Abstract
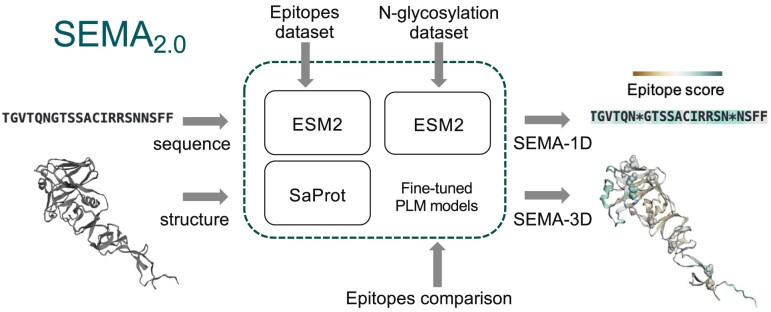
Prediction of conformational B-cell epitopes is a crucial task in vaccine design and development. In this work, we have developed SEMA 2.0, a user-friendly web platform that enables the research community to tackle the B-cell epitopes prediction problem using state-of-the-art protein language models. SEMA 2.0 offers comprehensive research tools for sequence- and structure-based conformational B-cell epitopes prediction, accurate identification of N-glycosylation sites, and a distinctive module for comparing the structures of antigen B-cell epitopes enhancing our ability to analyze and understand its immunogenic properties. SEMA 2.0 website https://sema.airi.net is free and open to all users and there is no login requirement. Source code is available at https://github.com/AIRI-Institute/SEMAi
Graphical Abstract
Graphical Abstract.
Introduction
Antigen-antibody interactions play a significant role in the immune response. The specific, discontinuous binding sites on the antigen structure that are recognized by antibodies produced by B-cells are known as conformational B-cell epitopes. Predicting conformational B-cell epitopes in antigens is a critical task in immunology and vaccine development. AI-based approaches based on large pre-trained protein language models (PLMs) offer significant advantages in addressing this challenge. Recently, we have developed SEMA (Spatial Epitope Modelling with Artificial Intelligence), a web server that predicts conformational epitopes with high accuracy (1).
In this work, we introduce SEMA 2.0, a new version of the SEMA web platform featuring an updated dataset, improved underlying models, and novel functionality. Notably, large state-of-the-art pre-trained PLMs were used for sequence-based prediction of conformational B-cell epitopes. For the sequence-based prediction, we used new ESM2 (Evolutionary Scale Modeling) models with 3 billion parameters which show improvements over ESM-1v models with 650 million parameters that we used in the previous version of SEMA (1–3). For the structure-based prediction of B-cell conformational epitopes, we utilized PLMs with a geometric modality, SaProt (Structure-aware Protein language model) in contrast to ESM-IF1 (ESM Inverse folding model) used in the previous version (4). This approach takes into account the spatial arrangement of the target antigen as well as its primary sequence, allowing for the prediction of B-cell conformational epitopes while considering the antigen’s conformational and multimeric states.
Several web servers and models for the conformational B-cell epitope predictions were recently published. Among them BepiPred 3.0 (5), SEPPA-3.0 (6), DiscoTope 3.0 (7). The major advantage of the SEMA 2.0 web server, is leveraging large pre-trained PLMs to achieve high performance metrics together with additional functionality. In particular, we have developed a model capable of identifying local structural similarities within two antigen structures aiding in the identification of similar conformational B-cell epitopes that may mimic immune responses. Post-translational modifications (PTMs), such as N- glycosylation are known to significantly affect the immunogenic properties of the antigen (6,8). To take into account this SEMA 2.0 includes a pre-trained model designed to predict N-glycosylation sites alongside conformational B-cell epitopes.
Overall, this work introduces a comprehensive web server for B-cell conformational epitope prediction, enriching the field of computational immunology with new functionalities and broad applicability.
