In the title compound, the pyridyl ring and the Bdan (dan = 1,8-diaminonaphto) group subtend a dihedral angle of 24.57 (5)°. In the crystal, the molecules make  (28) hydrogen-bonding networks around the fourfold inversion axis, giving a cyclic tetramer. The molecules form columnar stacks along the c axis.
(28) hydrogen-bonding networks around the fourfold inversion axis, giving a cyclic tetramer. The molecules form columnar stacks along the c axis.
Keywords: crystal structure, pyridine derivative, tetrameric structure, dan
Abstract
The title compound, C15H12BN3, is a type of diazaborinane featuring substitution at 1, 2, and 3 positions in the nitrogen–boron six-membered heterocycle. It is comprised of two almost planar units, the pyridyl ring and the Bdan (dan = 1,8-diaminonaphtho) group, which subtend a dihedral angle of 24.57 (5)°. In the crystal, the molecules are linked into R44(28) hydrogen-bonding networks around the fourfold inversion axis, giving cyclic tetramers. The molecules form columnar stacks along the c axis.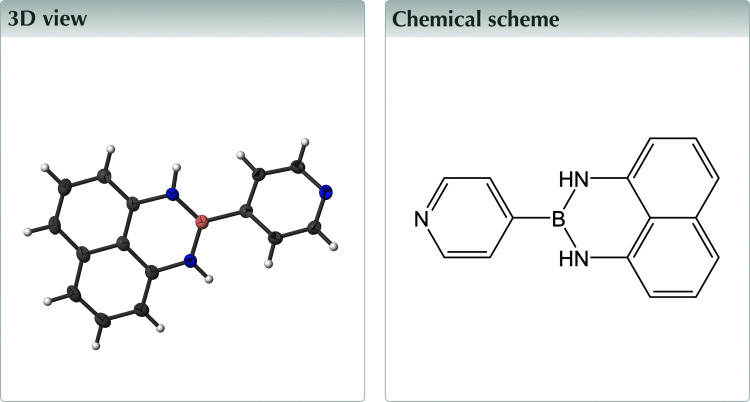
Structure description
The title compound, C15H12BN3, is a type of diazaborinane that is substituted at the 1, 2, and 3 positions in the nitrogen–boron six-membered heterocycle. Recently, diazaborinanes have been found to stabilize organic radicals (LaPorte et al., 2023 ▸).
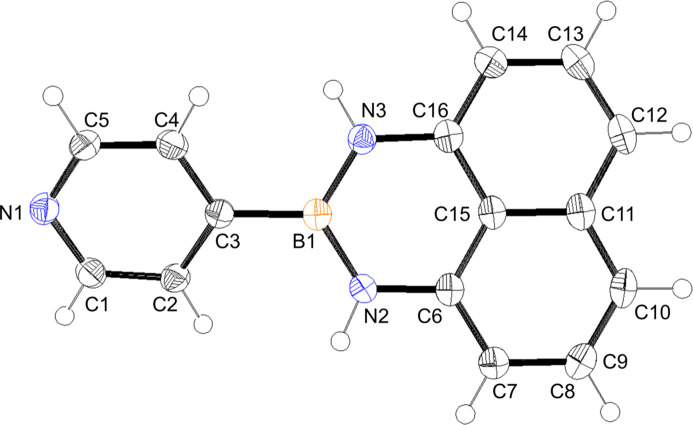
The title molecule (Fig. 1 ▸) is comprised of two almost planar units, the N1/C1–C5 pyridyl ring and the N2/N3/C6–C15/B1 group, which subtend a dihedral angle of 24.57 (5)°. This is slightly larger than those in related compounds that have almost planar structures (Akerman et al., 2011 ▸; Slabber et al., 2011 ▸).
Figure 1.
The title compound with the atom-numbering scheme. Displacement ellipsoids are drawn at the 50% probability level and H atoms are shown as small spheres of arbitrary radii.
