Abstract
Bladder cancer (BlCa) is an extensively heterogeneous disease that leads to great variability in tumor evolution scenarios and lifelong patient surveillance, emphasizing the need for modern, minimally invasive precision medicine. Here, we explored the clinical significance of copy number alterations (CNAs) in BlCa. CNA profiling was performed in 15 patient-derived xenografts (PDXs) and validated in The Cancer Genome Atlas BlCa (TCGA-BLCA; n = 408) and Lindgren et al. (n = 143) cohorts. CDKN2A copy number loss was identified as the most frequent CNA in bladder tumors, associated with reduced CDKN2A expression, tumors of a papillary phenotype, and prolonged PDX survival. The study’s screening cohort consisted of 243 BlCa patients, and CDKN2A copy number was assessed in genomic DNA and cell-free DNA (cfDNA) from 217 tumors and 189 pre-treatment serum samples, respectively. CDKN2A copy number loss was correlated with superior disease-free and progression-free survival of non-muscle-invasive BlCa (NMIBC) patients. Moreover, a higher CDKN2A index (CDKN2A/LEP ratio) in pre-treatment cfDNA was associated with advanced tumor stage and grade and short-term NMIBC progression to invasive disease, while multivariate models fitted for CDKN2A index in pre-treatment cfDNA offered superior risk stratification of T1/high-grade and EORTC high-risk patients, enhancing prediction of treatment outcome. CDKN2A copy number status could serve as a minimally invasive tool to improve risk stratification and support personalized prognosis in BlCa.
Keywords: MT: Regular Issue, PDX mouse model, copy number variation, CNV, cfDNA, ctDNA, p16INK4A, p14ARF, liquid biopsy, DNA-seq, prognosis
Graphical abstract
Copy number loss of CDKN2A serves as a powerful independent indicator of non-muscle-invasive bladder cancer progression to higher/invasive stages following treatment. Our findings exploit the CDKN2A index in pre-treatment cfDNA as a potent, minimally invasive alternate that could further stratify T1HG and EORTC high-risk groups for short-term progression, ameliorating patient welfare and supporting personalized treatment/monitoring.
Introduction
Bladder cancer (BlCa) represents the third most prevalent malignancy, causing considerable morbidity and mortality rates among men in European countries.1 The predominant histological type, urothelial bladder carcinoma (UBC; 90%), progresses through two distinct pathways that place significant challenges for clinical management. Non-muscle-invasive BlCa (NMIBC; 75%), although not considered life-threatening per se, exhibits high propensity rates for recurrence and progression to invasive stages, whereas muscle-invasive BlCa (MIBC; 25%) displays poor 5-year survival rates by frequently becoming metastatic and lethal.2,3,4 Although several recent advancements in BlCa therapy have improved the response rates and patient survival, disease prognosis remains inadequate, and personalized treatment persists as a major medical need.5,6 Notably, due to the high clinical heterogeneity, BlCa monitoring relies on procedure-based, lifelong follow-up strategies with medical imaging and invasive interventions, mainly cystoscopies, which have a severe impact on patient quality of life and impose the highest lifetime costs per patient to treat on healthcare systems.7 Hence, novel molecular markers and minimally invasive liquid biopsy tools that can facilitate personalized prognosis and tailored therapeutics are of utmost clinical importance.8
Over the past decade, substantial progress has been achieved in understanding and elucidating the highly heterogeneous molecular landscape of bladder tumors.9,10 Besides single-point mutations, gene-level copy number alterations (CNAs) are tightly intertwined with gene expression and tumor-related pathway regulation, being repeatedly associated with urothelial carcinoma development and progression11,12,13 To date, recent technological advances have allowed researchers to sensitively profile CNAs in both tumor genomic DNA (gDNA) and circulating cell-free DNA (cfDNA), providing a potent approach for the individual characterization of a patient’s genomic spectrum and modern, minimally invasive disease prognosis and monitoring.14,15,16 However, there is still limited understanding and exploitation in the clinical setting for BlCa patients.
In the present study, we explored CNAs landscape in urothelial bladder tumors, utilizing both patient-derived xenograft (PDX) mouse models and BlCa patients, highlighting, for the first time, the copy number loss of CDKN2A (cyclin-dependent kinase inhibitor 2A) as a potent, minimally invasive tool to improve patient risk stratification and support personalized disease prognosis. In this regard, CNA profiling of 15 PDXs and 2 external validation cohorts (The Cancer Genome Atlas [TCGA]-BLCA, n = 408;17 Lindgren et al., n = 14318) revealed CDKN2A copy number loss as the most frequent alteration of bladder tumors, correlated with reduced CDKN2A expression, tumors of a papillary phenotype, and favorable PDX survival. To support expansion into clinical practice, CNAs of CDKN2A were analyzed in gDNA of bladder tumors and pre-treatment cfDNA in a screening cohort of 243 BlCa patients. Our findings highlighted that CDKN2A copy number loss, both in tumors and cfDNA, serves as a powerful independent predictor of NMIBC progression to invasive disease following treatment.
Results
Profiling of CNAs in BlCa PDXs
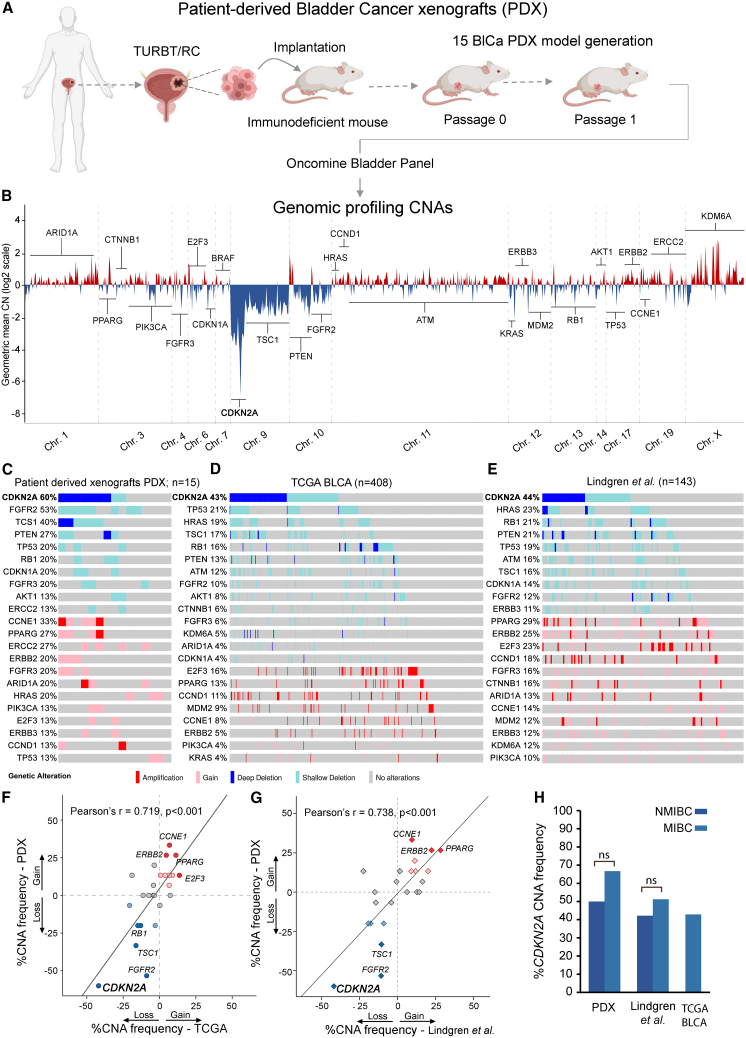
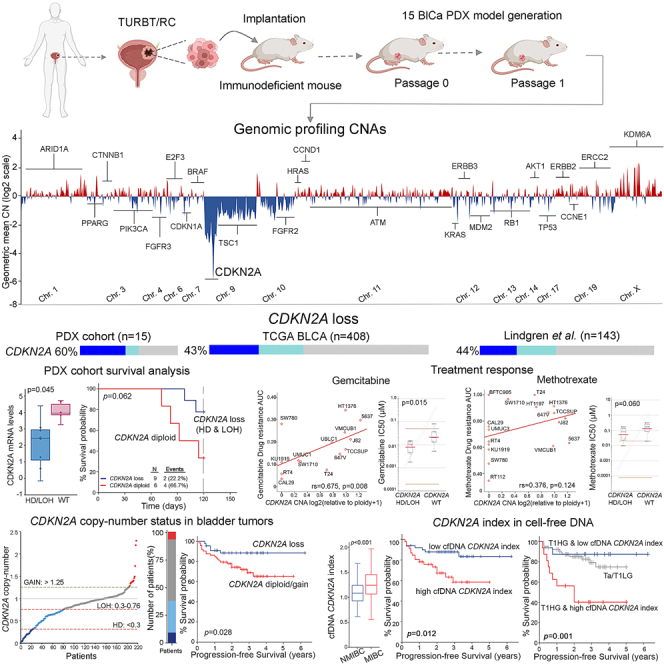
Bladder tumor specimens were grafted in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice to successfully generate 15 BlCa-PDX models (6 NMIBC, 9 MIBC). CNA profiling of PDX models was performed by DNA-sequencing (DNA-seq) (Figure 1A) and highlighted a wide chromosome 9 loss, with the most deep deletion spanning the CDKN2A gene locus on 9p21 (Figure 1B). Downstream analysis revealed that the most frequent focal homozygous deletion (HD) and loss of heterozygosity (LOH) events, were identified in the CDKN2A (60%), FGFR2 (53%), TSC1 (40%), PTEN (27%), RB1 (20%), TP53 (20%), and CDKN1A (20%) genes, while gains and focal amplifications were observed in the CCNE1 (33%), ERCC2 (27%), PPARG (27%), ARID1A (20%), HRAS (20%), FGFR3 (20%), ERBB2 (20%), CCND1 (13%), and E2F3 (13%) genes (Figure 1C). PDX analysis demonstrated the loss (HD/LOH) of the CDKN2A gene locus as the most frequent CNA, which was homozygous deleted in 7 (HD: 46.7%) and heterozygous deleted in 2 (LOH: 13.3%) PDX models (Figure S1).