Materials and methods
Datasets
Epitopes dataset
The epitopes dataset was generated as described in the (1) using the Protein Data Bank (PDB) database as released on 28 December 2023. In brief, for the amino acid residues of the antigen within the 8 Å of the interacting antibody the contact number was calculated. Amino acid residues beyond 16 Å of the interaction site were masked. Additionally, the training set was filtered out from the homologous sequences in the test set using the BLAST tool (9). For this purpose sequences with more than 30% identity and the E-value lower 0.05 were excluded from the training set. The final train and test sets consist of 1544 and 101sequences respectively (Supplementary Tables S1 and S2).
N-glycosylation dataset
To train the model for N-glycosylation sites prediction, we used a dataset collected by Wang et al. (10). We used a binary classification to mark residues with N-glycans and without. In total, the dataset included 8,963 samples and was divided into training and test sets in a ratio of 9:1.
SEMA-1D ensemble
In SEMA-1D we used an ensemble of 5 ESM-1v models with 650 million parameters. For SEMA-1D 2.0, we replaced the ESM-1v models with more powerful ESM-2 models (2) with 3 billion parameters each. We used exactly the same training hyperparameters as in (1). We also trained an ensemble of 5 ESM-2 models with 650 million parameters for comparison.
SEMA-3D ensemble
The previous version of the SEMA-3D ensemble consisted of 5 ESM-IF1 geometric models. In SEMA-3D 2.0, we used an ensemble of 5 pre-trained protein bimodal SaProt models with 650 million parameters each (4). SaProt is a transformer model, with architecture similar to ESM2. Structure information of each residue is encoded in one of 20 tokens from 3Di Foldseek vocabulary (11). Foldseek finds nearest neighbours for each residue and derives multiple features: seven angles, the Euclidean CA distance and two sequence distance features from the six CA coordinates of the two backbone fragments. It utilizes vector quantized variational autoencoder (VQ-VAE) to encode these relative structure features into one of 20 3Di tokens (11). This makes it possible to predict epitopes based on both structure- and sequence- modalities. To obtain 3Di tokens, the input protein structures derived from the PDB database were processed by FoldSeek. Models in SEMA-3D 2.0 were trained on the same dataset as SEMA-1D 2.0 using structures derived from the PDB database as an input. Training hyperparameters for models in SEMA-3D were the same as for the previous version and described in (1). Additionally, inference of the SEMA-3D model can be done in multichain mode. Within this mode, the protein subunits are presented as a single sequence, while the structure-aware dictionary is calculated in the same way as in the SEMA-3D monomer.
Model for structural comparison of antigens
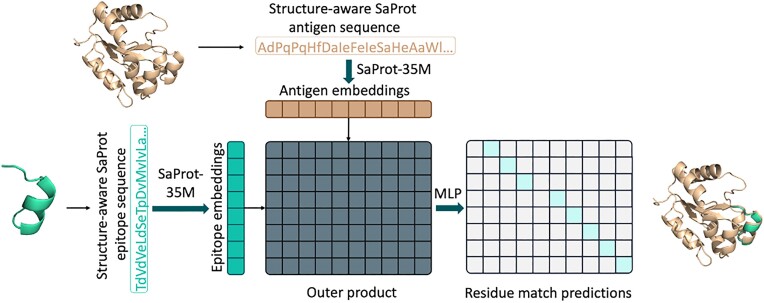
The architecture of the model that allows the conducting local structure comparison of proteins is shown in Figure 1. The model is trained to identify local structural similarities within proteins, based on the non-linear transformation of multiplication of the embeddings of PLM with geometric modalities.
Figure 1.
Training of the model to detect structural similarities between epitopes and antigen structures. Within the pipeline, both epitope-like structures and target antigen structures are used to generate structurally aware SaProt sequences, which are then passed into the SaProt-35M encoder. The outer product of encoder embeddings is then fed into a 2-layer MLP (multi-layer perceptron) network to predict matches between individual residues. Mint green elements of the predictions matrix correspond to values of 1 (match), and grey elements—to values of 0 (no match). This prediction is used to identify structurally similar regions.
