Abstract
Glycoproteins homologous to the type I membrane glycoprotein B (gB) of herpes simplex virus 1 (HSV-1) are the most highly conserved glycoproteins within the family Herpesviridae and are present in members of each herpesvirus subfamily. In the alphaherpesvirus pseudorabies virus (PrV), gB is required for entry into target cells and for direct viral cell-to-cell spread. These processes, though related, appear to be distinct, and thus it was interesting to analyze whether they require different functions of gB. To this end, we established cell lines stably expressing different carboxy-terminally truncated versions of PrV gB by deleting either (i) one predicted intracytoplasmic α-helical domain encompassing putative YQRL and dileucine internalization signals, (ii) two predicted intracytoplasmic α-helical domains, (iii) the complete intracytoplasmic domain, or (iv) the intracytoplasmic domain and the transmembrane anchor region. Confocal laser scanning microscopy showed that gB derivatives lacking at least the last 29 amino acids (aa) localize close to the plasma membrane, while the full-length protein accumulates in intracellular aggregations. Trans-complementation studies with a gB-deleted PrV (PrV-gB−) demonstrated that the 29-aa truncated form lacking the putative internalization signals and the C-terminal α-helical domain (gB-008) was efficiently incorporated into PrV-gB− virions and efficiently complemented infectivity and cell-to-cell spread. Moreover, gB-008 exhibited an enhanced fusogenic activity. In contrast, gB proteins lacking both α-helical domains (gB-007), the complete intracytoplasmic domain, or the intracytoplasmic domain and transmembrane anchor were only inefficiently or not at all incorporated into PrV-gB− virions and did not complement infectivity. However, gB-007 was able to mediate cell-to-cell spread of PrV-gB−. Similar phenotypes were observed when virus recombinants expressing gB-008 or gB-007, respectively, instead of wild-type gB were isolated and analyzed. Thus, our data show that internalization of gB is not required for gB incorporation into virions nor for its function in either entry or cell-to-cell spread. Moreover, they indicate different requirements for gB in these membrane fusion processes.
Herpesvirus glycoproteins play important roles in virus infectivity. They are involved in mediating attachment and entry of free virions, virus maturation and egress, and direct viral cell-to-cell spread from infected to adjacent noninfected cells (42). In the alphaherpesviruses, four glycoproteins, gB, gD, and the gH-gL complex, have been shown to be required for entry of wild-type viruses. These proteins are also required for cell-to-cell spread of most alphaherpesviruses, indicating a close relationship between the membrane fusion processes during entry and cell-to-cell spread. However, in pseudorabies virus (PrV), the essential gD is required for entry but dispensable for cell-to-cell spread (31, 36), and varicella-zoster virus (VZV) completely lacks a gD homolog (8). Thus, although there are similarities between entry and cell-to-cell spread, in PrV, these processes can clearly be differentiated based on the requirement for gD.
Whereas the function of gD has recently at least partially been clarified by the identification of gD-binding cellular virus receptors (11, 46, 48), the role of gB and the gH-gL complex in entry is still unclear. It has been proposed that herpesvirus gB encompasses a region near the transmembrane domain which might act as a fusion peptide (37) similar to those present in the fusion-inducing glycoproteins of, e.g., influenza virus and human immunodeficiency virus (13). However, proof for this assumption is lacking.
gB homologs have been found in every herpesvirus analyzed (32), and they constitute the most highly conserved glycoprotein species in members of the family Herpesviridae. Most gB homologs form disulfide-linked homodimers which are posttranslationally cleaved into two subunits per monomer. Although this proteolytic cleavage is reminiscent of the activation of other viral fusion proteins (influenza virus, paramyxovirus, human immunodeficiency virus), proteolytic cleavage is not required for function of bovine herpesvirus 1 (BHV-1) gB (22) and herpes simplex virus type 1 (HSV-1) gB is not cleaved at all (41). Thus, the importance of the cleavage for gB function is unclear.
PrV gB is a type I membrane glycoprotein of 913 amino acids (aa), including a 58-aa putative signal peptide; three C-terminally located hydrophobic domains, of which the last one probably represents the transmembrane region; and a 93-aa intracytoplasmic C-terminal tail (38). Within the last 65 aa of this tail, two α-helical domains and putative dileucine and YQRL endocytosis signals (25) are located. PrV gB is proteolytically cleaved in the Golgi apparatus (47) into two subunits of 69 and 58 kDa (23, 49), respectively, which comprise roughly the amino-terminal and carboxy-terminal halves of the molecule (49) and remain linked via disulfide bonds. PrV mutants lacking gB could only be isolated on gB-expressing complementing cells, indicating an essential requirement for gB in viral replication (31, 36). In particular, in the absence of gB, extracellular virions are unable to penetrate into target cells unless membrane fusion is experimentally induced by polyethylene glycol (36). Moreover, in the absence of gB, PrV is unable to directly spread from cell to cell, which parallels the situation in other herpesviruses.
We wanted to functionally dissect the gB protein in order to identify specific domains which may differentiate PrV gB function in entry and cell-to-cell spread. To this end, we started by constructing different C-terminally truncated PrV gB versions. The intracytoplasmic tail of HSV-1 gB has been shown to be involved in the regulation of cell fusion, in that mutations within this part of the molecule can result in syncytial phenotypes (1, 4, 6) without loss of function (14). A similar regulatory function has been proposed to reside in the cytoplasmic tail of gB of human cytomegalovirus (HCMV) (3, 44).
MATERIALS AND METHODS
Viruses and cells.
All virus mutants are based on PrV strain Ka (PrV-Ka) (17). PrV-1112 carries a lacZ expression cassette inserted into the nonessential gG locus (27). In a multitude of in vitro and in vivo experiments, it was biologically identical to wild-type PrV.
For transient or stable expression, rabbit kidney cells (RK13) were transfected with 5 μg of plasmid DNA by using SuperFect Reagent (Qiagen, Hilden, Germany) under transfection conditions suggested by the manufacturer. To establish stable recombinants, cells were selected in medium containing 0.5 mg of G418 (Life Technologies, Karlsruhe, Germany) per ml.
Immunodetection.
Western blotting (immunoblotting) and radioimmunoprecipitation were performed as described previously (19, 23). Monoclonal antibody (MAb) b43-b5, directed against the amino-terminal subunit of PrV gB, was used in Western blot analysis. Anti-gB MAbs a80-c16 and A20-c26 were used for radioimmunoprecipitation and immunofluorescence, respectively. Anti-gE MAb A9-b15-28 was used as a control.