Figure 1.
CNA analysis of PDX mouse models
(A) Study design of PDX generation and CNA profiling. (B) CNA profiles of PDXs across the genome. Copy number losses are shown in blue and copy number gains in red. (C–E) Oncoplots of CNAs in PDX tumors (C) and the TCGA-BLCA (D) and Lindgren et al. (E) cohorts. (F and G) Pearson correlation dot plot of CNA frequency (%) between the PDX and TCGA-BLCA (F) and between the PDX and Lindgren et al. (G) cohorts. (H) Bar graph of CDKN2A CNA frequency (%) in the NMIBC and MIBC of PDX models and the Lindgren et al. and TCGA-BLCA cohorts. Oncoplots were generated by the cBioPortal Oncoprinter tool.
To confirm our data, the TCGA-BLCA and Lindgren et al. cohorts were analyzed as validation cohorts. TCGA-BLCA analysis confirmed CDKN2A copy number loss (HD/LOH, 43%) as the most frequent CNA in bladder tumors, followed by TP53 (21%), HRAS (19%), TSC1 (17%), RB1 (16%), PTEN (13%), ATM (12%), and FGFR2 (10%) focal losses, while the most commonly amplified genes were E2F3 (16%), PPARG (13%), CCND1 (11%), and MDM2 (9%) (Figure 1D). Similarly, in the Lindgren et al. cohort, CDKN2A copy number loss (44%) was the most common alteration, followed by focal losses of HRAS (23%), RB1 (21%), PTEN (21%), and TP53 (19%) and gains of PPARG (29%), ERBB2 (25%), E2F3 (23%), and CCND1 (18%) (Figure 1E). The high concordance between PDX and validation datasets (PDX vs. TCGA-BLCA: r = 0.719, p < 0.001; PDX vs. Lindgren et al.: r = 0.738, p < 0.001; Figures 1F and 1G) confirmed that PDX models mirrored the CNA landscape of primary bladder tumors. Interestingly, CDKN2A displayed the highest HD rates (PDX: 46.7%, TCGA-BLCA: 22.6%, Lindgren et al., 21.3%), demonstrating the frequent complete loss of the CDKN2A gene locus in urothelial malignancy, while a similar distribution of CDKN2A deletions was observed between NMIBC and MIBC patients (Figures 1C–1E, 1H, and S1).
CDKN2A analysis in PDX models
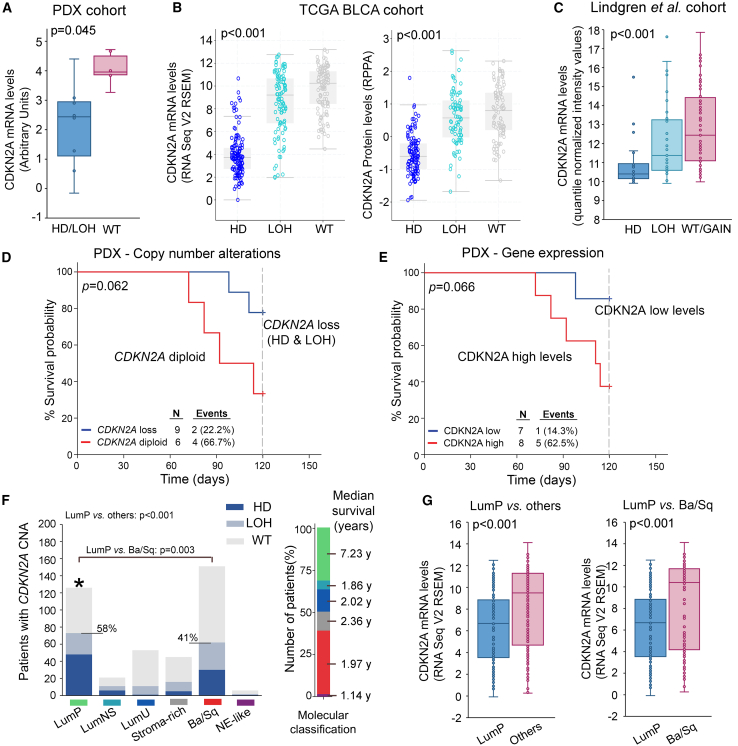
CDKN2A copy number loss (HD/LOH) exhibited profound effects on CDKN2A expression in PDX models and the TCGA-BLCA and Lindgren et al. cohorts, as significantly lower CDKN2A mRNA and protein levels were detected in bladder tumors harboring CDKN2A HD/LOH compared to wild-type (WT) tumors (Figures 2A–2C). Focusing on the survival of PDX models, although marginally not significant (p = 0.062), CDKN2A loss was correlated with prolonged survival probability (Figure 2D). Survival analysis based on CDKN2A expression displayed similar results (p = 0.066), affirming that reduced CDKN2A expression and copy number loss were both correlated with superior PDX survival (Figure 2E). Moreover, the association of CDKN2A copy number status with the most recent molecular classification system of the TCGA-BLCA dataset (consensusMIBC 2020)12 unveiled the significant prevalence of CDKN2A copy number loss and lower CDKN2A expression in luminal papillary (LumP) tumors, which are characterized by superior prognosis compared to other subtypes (p < 0.001; Figures 2F and 2G).
Figure 2.
CDKN2A copy number status in bladder tumors
(A–C) Boxplots presenting the correlation of CDKN2A copy number loss (HD/LOH) with mRNA expression across PDX samples (A), mRNA and protein levels in the TCGA-BLCA cohort (B), and mRNA levels in the Lindgren et al. cohort (C). The p values were calculated by Mann-Whitney U (A) and Kruskal-Wallis (B and C) tests. (D and E) Kaplan-Meier survival curves of PDX mouse models according to CDKN2A copy number status (D) and CDKN2A mRNA levels (E). The p value was calculated by log rank test. (F) Bar graphs of CDKN2A copy number status distribution across the molecular subtypes (consensusMIBC 2020) of the TCGA-BLCA cohort. The p values were evaluated by Fisher’s exact test. (G) Boxplots presenting the correlation of CDKN2A expression between LumP vs. other subtypes and LumP vs. Ba/Sq subtypes. The p values were calculated by Mann-Whitney U test.
CDKN2A copy number loss in the bladder tumor molecular background and chemotherapy response
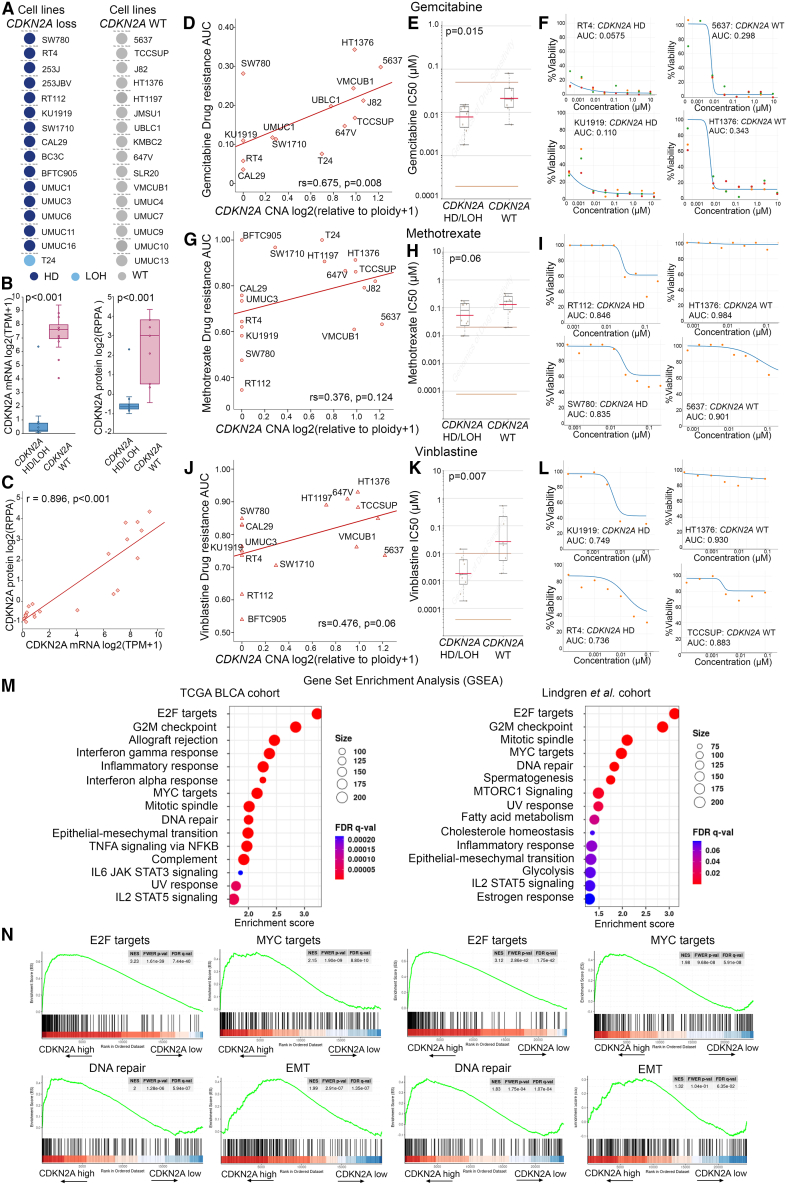
Our results demonstrated the association of CDKN2A copy number loss with tumors of papillary phenotype and superior disease prognosis, prompting us to get further insights into the role of CDKN2A copy number loss in the bladder tumor molecular landscape and treatment response. In this regard, we utilized Cancer Dependency Map (DepMap; https://depmap.org/portal/) to assess the correlation of CDKN2A copy number status with the sensitivity/resistance of 32 BlCa cell lines (Figure 3A) to the most commonly used systemic chemotherapy drugs in BlCa treatment. BlCa cell lines harboring CDKN2A copy number loss were confirmed to express significantly reduced CDKN2A mRNA (p < 0.001; Figure 3B) and protein levels (p < 0.001; Figures 3B and 3C). Moreover, a significantly lower area under the dose-response curve (AUC; gemcitabine: rs = 0.675, p = 0.008 [Figure 3D]; methotrexate: rs = 0.376, p = 0.124 [Figure 3G]; vinblastine: rs = 0.476, p = 0.060 [Figure 3J]) and IC50 (half-maximal inhibitory concentration) values (gemcitabine: p = 0.015 [Figure 3E]; methotrexate: p = 0.060 [Figure 3H]; vinblastine: p = 0.007 [Figure 3K]) were documented for BlCa cells with CDKN2A copy number loss compared to WT ones, highlighting their increased sensitivity to gemcitabine (Figures 3D–3F), methotrexate (Figures 3G–3I), and vinblastine. Indicative dose-response curves of cell lines with CDKN2A copy number loss (e.g., RT4, RT112, KU1919, and SW780) compared to WT cells (e.g., 5637, HT1376, and TCCSUP) for gemcitabine, methotrexate, and vinblastine are displayed in Figures 3F, 3I, and 3L, respectively.