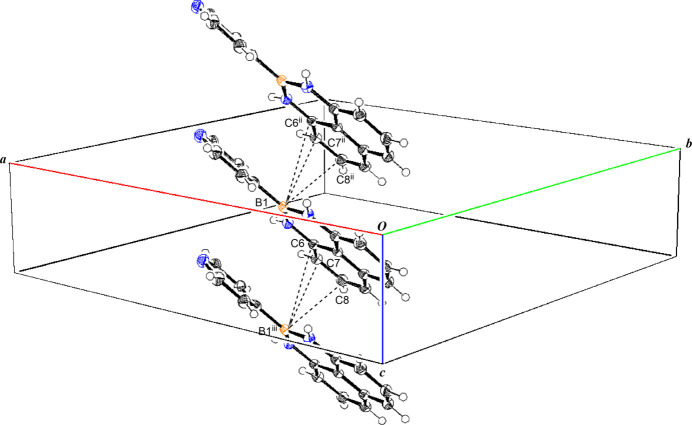
In the crystal, the molecules make 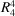 (28) hydrogen-bonding (Table 1 ▸) networks around the fourfold inversion axis, giving a cyclic tetramer as shown in Fig. 2 ▸. The formation of this tetrameric structure is thought to increase the dihedral angle. The molecules also stack along the c axis, as shown in Fig. 3 ▸, forming columnar stacks in which the B1⋯C6ii, B1⋯C7ii, and B1⋯C8ii distances are 3.656 (3), 3.513 (3) and 3.573 (3) Å, respectively [symmetry code:(ii) x, y, z − 1].
(28) hydrogen-bonding (Table 1 ▸) networks around the fourfold inversion axis, giving a cyclic tetramer as shown in Fig. 2 ▸. The formation of this tetrameric structure is thought to increase the dihedral angle. The molecules also stack along the c axis, as shown in Fig. 3 ▸, forming columnar stacks in which the B1⋯C6ii, B1⋯C7ii, and B1⋯C8ii distances are 3.656 (3), 3.513 (3) and 3.573 (3) Å, respectively [symmetry code:(ii) x, y, z − 1].
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯N1i | 0.93 (2) | 2.21 (2) | 3.113 (2) | 162 (2) |
Symmetry code: (i) 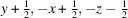 .
.
Figure 2.
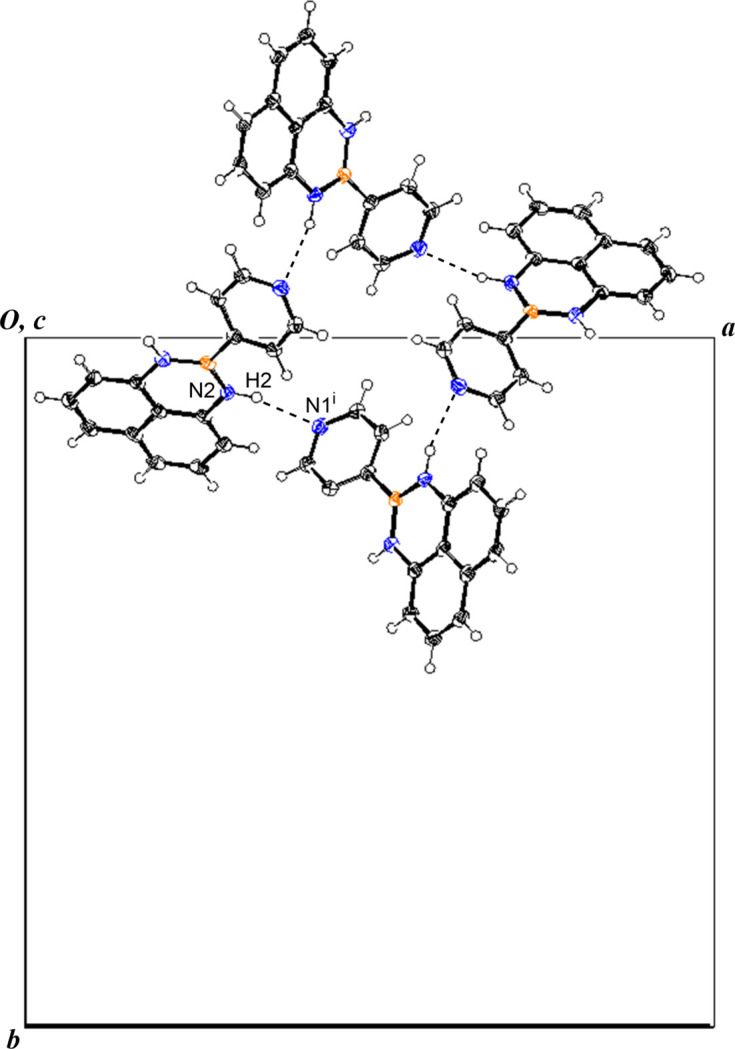
The hydrogen-bonding network of the title compound. [Symmetry code: (i) y + 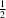 , −x +
, −x + 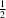 , −z −
, −z − 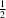 .]