Confocal laser scanning microscopy.
For indirect immunofluorescence analysis, cells expressing the various gB proteins were seeded on coverslips in a six-well tissue culture plate, grown to confluency, and fixed with 3% paraformaldehyde for 20 min. For permeabilization, they were subsequently incubated in 3% paraformaldehyde–0.3% Triton X-100 for 10 min. After repeated washing with phosphate-buffered saline (PBS), the monolayer was incubated with the anti-gB MAb A20-c26 diluted 1:10 in PBS for 1 h. After thorough washing, antimouse IgG-fluorescein isothiocyanate (FITC) conjugate (Dako, Hamburg, Germany) was added for 45 min. Cells were then washed twice and counterstained with 10−6 M propidium iodide in PBS–10% glycerol. Finally, samples were analyzed by confocal laser scanning microscopy (LSM 510; Zeiss, Oberkochen, Germany).
Southern blot analysis and sequencing.
Sequencing of double-stranded plasmid DNA by the dideoxy chain termination method (40) was performed as described previously (18). Southern blot analysis of BamHI-restricted and BamHI-EcoRI double-digested viral DNA was done by standard procedures (21, 39).
Plaque assay, penetration, and one-step growth.
The plaque assay was performed essentially as described previously (29). Assays of penetration kinetics by low-pH inactivation of extracellular virus and one-step growth analysis were done as described previously (21, 26).
Endocytosis assay.
RK13 cells grown on coverslips were transfected with pcDNA3 (InVitrogen, Groningen, The Netherlands) or plasmids expressing wild-type gB or gB-008 by using SuperFect reagent (Qiagen) and incubated at 37°C. Twenty hours after transfection, cells were cooled on ice for 10 min and rinsed with cold PBS. Cells were then incubated on ice for 45 min with anti-gB MAb A20-c26 diluted 1:10 in PBS–5% bovine serum albumin. After three washes with cold PBS, cells were shifted to 37°C by the addition of prewarmed medium containing 100 μM chloroquine and incubated for 30 min at 37°C in an incubator. Thereafter, cells were fixed with 3% paraformaldehyde for 15 min and subsequently permeabilized with 3% paraformaldehyde–0.3% Triton X-100 for 15 min. After repeated washing with PBS, anti-mouse IgG-FITC conjugate was added for 45 min. Cells were then washed twice and counterstained with 10−6 M propidium iodide in PBS–10% glycerol. Samples were analyzed by confocal laser scanning microscopy (Zeiss; LSM 510).
Construction of gB genes that specify C-terminal truncations.
Open reading frames (ORFs) of all gB derivatives except gB-005 were amplified by PCR with Pfx-Platinum polymerase (Life Technologies) from plasmid CMV-gB (12) with the upstream primer 5′-TAACGGATCCATGCCCGCTGGTGGCGG-3′ and the downstream primers 5′-CAGAATTCCTACAGGGCGTCGGGGTCC-3′ for the complete gB ORF, 5′-CCGAATTCCTACCCGCTGTTCTTCTTGCGCG-3′ for gB-008, 5′-CCGAATTCCTAGGCCTCGTCCACGTCGCCTTC-3′ for gB-007, and 5′-CCGAATTCCTAGCGCGAGATGTGCCGGTAGGC-3′ for gB-006, respectively. In-frame start and stop codons are indicated by boldface letters. BamHI- and EcoRI restriction sites which were introduced for convenient cloning are underlined. After digestion with EcoRI and BamHI, PCR fragments were directly ligated into pcDNA3 (In Vitrogen). The plasmid encoding gB-005 was created by insertion of a BamHI-BstYI fragment excised from pMTgII (36) into pcDNA3.
Construction of PrV mutants.
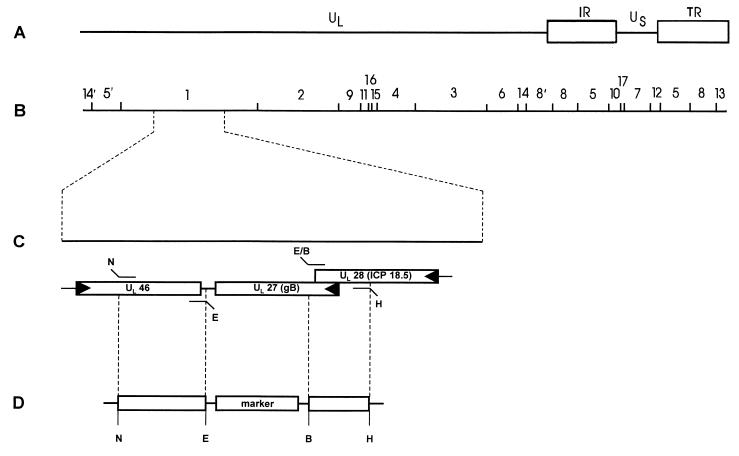
To generate a gB-null PrV mutant, a recombination vector based on pBluescript II SK− (Stratagene, Amsterdam, The Netherlands) was created carrying upstream ICP18.5 and downstream UL46 sequences for homologous recombination which were obtained by PCR from viral DNA.
The primers for the ICP18.5 fragment were 5′-CACAAAGCTTGGCCTCCTGCCGCACCTGAAG-3′ (nucleotides [nt] 2137 to 2157 of GenBank accession no. X14573) (30) and 5′-CACAGAATTCGGATCCTGAAGTTGCGCCCCTGC-3′ (nt 702 to 718 of GenBank accession no. M17321) (38). The primers for the UL46 fragment were 5′-ACGCGGCCGCACTGCCGCTTCCACCACCTGATC-3′ (nt 3496 to 3516 of GenBank accession no. AJ010303) (5) and 5′-ACGAATTCACAATAAACACGCACGCCGTGTGT-3′ (nt 4612 to 4628 of GenBank accession no. AJ010303) (5).
Green fluorescent protein (GFP) (16) or lacZ expression cassettes (27) were introduced between the recombination sequences to simplify selection and characterization of recombinants (Fig. 1). The resulting plasmids were named P021 and P022, respectively, and were transferred with full-length PrV-Ka DNA into RK13-gB cells by cotransfection with SuperFect Reagent as described above.
FIG. 1.