Figure 3.
In silico analysis of CDKN2A copy number loss in the bladder tumor molecular background and chemotherapy response
(A) CDKN2A copy number status of BlCa cell lines in the DepMap portal (https://depmap.org/portal/). (B) Boxplots presenting the correlation of CDKN2A copy number loss (HD/LOH) with mRNA and protein expression across BlCa cells. (C) Spearman correlation of CDKN2A mRNA and protein levels in BlCa cells. (D–L) Correlation of CDKN2A CNA with area under the dose-response curve (AUC) and IC50 values of BlCa cell lines for gemcitabine (D and E), methotrexate (G and H), and vinblastine (J and K). The p values were calculated by Mann-Whitney U test (B, E, H, and K) and Spearman analysis (C, D, G, and J). Shown are indicative dose-response curves of BlCa cells with CDKN2A copy number loss (e.g., RT4, RT112, KU1919, and SW780) compared to WT cells (e.g., 5637, HT1376, and TCCSUP) for gemcitabine (F), methotrexate (I), and vinblastine (L). (M) Dot plot of GSEA Hallmark analysis of the CDKN2A-related enriched gene sets in the TCGA-BLCA and Lindgren et al. cohorts. The size of the dots represents the number of genes, and the color of the dots represents the false discovery rate (FDR) q value. (N) Enrichment plots for the top four gene sets enriched in GSEA Hallmark analysis in the TCGA-BLCA and Lindgren et al. cohorts.
Moreover, gene set enrichment analysis (GSEA) was performed, utilizing the TCGA-BLCA and Lindgren et al. datasets. Intriguingly, apart from highlighting the role of CDKN2A in G2/M checkpoint state, our analysis revealed the significant CDKN2A-related enrichment of hallmark oncogenic gene sets, including E2F and MYC targets as well as DNA repair and epithelial-to-mesenchymal transition (EMT), underlying the potential involvement of CDKN2A in BlCa progression (Figures 3M and 3N).
CDKN2A copy number loss was associated with prolonged event-free survival of NMIBC patients
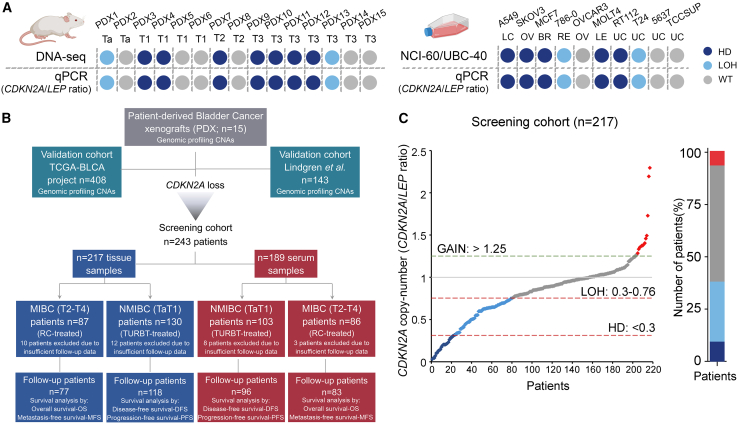
Motivated by our findings, we studied the clinical value of CDKN2A copy number status in a screening cohort of 243 BlCa patients, analyzing both patient tumors (n = 217) and pre-treatment serum samples (n = 189). Our qPCR-based quantification of CDKN2A copy number status displayed 100% concordance with both the DNA-seq data of the 15 PDX models and the known CDKN2A status (NCI-60/UBC-40 cell line panels) of 10 different cancer cell lines (Figures 4A and S2).19,20 In our screening cohort, CDKN2A copy number loss was observed in 37.8% (HD: n = 20, LOH: n = 62) of the patient bladder tumors, while CDKN2A losses were equally distributed between NMIBC (39.2%) and MIBC (35.6%) patients (Figure 4C). Figure 4B presents the REporting recommendations for tumour MARKer prognostic studies (REMARK) diagram of our clinical study, while detailed clinicopathological features of the patients are summarized in Table 1.
Figure 4.
CDKN2A copy-number status screening in BlCa patients
(A) Comparison of CDKN2A copy number status between DNA-seq and qPCR in PDX models (left) and NCI-60/UBC-40 cell line panels and qPCR in 10 selected cell lines (right). (B) REMARK diagram of the study. (C) Distribution of CDKN2A copy number status in tumors of our BlCa screening cohort.
Table 1.
Detailed clinicopathological features of the screening cohort
| Variable | No. of patients (n = 243) |
|
|---|---|---|
| BlCa gDNA cohort tissue (n = 217) |
BlCa cfDNA cohort serum (n = 189) |
|
| Disease | ||
| NMIBC (Ta, T1) | 130 (59.9%) | 103 (54.5%) |
| MIBC (T2–T4) | 87 (40.1%) | 86 (45.5%) |
| Tumor stage | ||
| pTa | 68 (31.3%) | 53 (28.0%) |
| pT1 | 62 (28.6%) | 50 (26.5%) |
| pT2 | 35 (16.1%) | 33 (17.5%) |
| pT3/T4 | 51 (23.5%) | 53 (28.0%) |
| Grade (WHO 2004) | ||
| Low | 80 (36.9%) | 61 (32.3%) |
| High | 137 (63.1%) | 128 (67.7%) |
| Grade (WHO 1973) | ||
| 1 | 21 (9.7%) | 20 (10.6%) |
| 2 | 74 (34.1%) | 52 (27.5%) |
| 3 | 122 (56.2%) | 117 (61.9%) |
| Gender | ||
| Male | 181 (83.4%) | 154 (81.5%) |
| Female | 36 (16.6%) | 35 (18.5%) |
| NMIBC (TaT1) | ||
| EORTC risk group | ||
| Low risk | 21 (16.2%) | 18 (17.5%) |
| Intermediate risk | 39 (30.0%) | 29 (28.2%) |
| High risk | 70 (53.8%) | 56 (54.3%) |
| Disease monitoring | ||
| Follow-up patients | 118 | 96 |
| Recurrence/progression | 49 (41.5%)/26 (22.0%) | 41 (42.7%)/23 (24.0%) |
| Mean DFS | 46.57 months | 45.71 months |
| Mean PFS | 59.77 months | 58.44 months |
| Excluded from follow-up | 12 | 7 |
| MIBC (T2–T4) | ||
| Disease monitoring | ||
| Follow-up patients | 77 | 83 |
| Progression/death | 49 (63.6%)/43 (55.8%) | 54 (65.1%)/48 (57.8%) |
| Mean DFS | 50.82 months | 48.62 months |
| Mean overall survival | 58.43 months | 55.53 months |
| Excluded from follow-up | 10 | 3 |
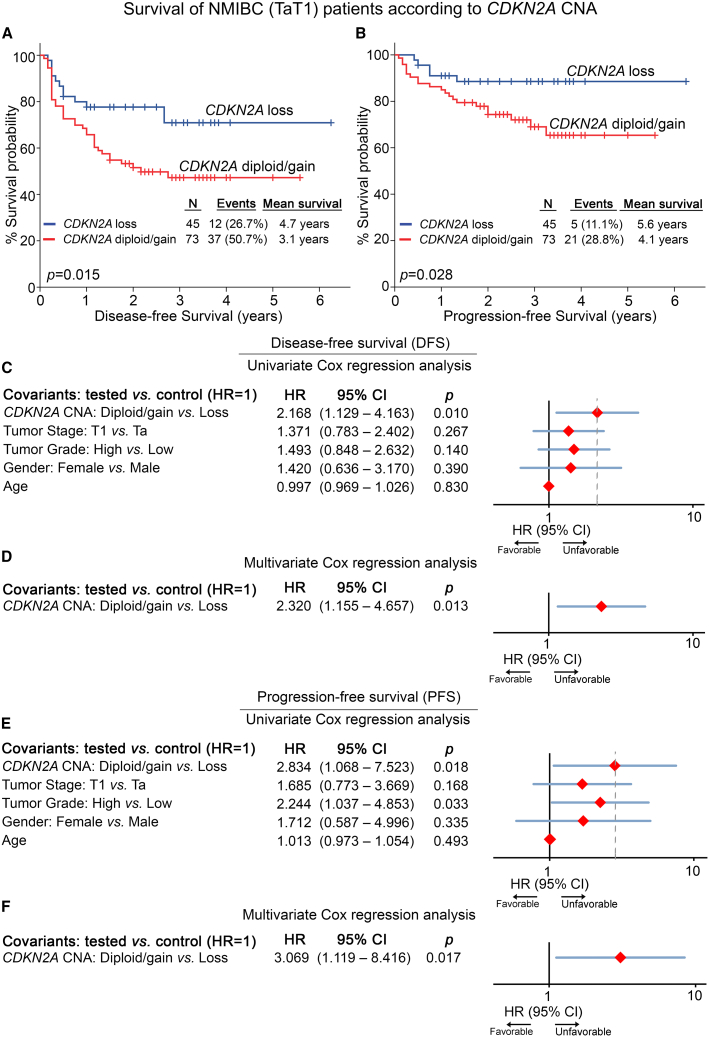
