2 gene of the AD169 strain of HCMV was used. The promoter for expression of the IE86 or IE72 protein contains the cis-acting repressive sequence (CRS) within the minimal CMV promoter (PminCMV). The replication-defective E1a− and E1b− recombinant adenovirus vector Ad-GFP was obtained from Beverly Davidson (University of Iowa). The titers of the various recombinant adenovirus vectors were determined by plaque assay on 293 cells. For transduction experiments, recombinant adenovirus vectors were added to 1 ml of Dulbecco's minimal essential medium containing 3 μl of Lipofectamine reagent (Life Technologies), which we found aids in the attachment of adenovirus to U373 cells. After 1 h at 37°C, the inoculum was removed and the cells were maintained in either low-serum medium for serum-starved cells or high-serum medium for growth-stimulated cells.
Enzymes.
All restriction enzymes and DNA-modifying enzymes were purchased from either New England Biolabs, Inc. (Beverly, Mass.) or Bethesda Research Laboratories, Inc. (Gaithersburg, Md.). T4 DNA ligase, the Klenow fragment of Escherichia coli DNA polymerase I, and calf intestinal phosphatase were acquired from Boehringer Mannheim Biochemicals (Indianapolis, Ind.). Taq DNA polymerase was obtained from Promega (Madison, Wis.). All enzymes were used according to the manufacturers' specifications.
Sequencing.
Genomic DNA from LXSN-IE86 cells was isolated and used in a PCR with one of seven primer pairs which span the IE2 cDNA. All oligonucleotides were purchased from Life Technologies. The primer pairs were designed to generate products no more than 350 bp long. The PCR products were gel purified and sequenced twice by the University of Iowa DNA core facility. Mutations in the IE2 gene were confirmed by additional PCR using LXSN-IE86 genomic DNA followed by a third sequencing.
Plasmid construction.
To mutate the CRS in the IE86 protein expression plasmid, pSVIE2 (38), two sets of PCR primers were used to amplify DNA from the MIE promoter while inducing site-specific mutations within the CRS. Primer pair 5′-CAAACCCCGACACGTAC-3′ and 5′-GAGCCTAGGTAGTGAACCGTCAG-3′ was used to amplify a 621-bp product from pSVIE2. Primer pair 5′-CGGTTCACTACCTAGGCTCTGCTT-3′ and 5′-ATGTCCAACATTACCGCCAT-3′ was used to amplify a 1,072-bp product from pSVIE2. After digestion of the 621-bp PCR product with AvrII and Bsp120I and digestion of the 1,072-bp PCR product with AvrII and SpeI, the two DNA fragments were ligated to pSVIE2 or pSVIE2HL, which were digested with Bsp120I and SpeI, to generate plasmids pSVIE2crs− and pSVIE2HLcrs−, respectively. The 5′ end of the CRS was changed from 5′-AGCTCGTTTAGTGAACCGT-3′ to 5′-AGCctaggTAGTGAACCGT-3′. (The CRS is in boldface, and the mutated bases are in lowercase letters.)
To generate the plasmid pGFP, plasmid pΔ-300/-1108SVGFP (39) was digested with BamHI and SalI, and the DNA fragment ends were made blunt with the Klenow fragment of E. coli DNA polymerase I and then ligated. To generate pGFP-IE86 and pGFP-IE86HL, plasmids pSVIE2crs− and pSVIE2HLcrs− were digested with SalI, treated with the Klenow fragment of DNA polymerase I, and then digested with restriction endonuclease NsiI. The DNA fragments were gel purified and ligated to pGFP, which was digested with EcoRI, treated with the Klenow fragment of DNA polymerase I, and then digested with NsiI. All plasmids were sequenced by the University of Iowa DNA core facility.
Western blots.
Adenovirus vector-transduced cells or transiently transfected 293T cells were collected and lysed by sonication in phosphate-buffered saline (PBS), pH 7.4. Protein concentrations were determined by a Bradford assay (Bio-Rad, Richmond, Calif.), and equal amounts of protein lysate were added to 2× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis loading buffer containing 200 mM Tris-HCl (pH 7.8), 8% SDS, 0.02% bromophenol blue, and 20% β-mercaptoethanol. After being boiled for 5 min, the denatured polypeptides were fractionated by electrophoresis in an SDS–9% polyacrylamide gel and transferred to nitrocellulose. Viral proteins were detected with monoclonal antibody 810 (Chemicon, Temecula, Calif.), which recognized the IE72, IE86, and IE86HL proteins. The enhanced chemiluminescence detection reagents and secondary peroxidase-labeled antibody (Pierce, Rockford, Ill.) were used according to the manufacturer's directions.
Northern blots.
Cytoplasmic RNA was isolated from HCMV-infected cells treated with cycloheximide (CHX) for 6 h as described previously (23, 35). The RNAs were fractionated by electrophoresis in denaturing 1.5% agarose gels containing 6% formaldehyde and blotted onto Nytran (Schleicher and Schuell, Keene, N.H.) as described previously (40). The primer pair 5′-GAGAAGGGAATGATGGGAGCC-3′ and 5′-CTAAATCGTCATGGGCCGTC-3′ was used to generate a 301-bp PCR product from plasmid pXbaI-A (57) containing the UL13 open reading frame. The PCR product was gel purified and 32P labeled with Rediprime II (Amersham) for use as a UL13-specific probe. Hybridization of the 32P-labeled probe was done at 68°C overnight as described previously (40).
RNase protection assay.
Riboprobe synthesis was performed as described previously (22). Twenty micrograms of cytoplasmic RNA was hybridized to the 32P-labeled riboprobes at 25°C overnight. The specific activities of the riboprobes were similar. After digestion with 100 U of RNase T1 (Boehringer Mannheim) at 37°C for 1 h and then with 65 μg of proteinase K at 37°C for 15 min, the protected RNAs were fractionated in a denaturing 6% polyacrylamide–7 M urea gel. Signals were detected by autoradiography on Hyperfilm MP (Amersham).
Flow cytometry.
Cell sorting was carried out on a Coulter (Opalocka, Fla.) EPICS 753 cell sorter, and fluorescent cell scanning was performed on a Becton Dickinson FACScan (San Jose, Calif.) using CellQuest DNA analysis software (Verity Software House Inc., Topsham, Maine). The cells were trypsinized and washed twice in PBS, pH 7.4. After suspension in PBS, the cells were sorted according to forward and side scatter (mock infected) or fluorescence due to GFP expression. Twenty thousand cells positive for GFP were plated in high-serum medium. After 6 days, CFU were identified by fixation with 10% formalin and staining with 0.3% methylene blue dye. GFP-positive cells were also suspended in a hypotonic propidium iodide solution containing 0.1% Triton X-100, 3.4 mM sodium citrate, and 50 μg of propidium iodide/ml at 4°C for 30 min. The ModFit software program (Becton Dickinson) was used to calculate the cell cycle profile of propidium iodide-stained cells according to the manufacturer's specifications.