A specific dataset comprising protein structures and epitope-like regions from homologous proteins was prepared. For this purpose, AlphaFold-predicted structures from the SwissProt database (12) were clustered based on sequence homology >45%. Clustering was performed using MMSeqs (13) software with the coverage mode set to 0, and a coverage threshold of 0.7 established. Within each cluster, the central structure was selected as the reference. To select discontinuous epitope-like protein fragments, amino acid residues on the protein structure’s surface were randomly selected that have epitope score predicted by SEMA 2.0 higher than threshold value. Protein fragments within a 5.0 Å radius of surface-exposed amino acid residues were included to the discontinuous epitope-like entity. Each fragment had a minimum length of five amino acid residues. Such entities were included in the dataset if the average p-LDDT value, as predicted by AlphaFold for the structure, exceeded 70, and at least five polar and solvent-exposed amino acid residues were present within the structure. This analysis was conducted using PyMOL scripts (version 2.5.4). Thus for each pair of homologous proteins within the cluster, discontinuous epitope-like protein regions in size from 20 to 70 residues were selected.
The model was trained to predict matching residues between the reference structure and discontinuous epitope-like protein regions. The matching residues were represented as a binary matrix, indicating residue-wise matches between the reference and epitope structures. The prediction of each element in the alignment matrix was treated as a distinct binary classification task, for which the training involved minimizing the binary cross-entropy loss function. The training dataset comprised 28 688 samples, with 3156 samples randomly selected for model validation. The following set of hyper-parameters was used to train the model: SaProt-35M encoder, AdamW optimizer, 1 × 10−4 learning rate with linear scheduler, trained for six epochs. To filter out noise in the model output we include only those pairs of matching residues that have at least two additional matching amino acid residues within a 12 Å distance.
N-glycosylation model
The N-glycosylation prediction model was obtained by adding a fully-connected linear layer on the top layer of the ESM-2 pre-trained model. The fully-connected layer takes input from the last layer of the respective pre-trained PLM and returns a one-dimensional vector of logits. Logits were used for the classification of Asparagine amino acid residue on whether it was N-glycosylated or not. We used the Adam optimizer and binary cross-entropy (BCE) loss function to train the model. The model was trained for two epochs with a starting learning rate of 1e−5.
Results
Updated SEMA-1D and SEMA-3D ensembles
The previous version of SEMA-1D has been updated by utilizing larger pre-trained PLMs. These models underwent training using either the previous training set or a new training set. The primary distinction from the previously published training set (1) lies in the inclusion of additional labeled antigen samples from newly released structures of the PDB database. In contrast to the previous version, a stricter filter was applied to exclude homologous sequences (with similarity >30%) from the training set that is present in the test set samples.
Following (1), the new SEMA-1D and SEMA-3D, as well as the final ensemble, were evaluated using two test sets: masked and unmasked. The masked test set included tasks to predict epitope residues within a distance of 16 Å from known epitopes, thereby avoiding potentially uncharacterized antigen regions due to the absence of experimental data. Conversely, the unmasked test set involved tasks to predict epitope residues across the entire length of the antigen sequence.
As shown in Table 1, implementing a more stringent exclusion of homologous sequences from the training set leads to a slight decrease in the ROC AUC metric for ESM-1v. However, the utilization of larger models from the ESM-2 family markedly enhances model performance. Specifically, the ESM-2 model with 3 billion parameters achieves an ROC AUC of 0.76 on the unmasked test set and 0.73 on the masked test sets.
Table 1.