Kaplan-Meier and Cox regression analyses were performed to evaluate the clinical significance of CDKN2A copy number loss for the post-treatment outcome of BlCa patients (Figure 5; Tables S1 and S2). Due to the diverse disease courses, survival analysis was performed separately in the NMIBC and MIBC patients, using tumor relapse and progression as well as metastasis/progression and mortality as clinical endpoints, respectively. NMIBC patients harboring CDKN2A copy number loss (HD/LOH) displayed superior disease-free survival (DFS) (p = 0.015; Figure 5A) and progression-free survival (PFS) (p = 0.028; Figure 5B) compared to CDKN2A diploid/gain patients, supporting the favorable prognostic value of CDKN2A loss observed in the PDX mouse models. In this regard, univariate Cox proportional regression analysis verified the significantly higher risk for short-term disease recurrence (hazard ratio [HR] = 2.168, p = 0.010; Figure 5C) and progression (HR = 2.834, p = 0.018; Figure 5E) of the CDKN2A diploid/gain NMIBC patients. Moreover, CDKN2A copy number status was highlighted as an independent predictor of NMIBC relapse (HR = 2.320, p = 0.013; Figure 5D) and progression to invasive disease stages (HR = 3.069, p = 0.017; Figure 5F) by multivariate Cox regression models adjusted for tumor stage, grade, patient gender, and age. Separate evaluation of HD vs. LOH CDKN2A status revealed that TaT1 patients harboring HD presented significantly extended PFS intervals (p = 0.048; Figure S3), and none of the CDKN2A HD patients displayed disease progression within their follow-up time compared to LOH and diploid/gain patients. Finally, the analysis did not reveal any significant predictive value of CDKN2A copy number status for MIBC (Figure S4; Table S2), highlighting its specific nature for NMIBC outcome.
Figure 5.
CDKN2A copy number loss is associated with superior event-free survival of NMIBC patients
(A and B) Kaplan-Meier survival curves for disease-free survival (DFS; A) and progression-free survival (PFS; B) of the NMIBC cohort according to CDKN2A copy number status. The p values were calculated by log rank test. (C–F) Forest plots of the univariate and multivariate Cox regression analysis for DFS (C and D) and PFS (E and F) of NMIBC patients. Internal validation was performed by bootstrap Cox proportional regression analysis based on 1,000 bootstrap samples. HR, hazard ratio; 95% CI, 95% confidence interval of the estimated HR.
Clinical utility of CDKN2A copy number status in pre-treatment cfDNA
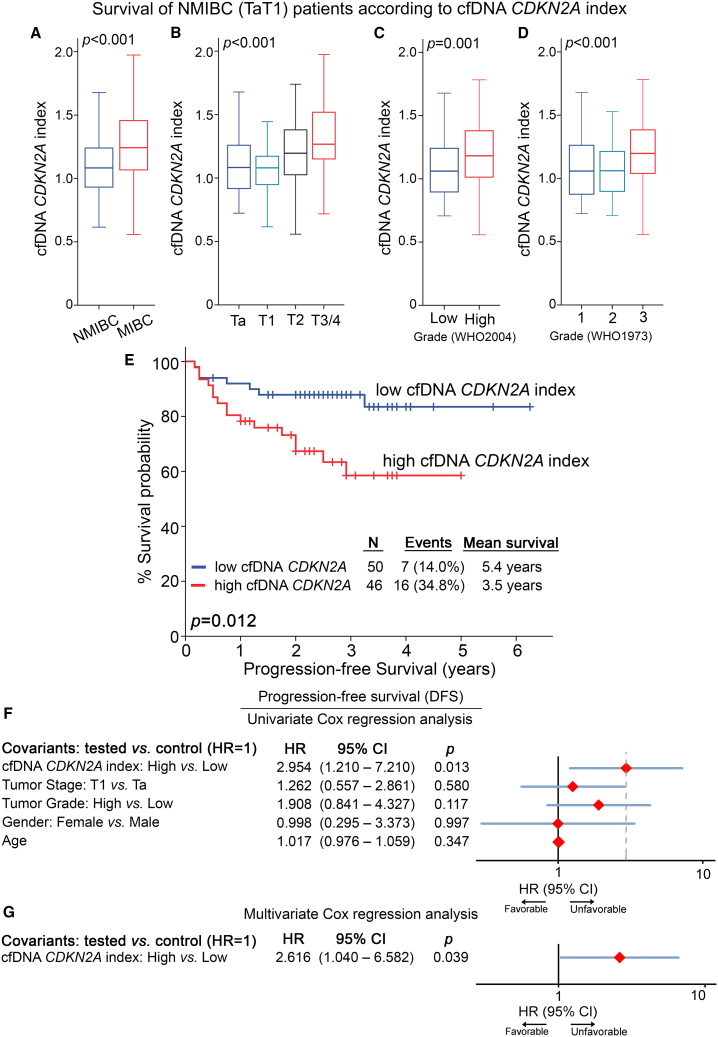
The powerful prognostic significance of CDKN2A copy number status in bladder tumors prompted us to exploit its clinical value in patient pre-treatment cfDNA. The analysis highlighted the association of a higher CDKN2A index (CDKN2A/LEP ratio) in pre-treatment cfDNA with muscle-invasive (T2–T4) compared to superficial tumors (TaT1) (p < 0.001; Figure 6A) as well as with higher pathological tumor stages (p < 0.001; Figure 6B) and grade (p ≤ 0.001; Figures 6C and 6D).
Figure 6.
High CDKN2A index in pre-treatment cfDNA is associated with worse prognosis
(A–D) Boxplots presenting the correlation of the CDKN2A index in pre-treatment cfDNA with patient tumor stage (A and B) and grade (C and D). The p values (two-sided) were calculated by Mann-Whitney U (A and C) and Kruskal-Wallis (B and D) tests. (E) Kaplan-Meier survival curve for the PFS of the NMIBC cohort according to CDKN2A index in pre-treatment cfDNA. The p value was calculated by log rank test. (F and G) Forest plots of the univariate and multivariate Cox regression analysis for the PFS of NMIBC patients. Internal validation was performed by bootstrap Cox proportional regression analysis based on 1,000 bootstrap samples.
Moreover, Kaplan-Meier (Figures 6 and S5) and Cox regression (Figure 6; Tables S3 and S4) analyses were performed to evaluate the clinical significance of the CDKN2A index in pre-treatment cfDNA for disease course. In line with the results in tumor specimens, both Kaplan-Meier curves (p = 0.012; Figure 6E) and univariate Cox regression analysis (HR = 2.954; p = 0.013; Figure 6F) clearly illustrated the notably higher risk for short-term progression to invasive disease of TaT1 patients with a higher CDKN2A index. Finally, multivariate Cox models, adjusted for tumor stage, grade, patient gender, and age, clearly confirmed the worse prognosis of NMIBC patients with a higher cfDNA CDKN2A index independent of the clinically used disease markers (HR = 2.616, p = 0.039; Figure 6G). Similar to tumor CDKN2A status, the survival analysis did not highlight any statistically significant correlation with MIBC outcome (Table S4).
Analysis of CDKN2A copy number status in pre-treatment cfDNA improves the prognostic value of the established and clinical used disease markers
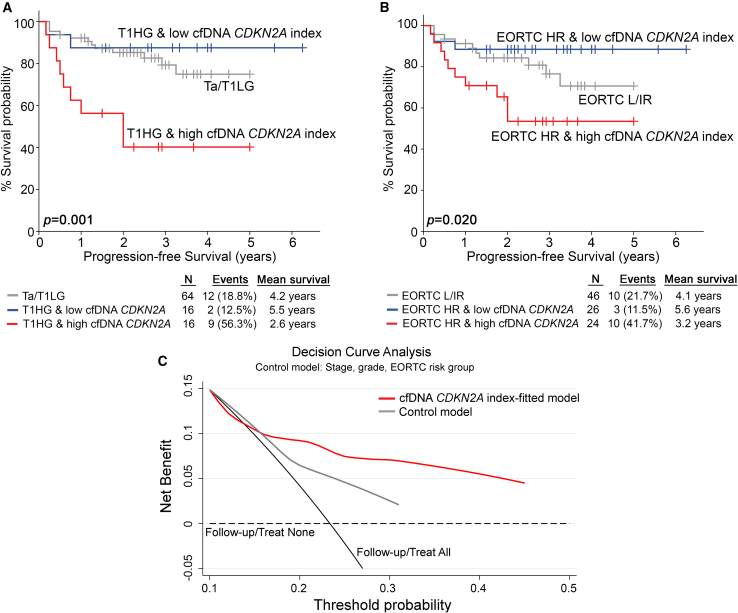
The independent clinical value of CDKN2A index in cfDNA for NMIBC outcome prompted us to evaluate its ability to strengthen the prognostic significance of the established disease markers. Multivariate models integrating cfDNA CDKN2A index with disease clinical markers clearly offered superior risk stratification and positive prediction of NMIBC progression to muscle-invasive disease (Figure 7). In this regard, the elevated CDKN2A index in pre-treatment cfDNA effectively distinguished T1HG patients at higher risk for progression (p = 0.001; Figure 7A). Similarly, a higher cfDNA CDKN2A index was associated with short-term progression among European Organisation of Research and Treatment of Cancer (EORTC) high-risk patients (p = 0.020; Figure 7B). Ultimately, decision curve analysis (DCA) analysis of cfDNA CDKN2A index-fitted multivariate prediction models, combining CDKN2A index with tumor stage, grade, and EORTC risk group clearly demonstrated the superior net benefit for the prediction of short-term progression of NMIBC patients compared to the control model of the established clinical markers alone (Figure 7C).