.]
Figure 3.
The stacking structure along the c axis. [Symmetry codes:(ii) x, y, z − 1; (iii) x, y, z + 1.]
Synthesis and crystallization
The title compound was prepared according to the literature method (Hashimoto & Okuno, 2024 ▸). Single crystals of sufficient quality were obtained by recrystallization from chloroform solution.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C15H12BN3 |
| M r | 245.09 |
| Crystal system, space group | Tetragonal, I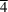
|
| Temperature (K) | 100 |
| a, c (Å) | 21.5659 (3), 5.0863 (1) |
| V (Å3) | 2365.58 (8) |
| Z | 8 |
| Radiation type | Cu Kα |
| μ (mm−1) | 0.65 |
| Crystal size (mm) | 0.20 × 0.05 × 0.05 |
| Data collection | |
| Diffractometer | XtaLAB Synergy R, DW system, HyPix |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2024 ▸) |
| Tmin, Tmax | 0.807, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 7772, 2268, 2125 |
| R int | 0.036 |
| (sin θ/λ)max (Å−1) | 0.626 |
| Refinement | |
| R[F2 > 2σ(F2)], wR(F2), S | 0.031, 0.083, 1.06 |
| No. of reflections | 2268 |
| No. of parameters | 180 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.15, −0.16 |
| Absolute structure | Flack x determined using 840 quotients [(I+)−(I−)]/[(I+)+(I−)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | −0.2 (3) |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314624006151/bx4031sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314624006151/bx4031Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314624006151/bx4031Isup3.cml
CCDC reference: 2364937
Additional supporting information: crystallographic information; 3D view; checkCIF report
full crystallographic data
\ 2-(Pyridin-4-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]\ diazaborinine . Crystal data
| C15H12BN3 | Dx = 1.376 Mg m−3 |
| Mr = 245.09 | Cu Kα radiation, λ = 1.54184 Å |
| Tetragonal, I4 | Cell parameters from 5132 reflections |
| a = 21.5659 (3) Å | θ = 2.9–75.0° |
| c = 5.0863 (1) Å | µ = 0.65 mm−1 |
| V = 2365.58 (8) Å3 | T = 100 K |
| Z = 8 | Block, clear colourless |
| F(000) = 1024 | 0.2 × 0.05 × 0.05 mm |
\ 2-(Pyridin-4-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]\ diazaborinine . Data collection
| XtaLAB Synergy R, DW system, HyPix diffractometer | 2268 independent reflections |
| Radiation source: Rotating-anode X-ray tube, Rigaku (Cu) X-ray Source | 2125 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.036 |