Transient transfections.
Cells, synchronized by serum starvation for 24 h prior to transfection, were overlaid with 2 ml of Dulbecco's minimal essential medium without fetal calf serum. Transient transfection was done by the calcium precipitation method of Graham and van der Eb (17). Low- or high-serum medium was added for an additional 24 h, depending on the experiment.
RESULTS
Effect of HCMV on the cell cycle of permissive U373 cells.
HCMV infection of serum-starved permissive HFF induces a cell cycle halt primarily at the G1/S transition point (7, 11, 31, 41). Our investigation of HFF infected with RΔ-582/-1108gfp gave similar results (data not shown). The U373 cells are also an HCMV-permissive cell line. There is controversy over the stage at which the cell cycle is halted by HCMV and which virus-specified proteins are involved in halting cell cycle progression. We investigated the effects of HCMV infection on the cell cycle of synchronized U373 cells. To synchronize the cells, they were incubated in low-serum medium (0.2%) for 48 h and then either mock infected or infected with the recombinant virus RΔ-582/-1108gfp (5 PFU/cell), which expresses GFP soon after infection. The cells were maintained in low-serum medium for an additional 24 h. To determine if the cells would complete the cell cycle, high-serum (10%) medium was added for 48 h. The cells were then sorted by FACS for side and forward scatter (mock infected) or for GFP expression (RΔ-582/-1108gfp infected). Since we used a recombinant HCMV that expresses the humanized GFP with early kinetics, we isolated and studied only the infected cells. FACS was necessary because we found that the efficiency with which HCMV Towne strain infects cells depends on the cell type. Using 5 PFU of the recombinant virus RΔ-582/-1108gfp/cell, we found that 93% of HFF were fluorescent by 48 h. Only 72% of U373 cells, 73% of human SMCs, 23% of HAECs, and 17% of HPVAECs were fluorescent by 48 h postinfection (p.i.) (data not shown). Since HCMV Towne strain was not adapted to grow in endothelial cells, the low infectivity of endothelial cells was expected. The DNA content of the sorted cells was determined by staining them with propidium iodide followed by FACS scanning as described in Materials and Methods.
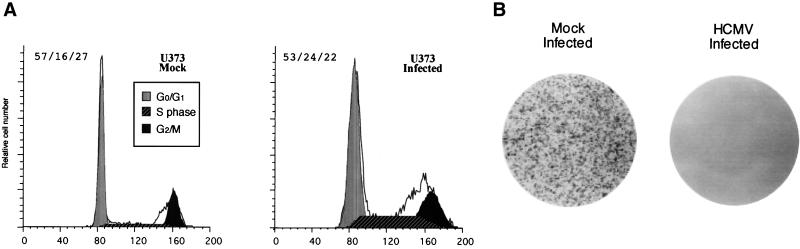
An increase in the S phase of the cell cycle of the infected cells relative to that of the mock-infected cells (24 versus 16%, respectively) was repeatedly detected with the U373 cells. In contrast, there was little change in G0/G1 (Fig. 1A). These observations suggested that the infected cells were halted in the S phase of the cell cycle.
FIG. 1.
Progression of permissive U373 cells into the S phase and inhibition of cell division after HCMV infection. (A) Cell cycle analysis of recombinant virus RΔ-582/-1108gfp-infected U373 cells. U373 cells, serum starved for 48 h, were mock infected or infected with 5 PFU of recombinant virus/cell. The cells were maintained in low-serum medium for an additional 24 h and then in high-serum medium for 48 h. Mock-infected U373 cells were sorted by forward and side scatter. Infected U373 cells were sorted by fluorescence. After propidium iodide (PI) staining, the DNA content of an equal number of cells was determined using a FACS scanner. The PI profile is shown as the solid histogram line. The PI histogram cell cycle analysis was determined by use of the ModFit program as described in Materials and Methods. The quantitation of cell cycle phases is given as percent G0/G1/percent S/percent G2/M. (B) Colony-forming assay using mock-infected or RΔ-582/-1108gfp-infected U373 cells. The sorted cells (20,000) were replated in high-serum medium, incubated at 37°C for 6 days, fixed with formalin, and stained with methylene blue as described in Materials and Methods. A result representative of several experiments is shown.
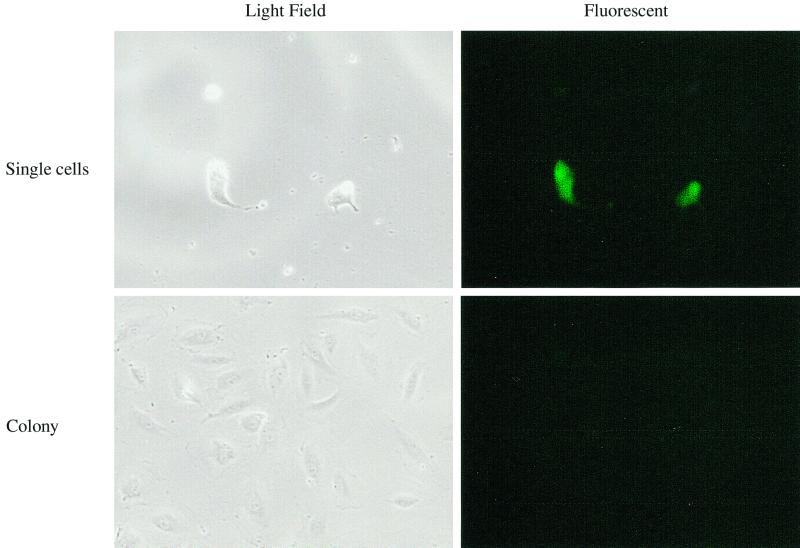
To demonstrate whether HCMV infection inhibited cell division, U373 cells were synchronized by serum starvation and infected with the recombinant virus RΔ-582/-1108gfp. RΔ-582/-1108gfp-infected U373 cells were sorted for GFP expression 48 h after infection and replated in high-serum medium. The mock-infected U373 cells grew to a confluent monolayer by 6 days after plating, but the infected GFP-positive cells were not able to divide during the 6-day period (Fig. 1B). Individual HCMV-infected, GFP-expressing U373 cells were present in the dishes during the 6-day period (data not shown). The morphology of these isolated cells at 6 days p.i. was typical of HCMV-infected cells. Unlike the infected U373 cells, there were a few colonies that arose after infection of SMCs, HAECs, and HPVAECs (all less than 10 colonies from 20,000 cells). All of the cells present in these colonies 6 days p.i. were GFP negative. A representative GFP fluorescence is shown in Fig. 2. The GFP-negative cells were either resistant to HCMV infection or unable to express early viral genes after infection. These results may explain the persistent HCMV infection of aortic endothelial cells reported by Fish et al. (14). The effect of HCMV infection on cell division was more profound than the cell cycle results, since this assay looks at cells over several days and is a cumulative event. We conclude that cell division in all HCMV-infected cells was inhibited.