Performance comparison (ROC AUC) of SEMA 1.0 and SEMA 2.0 models based on the PLM model using the training and test sets from work (1) (old dataset) and updated training set (new dataset)
| Old dataset | New dataset | |||||
|---|---|---|---|---|---|---|
| Model | Modality | Masked test | Unmasked test | Masked test | Unmasked test | |
| SEMA 1.0 | ESM-1v | 1D | 0.715 | 0.748 | 0.691 | 0.723 |
| ESM-IF1 | 3D | 0.726 | 0.756 | 0.726 | 0.752 | |
| SEMA 2.0 | ESM-2 650M | 1D | 0.728 | 0.768 | 0.715 | 0.747 |
| ESM-2 3B | 1D | 0.723 | 0.766 | 0.731 | 0.766 | |
| SaProt | 3D | 0.743 | 0.781 | 0.731 | 0.761 | |
| SaProt+ESM-2 650M | 1D/3D | 0.752 | 0.79 | 0.740 | 0.771 | |
| SaProt+ESM-2 3B | 1D/3D | 0.751 | 0.791 | 0.744 | 0.777 | |
For the new SEMA-3D ensemble, we utilized the state-of-the-art PLM that incorporates both sequence and geometric modalities, SaProt (4). The application of SaProt has led to enhanced performance metrics compared to the previous version of the SEMA-3D ensemble, which was based on the ESM-IF1 models (14), as detailed in Table 1.
Moreover, we demonstrate that the final ensembling of SEMA-1D and SEMA-3D predictions significantly improves the ROC AUC metrics on both masked and unmasked test sets, as shown in Table 1.
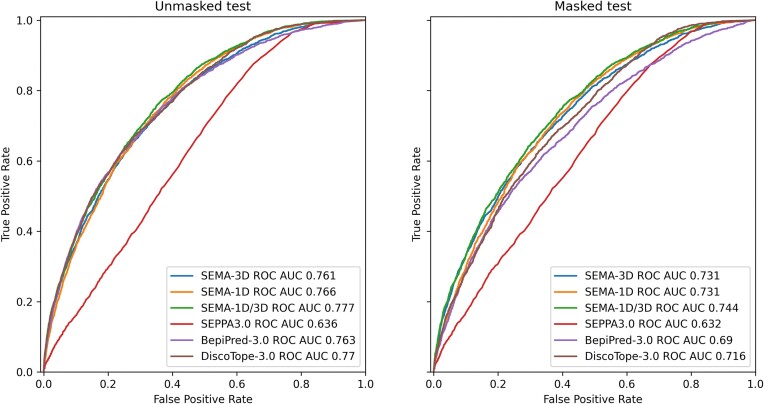
To benchmark the performance of the SEMA-1D and SEMA-3D ensembles against other commonly used tools, we evaluated the metrics for DiscoTope-3.0 (7), BepiPred-3.0 (5) and SEPPA 3.0 (6) using the same test sets (Table 2 and Figure 2). DiscoTope-3.0 and BepiPred-3.0 models exhibited similar performance on the unmasked test sets compared to all SEMA 2.0 models. However, the SEMA-1D/3D ensemble demonstrates superior performance on the masked test set. The masked test set was designed to focus on protein regions in close proximity to experimentally determined antibody contacts while excluding uncharacterized protein regions. Specifically, it includes only antigen residues located within a proximity of 16 Å from the known antibody binding site, with a positive label assigned for antigen residues located within 4.5 Å from the targeting antibody residues, and a negative label if it is located further away. This test set enables a more rigorous evaluation of the model’s ability to delineate the boundaries of the conformational epitope, while excluding distant regions for which the experimental annotation might be missing. The superior performance of SEMA 2.0 within this test set highlights the additional advantages of our model.
Table 2.