| Detector resolution: 10.0000 pixels mm-1 | θmax = 75.0°, θmin = 2.9° |
| ω scans | h = −25→26 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2024) | k = −26→27 |
| Tmin = 0.807, Tmax = 1.000 | l = −5→6 |
| 7772 measured reflections |
\ 2-(Pyridin-4-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]\ diazaborinine . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.031 | w = 1/[σ2(Fo2) + (0.0502P)2 + 0.1789P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.083 | (Δ/σ)max < 0.001 |
| S = 1.06 | Δρmax = 0.15 e Å−3 |
| 2268 reflections | Δρmin = −0.15 e Å−3 |
| 180 parameters | Absolute structure: Flack x determined using 840 quotients [(I+)-(I-)]/[(I+)+(I-)] (Parsons et al., 2013) |
| 0 restraints | Absolute structure parameter: −0.2 (3) |
| Primary atom site location: structure-invariant direct methods |
\ 2-(Pyridin-4-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]\ diazaborinine . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. The positions of the N-bound H atoms were obtained from difference Fourier maps and were refined isotropically. The C-bound H atoms were placed at ideal positions and were refined as riding on their parent C atoms. Uiso(H) values of the H atoms were set at 1.2Ueq(parent atom for Csp2). |
\ 2-(Pyridin-4-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]\ diazaborinine . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| N3 | 0.20041 (7) | 0.03150 (8) | −0.0041 (4) | 0.0233 (4) | |
| C7 | 0.28868 (9) | 0.15497 (9) | 0.5132 (4) | 0.0252 (4) | |
| H7 | 0.3324 | 0.1604 | 0.5071 | 0.030* | |
| C1 | 0.39265 (9) | −0.01777 (9) | −0.5063 (4) | 0.0270 (4) | |
| H1 | 0.4315 | −0.0032 | −0.5689 | 0.032* | |
| N2 | 0.29394 (8) | 0.07934 (7) | 0.1587 (3) | 0.0217 (3) | |
| C13 | 0.10022 (9) | 0.05895 (9) | 0.1820 (4) | 0.0267 (4) | |
| H13 | 0.0797 | 0.0324 | 0.0611 | 0.032* | |
| C10 | 0.15895 (9) | 0.14042 (9) | 0.5376 (4) | 0.0255 (4) | |
| C15 | 0.19455 (9) | 0.10642 (8) | 0.3491 (4) | 0.0219 (4) | |
| C5 | 0.31573 (9) | −0.08947 (9) | −0.5129 (4) | 0.0273 (4) | |
| H5 | 0.2989 | −0.1270 | −0.5796 | 0.033* | |
| C6 | 0.26012 (9) | 0.11421 (8) | 0.3414 (4) | 0.0218 (4) | |
| C14 | 0.16387 (9) | 0.06513 (9) | 0.1728 (4) | 0.0231 (4) | |
| C11 | 0.09363 (10) | 0.13134 (9) | 0.5452 (4) | 0.0288 (5) | |
| H11 | 0.0693 | 0.1527 | 0.6721 | 0.035* | |
| B1 | 0.26579 (10) | 0.03753 (10) | −0.0181 (4) | 0.0220 (4) | |