Construction of PrV mutants. (A) Schematic diagram of the PrV genome with BamHI restriction sites (B). The PrV genome consists of a unique long (UL) and a unique short (US) region. The latter is flanked by inverted repeats (IR, internal repeat; TR, terminal repeat). (C) Enlargement of BamHI fragment 1, which includes the relevant ORFs with transcriptional orientation indicated by arrows. Fragments were amplified by two-step PCR utilizing the primers as shown and inserted via restriction sites (E, EcoRI; B, BamHI; H, HindIII; N, NotI) into the recombination plasmid (D) based on pBluescript II SK−.
To obtain a gB rescuant and PrV mutants expressing truncated gB proteins, ORFs for wild-type gB, gB-008, and gB-007 were introduced into plasmid P021 (see above) instead of the marker gene cassette by using BamHI and EcoRI restriction sites. After cotransfection of the resulting plasmids and DNA of PrV-ΔgBGFP into RK13 cells, nonfluorescent plaques were picked and purified to homogeneity. Plaque isolates were designated as PrV-ΔgBGFPR, PrV-008, and PrV-007, respectively, and characterized by Southern blotting and radioimmunoprecipitation.
RESULTS
Isolation of PrV-ΔgBβ and PrV-ΔgBGFP.
Previously, we isolated a gB-deficient PrV mutant which carried a lacZ expression cassette inserted into a partially deleted UL27 (gB) gene (22, 36). Since in this mutant ca. one-third of the gB gene with 5′ and 3′ sequences of the UL27 ORF was still present, the rescue frequency on complementing gB-expressing cells was relatively high. To reduce rescue, a PrV mutant lacking most of the UL27 ORF was isolated. To this end, a recombination plasmid was constructed by PCR which contained of the 3′-end of the UL28 gene which resides upstream from and partially overlaps with the UL27 gene. Therefore, the very 5′ end of the UL27 ORF had to be retained not to affect the UL28 gene. However, at the 3′ end, no UL27-specific sequences were left, and the UL28 part was directly fused to the downstream UL46 gene (Fig. 1). In PrV, UL46 has been shown to be located adjacent to UL27 (5) due to an internal inversion within the UL region (2) encompassing the UL27 to UL44 genes (9). To facilitate isolation of virus mutants, lacZ or GFP expression cassettes were introduced. The respective virus mutants PrV-ΔgBβ and PrV-ΔgBGFP were plaque purified and propagated on cell line RK13-gB, which constitutively expresses the UL27 gene under control of the HCMV immediate-early promoter-enhancer (see below). On these cells, rescue-free gB-negative PrV stocks could be obtained.
Construction of cell lines expressing C-terminally truncated gB.
The C-terminal 98 aa of PrV gB contain two extended α-helical domains, designated I and II (10, 38) (Fig. 2). Domain II encompasses putative YQRL and dileucine endocytosis signals (25) (Fig. 2). To analyze the function of the cytoplasmic tail as a whole and of the different domains, C-terminally truncated gB derivatives were constructed by PCR by insertion of stop codons before the second α-helical domain (gB-008), at the 4th aa of the first α-helical domain (gB-007), and at the end of the putative transmembrane region (gB-006). A gB lacking the transmembrane region and the cytoplasmic tail was expressed from a BamHI-BstYI fragment excised from pMTgII (gB-005) (36). All gB derivatives were cloned into pcDNA3, which allows expression under control of the HCMV immediate-early promotor-enhancer, as was the PCR product containing the complete UL27 ORF (gB). After transfection into RK13 cells, stably expressing cell lines were established and analyzed.
FIG. 2.
Predicted amino acid sequence of the carboxy-terminal portion of PrV gB (10). C-terminal amino acid sequences of gB and truncated derivatives are shown. The hydrophobic stretch including the transmembrane domain is shaded grey, relevant α-helices are underlined and marked I and II, and the YQRL and dileucine endocytosis motifs are boxed.
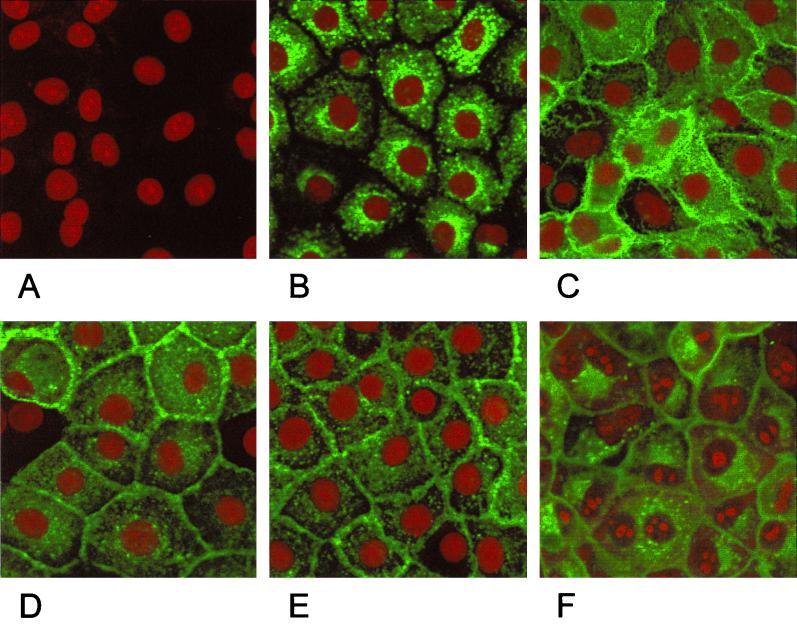
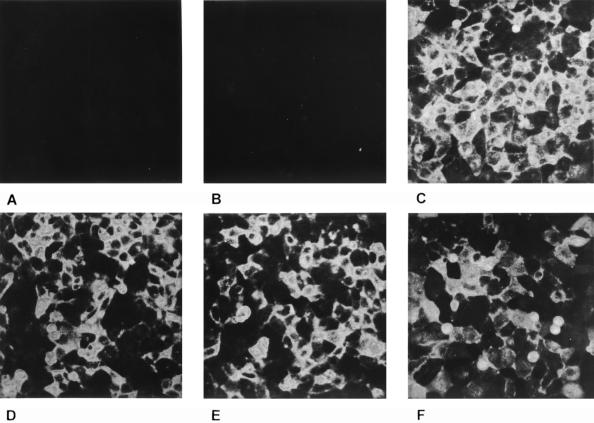
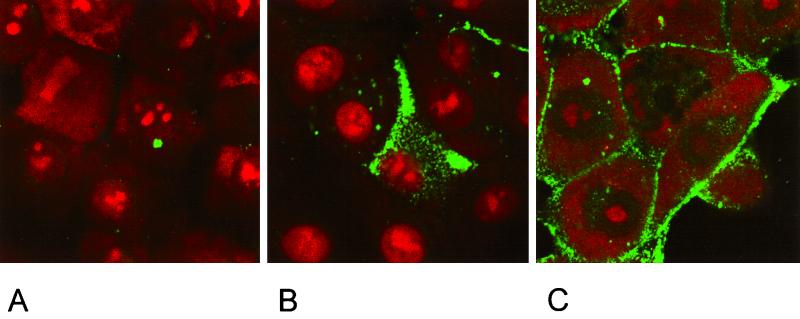
In indirect immunofluorescence using a gB-specific MAb, cells expressing full-length gB showed a punctate staining in the cytoplasm (Fig. 3B). No membrane fluorescence was observed in these cells (Fig. 4B). In contrast, cells expressing the C-terminally truncated gB-008, gB-007, and gB-006 showed a pattern of gB expression which suggested membrane localization of the protein (Fig. 3C, D, and E). This could be verified by immunofluorescence assays on nonpermeabilized cells (Fig. 4). Without permeabilization, only cells expressing gB-008 (Fig. 4C), gB-007 (Fig. 4D), and gB-006 (Fig. 4E) yielded a positive staining, whereas RK13-gB cells expressing wild-type gB (Fig. 4B) failed to react. Cells expressing gB-005 which lacks the putative transmembrane region showed a faint membrane-associated fluorescence (Fig. 4F) in addition to a punctate staining in the cytoplasm (Fig. 3F).