Quality comparison of SEMA2.0 and other models
| Model | AUC | Threshold | PPV | Sensivity | MCC | |
|---|---|---|---|---|---|---|
| Masked test | SEMA-1D | 0.731 | 0.362 | 0.613 | 0.67 | 0.278 |
| SEMA-3D | 0.731 | 0.71 | 0.615 | 0.666 | 0.276 | |
| SEMA-1D/3D | 0.744 | 0.483 | 0.617 | 0.677 | 0.288 | |
| DiscoTope-3.0 | 0.716 | 0.164 | 0.602 | 0.651 | 0.249 | |
| BepiPred-3.0 | 0.69 | 0.148 | 0.595 | 0.638 | 0.23 | |
| SEPPA3.0 | 0.632 | 0.048 | 0.581 | 0.599 | 0.179 | |
| Unmasked test | SEMA-1D | 0.766 | 0.343 | 0.561 | 0.696 | 0.218 |
| SEMA-3D | 0.761 | 0.637 | 0.561 | 0.689 | 0.215 | |
| SEMA-1D/3D | 0.777 | 0.479 | 0.564 | 0.703 | 0.229 | |
| DiscoTope-3.0 | 0.77 | 0.164 | 0.568 | 0.694 | 0.229 | |
| BepiPred-3.0 | 0.763 | 0.102 | 0.56 | 0.693 | 0.215 | |
| SEPPA3.0 | 0.636 | 0.048 | 0.542 | 0.611 | 0.137 |
For all models, we used same the set of proteins as in (1). Performance of the models was assessed using AUC, Matthews correlation coefficient (MCC), positive predictive value (PPV) and sensitivity metrics. To calculate MCC, PPV and sensitivity we converted prediction values to a binary values applying a threshold. Threshold was set as an optimal cut-off provided by ROC AUC analysis corresponding to the highest true positive rate together with the lowest false positive rate.
Figure 2.
ROC AUC metrics calculated on masked (left) and unmasked (right) test sets from (1).
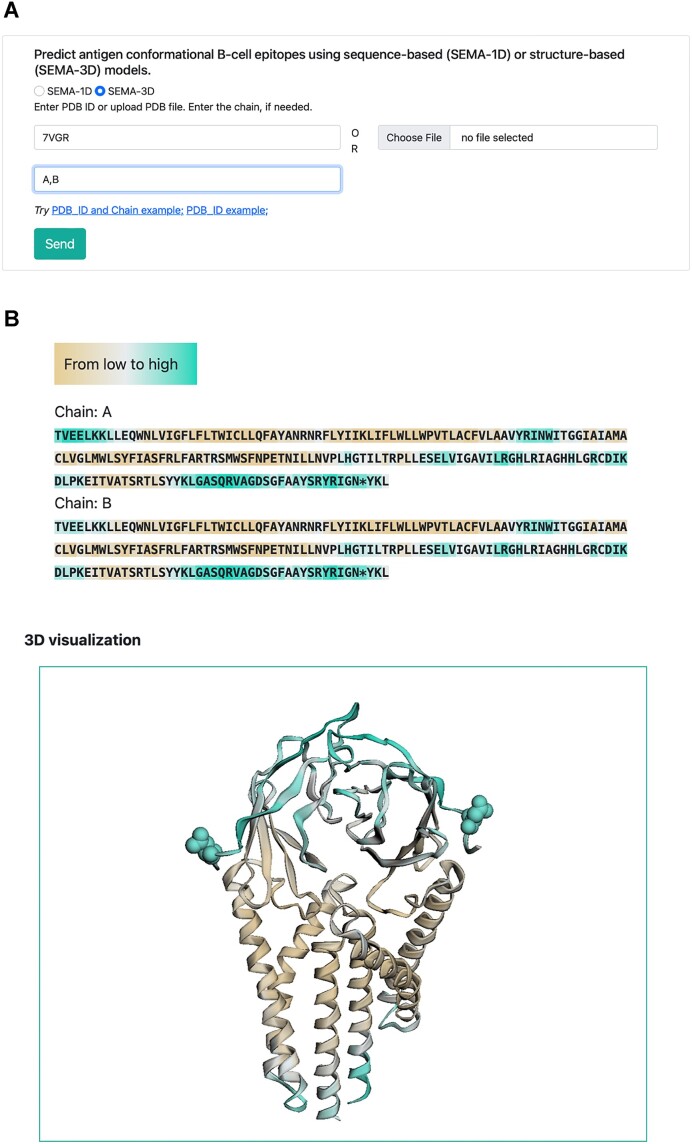
The special interest of the SEMA-3D model could be in the prediction of the conformational B-cell epitopes of the antigen structures that fold in the multimeric state. SEMA-3D can also predict epitopes for dimers, e.g. SARS-CoV-2 M protein (Figure 3) with similar ROC AUC scores, 0.906 for the dimer versus 0.884 for the monomers separately.