| C3 | 0.30411 (9) | −0.00146 (9) | −0.2242 (4) | 0.0220 (4) | |
| C4 | 0.28147 (9) | −0.05701 (9) | −0.3271 (4) | 0.0262 (4) | |
| H4 | 0.2426 | −0.0727 | −0.2700 | 0.031* | |
| C12 | 0.06564 (9) | 0.09191 (9) | 0.3700 (4) | 0.0286 (5) | |
| H12 | 0.0219 | 0.0868 | 0.3758 | 0.034* | |
| N1 | 0.37087 (8) | −0.07118 (8) | −0.6036 (3) | 0.0268 (4) | |
| C2 | 0.36184 (9) | 0.01756 (9) | −0.3191 (4) | 0.0246 (4) | |
| H2A | 0.3801 | 0.0547 | −0.2555 | 0.030* | |
| C9 | 0.19006 (10) | 0.18127 (9) | 0.7109 (4) | 0.0277 (5) | |
| H9 | 0.1670 | 0.2039 | 0.8380 | 0.033* | |
| C8 | 0.25310 (10) | 0.18841 (9) | 0.6969 (4) | 0.0279 (4) | |
| H8 | 0.2732 | 0.2164 | 0.8132 | 0.033* | |
| H3 | 0.1804 (12) | 0.0058 (12) | −0.125 (6) | 0.042 (7)* | |
| H2 | 0.3364 (11) | 0.0870 (10) | 0.167 (5) | 0.028 (6)* |
\ 2-(Pyridin-4-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]\ diazaborinine . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N3 | 0.0236 (8) | 0.0238 (8) | 0.0225 (8) | 0.0017 (6) | −0.0004 (7) | −0.0003 (7) |
| C7 | 0.0272 (9) | 0.0252 (9) | 0.0233 (10) | 0.0016 (8) | −0.0001 (8) | 0.0025 (8) |
| C1 | 0.0229 (9) | 0.0263 (9) | 0.0317 (10) | 0.0004 (8) | 0.0016 (9) | 0.0004 (9) |
| N2 | 0.0203 (8) | 0.0225 (7) | 0.0223 (8) | 0.0012 (6) | 0.0012 (7) | 0.0008 (7) |
| C13 | 0.0254 (10) | 0.0260 (9) | 0.0287 (10) | −0.0001 (8) | 0.0002 (9) | 0.0042 (9) |
| C10 | 0.0313 (10) | 0.0235 (9) | 0.0216 (10) | 0.0058 (8) | 0.0042 (8) | 0.0068 (8) |
| C15 | 0.0262 (9) | 0.0202 (8) | 0.0193 (9) | 0.0045 (7) | 0.0022 (8) | 0.0053 (7) |
| C5 | 0.0272 (10) | 0.0245 (9) | 0.0301 (10) | −0.0008 (8) | −0.0020 (9) | −0.0043 (8) |
| C6 | 0.0270 (9) | 0.0200 (8) | 0.0183 (9) | 0.0027 (7) | 0.0016 (8) | 0.0050 (8) |
| C14 | 0.0271 (9) | 0.0209 (9) | 0.0214 (9) | 0.0025 (7) | 0.0018 (8) | 0.0057 (8) |
| C11 | 0.0301 (10) | 0.0283 (10) | 0.0278 (11) | 0.0088 (8) | 0.0094 (9) | 0.0068 (8) |
| B1 | 0.0254 (10) | 0.0194 (9) | 0.0212 (10) | 0.0027 (8) | 0.0001 (9) | 0.0041 (9) |
| C3 | 0.0227 (9) | 0.0228 (9) | 0.0204 (9) | 0.0030 (7) | −0.0033 (7) | 0.0017 (7) |
| C4 | 0.0223 (9) | 0.0274 (9) | 0.0290 (10) | −0.0008 (8) | −0.0003 (8) | 0.0000 (9) |
| C12 | 0.0248 (9) | 0.0290 (10) | 0.0319 (11) | 0.0044 (8) | 0.0051 (9) | 0.0092 (9) |
| N1 | 0.0272 (9) | 0.0263 (8) | 0.0270 (9) | 0.0038 (7) | −0.0015 (7) | −0.0034 (7) |
| C2 | 0.0253 (9) | 0.0210 (9) | 0.0276 (10) | −0.0009 (8) | −0.0009 (8) | −0.0012 (8) |