Figure 7.
Evaluation of the CDKN2A index in pre-treatment cfDNA improves risk stratification and prediction of NMIBC progression to muscle-invasive disease
(A and B) Kaplan-Meier survival curves for the PFS of NMIBC patients according to CDKN2A index in pre-treatment cfDNA combined with tumor stage/grade (A) and EORTC risk group (B). The p values were calculated by log rank test. (C) DCA curves of “cfDNA CDKN2A index-fitted” and “control” multivariate prognostic models for the PFS of NMIBC patients. Net benefit is plotted against various ranges of threshold probabilities.
Discussion
Despite the significant progress in BlCa therapy, the high rates of tumor recurrence/mortality and drug resistance remain major obstacles in efficient clinical management. An improved understanding of the genetic mechanisms underlying BlCa progression could be exploited to ameliorate non-invasive molecular diagnostics and tailored therapeutics.21,22,23 Here, utilizing PDX mouse models and BlCa patient cohorts, we performed, for the first time, an integrative analysis of CNAs in bladder tumors and unveiled the clinical utility of both tumor and cfDNA copy number status of CDKN2A in improving personalized prognosis and prediction of BlCa patient treatment outcome.
DNA-seq of PDX models highlighted that CDKN2A displays the highest HD and LOH rates in UBC, which were further confirmed in the TCGA-BLCA and Lindgren et al. validation cohorts. Transcriptional analysis in both PDX models and validation cohorts revealed that CDKN2A copy number loss (HD/LOH) diminished CDKN2A expression and was associated with low-risk LumP tumors and prolonged PDX survival. Additionally, CDKN2A copy number loss was correlated with higher sensitivity of BlCa cells in gemcitabine, methotrexate, and vinblastine, while GSEA of the TCGA-BLCA and Lindgren et al. cohorts highlighted the CDKN2A-related enrichment of hallmark oncogenic gene sets, including E2F and MYC targets as well as the G2M checkpoint, DNA repair, and EMT. Prompted by our results, we developed an in-house qPCR assay for the analysis of CDKN2A copy number status in our screening BlCa cohort, confirming the high prevalence of CDKN2A copy number loss in urothelial cancer. The survival analysis of our screening cohort demonstrated the superior prognosis and treatment outcome of TaT1 patients with CDKN2A deletions compared to CDKN2A diploid/gain patients, independent of the established disease markers and patient clinicopathological data, further validating the favorable prognostic utility of CDKN2A copy number loss observed in the PDX mouse models.
While cystoscopies are currently the gold-standard clinical tool for BlCa management, sampling restrictions may lead to tumor understaging and inadequate representation of disease heterogeneity, all while subjecting patients to invasive procedures.24 In this regard, liquid biopsies hold great promise as new, minimally invasive tool, with significant implications in clinical practice. Given the potent clinical utility of CDKN2A copy number status in NMIBC prognosis, we further interrogated its value in pre-treatment patients’ circulation. Notably, NMIBC patients with a high CDKN2A index in pre-treatment cfDNA were strongly associated with advanced tumor stage and grade, along with a significantly higher risk for short-term disease progression following transurethral tumor resection, independent of patients’ clinicopathological data. In this regard, cfDNA CDKN2A-fitted multivariate models significantly ameliorated risk stratification and prognosis of NMIBC, resulting in the advanced positive prediction of disease progression within the clinically heterogeneous group of T1HG and EORTC-high risk patients. The superior risk stratification of NMIBC patients using cfDNA CDKN2A-fitted models could be applied in clinical practice and support personalized treatment/monitoring decision-making. In this regard, NMIBC patients at higher risk for progression to higher/invasive stages could be considered candidates for HR-like treatment/management by being subjected to prolonged intravesical bacillus Calmette-Guérin (BCG) administration (induction plus maintenance BCG for 3 years) or to immediate radical cystectomy (RC) in shared decision-making, also raising patients’ awareness. Moreover, a risk-based follow-up strategy for NMIBC patients could also be utilized to reduce healthcare systems’ economic burden and raise patients’ quality of life.
CDKN2A encodes for the cycle-dependent kinases p14ARF/p16INK4A, while its activation has been closely associated with senescence.25 In a clinical approach, the value of CDKN2A in BlCa prognostication has proven to be controversial. CDKN2A deletion and/or decreased p16INK4A expression has been documented to predict BlCa progression and lead to immune checkpoint therapy resistance.26,27 However, new studies have highlighted a multifaceted role of CDKN2A in BlCa development. In this regard, and in line with our findings, CDKN2A copy number loss and/or reduced gene expression has been associated with luminal/papillary bladder tumors,17,18,28 while p14ARF/p16INK4A overexpression has been correlated with inferior survival expectancy of NMIBC patients and poor chemotherapy response of MIBC patients.29,30,31,32
Mechanistically, p14ARF and p16INK4A inhibit cell cycle progression and were initially described as potent tumor suppressors;33 however, recent evidence has unveiled their pro-survival functions. In particular, p14ARF-induced stabilization of FAK and SLUG, contributes to cervical and prostate cancer cell growth/proliferation, respectively, while human papillomavirus (HPV)-associated tumors induce p16INK4A expression to sustain their survival.34,35,36 Moreover, senescent cancer cells are characterized by high levels of p16INK4A,37,38 and senolytic treatment strategies targeting p16INK4A-high senescent cells efficiently increase the chemotherapy response and survival of glioblastoma- and breast cancer-bearing mice39,40 Although the role of CDKN2A has not yet been clarified in bladder tumors, future studies will pave the way to unveil the causal link between its function and the strong clinical value in NMIBC prognosis and post-treatment outcome.
In conclusion, our study aimed to evaluate CNA emergence and clinical application in BlCa. Following the establishment and CNA profiling of PDX mouse models, CDKN2A copy number loss was highlighted as the most frequent CNA in bladder tumors and correlated with reduced CDKN2A expression, LumP tumors, and prolonged survival of the PDX models. The analysis of bladder tumors and pre-treatment cfDNA of the BlCa patient cohort clearly highlighted CDKN2A copy number status as a powerful independent indicator of NMIBC patient prognosis. Finally, we demonstrated that cfDNA CDKN2A index-fitted multivariate models offered superior risk stratification of NMIBC patients and improved prediction of post-treatment outcome compared to the clinically used disease markers, endorsing CNA analysis of CDKN2A in pre-treatment cfDNA for modern non-invasive precision medicine of BlCa patients.
Materials and methods
PDX mouse models
Primary bladder tumor samples (up to 5 mm, depending on tumor availability) were grafted subcutaneously in 6-week-old NSG mice (stock number 005557) obtained from The Jackson Laboratory (Bar Harbor, ME, USA). Mice were anesthetized with an intraperitoneal injection of a ketamine:xylazine solution (90 mg/kg ketamine and 10 mg/kg xylazine).41 Tumors were allowed to reach a size up to 1 cm in diameter, and then mice were euthanized by cervical dislocation. During this period, tumor dimensions were measured weekly with calipers. Mice were closely monitored and sacrificed when tumors reached a size of 1 cm or displayed physical signs of discomfort, such as severe body weight loss or ulceration. The growth time from implantation varied from 1 to 7 months. Grafted tumors that failed to grow within 8 months (∼30%) were considered “no take.”42 Harvested PDX tumors were snap frozen for future implantation and/or stored at −80°C for subsequent molecular analysis. Tumor segments for future implantation were placed in cryovials containing 1.5 mL of cryopreservation medium: 5% DMSO/95% fetal bovine serum (Sigma-Aldrich). Cryotubes were put in a freezing container containing isopropanol, placed in an −80°C freezer overnight, and transferred to a liquid nitrogen tank the next day. Mice were housed in individually ventilated cages under specific pathogen-free conditions at the Biomedical Research Foundation of the Academy of Athens (BRFAA) animal facility. All experiments were performed in compliance with national and international legislation and were approved by the BRFAA Committee of Ethics on Animal Experiments and the Attica Prefecture.