FIG. 3.
Subcellular localization of C-terminally truncated gB forms. Normal RK13 cells (A) and cells expressing wild-type gB (B), gB-008 (C), gB-007 (D), gB-006 (E), or gB-005 (F) were grown to confluency, fixed with 3% paraformaldehyde–0.3% Triton X-100, and incubated with anti-gB MAb A20-c26. Confocal laser scanning microscopy was performed after incubation with FITC-conjugated secondary antibodies and staining of nuclear DNA with propidium iodide.
FIG. 4.
Surface expression of C-terminally truncated gB. Normal RK13 cells (A) and cells expressing wild-type gB (B), gB-008 (C), gB-007 (D), gB-006 (E), or gB-005 (F) were grown to confluency, fixed with 3% paraformaldehyde, and incubated with the anti-gB MAb A20-c26. Fluorescence microscopy was performed after incubation with FITC-conjugated secondary antibodies.
Characterization of C-terminally truncated gB derivatives.
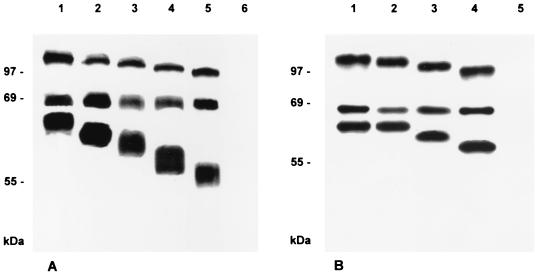
To check for correct expression of the respective gB protein, proteins were metabolically radiolabeled with Tran-S35-Label (ICN, Eschwege, Germany), immunoprecipitated, and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide) under reducing conditions. As shown in Fig. 5A, lane 1, from RK13-gB cells, the two gB subunits of 69 and 58 kDa were precipitated in addition to the uncleaved precursor of ca. 120 kDa. In cells expressing C-terminally truncated gB proteins, as expected, the N-terminal 69-kDa subunit remained intact, whereas the uncleaved precursor and the C-terminal 58-kDa subunit became successively smaller, paralleling the size of the deletion (Fig. 5A, lanes 2 to 5). As demonstrated by radioimmunoprecipitation (Fig. 5A) and fluorescence-activated cell sorter (FACS) analysis of permeabilized cells (data not shown), gB expression levels of the selected cell clones were comparable. To ascertain that wild-type gB is indeed endocytosed from the cell surface, an endocytosis assay was performed. As shown in Fig. 6B, wild-type gB is predominantly detected intracellularly after incubation on ice with an anti-gB MAb followed by a chase period at 37°C and detection by secondary antibody. This parallels previous results on gB endocytosis (43). In contrast, gB-008 (Fig. 6C) remains primarily membrane associated. No gB-specific signal was detected after transfection with control pcDNA3 (Fig. 6A).
FIG. 5.
Immunoprecipitation of C-terminally truncated gB. Lysates from metabolically radiolabeled RK13 cells expressing wild-type gB (A, lane 1), gB-008 (A, lane 2), gB-007 (A, lane 3), gB-006 (A, lane 4), or gB-005 (A, lane 5) and from normal RK13 cells (A, lane 6) or RK13 cells infected with PrV-1112 (B, lane 1), PrV-ΔgBGFPR (B, lane 2), PrV-008 (B, lane 3), PrV-007 (B, lane 4), or PrV-ΔgBβ (B, lane 5) were precipitated with the gB-specific MAb a80-c16. Precipitates were analyzed by SDS-PAGE under reducing conditions, and labeled protein was visualized by autoradiography.
FIG. 6.
Endocytosis assay. RK13 cells were transfected with pcDNA3 (A) or plasmids expressing wild-type gB (B) or gB-008 (C) for 20 h prior to an indirect immunofluorescence endocytosis assay (43) with anti-gB MAb A20-c26.
Plaque formation by PrV-gB− on cells expressing C-terminally truncated gB derivatives.