Figure 3.
Example of SEMA-3D web server input (A) and output (B) for the SARS-CoV-2 M protein dimer (PDB ID: 7VGR, chains A and B). The protein sequence is color-coded based on predicted values: brown indicates a low epitope score of zero, while cyan denotes that the epitope score exceeds the threshold, classifying the protein region as an epitope. Amino acids predicted to undergo N-glycosylation are marked with an asterisk in the sequence and depicted as spheres in the 3D visualization.
N-glycosylation prediction model
We integrated within the SEMA 2.0 pre-trained model to predict the N-glycosylation sites. For this purpose, we fine-tuned the ESM-2 model with 650 million parameters to solve the binary classification task. The fine-tuned model demonstrated similar high metrics as the MuSiteDeep model (10) with a ROC AUC value of 0.99.
Structural comparison of antigens
In SEMA 2.0, we add an additional model to identify local similarities between two input antigen structures. This model (Figure 1) is based on the neural network that conducts a comparison of representations from the PLM with geometric modalities SaProt (4) for amino acid residues of two proteins to predict the presence of structural similarities.
The hyperparameters of the model were experimentally derived to achieve the highest PRC AUC and ROC AUC metrics on the held-out portion of the dataset. Our best-performing model achieved 0.949 PRC AUC (0.998 ROC AUC) on the validation set of homologous protein fragments.
Although the model was trained on pairs of small epitope-like structures and larger antigens, our test cases show that the model is generalized to capture structural similarity across two full-length antigens.
As a test case, we performed a comparison of RBD domains of SARS-CoV and SARS-CoV-2. As expected, the algorithm allows to correctly assign the structurally similar regions (Figure 4). Other test cases included the identification of mimicking regions within synthetic immunogens and corresponding antigen structures. In both cases published by (15,16), the module allows to both identify epitopes as well correctly align the matched residues.
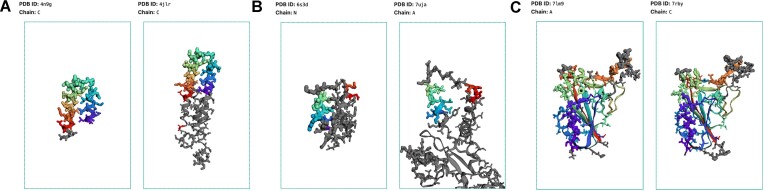
Figure 4.
(A) Examples of epitopes comparison model inference for the RSV-Presenting Epitope Scaffold (PDB ID 4N9G, chain C) and RSV immunogen (PDB ID 4JLR, chain C) (B) De novo protein encoding RSV epitopes from (16) (PDB ID 6S3D, chain N) and RSV antigen (PDB ID 7UJA, chain A). (C) SARS-CoV-2 S-protein RBD (PDB ID 7LM9, chain A) and SARS-CoV RBD (PDB ID 7RBY, chain C). Epitope comparison analysis output for SARS-CoV2 S-protein RBD and (PDB ID 7B3O, chain E; PDB ID 7LM9, chain A). Similar regions of the tertiary structures are depicted in the same colors (similarity score > 2), whereas regions with no similarity are shown in grey. Epitope residues are represented as sticks, with their radii corresponding to the epitope scores.
Usage
SEMA web server
The SEMA 2.0 web interface starts from the home page, which offers a concise description of the instruments. From there, users can access more detailed instructions via the ‘About’ tab or start their analysis directly. SEMA facilitates two types of analyses: the prediction of conformational B-cell epitopes based on either the primary protein sequence or tertiary structure (accessible through the ‘Predict epitopes’ tab), and the structural comparison of antigen epitopes (found under the ‘Compare epitopes’ tab).