FIG. 2.
HAECs after infection with recombinant virus RΔ-582/-1108gfp. Serum-starved HAECs were infected with 5 PFU of recombinant virus/cell. The medium was replaced at 24 h p.i. with high-serum medium. The infected cells were sorted by fluorescence, and 20,000 cells were replated in high-serum medium. After 8 days, the cells were analyzed for fluorescence using an inverted fluorescent microscope. The same field of view was photographed using light field and fluorescent illumination. A representative field of view is shown. Similar results were obtained with SMCs and HPVAECs.
Expression of HCMV IE86 protein using Tet-inducible replication-defective adenovirus vectors.
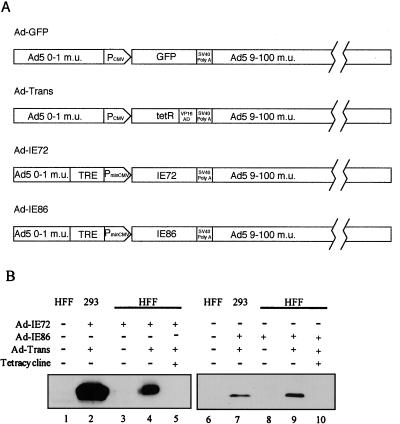
Since it is difficult to transfect the majority of U373 cells in culture, we used replication-defective adenovirus vectors to express either the IE72 or the IE86 protein and determined the effects of these viral IE proteins on cell cycle progression. The structures of the adenovirus vectors are shown in Fig. 3A. All of the vectors have the same adenovirus backbone. Replication-defective adenovirus which expresses the humanized GFP downstream of the CMV MIE promoter served as a control (Fig. 3A). Ad-Trans is a recombinant adenovirus vector which expresses the TetR-VP16 fusion protein which binds to the tetracycline repressor element (TRE) in the absence of tetracycline and activates the minimal CMV MIE promoter (PminCMV) of the recombinant adenovirus vectors Ad-IE72 and Ad-IE86. PminCMV has the CRS element, but the strong viral enhancer was removed (Fig. 3A). It has been reported that both the IE72 and IE86 proteins are required to rescue replication of an E1a and E1b knockout adenovirus (56), but it requires 4 to 5 days to see adenovirus cytopathic effect. Because E1a and E1b are both removed from the replication-defective recombinant adenoviruses, the expression of the IE72 protein or the IE86 protein alone was not sufficient to allow for detectable adenovirus vector replication within the assay period (data not shown).
FIG. 3.
Replication-defective adenovirus vectors for expression of IE72 or IE86 protein. (A) Diagram of replication-defective adenovirus vectors. Ad-GFP expresses the humanized GFP. Ad-Trans expresses the TetR-VP16 fusion protein, which binds to the TRE and activates the minimal CMV promoter (PminCMV). The Ad-IE72 and Ad-IE86 replication-defective adenovirus vectors express the IE72 or IE86 protein under control of the TRE-PminCMV promoter. (B) Western blot analysis of IE72 and IE86 protein expression detected by monoclonal antibody 810. Lanes: 1, 3 to 6, and 8 to 10, HFF; 2 and 7, 293 cells; 1 and 6, no adenovirus vector treatment; 2, Ad-IE72 plus Ad-Trans; 3, Ad-IE72 alone; 4, Ad-IE72 plus Ad-Trans; 5, Ad-IE72 plus Ad-Trans in the presence of 10 μg of tetracycline/ml; 7, Ad-IE86 plus Ad-Trans; 8, Ad-IE86 alone; 9, Ad-IE86 plus Ad-Trans; 10, Ad-IE86 plus Ad-Trans in the presence of 10 μg of tetracycline/ml.
To confirm the expression of the IE72 and IE86 proteins in HFF or 293 cells transduced with Ad-IE72 or Ad-IE86 with or without Ad-Trans in the presence or absence of tetracycline, Western blot analysis was done as described in Materials and Methods. The IE72 and IE86 proteins were detected in HFF or 293 cells (Fig. 3B, lanes 4 and 9 and lanes 2 and 7, respectively). Viral IE protein expression was inhibited by the presence of 10 μg of tetracycline/ml (Fig. 3B, lanes 5 and 10). The lower levels of the IE86 protein were possibly due to the IE86 protein repressing the expression of the TetR-VP16 fusion protein because of the presence of the CRS in the CMV promoter of this recombinant adenovirus vector.
Expression of a functional Ad-IE86 protein.
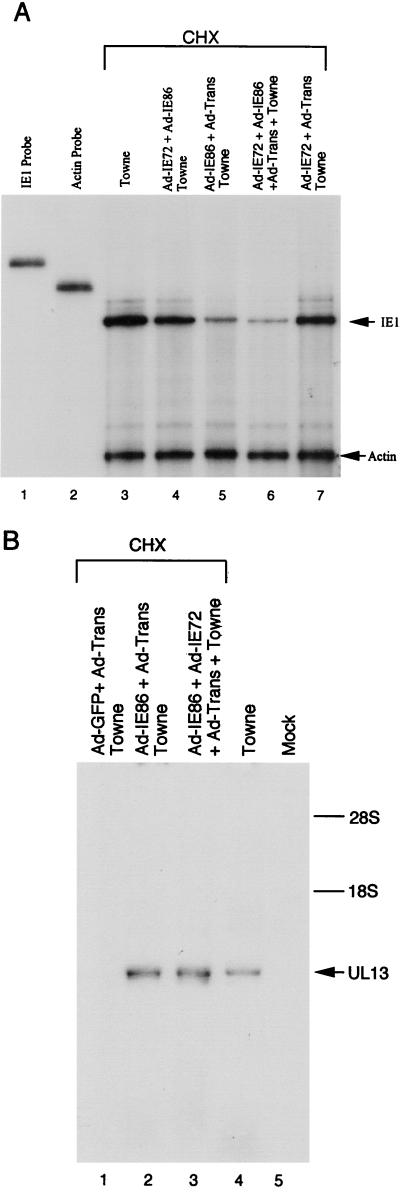
To demonstrate the function of the IE86 protein in the context of the viral genome, we tested the effects of the IE86 protein on the MIE promoter. HFF were either mock transduced or transduced with 10 PFU of Ad-IE72 plus Ad-IE86, Ad-IE86 plus Ad-Trans, both Ad-IE72 and Ad-IE86 plus Ad-Trans, or Ad-IE72 plus Ad-Trans per cell. After 24 h, the cells were infected with HCMV Towne strain (5 PFU/cell) in the presence of CHX. CHX was used to eliminate the effects of the HCMV-encoded IE72 and IE86 proteins. RNA was isolated 6 h p.i. and used in RNase protection assays with 32P-labeled antisense RNA probes. The probes protected actin and HCMV IE1 RNA. To distinguish viral RNA from the HCMV genome from viral RNA from the adenovirus IE1 cDNA, we used a probe that detects IE1 intron-containing RNA. When normalized to the internal actin control, IE86 protein expression prior to HCMV infection led to a four- to fivefold reduction in the amount of steady-state IE1 precursor RNA (Fig. 4A, lanes 5 and 6). High levels of IE72 protein expression had no significant effect on the steady-state levels of IE1 precursor RNA (Fig. 4A, lane 7). These observations demonstrated for the first time that adenovirus vector-expressed IE86 protein can function to repress the MIE promoter in the context of the viral genome, but the IE72 protein had no effect on the MIE promoter.