| C9 | 0.0383 (11) | 0.0243 (10) | 0.0205 (10) | 0.0092 (8) | 0.0042 (9) | 0.0014 (8) |
| C8 | 0.0387 (11) | 0.0223 (9) | 0.0226 (10) | 0.0035 (8) | −0.0019 (9) | 0.0010 (8) |
\ 2-(Pyridin-4-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]\ diazaborinine . Geometric parameters (Å, º)
| N3—C14 | 1.399 (3) | C10—C9 | 1.416 (3) |
| N3—B1 | 1.418 (3) | C15—C6 | 1.425 (3) |
| N3—H3 | 0.93 (3) | C15—C14 | 1.426 (3) |
| C7—H7 | 0.9500 | C5—H5 | 0.9500 |
| C7—C6 | 1.384 (3) | C5—C4 | 1.389 (3) |
| C7—C8 | 1.408 (3) | C5—N1 | 1.335 (3) |
| C1—H1 | 0.9500 | C11—H11 | 0.9500 |
| C1—N1 | 1.339 (3) | C11—C12 | 1.372 (3) |
| C1—C2 | 1.389 (3) | B1—C3 | 1.578 (3) |
| N2—C6 | 1.401 (2) | C3—C4 | 1.396 (3) |
| N2—B1 | 1.411 (3) | C3—C2 | 1.397 (3) |
| N2—H2 | 0.93 (2) | C4—H4 | 0.9500 |
| C13—H13 | 0.9500 | C12—H12 | 0.9500 |
| C13—C14 | 1.380 (3) | C2—H2A | 0.9500 |
| C13—C12 | 1.405 (3) | C9—H9 | 0.9500 |
| C10—C15 | 1.431 (3) | C9—C8 | 1.370 (3) |
| C10—C11 | 1.423 (3) | C8—H8 | 0.9500 |
| C14—N3—B1 | 123.02 (17) | C13—C14—N3 | 122.15 (18) |
| C14—N3—H3 | 118.1 (17) | C13—C14—C15 | 120.03 (18) |
| B1—N3—H3 | 118.8 (17) | C10—C11—H11 | 119.9 |
| C6—C7—H7 | 119.9 | C12—C11—C10 | 120.23 (18) |
| C6—C7—C8 | 120.13 (18) | C12—C11—H11 | 119.9 |
| C8—C7—H7 | 119.9 | N3—B1—C3 | 120.36 (18) |
| N1—C1—H1 | 118.0 | N2—B1—N3 | 117.04 (18) |
| N1—C1—C2 | 123.90 (18) | N2—B1—C3 | 122.60 (17) |
| C2—C1—H1 | 118.0 | C4—C3—B1 | 121.57 (17) |
| C6—N2—B1 | 122.82 (16) | C4—C3—C2 | 115.74 (18) |
| C6—N2—H2 | 112.8 (15) | C2—C3—B1 | 122.68 (17) |
| B1—N2—H2 | 124.4 (15) | C5—C4—C3 | 120.12 (18) |
| C14—C13—H13 | 119.9 | C5—C4—H4 | 119.9 |
| C14—C13—C12 | 120.1 (2) | C3—C4—H4 | 119.9 |
| C12—C13—H13 | 119.9 | C13—C12—H12 | 119.3 |
| C11—C10—C15 | 118.62 (19) | C11—C12—C13 | 121.46 (18) |
| C9—C10—C15 | 118.83 (18) | C11—C12—H12 | 119.3 |
| C9—C10—C11 | 122.54 (18) | C5—N1—C1 | 116.03 (17) |
| C6—C15—C10 | 119.37 (18) | C1—C2—C3 | 120.15 (17) |
| C6—C15—C14 | 121.14 (17) | C1—C2—H2A | 119.9 |
| C14—C15—C10 | 119.49 (17) | C3—C2—H2A | 119.9 |
| C4—C5—H5 | 118.0 | C10—C9—H9 | 119.7 |
| N1—C5—H5 | 118.0 | C8—C9—C10 | 120.51 (19) |
| N1—C5—C4 | 124.06 (18) | C8—C9—H9 | 119.7 |
| C7—C6—N2 | 121.89 (17) | C7—C8—H8 | 119.4 |
| C7—C6—C15 | 119.97 (17) | C9—C8—C7 | 121.19 (19) |
| N2—C6—C15 | 118.14 (16) | C9—C8—H8 | 119.4 |
| N3—C14—C15 | 117.81 (16) | ||
| N3—B1—C3—C4 | −23.9 (3) | C11—C10—C15—C6 | −179.01 (17) |