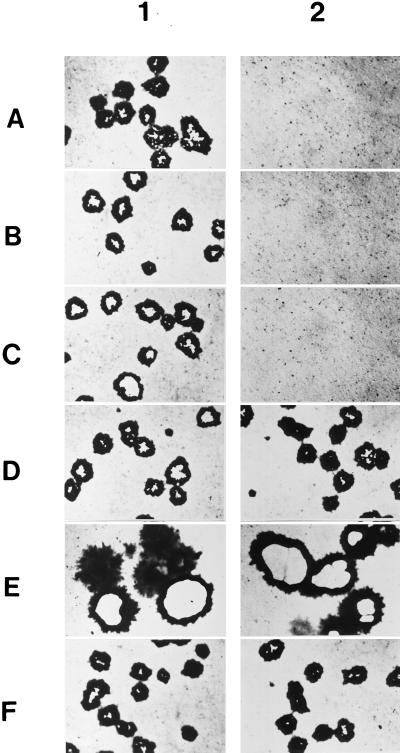
PrV gB is essential for direct viral cell-to-cell spread resulting in plaque formation. Thus, PrV-gB− does not form plaques on noncomplementing cells, but, after phenotypic complementation by propagation on gB-expressing cells, it is able to complete a single replicative cycle. To test for function of the C-terminally truncated gB derivatives in cell-to-cell spread, RK13-008, RK13-007, RK13-006, and RK13-005 cells were infected by phenotypically complemented PrV-ΔgBβ (Fig. 7, column 2) under plaque assay conditions and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) 2 days after infection. For comparison, RK13 and RK13-gB cells were similarly infected (Fig. 7A and F). As shown in Fig. 7, RK13-008 cells expressing gB lacking the putative endocytosis signals and second α-helical domain (Fig. 7E) and RK13-007 cells expressing gB lacking in addition the first α-helical domain (Fig. 7D) complemented plaque formation. Interestingly, on cells expressing gB-008, plaque size was increased approximately twofold over wild-type gB-expressing cells (compare Fig. 7E and F). In contrast, infection of RK13-006 cells which express a gB which lacks the complete cytoplasmic tail (Fig. 7C) did not result in plaque formation by PrV-ΔgBβ. As expected, RK13-005 cells which express gB lacking the transmembrane domain (Fig. 7B) and RK13 cells not expressing any gB (Fig. 7A) also did not complement plaque formation. As a control, all cells were infected with wild-type-like PrV-1112, which produced plaques on all cell lines (Fig. 7, column 1). On RK13-008 cells, plaque size was increased approximately twofold (Fig. 7E).
FIG. 7.
Infection of gB-expressing cells. Cells were infected with wild-type-like PrV-1112 (panel 1) or PrV-ΔgBβ (panel 2) and stained with X-Gal 2 days after infection. Whereas plaques developed on all cell lines infected with PrV-1112, only single infected cells were observed on RK13 (A), RK13-005 (B), and RK13-006 cells (C) infected with PrV-ΔgBβ. Note the increased plaque size on RK13-008 (E) and the formation of wild-type-like plaques on RK13-007 cells (D) following infection with PrV-ΔgBβ, which are similar to those produced by gB-negative PrV on full-length gB-expressing RK13-gB cells (F).
Incorporation of C-terminally truncated gB into virions.
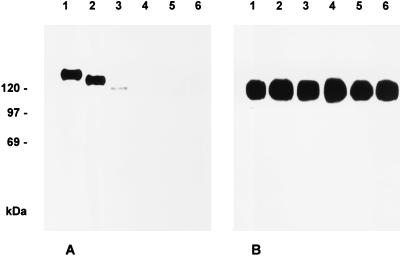
PrV gB is required for infectivity of free virions. Virus particles lacking gB are unable to enter target cells unless membrane fusion is induced experimentally by, e.g., polyethylene glycol. To test for functional complementation of the entry defect by the C-terminally truncated gB proteins, incorporation of mutated gBs into the virion was first analyzed. To this end, cell lines expressing mutated gB proteins were infected for 2 days at a multiplicity of infection (MOI) of 10 with phenotypically complemented PrV-ΔgBβ, and progeny virus particles were purified from the supernatant by sucrose gradient centrifugation and analyzed by Western blotting. As shown in Fig. 8A, wild-type gB and gB-008 and gB-007 were detected in purified virion preparations, although the signal from gB-007 virions was significantly weaker. In contrast, in virus progeny from cells expressing gB-006 or gB-005, no gB was found. As a control, gE was present in similar amounts in all virus preparations (Fig. 8B). Thus, absence of the putative endocytosis signals and both C-terminal α-helical domains does not totally preclude incorporation of gB into virions, but incorporation was highly inefficient in the absence of both α-helical domains.
FIG. 8.
Incorporation of C-terminally truncated gB into virions. RK13-gB (lane 1), RK13-008 (lane 2), RK13-007 (lane 3), RK13-006 (lane 4), RK13-005 (lane 5), and normal RK13 cells (lane 6) were infected with PrV-ΔgBβ at an MOI of 10. Samples of sucrose gradient-purified progeny virions were subjected to SDS-PAGE under nonreducing conditions followed by Western blot analysis with anti-gB MAb b43-b5 (A) or anti-gE MAb A9-b15-28 (B).
Complementation of infectivity of PrV-gB− by C-terminally truncated gB derivatives.
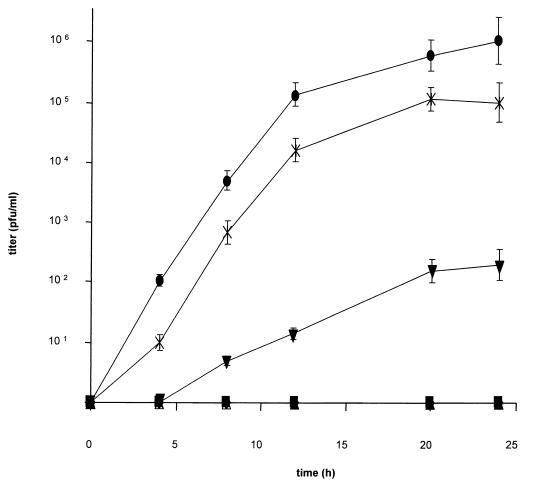
To assay functional complementation of the entry defect, one-step growth kinetics of phenotypically complemented PrV-ΔgBβ were established on cells expressing the different C-terminally truncated gB versions. As shown in Fig. 9, RK13 cells expressing gB-008 complemented infectivity of PrV-gB−, although ca. 10-fold less efficiently than RK13-gB cells. Thus, neither the putative endocytosis signals nor the C-terminal α-helical domain II is required for function of gB in entry. In contrast, gB-007 lacking in addition α-helical domain I complemented infectivity only to a very low but detectable extent, which parallels the inefficient incorporation into virus particles (see above). Cells expressing gB-006 with a complete deletion of the intracytoplasmic domain or gB-005 which lacks the transmembrane domain did not show any complementation, and neither did parental RK13 cells.
FIG. 9.