FIG. 4.
IE86 protein regulatory functions in the context of the HCMV genome. (A) Repression of the MIE promoter. HFF were transduced with different replication-defective adenovirus vectors and then 24 h later were infected with 5 PFU of HCMV Towne strain/cell in the presence of CHX. Cytoplasmic RNA was isolated at 6 h p.i. and used in an RNase protection assay with 32P-labeled antisense IE1 probe. 32P-labeled antisense actin probe was used for an RNA-loading control. Lanes: 1, undigested IE1 probe; 2, undigested actin probe; 3, no adenovirus; 4, Ad-IE72 plus Ad-IE86; 5, Ad-IE86 plus Ad-Trans; 6, both Ad-IE72 and Ad-IE86 plus Ad-Trans; 7, Ad-IE72 plus Ad-Trans; 3 to 7, infected with HCMV Towne strain. (B) Activation of an early viral promoter. HFF were transduced with different replication-defective adenovirus vectors and then 24 h later were infected with 5 PFU of HCMV Towne strain/cell in the presence of CHX. A HCMV Towne-infected control was not transduced with adenovirus vectors and not treated with CHX. Cytoplasmic RNA was isolated 6 h after infection. Northern blot hybridization was done with a 32P-labeled UL13 probe. Lanes: 1, Ad-GFP plus Ad-Trans; 2, Ad-IE86 plus Ad-Trans; 3, both Ad-IE72 and Ad-IE86 plus Ad-Trans; 4 to 5, no adenovirus vectors; 1 to 3, infected with HCMV Towne strain in the presence of CHX; 4, infected with HCMV Towne strain without CHX. The locations of the 28S and 18S ribosomal RNAs are indicated.
To determine if the transduced IE86 protein can activate a viral early promoter in the context of the viral genome, HFF were mock transduced or transduced with 10 PFU of the adenovirus vectors/cell and infected with HCMV Towne strain 24 h later as described above. RNA was isolated 6 h p.i., fractionated by denaturing agarose gel electrophoresis, and transferred to a Nytran membrane. Since preliminary gene microarray analysis indicated that the UL13 promoter was highly activated by the IE86 protein, a random-primed 32P-labeled UL13-specific probe was used. Preexpression of the IE86 protein or both the IE86 and IE72 proteins transactivated the UL13 promoter in the presence of CHX (Fig. 4B, lanes 2 and 3). The IE86 protein was necessary for promoter activation, but the IE72 protein did not synergize the early promoter activation. Transduction with Ad-GFP plus Ad-Trans had no effect (Fig. 4B, lane 1). These data indicated for the first time that the IE86 protein can activate an early viral promoter in the context of the viral genome.
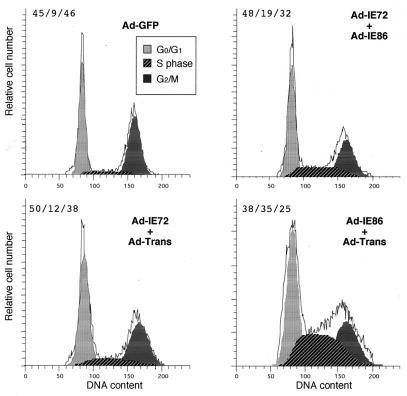
Ad-IE86 protein effects on the S phase.
Weibusch and Hagemeier (59) reported that the IE86 protein inhibited cell cycle progression of U373 cells at the G1/S transition point; however, several reports indicated that the viral IE86 protein had a role in increasing and interacting with factors involved in the S phase of the cell cycle (6, 19, 50). Since transfection of U373 cells is inefficient (10% or less), we used replication-defective recombinant adenovirus vectors that can transduce more than 95% of the U373 cells as determined by GFP fluorescence with Ad-GFP (10 PFU/cell) (data not shown). We determined the effect of the IE86 protein on U373 cells by using adenovirus vectors. Permissive U373 cells were synchronized by maintaining the cells in low-serum medium for 48 h, and then the cells were transduced with a total of 10 PFU of either Ad-GFP, Ad-IE72 plus Ad-IE86, Ad-IE72 plus Ad-Trans, or Ad-IE86 plus Ad-Trans per cell. The cells were maintained in low-serum medium for an additional 24 h, and then high-serum medium with nocodazole was added for an additional 48 h. The cells were harvested and stained with propidium iodide for cell cycle analysis as described in Materials and Methods.
The cell cycle distribution of Ad-GFP-transduced cells was similar to that of mock-infected U373 cells (data not shown), with an S-phase component of 9 versus 8% (Fig. 5B). Cells transduced with Ad-IE72 plus Ad-IE86 had a modest increase in the S-phase component of the cell cycle (Fig. 5). This effect was probably due to a low level of IE86 protein expression in the absence of induction by Ad-Trans. Cells transduced with Ad-IE72 plus Ad-Trans had a slight increase in the S-phase component. In contrast, cells transduced with Ad-IE86 plus Ad-Trans had a significant increase to 35% of the cells in the S-phase component (Fig. 5). However, the cells were blocked in the S phase and did not progress through the S phase to accumulate in G2/M in the presence of nocodazole. These data suggest that the IE86 protein can increase the number of cells in the S phase independently of other HCMV-encoded genes.
FIG. 5.
Effect of the IE86 protein on the S phase of permissive U373 cells. U373 cells, serum starved for 48 h, were treated with either 10 PFU of Ad-GFP/cell or of 5 PFU each of either Ad-IE72 plus Ad-IE86, Ad-IE72 plus Ad-Trans, or Ad-IE86 plus Ad-Trans per cell. After 24 h, the cells were treated with nocodazole and stimulated with high-serum medium. After an additional 24 h, the cells were stained with propidium iodide (PI), and the DNA content of an equal number of cells was determined using a FACS scanner. The PI profile is shown as the solid histogram line. The PI histogram cell cycle analysis was determined by use of the ModFit program as described in Materials and Methods and is given as percent G0/G1/percent S/percent G2/M.
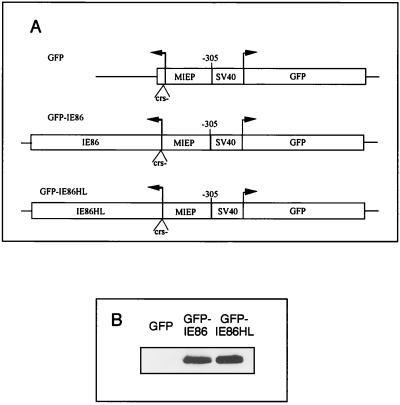
IE86 protein specifically halts cell cycle progression in the S phase.