| N3—B1—C3—C2 | 154.92 (19) | C11—C10—C15—C14 | 0.7 (3) |
| N2—B1—C3—C4 | 156.09 (19) | C11—C10—C9—C8 | 179.81 (19) |
| N2—B1—C3—C2 | −25.1 (3) | B1—N3—C14—C13 | 179.50 (19) |
| C10—C15—C6—C7 | −0.8 (3) | B1—N3—C14—C15 | −1.1 (3) |
| C10—C15—C6—N2 | 178.65 (16) | B1—N2—C6—C7 | 179.99 (18) |
| C10—C15—C14—N3 | −178.38 (17) | B1—N2—C6—C15 | 0.5 (3) |
| C10—C15—C14—C13 | 1.0 (3) | B1—C3—C4—C5 | 178.49 (18) |
| C10—C11—C12—C13 | 0.9 (3) | B1—C3—C2—C1 | −178.13 (18) |
| C10—C9—C8—C7 | −0.8 (3) | C4—C5—N1—C1 | −0.6 (3) |
| C15—C10—C11—C12 | −1.7 (3) | C4—C3—C2—C1 | 0.7 (3) |
| C15—C10—C9—C8 | 0.5 (3) | C12—C13—C14—N3 | 177.53 (17) |
| C6—C7—C8—C9 | 0.3 (3) | C12—C13—C14—C15 | −1.8 (3) |
| C6—N2—B1—N3 | −0.3 (3) | N1—C1—C2—C3 | −1.1 (3) |
| C6—N2—B1—C3 | 179.77 (16) | N1—C5—C4—C3 | 0.3 (3) |
| C6—C15—C14—N3 | 1.4 (3) | C2—C1—N1—C5 | 0.9 (3) |
| C6—C15—C14—C13 | −179.24 (18) | C2—C3—C4—C5 | −0.4 (3) |
| C14—N3—B1—N2 | 0.6 (3) | C9—C10—C15—C6 | 0.3 (3) |
| C14—N3—B1—C3 | −179.47 (17) | C9—C10—C15—C14 | −179.93 (16) |
| C14—C13—C12—C11 | 0.9 (3) | C9—C10—C11—C12 | 179.00 (18) |
| C14—C15—C6—C7 | 179.45 (17) | C8—C7—C6—N2 | −178.96 (17) |
| C14—C15—C6—N2 | −1.1 (3) | C8—C7—C6—C15 | 0.5 (3) |
\ 2-(Pyridin-4-yl)-2,3-dihydro-1H-naphtho[1,8-de][1,3,2]\ diazaborinine . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···N1i | 0.93 (2) | 2.21 (2) | 3.113 (2) | 162 (2) |
Symmetry code: (i) y+1/2, −x+1/2, −z−1/2.
References
- Akerman, M. P., Robinson, R. S. & Slabber, C. A. (2011). Acta Cryst. E67, o1873. [DOI] [PMC free article] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst.42, 339–341.
- Hashimoto, S. & Okuno, T. (2024). IUCrData, 9, x240362. [DOI] [PMC free article] [PubMed]
- LaPorte, A. J., Feldner, J. E., Spies, J. C., Maher, T. J. & Burke, M. D. (2023). Angew. Chem. Int. Ed.62, e202309566. [DOI] [PubMed]
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Rigaku OD (2024). CrysAlis PRO. Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Slabber, C. A., Grimmer, C., Akerman, M. P. & Robinson, R. S. (2011). Acta Cryst. E67, o1995. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2414314624006151/bx4031sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2414314624006151/bx4031Isup2.hkl
Supporting information file. DOI: 10.1107/S2414314624006151/bx4031Isup3.cml
CCDC reference: 2364937
Additional supporting information: crystallographic information; 3D view; checkCIF report