One-step growth. One-step growth kinetics of PrV-ΔgBβ in normal RK13 (⧫), RK13-005 (▴), RK13-006 (■), RK13-007 (▾), RK13-008 (×), and RK13-gB (●) cells were determined after infection at an MOI of 10. Titers are indicated in PFU per milliliter. Data are averages of three independent experiments. Vertical lines indicate standard deviations.
Growth properties of PrV recombinants expressing C-terminally truncated gB derivatives.
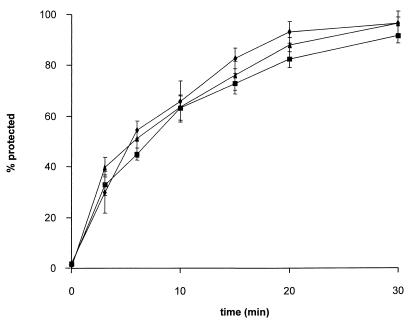
To test for the function of truncated gB derivatives within the viral background, PrV mutants were isolated by inserting mutated gB genes gB-008 and gB-007 into PrV-ΔgBGFP, thereby deleting the marker gene insert. In addition, PrV-ΔgBGFP was rescued by the wild-type gB gene. Expression of mutated forms of gB was verified by radioimmunoprecipitation of lysates of RK13 cells infected with wild-type, mutant, and rescued viruses (Fig. 5B). As expected, one-step growth kinetics and plaque formation did not differ compared to replication of PrV-ΔgBGFP on the cell lines stably expressing mutated gB proteins (data not shown). When constitutively expressed in RK13 cells, gB-008 exhibits a higher fusogenic activity, resulting in larger plaques (Fig. 7). Thus, we analyzed whether this increase in fusogenicity also influences early events during virus infection, i.e., penetration. As shown in Fig. 10, the penetration kinetics of PrV-008 were similar to those of the wild-type-like PrV-1112 and rescuant PrV-ΔgBGFPR, indicating different properties or an altered involvement of gB-008 in direct viral cell-to-cell spread and entry.
FIG. 10.
Penetration kinetics. The rate of entry of PrV-1112 (▴), PrV-ΔgBGFPR (■), and PrV-008 (⧫) into RK13 cells was determined by low-pH inactivation of extracellular virus at different time points after temperature shift. The percentage of plaques at a given time point compared to that in PBS-treated control plates is shown. Data are averages of three independent experiments. Vertical lines indicate standard deviations.
DISCUSSION
Glycoprotein B of PrV is essential for infection of target cells by free virions and for direct viral cell-to-cell spread. In this report, the function in these processes of different domains within the cytoplasmic tail of PrV gB was analyzed. To this end, mutated gB proteins were expressed and characterized.
While wild-type gB was mainly observed in intracellular accumulations and not at the plasma membrane, all gB derivatives containing truncations of at least the carboxy-terminal 29 aa of the cytoplasmic tail localized to the plasma membrane. This was true for gB expressed during virus infection as well as gB expressed without other viral proteins in stably transformed cell lines. Thus, within the last 29 aa of PrV gB, a signal appears to be located which precludes accumulation of gB in the plasma membrane. Within this region of gB, a dileucine motif and a YQRL motif, both potential signals for internalization (24, 25), are present. A similar YQRL endocytosis motif has been found in gB of HCMV (34) and VZV (T. Heinemann, N. Krudwig, and S. Hall, 24th Int. Herpesvirus Workshop, abstr. 6.019, 1999), which both have been shown to be internalized (34, 45; Heinemann et al., 24th Int. Herpesvirus Workshop). In contrast, HSV-1 gB has primarily been demonstrated at the plasma membrane (15, 35) despite the presence of a similar internalization motif.
As shown here, differences in intracellular localization alone do not appear to influence gB incorporation into and function in virions. The mutant gB protein lacking α-helical domain II, including the putative endocytosis motifs, is functional in complementing a gB-negative PrV mutant in trans after expression in a stably transformed cell line as well as after expression from the PrV genome, although it results in a ca. 10-fold reduction in titer. Thus, preferential presence in the plasma membrane is not required for function of wild-type gB, and internalization is not required for incorporation and function of C-terminally truncated gB.
However, removal of the last 29 aa of gB altered its fusogenicity (20), a result which parallels similar findings in HSV-1 (1, 6). Therefore, within the C-terminal α-helical domain, an element that regulates fusogenic activity of gB homologs appears to be located. Whether this domain is directly involved in controlling fusion or whether mutation of this region results in a conformational change altering the fusion capacity of gB is unclear. However, the conservation of this phenotype indicates a common pathway for fusion regulation or deregulation via gB. Interestingly, the increase in fusogenic properties of gB-008 did not influence gB function during penetration. Thus, the penetration kinetics of recombinant PrV expressing gB-008 were indistinguishable from those of wild-type PrV or a gB rescuant.
Truncation of the last 60 aa in gB-007, which results in elimination of both predicted α-helices, resulted in an inefficient incorporation of gB into virions, despite the presence of the mutant protein at the plasma membrane. This inefficient incorporation parallels and probably explains the drastically reduced infectivity of mutant viruses carrying this protein either supplied in trans by stably expressing cell lines or in cis when expressed from the genome of a corresponding virus recombinant. However, although only at a very limited level, but clearly above background, the mutated gB still rescued a certain level of infectivity from the gB-negative PrV mutant. In contrast, a similarly mutated HSV-1 gB did not exhibit any complementation capacity for a gB-deleted HSV-1 mutant (1, 7). Whether this is a virus-specific difference or can be explained by different assay systems is unclear at present.
Surprisingly, neither kinetics of plaque formation nor plaque sizes were found to be different when either wild-type gB or gB-007 was used for complementation. Thus, the absence of the last two α-helical domains, while severely impairing rescue of infectivity of free virions, did not interfere with gB function in direct viral cell-to-cell spread. This demonstrates different requirements for gB in membrane fusion during entry and during cell-to-cell spread. Obviously, efficient incorporation into virions is not required for normal levels of direct viral cell-to-cell spread. Based on the activity of complement-independent neutralizing antibodies, it has been postulated that different domains of gB may be involved in penetration and cell-to-cell spread (28). In PrV gB, α-helical domain II, which includes the internalization signals, appears to modulate and/or regulate the fusogenic activity of gB, whereas α-helical domain I could be responsible for efficient incorporation of gB into virions. Interestingly, neither of the two domains appears to be required for gB function in promoting fusion events during direct viral cell-to-cell spread.