Even though the adenovirus E1a, E1b, and E3 genes were deleted in the adenovirus vectors, it is possible that the IE86 protein indirectly halted the cell cycle via adenovirus proteins, such as those encoded from the E4 region. The adenovirus E4 gene products were reported to block the cell cycle in the S phase (60). Therefore, it was necessary to exclude the effects of the adenovirus E4 gene products. Because transient transfections are inherently inefficient and indicator genes on two separate plasmids are not always taken up by the same cell, we constructed plasmids which have the CMV MIE promoter with the CRS mutated driving expression of either the IE86 protein or a mutant IE86 protein and a divergent simian virus 40 (SV40) promoter driving expression of humanized GFP on the same plasmid (Fig. 6A). The CRS of the MIE promoter was mutated, because the wild-type IE86 protein represses the MIE promoter by binding to the CRS (36, 37). The IE86HL protein has the amino acids histidine 446 and histidine 452 in a putative zinc finger domain converted to leucines as described previously (37). The divergent SV40 promoter driving expression of GFP allows for the selection of transfected cells by FACS scanning. Western blots demonstrated that the IE86 protein and the IE86HL proteins were expressed at equal levels in 293T cells (Fig. 6B).
FIG. 6.
Expression of wild-type and mutant IE86 proteins. (A) Diagram of plasmids used for GFP, IE86, or IE86HL protein expression. An SV40 promoter drives expression of GFP in a direction opposite to that of the MIE promoter (MIEP) driving expression of either no viral gene (GFP), HCMV IE86 (GFP-IE86), or a mutant form of IE86 (GFP-IE86HL). The CRS of the MIE promoter was mutated (crs−) to allow equal expression of both the IE86 and IE86HL proteins. (B) Western blot analysis of IE86 and IE86HL protein expression detected by monoclonal antibody 810. The 293T cells were transiently transfected with 10 μg of either plasmid GFP, GFP-IE86, or GFP-IE86HL.
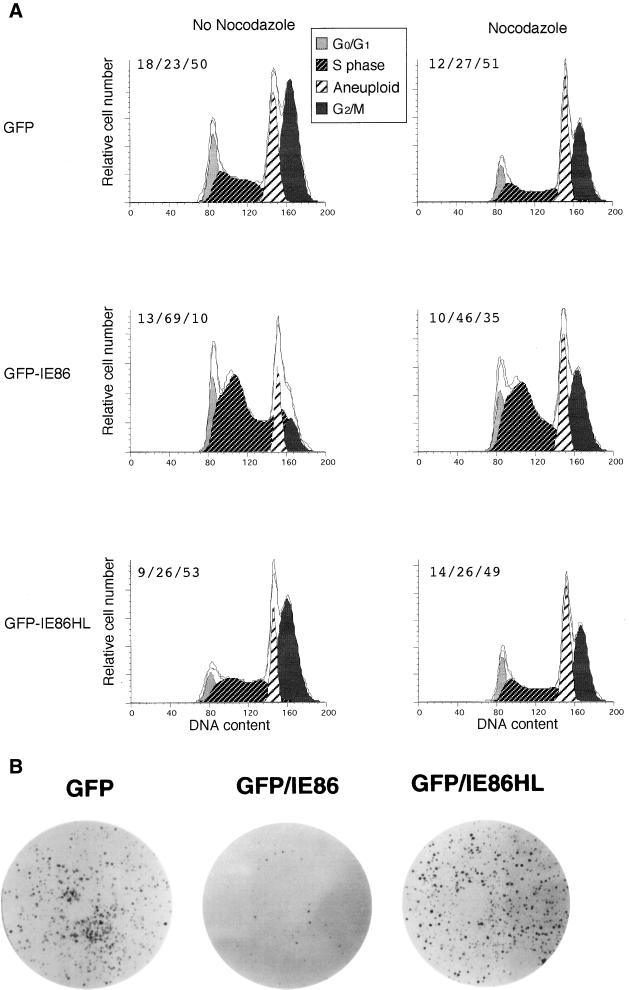
To determine the effect of the wild-type IE86 protein in the absence of any other viral proteins known to halt the cell cycle, 293T cells were transiently transfected with plasmids that express either GFP alone, GFP plus the IE86 protein, or GFP plus the IE86HL protein. We used 293T cells because we obtained transfection efficiencies as high as 40% as determined by FACS, but transfection efficiencies of only 10% or 1% were obtained with U373 cells or HFF, respectively (data not shown). In addition, the HFF did not remain viable after transfection plus treatment with nocodazole. The adenovirus E1a and E1b proteins are expressed endogenously in the 293T cells, but they were not expected to play a role in halting the cell cycle, since these viral proteins are involved in cell cycle progression. To synchronize the 293T cells, they were serum starved for 48 h prior to transfection with 10 μg of the GFP-expressing plasmids. The transiently transfected cells were kept in low-serum medium for an additional 24 h. To determine the effects on cell cycle progression, the cells were fed high-serum medium with or without nocodazole. After an additional 48 h, the cells were sorted by FACS, stained with propidium iodide, and then subjected to cell cycle analysis as described in Materials and Methods.
All of the sorted 293T cells had an aneuploid peak (Fig. 7A). The plasmid GFP and GFP-IE86HL gave similar cell cycle profiles, with 27 to 23% of the cells in the S phase with or without nocodazole treatment (Fig. 7A). In contrast, GFP-IE86-transfected cells had a significantly higher S-phase component of 46 and 69% with and without nocodazole, respectively. These data indicate that the expression of the wild-type IE86 protein can cause cells to accumulate in the S phase of the cell cycle.
FIG. 7.
Effect of HCMV IE86 protein on the S phase of nonpermissive 293T cells. (A) Cell cycle analysis in the presence or absence of nocodazole. Serum-starved 293T cells were transiently transfected with 10 μg of either plasmid GFP, GFP-IE86, or GFP-IE86HL. The cells were stimulated with high-serum medium with or without nocodazole at 24 h after transfection. At 48 h, the GFP+ cells were sorted by FACS, stained with propidium iodide (PI), and then FACS scanned. The PI histogram cell cycle profile was determined by using ModFit software as described in Materials and Methods. The quantitation of cell cycle phases is given as percent G0/G1/percent S/percent G2/M. (B) Wild-type IE86 protein inhibition of cell division. After 24 h, GFP-positive cells from each of the transient transfections described above were collected by FACS, and 20,000 GFP-positive cells were replated in high-serum medium. After 6 days at 37°C, the cells were fixed with formalin and stained with methylene blue as described in Materials and Methods. A result representative of several experiments is shown.