Screening cohort
The screening cohort of the study consisted of 243 patients diagnosed with primary BlCa at “Laiko” General Hospital (Athens, Greece). Fresh-frozen bladder tumors were obtained following transurethral resection of bladder tumor (TURBT) for NMIBC patients (TaT1) or RC for MIBC patients (T2–T4), while blood samples were collected prior to surgery. NMIBC patients received adjuvant treatment according to European Association of Urology (EAU) guidelines, while none of the patients received neoadjuvant therapy prior to surgery. Risk-group stratification of NMIBC patients was carried out according to EORTC guidelines. Based on EAU guidelines, post-treatment follow-up involved cystoscopy and urinary cytology for NMIBC patients or renal ultrasound and thoracoabdominal computed tomography CT/MRI every 3 and 6 months, respectively, for MIBC. If symptoms were observed, then supplemental CT/MRI, kidney ultrasound, and brain MRI/bone scans were performed. Tumor relapse (recurrence of an equal/lower pathologic stage) and progression (recurrence of a higher/invasive stage) of NMIBC was diagnosed by TURBT after positive cystoscopy results, while recurrence/metastasis of MIBC was affirmed by CT. During a median follow-up time of 35 months (95% confidence interval [CI]: 31.36–38.64), 221 patients were successfully followed up, whereas 22 patients were excluded due to insufficient monitoring data. Study approval was granted by the Ethics Committee of “Laiko” General Hospital. The study was conducted in compliance with the ethics standards of the 1975 Declaration of Helsinki, as revised in 2008. Before sample collection, all patients provided signed informed consent.
Validation cohorts
The TCGA (TCGA-BLCA project) and Lindgren et al. BlCa cohorts were utilized as validation cohorts of the study.17,18 TCGA-BLCA consists of 412 patients diagnosed with UBC (n = 409, including 406 T2–T4 patients), papillary adenocarcinomas (n = 1), epithelial carcinomas (n = 1), and squamous cell carcinomas (n = 1). Affymetrix SNP6.0 arrays were used to assess CNAs, while mRNA expression profiles were generated by paired-end whole-transcriptome RNA sequencing (Illumina HiSeq platform). CNA and normalized RNA expression data along with the clinical data of the TCGA-BLCA project are available and can be explored through the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/projects/TCGA-BLCA) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org). For the molecular classification of the TCGA-BLCA cohort, we utilized the unified consensus subtyping system (consensusMIBC 2020) proposed by Kamoun et al. that differentiates patients into 6 biologically relevant molecular classes; namely, LumP; luminal nonspecified; luminal unstable; stroma-rich, basal/squamous; and neuroendocrine-like.12 The Lindgren et al. cohort includes 145 bladder tumor specimens [NMIBC = 102; MIBC = 42; Carcinoma in situ (Cis or Tis) = 1], submitted to genome tiling array [32K bacterial artificial chromosome (BAC) array], and gene expression array (Illumina HumanHT-12 v.3.0 expression beadchip) analysis for CNA and mRNA expression profiling, respectively. Publicly available clinical, can, and normalized RNA expression data were downloaded from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32549).
Cell lines
A total of 10 human cell lines were used in the study: T24, TCCSUP, HTB-9, RT112 (BlCa), MCF-7 (breast cancer), OVCAR-3, SK-OV-3 (ovarian cancer), A549 (lung adenocarcinoma), MOLT-4 (acute T lymphoblastic leukemia), and 786-O (renal cell carcinoma). Each cell line was cultured according to the American Type Culture Collection instructions.
DNA extraction from tumors/cell lines and total RNA purification from PDX samples
gDNA from cell lines and patients/PDX tumor samples was isolated using TRI-Reagent (Molecular Research Center, Cincinnati, OH, USA) following homogenization of 5–10 × 106 cells or 40–100 mg of fresh-frozen tissue specimens, respectively. Prior to gDNA extraction, total RNA was also extracted from PDX samples using the phenol-chloroform method. According to the manufacturer’s instructions, gDNA was dissolved and pH adjusted in 8 mM NaOH and 0.1 M HEPES buffer, total RNA was dissolved in RNA storage solution (Invitrogen, Carlsbad, CA, USA), and both analytes were stored at −80°C until analysis. The resulting DNA/RNA quality and concentration were assessed spectrophotometrically using a BioSpec Nano UV-visible spectrophotometer (Shimadzu, Kyoto, Japan).
cfDNA extraction from serum samples
5 mL of venous blood was sampled and allowed to clot at room temperature for 30 min before centrifugation at 1,200 × g for 20 min at 4°C. The serum supernatant was collected without disturbing the cellular layer and then stored at −80°C until analysis. Prior to cfDNA extraction, hemolyzed serum samples were excluded from the analysis by measuring spectrophotometrically free hemoglobin levels using the Harboe method and Allen correction. 500 μL of serum sample was used for total cfDNA extraction with the NucleoSpin cfDNA XS Kit (Macherey-Nagel, Düren, Germany) according to manufacturer’s instructions. Total cfDNA quantification was performed fluorometrically on the Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA) with the dsDNA HS Assay Kit (Life Technologies), while cfDNA fragment quantification and size distribution were assessed by capillary electrophoresis using the Agilent 2100 bioanalyzer (Agilent Technologies) with the Agilent High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s instructions.
DNA-seq
For each PDX sample, 10 ng of gDNA was amplified using the Oncomine Bladder Panel (Thermo Scientific) to construct DNA libraries using the Ion AmpliSeq Library Kit (Thermo Scientific). FuPa reagent was utilized to partially digest the primers, and the produced libraries were indexed with a unique adapter using the Ion Xpress Barcode Adapter Kit (Thermo Scientific). All barcoded libraries were purified using the Agencourt AMPure XP Beads (Beckman Coulter), quantified with a Qubit 4.0 fluorometer (Thermo Scientific), diluted to 50 pM, and pooled in equimolar proportions. Template preparation, enrichment, and chip loading were carried out on the Ion Chef system (Thermo Scientific). Sequencing was performed on S5XL, on a 530 chip, using the Ion 510 & Ion 520 & Ion 530 Kit-Chef Kit (Thermo Scientific). All procedures were performed according to the manufacturer’s instructions.
Bioinformatics analysis
Base calling, demultiplexing, and alignment to the hg19 reference genome (GRCh37) of the sequencing raw data were performed on the Torrent Suite 5.10 software (Thermo Scientific) using default parameters. Variant calling was performed by the VariantCaller v.5.8.0.19 plug-in and coverage analysis by the coverageAnalysis v.5.8.0.8 plug-in in Torrent Suite 5.10. The data were further analyzed on Ion Reporter Software 5.18 for the detection and annotation of low-frequency somatic variants (SNPs, insertions or deletions, and CNAs) using the Oncomine Bladder – 530 – w4.2 – DNA – Single Sample workflow (v.4.2) and the Oncomine Extended (5.18) filter set. Quality control and coverage metrics of DNA-seq are included in Table S5.
In silico analysis
The GSEA software (Broad Institute, MA, USA) and MSigDB v.6.2 Hallmark gene set collection were used to identify the enrichment of CDKN2A-related gene sets in the TCGA-BLCA and Lindgren et al. cohorts. The GSEA software was assessed through Gene Enrichment Identifier (https://yoavshaul-lab.shinyapps.io/gsea-geni/).43 Gene sets with absolute normalized enrichment score (NES) > 1.8 and false discovery rate (FDR) q < 0.05 were considered significant and reported.
CNA and mRNA expression profiling as well as response data to gemcitabine, methotrexate, and vinblastine of BlCa cell lines were obtained through the DepMap portal (https://depmap.org/portal) of the Broad Institute. The IC50 values represent the drug concentration that reduced cell viability by 50%, while the area under the AUC is calculated by plotting percent viability vs. drug concentration.
First-strand cDNA synthesis and CDKN2A expression quantification
Reverse transcription was performed in a 20-μL reaction by 50 U MMLV reverse transcriptase (Invitrogen), 40 U recombinant ribonuclease inhibitor (Invitrogen), and 5 μM oligo-deoxythymidine primer, using as template 1 μg of total RNA. Reverse transcription was performed at 37°C for 60 min, whereas enzyme was inactivated at 70°C for 15 min.
CDKN2A expression levels were quantified by SYBR-Green fluorescence-based quantitative PCR (qPCR). Based on published sequences, specific CDKN2A (GenBank: NM_058195.4, NM_000077.5; F: 5′-GAAGGTCCCTCAGACATCCCC-3′, R: 5′-CCCTGTAGGACCTTCGGTGAC-3′) and HPRT1 (GenBank: NM_000194.3; F: 5′-TGGAAAGGGTGTTTATTCCTCAT-3′, R: 5′- ATGTAATCCAGCAGGTCAGCAA-3′) primers were designed to produce specific 120-bp CDKN2A and 151-bp HPRT1 amplicons. The qPCR assays performed in the QuantStudio 5 Real-Time PCR System (Applied Biosystems) in 10-μL reactions consisted of Kapa SYBR Fast Universal 2× qPCR MasterMix (Kapa Biosystems, Woburn, MA, USA), 100 nM of each specific qPCR primer, and 10 ng of cDNA template. Polymerase activation was performed at 95°C for 3 min, followed by 40 cycles of a denaturation step at 95°C for 15 s and a primer annealing and extension step at 60°C for 1 min. Following amplification, melting curve analysis and agarose gel electrophoresis of the products were used for the discrimination of specific amplicons. CDKN2A expression was quantified by the 2−ΔΔCT relative quantification method, using HPRT1 as an internal control for normalization purposes.