Further truncation of the carboxy-terminal end of gB until the hydrophobic membrane anchor (gB-006) or removal of the complete hydrophobic stretch including the membrane anchor (gB-005) resulted in gB species that support neither infectivity nor plaque formation. Thus, within the 28 aa present in gB-007 and absent in gB-006, a region is located which is important for gB function in direct viral cell-to-cell spread. This is interesting, since gB-006 is still predominantly found at the plasma membrane and thus may participate in membrane fusion processes. In contrast, gB-005 is secreted in large amounts from expressing cells concurring with the deletion of the predicted transmembrane anchor. Nevertheless, a significant amount of gB-005 is detectable near the plasma membrane by immunofluorescence. Whether this is caused by interstitial accumulation of released gB, membrane-association of gB via hydrophobic segments despite absence of the predicted membrane anchor, or the interaction of soluble gB with a cellular membrane component is unclear. However, neither of our gB-expressing cell lines exhibits the phenotype seen for gD-expressing cells, i.e., interference with viral infection. For gD-mediated interference, sequestration of cellular receptors has been proposed as the most likely explanation, although formal proof of this hypothesis is still lacking (33). The absence of interference in our gB-expressing cells despite the predominantly plasma membrane localization of the mutated proteins argues against the existence of cellular gB receptors which are saturable by membrane-bound or membrane-associated gB, at least in a wild-type PrV infection.
In summary, we identified several distinct domains within the cytoplasmic tail of PrV gB which are individually involved in different processes during virus infection, including intracellular trafficking, incorporation of gB into virions, and membrane fusion during entry and direct cell-to-cell spread. Whereas the α-helical domain II including dileucine and YQRL internalization signals is not required for virion localization of gB or its function during entry or cell-to-cell spread, in the absence of α-helical domain I, incorporation into virions and function during entry are inefficient. Surprisingly, gB function in direct cell-to-cell spread was not affected by this deletion. In the absence of the complete cytoplasmic tail, however, function of gB in either entry or direct cell-to-cell spread was abolished.
ACKNOWLEDGMENTS
This study was supported by the Deutsche Forschungsgemeinschaft (grant Me 854/4-1).
We thank N. Osterrieder for help with the confocal laser scanning analyses.
REFERENCES
- 1.Baghian A, Huang L, Newman S, Jayachandra S, Kousoulas K G. Truncation of the carboxy-terminal 28 amino acids of glycoprotein B specified by herpes simplex virus type 1 mutant amb1511-7 causes extensive cell fusion. J Virol. 1993;67:2396–2401. doi: 10.1128/jvi.67.4.2396-2401.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Porat T, Veach R A, Ihara S. Localization of the regions of homology between the genomes of herpes simplex virus type 1 and pseudorabies virus. Virology. 1983;127:194–204. doi: 10.1016/0042-6822(83)90383-5. [DOI] [PubMed] [Google Scholar]
- 3.Bold S, Ohlin M, Garten W, Radsak K. Structural domains involved in human cytomegalovirus glycoprotein B-mediated cell-cell fusion. J Gen Virol. 1996;77:2297–2302. doi: 10.1099/0022-1317-77-9-2297. [DOI] [PubMed] [Google Scholar]
- 4.Bond V C, Person S, Warner S C. The isolation and characterization of mutants of herpes simplex virus type 1 that induce cell fusion. J Gen Virol. 1982;61:245–254. doi: 10.1099/0022-1317-61-2-245. [DOI] [PubMed] [Google Scholar]
- 5.Bras F, Dezélée S, Simonet B, Nguyen X, Vende P, Flamand A, Masse M J. The left border of the genomic inversion of pseudorabies virus contains genes homologous to the UL46 and UL47 genes of herpes simplex virus type 1, but no UL45 gene. Virus Res. 1999;60:29–40. doi: 10.1016/s0168-1702(98)00146-4. [DOI] [PubMed] [Google Scholar]
- 6.Bzik D J, Fox B A, DeLuca N A, Person S. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology. 1984;137:185–190. doi: 10.1016/0042-6822(84)90022-9. [DOI] [PubMed] [Google Scholar]
- 7.Cai W, Person S, Warner S C, Zhou J, DeLuca N A. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J Virol. 1987;61:714–721. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison A J, Scott J E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986;67:1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- 9.Dezélée B, Bras F, Vende P, Simonet B, Nguyen X, Flamand A, Masse M J. The Bam HI fragment 9 contains genes homologous to the UL24, UL25, UL26 and UL26.5 genes of herpes simplex virus type 1. Virus Res. 1996;42:27–39. doi: 10.1016/0168-1702(96)01293-2. [DOI] [PubMed] [Google Scholar]
- 10.Garnier J, Gibrat J-F, Robson B. GOR secondary structure prediction method, version IV. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 11.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 12.Gerdts V, Jöns A, Mettenleiter T C. Potency of an experimental DNA vaccine against Aujeszky's disease in pigs. Vet Microbiol. 1999;66:1–13. doi: 10.1016/s0378-1135(98)00300-9. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 14.Huff V, Cai W, Glorioso J C, Levine M. The carboxy-terminal 41 amino acids of herpes simplex virus type 1 glycoprotein B are not essential for production of infectious virus particles. J Virol. 1988;62:4403–4406. doi: 10.1128/jvi.62.11.4403-4406.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson D C, Spear P G. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J Virol. 1982;43:1102–1112. doi: 10.1128/jvi.43.3.1102-1112.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jöns A, Mettenleiter T C. Green fluorescent protein expressed by recombinant pseudorabies virus as an in vivo marker for viral replication. J Virol Methods. 1997;66:283–292. doi: 10.1016/s0166-0934(97)00065-7. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan A S, Vatter A. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959;13:78–92. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 18.Klupp B G, Mettenleiter T C. Sequence and expression of the glycoprotein gH gene of pseudorabies virus. Virology. 1991;182:732–741. doi: 10.1016/0042-6822(91)90614-h. [DOI] [PubMed] [Google Scholar]