To determine the effect of wild-type IE86 protein on cell division, 293T cells were serum starved for 48 h and then transfected with 10 μg of either plasmid GFP, GFP-IE86, or GFP-IE86HL. After an additional 24 h in low-serum medium, the cells were fed high-serum medium. Forty-eight hours later, the cells were sorted by FACS for GFP fluorescence. The sorted cells (20,000) were replated in high-serum medium. Cells expressing GFP alone or GFP plus the IE86HL protein grew when replated on the dishes in high-serum medium (Fig. 7B). GFP-positive cells that also expressed the IE86 protein were restricted in their ability to grow and were unable to generate colonies after 6 days. Single GFP-expressing cells were present that did not give rise to colonies after 6 days of incubation (data not shown). The few colonies that did develop either had lost the plasmid or were not transfected originally. We conclude that the wild-type IE86 protein can cause 293T cells to accumulate in the S phase of the cell cycle and can inhibit cell division.
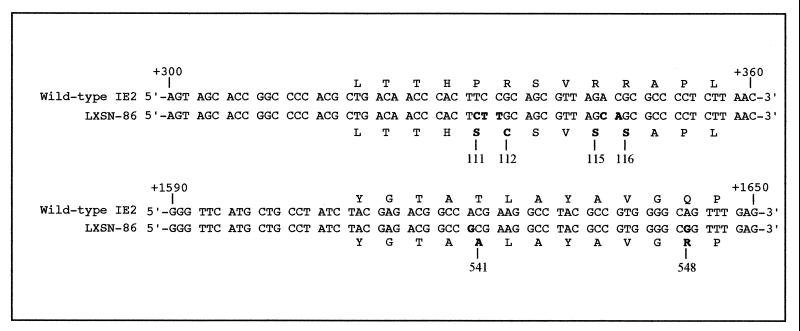
LXSN-IE86 cell line contains a mutant IE2 gene.
Recently, Bonin and McDougall (5) isolated an IE86 protein-expressing HF cell line (LXSN-IE86) using a retroviral vector with selection for neomycin resistance. If wild-type IE86 protein blocks cell division, then it is possible that the integrated IE2 gene was mutated. Therefore, we sequenced the IE2 gene in the LXSN-IE86 cells. We utilized seven sets of PCR primers to amplify overlapping 250- to 350-bp fragments of the IE2 gene in DNA isolated from the LXSN-IE86 cells. Upon sequencing the integrated viral gene, we found several mutations within exon 5 of the viral protein (Fig. 8). The carboxyl-terminal mutations were confirmed by independent sequencing of the same cell line by the Greaves Laboratory (R. Greaves [Cambridge University], personal communication). Since the mutant IE86 protein in LXSN-IE86 cells did not block cell cycle progression (5), the mutations may indicate important residues involved in an IE86 protein-induced cell cycle halt. How the wild-type protein blocks cell cycle progression requires further investigation.
FIG. 8.
Sequence comparison between the IE2 gene from HCMV Ad169 and that from LXSN-IE86 cells. The nucleotide sequence comparison of +300 to +360 and +1590 to +1650 relative to the transcription start site of the MIE promoter is shown. Mutant nucleotides and amino acids are in boldface. The mutant amino acid numbers are indicated. Sequencing was performed as described in Materials and Methods.
DISCUSSION
HCMV infects quiescent, differentiated cells that do not have available factors required for viral DNA synthesis. To create a viral replication factory, the virus must activate the appropriate biosynthetic pathways for DNA synthesis. After HCMV infection, the levels of factors involved in cellular DNA synthesis are increased, including proliferating cell nuclear antigen, thymidine kinase, DNA polymerase α, ornithine decarboxylase, and certain cyclins (6, 7, 11, 61). HCMV infection of permissive cells induces a cell cycle halt. Several reports suggest a cell cycle halt at the G1/S transition point in HFF (7, 11, 31, 41). However, other reports suggest that HCMV infection can induce cell cycle halts at other stages of the cell cycle (26, 31, 43). Determining which viral gene products stimulate G0 cells to enter an aborted cell cycle is important for understanding the biology of the virus-cell interaction.
We investigated the effects of HCMV infection on the permissive U373 cell line as well as other permissive cells. HCMV-infection of serum-starved U373 cells caused an increase in the number of cells present in the S phase. Since the viral genome is small compared to the cellular genome, viral DNA replication alone would not account for the changes seen in the S phase. The infected U373 cells, as well as the other permissive cells tested, were unable to complete cell division even though some cells made it to G2/M. The differences between the locations of the cell cycle halt in HFF and U373 cells may depend on the type of cell (primary versus transformed). Alternatively, the UL69 tegument protein, which halts the cell cycle in G1, functions more efficiently in HFF than in U373 cells.
In a similar experiment, Weibusch and Hagemeier (59) used a multiplicity of infection that infected only 45% of the U373 cells, which confounds the cell cycle analysis by including noninfected cells. They detected a threefold increase in the G1 population of infected versus mock-infected U373 cells. In contrast, we used a recombinant HCMV which expresses the humanized GFP with early kinetics and isolated only the productively infected cells. FACS was necessary because we found that infection of permissive cells with HCMV Towne strain at a multiplicity of infection of 5 PFU/cell depends on the cell type and can range from 17 (HPVAECs) to 93% (HFF). In addition, the cell cycle analysis after serum stimulation is critical. In our pilot studies with uninfected serum-starved U373 cells, we determined that at 24 h after the addition of high-serum medium in the presence of nocodazole, 18% of the cells were still in G0/G1, 66% of the cells were in the S phase, and only 15% of the cells progressed into G2/M. When serum-starved cells were harvested at 48 h after the addition of high-serum medium in the presence of nocodazole, only 5% of the cells were in G0/G1, 27% of the cells were in the S phase, and 67% of the cells progressed into G2/M (data not shown). Since we harvested the RΔ-582/-1108gfp-infected U373 cells 48 h after release from serum starvation, the cells recovered from the serum starvation and progressed into the cell cycle. This apparent lag in the recovery of U373 cells may in part explain why the observations of Weibusch and Hagemeier (59) are different than our results.
To determine the effects of the IE86 protein on the cell cycle, we used replication-defective adenovirus vectors expressing IE86 protein to transduce U373 cells. The adenovirus-transduced IE86 protein was functional in its ability to activate an early viral promoter and to repress the MIE promoter in the context of the viral genome. Transduction of functional IE86 protein into U373 cells led to a fourfold increase in the number of cells present in the S phase relative to control transduced cells (Ad-GFP). The IE72 protein did not have a significant effect on the S phase at 10 PFU of Ad-IE72 plus Ad-Trans vectors/cell. The IE86 protein in the absence of other viral proteins induced more cells to enter the S phase than in HCMV-infected U373 cells. This finding is consistent with the idea that HCMV DNA replication alone could not account for an increase in DNA content seen in the HCMV-infected U373 cells. Since the virion-associated UL69 protein of HCMV was not present in the transduced U373 cells, the cells did not demonstrate a G1/S halt. The cells progressed into the S phase, and then they were halted.