Epitope prediction
The epitope prediction tool utilizes both sequence-based (SEMA-1D) and structure-based (SEMA-3D) approaches. The models in these ensembles have been fine-tuned to predict the propensity of amino acid residues to interact with the Fab regions of immunoglobulins. Users can select between the SEMA-1D or SEMA-3D models by clicking on the corresponding radio buttons.
SEMA-1D accepts an amino acid sequence as input data, which can be submitted in two ways: by pasting or typing a sequence of interest. The output includes a predicted epitope score and an N-glycosylation label for each residue within the amino acid sequence. Users can download results in CSV format.
The results are visualized as a color-coded sequence, where the background color indicates the predicted epitope score, ranging from brown (non-epitope) to cyan (epitope). Predicted N-glycosylated amino acids are marked with an asterisk (refer to Figure 3, top part).
SEMA-3D processes tertiary structure and sequence as input data. Users can submit input data by either specifying a target chain or the entire structure in two ways: (i) entering a PDB ID, by which the corresponding structures will be downloaded from the PDB database or (ii) uploading a custom PDB file. In both scenarios, users have the option to specify a particular chain for analysis; if no chain is specified, the analysis will be conducted on the entire structure.
The SEMA-3D output includes a predicted epitope score and an N-glycosylation label for each residue in the amino acid sequence (as illustrated in Figure 3). Users can download a zip archive containing a file with results in CSV format and an original PDB file. The visualization displays both the color-coded sequence and the tertiary structure of the protein, using the same color gradient to indicate epitope scores. Predicted N-glycosylated amino acids (AAs) are marked with an asterisk in the sequence and represented as spheres in the 3D structure.
Epitopes comparison
The epitope comparison tool predicts conformational B-cell epitopes of two antigens and highlights regions of local structural similarity within the protein structures. This functionality enables the identification of structurally similar epitopes across two antigen structures.
Users can input tertiary structures, with the option to focus on a target chain for the proteins being studied. Each structure can be submitted in one of two ways: (i) by typing the PDB ID and chain, wherein the corresponding structures will be extracted from the PDB file downloaded from the PDB database or (ii) by uploading a custom PDB file and specifying a chain.
The output includes predicted similarity and SEMA-3D epitope scores for each residue in the amino acid sequences of both proteins under study (Figure 4). The similarity scores indicate the degree of local structural similarity between residual pairs from the first and second input proteins. Users can download a zip archive file including epitopes prediction results for each protein in CSV format, similarity score for residuals pairs (for pairs with similarity scores greater than 2) in CSV format, and original PDB files.
The visualization of results shows the tertiary structures of the proteins under study, colored according to their similarity score. This indicates that parts of the tertiary structures with structural similarities are highlighted in matching colors (for similarity scores >2), whereas dissimilar parts are displayed in grey. Epitopes are depicted as sticks, with their radii proportional to the epitope scores.
Web server implementation
SEMA 2.0 web server is based on the Unicorn HTTP server in combination with Nginx. The deep-learning framework was implemented in the Business logic layer by Python. We used PostgreSQL for the server’s database. The web interface is implemented with JavaScript libraries, jQuery and 3Dmol library.
Discussion
In this work, we present SEMA 2.0, a web server that utilizes cutting-edge PLMs specifically fine-tuned to predict conformational B-cell epitopes. Compared to its predecessor, SEMA 2.0 incorporates additional functionalities, including a model dedicated to predicting N-glycosylation post-translational modifications (PTMs) within protein sequences, support for multi-chain proteins in the SEMA-3D model, and a tool for evaluating the structural similarity of antigen epitopes.
In this study, we show that the substitution of ESM-1v models with the bigger pre-trained PLMs ESM-2 (2) and SaProt (4) results in a substantial increase in the quality of prediction. The application of the PLMs with geometric modalities allows us to efficiently take into account the structural properties of the antigen, which, in particular, can be used for antigens with multiple conformations or different oligomerization states.