CDKN2A CNA analysis
CNAs of CDKN2A in gDNA and cfDNA samples were analyzed by a SYBR Green-based qPCR assay using LEP as the diploid reference gene. According to published sequences and in silico analysis, specific primers for the CDKN2A (NCBI: 1029; NC_000009.12; F: 5′-ATGCCTGCTTCTACAAACCCAC-3′, R: 5′-GGAGCCCATACGCAACGAGAT-3′) and LEP (NCBI: 3952; NC_000007.14; F: 5′-CTCCACCCCATCCTGACCTTA-3′, R: 5′-AGGTTCTCCAGGTCGTTGGAT-3′) genes were designed for the amplification of specific 119-bp CDKN2A and 116-bp LEP amplicons. The qPCR assays were performed in 10-μL reactions that included Kapa SYBR Fast Universal 2× qPCR Master Mix (Kapa Biosystems), 200 nM of each PCR primer and 1 μL of gDNA or 300 nM of each PCR primer and 2 μL of cfDNA template in the 7500 Real-Time PCR System (Applied Biosystems). Polymerase activation was carried out at 95°C for 10 min, followed by 40 cycles of a denaturation step at 95°C for 30 s and a primer annealing and extension step at 60°C for 1 min. Thereafter, melting curve analysis and agarose gel electrophoresis were conducted to assess PCR amplicon specificity.
CDKN2A CNAs were calculated according to the 2−ΔΔCT relative quantification method, using LEP as the diploid reference gene.44 Standard curves for the identification of the assays’ limit of quantification and the determination of optimal cutoff values for CDKN2A homozygous/hemizygous deletions were constructed using diploid human gDNA as well as SK-OV-3 and RT112 cell line gDNA with known HD of CDKN2A (NCI-60/UBC-40 cell line panels19,20,45). All standard curve assays were performed in triplicate.
Statistical analysis
Statistical analysis was performed by SPSS Statistics 20 software (IBM, Armonk, NY, USA). Sapiro-Wilk and Kolmogorov-Smirnov tests were applied to test data’s normal distribution. The non-parametric Mann-Whitney U and Kruskal-Wallis tests were used to assess the correlation of CDKN2A copy number status and expression with categorical clinicopathological features, while the association between TCGA consensus classes and CDKN2A CNAs was calculated by Fisher’s exact tests. The similarity of CNA profiles between the PDX, TCGA-BLCA, and Lindgren et al. cohorts was evaluated by calculating the Pearson correlation of gene-level CNAs of the samples.
Kaplan-Meier survival curves using log rank test and Cox proportional regression analysis were implemented for patient survival analysis. The X-tile algorithm was applied for the optimal selection of the cutoff values of the cfDNA CDKN2A index (CDKN2A/LEP ratio). Internal validation was performed by bootstrap Cox proportional regression analysis based on 1,000 bootstrap samples. Finally, DCA was performed according to Vickers et al. by STATA 13 software (StataCorp, College Station, TX, USA) in order to evaluate the clinical benefit of the cfDNA CDKN2A index in patient treatment outcome.46,47
Data and code availability
Data supporting the findings of this study are available within the paper and its supplemental information. CNA and normalized RNA expression data along with the clinical data of the TCGA-BLCA project were acquired from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/projects/TCGA-BLCA) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org). Publicly available clinical, CNA, and normalized RNA expression data of the Lindgren et al. cohort were downloaded from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32549). CNA data for the NCI-60 cell lines used in the study can be retrieved from CellMiner (https://discover.nci.nih.gov/cellminer/), while CNA data for UBC-40 cell lines are available in the Earl et al. study.19 CNA, mRNA expression profiling, and drug response data of BlCa cell lines were obtained through the DepMap portal (https://depmap.org/portal) of the Broad Institute. All data are available from the corresponding authors upon reasonable request.
Acknowledgments
This research was co-financed by the European Regional Development Fund of the European Union and Greek national funds through the Operational Program Competitiveness, Entrepreneurship and Innovation under the call RESEARCH-CREATE-INNOVATE (project code: T2EDK-02196) to the National and Kapodistrian University of Athens (KE17358). The research work was supported by the Hellenic Foundation for Research and Innovation (HFRI) under the Third Call for HFRI PhD Fellowships (fellowship number 6123 to M.-A.P.).
Author contributions
M.-A.P., conceptualization and design, methodology, investigation, formal analysis, validation, visualization, data curation, and writing – original draft; K.-M.P. and K.P., investigation; P.L., G.K., Z.K., G.L., M.Z., and D.L., investigation and resources; D.C.S. and K.S., resources and writing—review and editing; A.K., investigation, resources, and writing – review and editing; A.S., resources, writing – review and editing, project administration, and funding acquisition; M.A., conceptualization and design, methodology, investigation, formal analysis, validation, visualization, data curation, writing – review and editing, project administration, and supervision. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omton.2024.200818.
Contributor Information
Andreas Scorilas, Email: ascorilas@biol.uoa.gr.
Margaritis Avgeris, Email: mavgeris@med.uoa.gr.
Supplemental information
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Cambier S., Sylvester R.J., Collette L., Gontero P., Brausi M.A., van Andel G., Kirkels W.J., Silva F.C.D., Oosterlinck W., Prescott S., et al. EORTC Nomograms and Risk Groups for Predicting Recurrence, Progression, and Disease-specific and Overall Survival in Non-Muscle-invasive Stage Ta-T1 Urothelial Bladder Cancer Patients Treated with 1-3 Years of Maintenance Bacillus Calmette-Guérin. Eur. Urol. 2016;69:60–69. doi: 10.1016/j.eururo.2015.06.045. [DOI] [PubMed] [Google Scholar]
- 3.Messing E.M., Tangen C.M., Lerner S.P., Sahasrabudhe D.M., Koppie T.M., Wood D.P., Jr., Mack P.C., Svatek R.S., Evans C.P., Hafez K.S., et al. Effect of Intravesical Instillation of Gemcitabine vs Saline Immediately Following Resection of Suspected Low-Grade Non-Muscle-Invasive Bladder Cancer on Tumor Recurrence: SWOG S0337 Randomized Clinical Trial. JAMA. 2018;319:1880–1888. doi: 10.1001/jama.2018.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witjes J.A., Bruins H.M., Cathomas R., Compérat E.M., Cowan N.C., Gakis G., Hernández V., Linares Espinós E., Lorch A., Neuzillet Y., et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur. Urol. 2021;79:82–104. doi: 10.1016/j.eururo.2020.03.055. [DOI] [PubMed] [Google Scholar]
- 5.Tran L., Xiao J.F., Agarwal N., Duex J.E., Theodorescu D. Advances in bladder cancer biology and therapy. Nat. Rev. Cancer. 2021;21:104–121. doi: 10.1038/s41568-020-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams S.B., Howard L.E., Foster M.L., Klaassen Z., Sieluk J., De Hoedt A.M., Freedland S.J. Estimated Costs and Long-term Outcomes of Patients With High-Risk Non-Muscle-Invasive Bladder Cancer Treated With Bacillus Calmette-Guérin in the Veterans Affairs Health System. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinton T.N., Chen Z., Wise H., Lenis A.T., Chavan S., Donoghue M.T.A., Almassi N., Chu C.E., Dason S., Rao P., et al. Genomic heterogeneity as a barrier to precision oncology in urothelial cancer. Cell Rep. 2022;41 doi: 10.1016/j.celrep.2022.111859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hoogstraten L.M.C., Vrieling A., van der Heijden A.G., Kogevinas M., Richters A., Kiemeney L.A. Global trends in the epidemiology of bladder cancer: challenges for public health and clinical practice. Nat. Rev. Clin. Oncol. 2023;20:287–304. doi: 10.1038/s41571-023-00744-3. [DOI] [PubMed] [Google Scholar]
- 9.Guo G., Sun X., Chen C., Wu S., Huang P., Li Z., Dean M., Huang Y., Jia W., Zhou Q., et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat. Genet. 2013;45:1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurst C.D., Platt F.M., Taylor C.F., Knowles M.A. Novel tumor subgroups of urothelial carcinoma of the bladder defined by integrated genomic analysis. Clin. Cancer Res. 2012;18:5865–5877. doi: 10.1158/1078-0432.CCR-12-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sjodahl G., Eriksson P., Patschan O., Marzouka N.A., Jakobsson L., Bernardo C., Lovgren K., Chebil G., Zwarthoff E., Liedberg F., Hoglund M. Molecular changes during progression from nonmuscle invasive to advanced urothelial carcinoma. Int. J. Cancer. 2020;146:2636–2647. doi: 10.1002/ijc.32737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamoun A., de Reyniès A., Allory Y., Sjödahl G., Robertson A.G., Seiler R., Hoadley K.A., Groeneveld C.S., Al-Ahmadie H., Choi W., et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2020;77:420–433. doi: 10.1016/j.eururo.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taber A., Christensen E., Lamy P., Nordentoft I., Prip F., Lindskrog S.V., Birkenkamp-Demtröder K., Okholm T.L.H., Knudsen M., Pedersen J.S., et al. Molecular correlates of cisplatin-based chemotherapy response in muscle invasive bladder cancer by integrated multi-omics analysis. Nat. Commun. 2020;11:4858. doi: 10.1038/s41467-020-18640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vandekerkhove G., Lavoie J.M., Annala M., Murtha A.J., Sundahl N., Walz S., Sano T., Taavitsainen S., Ritch E., Fazli L., et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat. Commun. 2021;12:184. doi: 10.1038/s41467-020-20493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soave A., Chun F.K.H., Hillebrand T., Rink M., Weisbach L., Steinbach B., Fisch M., Pantel K., Schwarzenbach H. Copy number variations of circulating, cell-free DNA in urothelial carcinoma of the bladder patients treated with radical cystectomy: a prospective study. Oncotarget. 