- 19.Klupp B G, Fuchs W, Weiland E, Mettenleiter T C. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J Virol. 1997;71:7687–7695. doi: 10.1128/jvi.71.10.7687-7695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klupp B G, Nixdorf R, Mettenleiter T C. Pseudorabies virus glycoprotein M inhibits membrane fusion. J Virol. 2000;74:6760–6768. doi: 10.1128/jvi.74.15.6760-6768.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopp A, Mettenleiter T C. Stable rescue of a glycoprotein gII deletion mutant of pseudorabies virus by glycoprotein gI of bovine herpesvirus 1. J Virol. 1992;66:2754–2762. doi: 10.1128/jvi.66.5.2754-2762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopp A, Blewett E, Misra V, Mettenleiter T C. Proteolytic cleavage of bovine herpesvirus 1 (BHV-1) glycoprotein gB is not necessary for its function in BHV-1 or pseudorabies virus. J Virol. 1994;68:1667–1674. doi: 10.1128/jvi.68.3.1667-1674.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukàcs N, Thiel H J, Mettenleiter T C, Rziha H-J. Demonstration of three major species of pseudorabies virus glycoproteins and identification of a disulfide-linked glycoprotein complex. J Virol. 1985;53:166–173. doi: 10.1128/jvi.53.1.166-173.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marks M, Ohno H, Kirchhausen T, Bonifacino J S. Protein sorting by tyrosine-based signals: adapting to the Ys and wherefores. Cell Biol. 1997;7:124–128. doi: 10.1016/S0962-8924(96)10057-X. [DOI] [PubMed] [Google Scholar]
- 25.Marsh M, McMahon H T. The structural era of endocytosis. Science. 1999;285:215–220. doi: 10.1126/science.285.5425.215. [DOI] [PubMed] [Google Scholar]
- 26.Mettenleiter T C. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology. 1989;171:623–625. doi: 10.1016/0042-6822(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 27.Mettenleiter T C, Rauh I. A glycoprotein gX-β-galactosidase fusion gene as insertional marker for rapid identification of pseudorabies virus mutants. J Virol Methods. 1990;30:55–65. doi: 10.1016/0166-0934(90)90043-f. [DOI] [PubMed] [Google Scholar]
- 28.Navarro D, Paz P, Pereira L. Domains of herpes simplex virus I glycoprotein B that function in virus penetration, cell-to-cell spread, and cell fusion. Virology. 1992;186:99–112. doi: 10.1016/0042-6822(92)90064-v. [DOI] [PubMed] [Google Scholar]
- 29.Nixdorf R, Schmidt J, Karger A, Mettenleiter T C. Infection of Chinese hamster ovary cells by pseudorabies virus. J Virol. 1999;73:8019–8026. doi: 10.1128/jvi.73.10.8019-8026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pederson N E, Enquist L W. The nucleotide sequence of a pseudorabies virus gene similar to ICP18.5 of herpes simplex virus type 1. Nucleic Acids Res. 1989;17:3597. doi: 10.1093/nar/17.9.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peeters B, de Wind N, Hooisma M, Wagenaar F, Gielkens A, Moormann R. Pseudorabies virus envelope glycoproteins gp50 and gII are essential for virus penetration, but only gII is involved in membrane fusion. J Virol. 1992;66:894–905. doi: 10.1128/jvi.66.2.894-905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pereira L. Function of glycoprotein B homologues of the family herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- 33.Petrovskis E A, Meyer A L, Post L E. Reduced yield of infectious pseudorabies virus and herpes simplex virus from cell lines producing viral glycoprotein gp50. J Virol. 1988;62:2196–2199. doi: 10.1128/jvi.62.6.2196-2199.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radsak K, Eickmann M, Mockenhaupt T, Bogner E, Kern H, Eis-Hübinger A, Reschke M. Retrieval of human cytomegalovirus glycoprotein B from infected cell surface for virus envelopment. Arch Virol. 1996;141:557–572. doi: 10.1007/BF01718317. [DOI] [PubMed] [Google Scholar]
- 35.Rasile L, Ghosh K, Raviprakash K, Ghosh H P. Effects of deletions in the carboxy-terminal hydrophobic region of herpes simplex virus glycoprotein gB on intracellular transport and membrane anchoring. J Virol. 1993;67:4856–4866. doi: 10.1128/jvi.67.8.4856-4866.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rauh I, Mettenleiter T C. Pseudorabies virus glycoproteins gII and gp50 are essential for virus penetration. J Virol. 1991;65:5348–5356. doi: 10.1128/jvi.65.10.5348-5356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reschke M, Reis B, Nöding K, Rohsiepe D, Richter A, Mockenhaupt T, Garten W, Radsak K. Constitutive expression of human cytomegalovirus glycoprotein B (gpUL55) with mutagenized carboxy-terminal hydrophobic domains. J Gen Virol. 1995;76:113–122. doi: 10.1099/0022-1317-76-1-113. [DOI] [PubMed] [Google Scholar]
- 38.Robbins A K, Dorney D J, Wathen M W, Whealy M E, Gold C, Watson R J, Holland L E, Weed S D, Levine M, Glorioso J C, Enquist L W. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J Virol. 1987;61:2691–2701. doi: 10.1128/jvi.61.9.2691-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarmiento M, Spear P G. Membrane proteins specified by herpes simplex viruses. IV. Conformation of the virion glycoprotein designated VP7(B2) J Virol. 1979;29:1159–1167. doi: 10.1128/jvi.29.3.1159-1167.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 43.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tugizov S, Wang Y, Quadri I, Navarro D, Maidji E, Pereira L. Mutated forms of human cytomegalovirus glycoprotein B are impaired in inducing syncytium formation. Virology. 1995;209:580–591. doi: 10.1006/viro.1995.1290. [DOI] [PubMed] [Google Scholar]
- 45.Tugizov S, Maidji E, Xiao J, Zheng Z, Pereira L. Human cytomegalovirus glycoprotein B contains autonomous determinants for vectorial targeting to apical membranes of polarized epithelial cells. J Virol. 1998;72:7374–7386. doi: 10.1128/jvi.72.9.7374-7386.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 47.Whealy M E, Card J P, Meade R P, Robbins A K, Enquist L W. Effect of brefeldin A on alphaherpesvirus membrane protein glycosylation and virus egress. J Virol. 1991;65:1066–1081. doi: 10.1128/jvi.65.3.1066-1081.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, Ponce de Leon M, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wölfer U, Kruft V, Sawitzky D, Hampl H, Wittmann-Liebold B, Habermehl K-O. Processing of pseudorabies virus glycoprotein gII. J Virol. 1990;64:3122–3125. doi: 10.1128/jvi.64.6.3122-3125.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]