The E4 gene of adenovirus has been implicated in inducing a G1/S and a G2 cell cycle halt (60). Because the E4 gene was present in the adenovirus vectors, it was possible that IE86 protein expression increased transcription from the adenovirus E4 gene. To eliminate the possibility of E4 gene involvement in the induction of the S phase and the cell cycle halt, plasmids were constructed which expressed GFP and a wild-type or a mutant IE86 protein. GFP alone or GFP plus an IE86 mutant protein failed to induce the S phase or a cell cycle halt in transfected 293T cells. In contrast, wild-type IE86 protein alone caused an accumulation of cells in the S phase and a halt in cell division. The adenovirus E1a and E1b proteins, expressed in the 293T cells, do not play a role, since they were not present in the replication-defective recombinant Ad-IE86 and Ad-Trans vectors which could induce the S phase and a cell cycle halt. Our results demonstrate that the expression of wild-type IE86 protein can cause cells to accumulate in the S phase of the cell cycle.
When Weibusch and Hagemeier (59) serum starved U373 cells prior to transfection followed by the addition of serum 42 h after transfection, they reported little change in the cell cycle between mock-transfected and IE86 protein-expressing cells. In fact, a modest increase in the S-phase component was seen (20% in the IE86 protein-expressing cells compared to 16% in the mock-infected cells) (59). When we selected the GFP-expressing cells and analyzed the cell cycle, we detected an increase in the S phase from 23% (GFP-mock) to 69% (GFP-IE86). Our plasmid constructs coexpressed GFP as well as the effector gene of interest in synchronized cells that were selected 48 h after transfection for GFP expression. The longer recovery period and the GFP selection allowed the cell cycle analysis to be performed on a uniform population. Differences in experimental design may explain the discrepancies in the effect of the IE86 protein on the stage of the cell cycle. However, the results of Weibusch and Hagemeier confirmed our original observation (Murphy and Stinski, 23rd Int. Herpesvirus Workshop) that wild-type IE86 protein can induce a cell cycle halt.
The ability of the IE86 protein to inhibit cell cycle progression might explain the difficulties we encountered in isolating recombinant adenovirus vectors expressing a wild-type IE2 gene product from an unregulated promoter (data not shown). In contrast, we and others (1, 2) have isolated a recombinant adenovirus vector expressing the wild-type IE1 gene product from an unregulated promoter. In addition, Greaves and Mocarski (18) were able to isolate an IE72 protein-expressing HF cell line. While, Bonin and McDougall (5) generated an IE86 protein-expressing HF cell line using a retroviral vector and a selectable gene product (G418), the IE86 protein encoded by the IE2 gene did not induce a halt in cell cycle progression. Upon sequencing the integrated viral gene, we found mutations within a critical carboxyl-terminal domain of the protein. These mutations were confirmed by independent sequencing of the same cell line by the R. Greaves Laboratory (Greaves, personal communication). When Weibusch and Hagemeier (59) mutated this region of the IE86 protein, they were unable to halt cell cycle progression. We propose that the use of a retroviral system compounded by selection for neomycin resistance resulted in the selection of an HF expressing a mutant IE86 protein. These mutations in the IE86 protein may have prevented the protein from inhibiting the cell cycle.
We observed a halt in the cell cycle of HCMV-infected permissive HFF, U373 cells, SMCs, HAECs, and HPVAECs, as well as nonpermissive 293T cells, transiently transfected with an IE86 protein-expressing plasmid. A halt in the S phase of the cell cycle may allow HCMV to utilize cellular factors for its own viral DNA replication. Weibusch and Hagemeier (59) have shown that the IE86 protein can affect the cell cycle in a p16INK4-, Rb-, p53-, or p21CIP-independent manner. The mechanism by which the IE86 protein halts cell cycle progression is presently not understood and requires further investigation.
ACKNOWLEDGMENTS
We thank members of the laboratory for helpful discussion and Jeffery Meier for critical reading of the manuscript. We are grateful to Philip Lashmit for assistance. We also thank the members of the flow cytometry facility at the University of Iowa.
This work was supported by grants AI-13562 (M.F.S.) and AI-21640 (J.A.N.) from the National Institutes of Health.
REFERENCES
- 1.Ahn J, Brignole E J, Hayward G S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J-H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn K, Angulo A, Ghazal P, Peterson P A, Yang Y, Fruh K. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc Natl Acad Sci USA. 1996;93:10990–10995. doi: 10.1073/pnas.93.20.10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alford C A, Britt W J. Cytomegalovirus. New York, N.Y: Raven Press, Ltd.; 1990. [Google Scholar]
- 5.Bonin L R, McDougall J K. Human cytomegalovirus IE2 86-kilodalton protein binds p53 but does not abrogate G1 checkpoint function. J Virol. 1997;71:5861–5870. doi: 10.1128/jvi.71.8.5861-5870.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 7.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 8.Caswell R, Hagemeier C, Chiou C-J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE2) protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcription regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 9.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiou C J, Zong J, Waheed I, Hayward G S. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J Virol. 1993;67:6201–6214. doi: 10.1128/jvi.67.10.6201-6214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drew W L, Mintz L, Hoo R, Finley T N. Growth of herpes simplex and cytomegalovirus in cultured human aveolar macrophages. Am Rev Respir Dis. 1979;119:287–291. doi: 10.1164/arrd.1979.119.2.287. [DOI] [PubMed] [Google Scholar]
- 13.Fish K N, Depto A S, Moses A V, Britt W, Nelson J A. Growth kinetics of human cytomegalovirus are altered in monocyte-derived macrophages. J Virol. 1995;69:3737–3743. doi: 10.1128/jvi.69.6.3737-3743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fish K N, Soderberg-Naucler C, Mills L K, Stenglein S, Nelson J A. Human cytomegalovirus persistently infects aortic endothelial cells. J Virol. 1998;72:5661–5669. doi: 10.1128/jvi.72.7.5661-5668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebert S, Schmolke S, Sorg G, Floss S, Plachter B, Stamminger T. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate early-mediated transactivation that is able to prevent viral replication. J Virol. 1997;71:7048–7060. doi: 10.1128/jvi.71.9.7048-7060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonczol E, Andrews P W, Plotkin S A. Cytomegalovirus replicates in differentiated but not in undifferentiated human embryonal carcinoma cells. Science. 1984;224:159–161. doi: 10.1126/science.6322309. [DOI] [PubMed] [Google Scholar]
- 17.Graham F L, van der Eb A J. A new technique for the assay of infectivity of adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 18.Greaves R F, Mocarski E S. Defective growth correlates with reduced accumulation of viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J Virol. 1998;72:366–379. doi: 10.1128/jvi.72.1.366-379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hagemeier C, Walker S M, Sissons P J, Sinclair J H. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J Gen Virol. 1992;73:2385–2393. doi: 10.1099/0022-1317-73-9-2385. [DOI] [PubMed] [Google Scholar]