2017;8:56398–56407. doi: 10.18632/oncotarget.17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rose Brannon A., Jayakumaran G., Diosdado M., Patel J., Razumova A., Hu Y., Meng F., Haque M., Sadowska J., Murphy B.J., et al. Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS. Nat. Commun. 2021;12:3770. doi: 10.1038/s41467-021-24109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson A.G., Kim J., Al-Ahmadie H., Bellmunt J., Guo G., Cherniack A.D., Hinoue T., Laird P.W., Hoadley K.A., Akbani R., et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell. 2017;171:540–556.e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindgren D., Sjödahl G., Lauss M., Staaf J., Chebil G., Lövgren K., Gudjonsson S., Liedberg F., Patschan O., Månsson W., et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earl J., Rico D., Carrillo-de-Santa-Pau E., Rodríguez-Santiago B., Méndez-Pertuz M., Auer H., Gómez G., Grossman H.B., Pisano D.G., Schulz W.A., et al. The UBC-40 Urothelial Bladder Cancer cell line index: a genomic resource for functional studies. BMC Genom. 2015;16:403. doi: 10.1186/s12864-015-1450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C. High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun H., Cao S., Mashl R.J., Mo C.K., Zaccaria S., Wendl M.C., Davies S.R., Bailey M.H., Primeau T.M., Hoog J., et al. Comprehensive characterization of 536 patient-derived xenograft models prioritizes candidates for targeted treatment. Nat. Commun. 2021;12:5086. doi: 10.1038/s41467-021-25177-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park S., Rong L., Owczarek T.B., Bernardo M.D., Shoulson R.L., Chua C.W., Kim J.Y., Lankarani A., Chakrapani P., Syed T., et al. Novel Mouse Models of Bladder Cancer Identify a Prognostic Signature Associated with Risk of Disease Progression. Cancer Res. 2021;81:5161–5175. doi: 10.1158/0008-5472.CAN-21-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pilala K.M., Papadimitriou M.A., Panoutsopoulou K., Barbarigos P., Levis P., Kotronopoulos G., Stravodimos K., Scorilas A., Avgeris M. Epigenetic regulation of MIR145 core promoter controls miR-143/145 cluster in bladder cancer progression and treatment outcome. Mol. Ther. Nucleic Acids. 2022;30:311–322. doi: 10.1016/j.omtn.2022.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caglic I., Panebianco V., Vargas H.A., Bura V., Woo S., Pecoraro M., Cipollari S., Sala E., Barrett T. MRI of Bladder Cancer: Local and Nodal Staging. J. Magn. Reson. Imag. 2020;52:649–667. doi: 10.1002/jmri.27090. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yurakh A.O., Ramos D., Calabuig-Fariñas S., López-Guerrero J.A., Rubio J., Solsona E., Romanenko A.M., Vozianov A.F., Pellin A., Llombart-Bosch A. Molecular and immunohistochemical analysis of the prognostic value of cell-cycle regulators in urothelial neoplasms of the bladder. Eur. Urol. 2006;50:506–515. doi: 10.1016/j.eururo.2006.03.027. discussion 515. [DOI] [PubMed] [Google Scholar]
- 27.Han G., Yang G., Hao D., Lu Y., Thein K., Simpson B.S., Chen J., Sun R., Alhalabi O., Wang R., et al. 9p21 loss confers a cold tumor immune microenvironment and primary resistance to immune checkpoint therapy. Nat. Commun. 2021;12:5606. doi: 10.1038/s41467-021-25894-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer G., Al-Ahmadie H., Schultz N., Hanrahan A.J., Ostrovnaya I., Balar A.V., Kim P.H., Lin O., Weinhold N., Sander C., et al. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J. Clin. Oncol. 2013;31:3133–3140. doi: 10.1200/JCO.2012.46.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raspollini M.R., Luque R.J., Menendez C.L., Bollito E., Brunelli M., Martignoni G., Montironi R., Cheng L., Blanca A., Baroni G., et al. T1 high-grade bladder carcinoma outcome: the role of p16, topoisomerase-IIα, survivin, and E-cadherin. Hum. Pathol. 2016;57:78–84. doi: 10.1016/j.humpath.2016.06.022. [DOI] [PubMed] [Google Scholar]
- 30.Breyer J., Wirtz R.M., Erben P., Worst T.S., Stoehr R., Eckstein M., Bertz S., Sikic D., Denzinger S., Burger M., et al. High CDKN2A/p16 and Low FGFR3 Expression Predict Progressive Potential of Stage pT1 Urothelial Bladder Carcinoma. Clin. Genitourin. Cancer. 2018;16:248–256.e2. doi: 10.1016/j.clgc.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Owczarek T.B., Kobayashi T., Ramirez R., Rong L., Puzio-Kuter A.M., Iyer G., Teo M.Y., Sánchez-Vega F., Wang J., Schultz N., et al. ARF Confers a Context-Dependent Response to Chemotherapy in Muscle-Invasive Bladder Cancer. Cancer Res. 2017;77:1035–1046. doi: 10.1158/0008-5472.CAN-16-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worst T.S., Weis C.A., Stöhr R., Bertz S., Eckstein M., Otto W., Breyer J., Hartmann A., Bolenz C., Wirtz R.M., Erben P. CDKN2A as transcriptomic marker for muscle-invasive bladder cancer risk stratification and therapy decision-making. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-32569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao R., Choi B.Y., Lee M.H., Bode A.M., Dong Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (p16(INK4a)) in Cancer. EBioMedicine. 2016;8:30–39. doi: 10.1016/j.ebiom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McLaughlin-Drubin M.E., Park D., Munger K. Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc. Natl. Acad. Sci. USA. 2013;110:16175–16180. doi: 10.1073/pnas.1310432110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vivo M., Fontana R., Ranieri M., Capasso G., Angrisano T., Pollice A., Calabrò V., La Mantia G. p14ARF interacts with the focal adhesion kinase and protects cells from anoikis. Oncogene. 2017;36:4913–4928. doi: 10.1038/onc.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie Y., Liu S., Lu W., Yang Q., Williams K.D., Binhazim A.A., Carver B.S., Matusik R.J., Chen Z. Slug regulates E-cadherin repression via p19Arf in prostate tumorigenesis. Mol. Oncol. 2014;8:1355–1364. doi: 10.1016/j.molonc.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohn R.L., Gasek N.S., Kuchel G.A., Xu M. The heterogeneity of cellular senescence: insights at the single-cell level. Trends Cell Biol. 2023;33:9–17. doi: 10.1016/j.tcb.2022.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soureas K., Papadimitriou M.A., Panoutsopoulou K., Pilala K.M., Scorilas A., Avgeris M. Cancer quiescence: non-coding RNAs in the spotlight. Trends Mol. Med. 2023;29:843–858. doi: 10.1016/j.molmed.2023.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Demaria M., O'Leary M.N., Chang J., Shao L., Liu S., Alimirah F., Koenig K., Le C., Mitin N., Deal A.M., et al. Cellular Senescence Promotes Adverse Effects of Chemotherapy and Cancer Relapse. Cancer Discov. 2017;7:165–176. doi: 10.1158/2159-8290.CD-16-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salam R., Saliou A., Bielle F., Bertrand M., Antoniewski C., Carpentier C., Alentorn A., Capelle L., Sanson M., Huillard E., et al. Cellular senescence in malignant cells promotes tumor progression in mouse and patient Glioblastoma. Nat. Commun. 2023;14:441. doi: 10.1038/s41467-023-36124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanaki Z., Voutsina A., Markou A., Pateras I.S., Potaris K., Avgeris M., Makrythanasis P., Athanasiadis E.I., Vamvakaris I., Patsea E., et al. Generation of Non-Small Cell Lung Cancer Patient-Derived Xenografts to Study Intratumor Heterogeneity. Cancers. 2021;13 doi: 10.3390/cancers13102446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rampias T., Karagiannis D., Avgeris M., Polyzos A., Kokkalis A., Kanaki Z., Kousidou E., Tzetis M., Kanavakis E., Stravodimos K., et al. The lysine-specific methyltransferase KMT2C/MLL3 regulates DNA repair components in cancer. EMBO Rep. 2019;20 doi: 10.15252/embr.201846821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayashi A., Ruppo S., Heilbrun E.E., Mazzoni C., Adar S., Yassour M., Rmaileh A.A., Shaul Y.D. GENI: A web server to identify gene set enrichments in tumor samples. Comput. Struct. Biotechnol. J. 2023;21:5531–5537. doi: 10.1016/j.csbj.2023.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papadimitriou M.A., Levis P., Kotronopoulos G., Stravodimos K., Avgeris M., Scorilas A. Preoperative Cell-Free DNA (cfDNA) in Muscle-Invasive Bladder Cancer Treatment Outcome. Clin. Chem. 2023;69:399–410. doi: 10.1093/clinchem/hvac218. [DOI] [PubMed] [Google Scholar]
- 45.Abaan O.D., Polley E.C., Davis S.R., Zhu Y.J., Bilke S., Walker R.L., Pineda M., Gindin Y., Jiang Y., Reinhold W.C., et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73:4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vickers A.J., Elkin E.B. Decision curve analysis: a novel method for evaluating prediction models. Med. Decis. Making. 2006;26:565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vickers A.J., Cronin A.M., Elkin E.B., Gonen M. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med. Inf. Decis. Making. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within the paper and its supplemental information. CNA and normalized RNA expression data along with the clinical data of the TCGA-BLCA project were acquired from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/projects/TCGA-BLCA) and the Memorial Sloan Kettering Cancer Center cBioPortal (http://www.cbioportal.org). Publicly available clinical, CNA, and normalized RNA expression data of the Lindgren et al. cohort were downloaded from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE32549). CNA data for the NCI-60 cell lines used in the study can be retrieved from CellMiner (https://discover.nci.nih.gov/cellminer/), while CNA data for UBC-40 cell lines are available in the Earl et al. study.19 CNA, mRNA expression profiling, and drug response data of BlCa cell lines were obtained through the DepMap portal (https://depmap.org/portal) of the Broad Institute. All data are available from the corresponding authors upon reasonable request.