- 22.Hermiston T W, Malone C L, Stinski M F. Human cytomegalovirus immediate-early two protein region involved in negative regulation of the major immediate-early promoter. J Virol. 1990;64:3532–3536. doi: 10.1128/jvi.64.7.3532-3536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho M. Cytomegalovirus: biology and infection. New York, N.Y: Plenum Publishing Corp.; 1991. [Google Scholar]
- 25.Ibanez C E, Schrier R, Ghazal P, Wiley C, Nelson J A. Human cytomegalovirus productively infects primary differentiated macrophages. J Virol. 1991;65:6581–6588. doi: 10.1128/jvi.65.12.6581-6588.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones T R, Wiertz E J H J, Sun L, Fish K N, Nelson J A. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci USA. 1996;93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jupp R, Hoffmann S, Stenberg R M, Nelson J A, Ghazal P. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J Virol. 1993;67:7539–7546. doi: 10.1128/jvi.67.12.7539-7546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo K, Kaneshima H, Mocarski E S. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc Natl Acad Sci USA. 1994;91:11879–11883. doi: 10.1073/pnas.91.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu M, Shenk T. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu M, Shenk T. Human cytomegalovirus UL69 protein induces cells to accumulate in G1 phase of the cell cycle. J Virol. 1999;73:676–683. doi: 10.1128/jvi.73.1.676-683.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukac D M, Alwine J C. Effects of human cytomegalovirus major immediate-early proteins in controlling the cell cycle and inhibiting apoptosis: studies with ts13 cells. J Virol. 1999;73:2825–2831. doi: 10.1128/jvi.73.4.2825-2831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lukac D M, Manuppello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lundquist C A, Meier J L, Stinski M F. A strong negative transcriptional regulatory region between the human cytomegalovirus UL127 gene and the major immediate-early enhancer. J Virol. 1999;73:9039–9052. doi: 10.1128/jvi.73.11.9039-9052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macias M P, Huang L, Lashmit P E, Stinski M F. Cellular and viral protein binding to a cytomegalovirus promoter transcription initiation site: effects on transcription. J Virol. 1996;70:3628–3635. doi: 10.1128/jvi.70.6.3628-3635.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macias M P, Stinski M F. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci USA. 1993;90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meier J L, Pruessner J A. The human cytomegalovirus major immediate-early distal enhancer region is required for efficient viral replication and immediate-early gene expression. J Virol. 2000;74:1602–1613. doi: 10.1128/jvi.74.4.1602-1613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meier J L, Stinski M F. Effect of a modulator deletion on transcription of the human cytomegalovirus major immediate-early genes in infected undifferentiated and differentiated cells. J Virol. 1997;71:1246–1255. doi: 10.1128/jvi.71.2.1246-1255.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morin J, Johann S, O'Hara B, Gluzman Y. Exogenous thymidine is preferentially incorporated into human cytomegalovirus DNA in infected human fibroblasts. J Virol. 1996;70:6402–6404. doi: 10.1128/jvi.70.9.6402-6404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E S. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salvant B S, Fortunato E A, Spector D H. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J Virol. 1998;72:3729–3741. doi: 10.1128/jvi.72.5.3729-3741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samaniego L A, Tevethia M J, Spector D J. The human cytomegalovirus 86-kilodalton immediate-early 2 protein: synthesis as a precursor polypeptide and interaction with a 75-kilodalton protein of probable viral origin. J Virol. 1994;68:720–729. doi: 10.1128/jvi.68.2.720-729.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86 kDa protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinzger C, Grefte A, Plachter B, Gouw A S H, The T H, Jahn G. Fibroblasts, epithelial cells, endothelial cells, and smooth muscle cells are the major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J Gen Virol. 1995;76:741–750. doi: 10.1099/0022-1317-76-4-741. [DOI] [PubMed] [Google Scholar]
- 47.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spector D H. Activation and regulation of human cytomegalovirus early genes. Intervirology. 1996;39:361–377. doi: 10.1159/000150507. [DOI] [PubMed] [Google Scholar]
- 49.Spector D J, Tevethia M J. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J Virol. 1994;68:7549–7553. doi: 10.1128/jvi.68.11.7549-7553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Speir E, Modali R, Huang E, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 51.Stenberg R M. The human cytomegalovirus major immediate-early gene. Intervirology. 1996;39:343–349. doi: 10.1159/000150505. [DOI] [PubMed] [Google Scholar]
- 52.Stinski M F. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J Virol. 1977;23:751–767. doi: 10.1128/jvi.23.3.751-767.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stinski M F, Macias M P, Malone C L, Thrower A R, Huang L. Regulation of transcription from the cytomegalovirus major immediate early promoter by cellular and viral proteins. In: Michelson S, Plotkin S A, editors. Multidisciplinary approach to understanding cytomegalovirus. New York, N.Y: Elsevier Science Publishers; 1993. pp. 3–12. [Google Scholar]
- 54.Stinski M F, Malone C L, Hermiston T W, Liu B. Regulation of human cytomegalovirus transcription. In: Wagner E K, editor. Herpesvirus transcription and its control. Boca Raton, Fla: CRC Press; 1991. pp. 245–260. [Google Scholar]
- 55.Streblow D N, Soderberg-Naucler C, Vieira J, Smith P, Wakabayashi E, Ruchti F, Mattison K, Altschuler Y, Nelson J A. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell. 1999;99:511–520. doi: 10.1016/s0092-8674(00)81539-1. [DOI] [PubMed] [Google Scholar]
- 56.Tevethia M J, Spector D J, Leisure K M, Stinski M F. Participation of two human cytomegalovirus immediate early gene regions in transcriptional activation of adenovirus promoters. Virology. 1987;161:276–285. doi: 10.1016/0042-6822(87)90119-x. [DOI] [PubMed] [Google Scholar]
- 57.Thomsen D R, Stinski M F. Cloning of the human cytomegalovirus genome as endonuclease XbaI fragments. Gene. 1981;16:207–216. doi: 10.1016/0378-1119(81)90077-9. [DOI] [PubMed] [Google Scholar]
- 58.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weibusch L, Hagemeier C. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J Virol. 1999;73:9274–9283. doi: 10.1128/jvi.73.11.9274-9283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wersto R P, Rosenthal E R, Seth P K, Eissa N T, Donahue R E. Recombinant, replication-defective adenovirus gene transfer vectors induce cell cycle dysregulation and inappropriate expression of cyclin proteins. J Virol. 1998;72:9491–9502. doi: 10.1128/jvi.72.12.9491-9502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamanishi K, Rapp F. Induction of host DNA synthesis and DNA polymerase by DNA-negative temperature-sensitive mutants of human cytomegalovirus. Virology. 1979;94:237–241. doi: 10.1016/0042-6822(79)90457-4. [DOI] [PubMed] [Google Scholar]