Additionally, we developed a model that can be applied to identify similar epitopes within different antigen structures, for instance, those corresponding to various viral or bacterial strains. It can also be applied for detecting regions that mimic the immune response in proteins without homology or overall structural similarity.
Supplementary Material
Contributor Information
Nikita V Ivanisenko, Bioinformatics Group, AIRI, Moscow, Russia; Laboratory of Computational Proteomics, Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia.
Tatiana I Shashkova, Bioinformatics Group, AIRI, Moscow, Russia.
Andrey Shevtsov, Bioinformatics Group, AIRI, Moscow, Russia; Regulatory Transcriptomics and Epigenomics Group, Research Center of Biotechnology RAS, Moscow, Russia.
Maria Sindeeva, Bioinformatics Group, AIRI, Moscow, Russia.
Dmitriy Umerenkov, Bioinformatics Group, AIRI, Moscow, Russia.
Olga Kardymon, Bioinformatics Group, AIRI, Moscow, Russia.
Data availability
The models are available via web interface https://sema.airi.net. The source code, including code for the dataset generation and models training, is available at GitHub (https://github.com/AIRI-Institute/SEMAi) and Zenodo (https://doi.org/10.5281/zenodo.11076971).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
No external funding.
Conflict of interest statement. None declared.
References
- 1. Shashkova T.I., Umerenkov D., Salnikov M., Strashnov P.V., Konstantinova A.V., Lebed I., Shcherbinin D.N., Asatryan M.N., Kardymon O.L., Ivanisenko N.V. SEMA: antigen B-cell conformational epitope prediction using deep transfer learning. Front. Immunol. 2022; 5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lin Z., Akin H., Rao R., Hie B., Zhu Z., Lu W., Smetanin N., Verkuil R., Kabeli O., Shmueli Y. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science. 2023; 379:1123–1130. [DOI] [PubMed] [Google Scholar]
- 3. Rives A., Meier J., Sercu T., Goyal S., Lin Z., Liu J., Guo D., Ott M., Zitnick C.L., Ma J. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2016239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su J., Han C., Zhou Y., Shan J., Zhou X., Yuan F. SaProt: protein language modeling with structure-aware vocabulary. 2023; bioRxiv doi:02 October 2023, preprint: not peer reviewed 10.1101/2023.10.01.560349. [DOI]
- 5. Clifford J.N., Høie M.H., Deleuran S., Peters B., Nielsen M., Marcatili P. BepiPred-3.0: Improved B-cell epitope prediction using protein language models. Protein Sci. 2022; 31:e4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhou C., Chen Z., Zhang L., Yan D., Mao T., Tang K., Qiu T., Cao Z. SEPPA 3.0—enhanced spatial epitope prediction enabling glycoprotein antigens. Nucleic Acids Res. 2019; 47:W388–W394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Høie M.H., Gade F.S., Johansen J.M., Würtzen C., Winther O., Nielsen M., Marcatili P. DiscoTope-3.0: Improved B-cell epitope prediction using inverse folding latent representations. Front. immunol. 2024; 15:1322712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rudd P.M., Elliott T., Cresswell P., Wilson I.A., Dwek R.A. Glycosylation and the immune system. Science. 2001; 291:2370–2376. [DOI] [PubMed] [Google Scholar]
- 9. Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC bioinformatics. 2009; 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang D., Liu D., Yuchi J., He F., Jiang Y., Cai S., Li J., Xu D. MusiteDeep: a deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020; 48:W140–W146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Kempen M., Kim S.S., Tumescheit C., Mirdita M., Lee J., Gilchrist C.L., Söding J., Steinegger M. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 2024; 42:243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022; 50:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hauser M., Steinegger M., Söding J. MMseqs software suite for fast and deep clustering and searching of large protein sequence sets. Bioinformatics. 2016; 32:1323–1330. [DOI] [PubMed] [Google Scholar]
- 14. Jing B., Eismann S., Suriana P., Townshend R.J., Dror R. Learning from protein structure with geometric vector perceptrons. 2020; arXiv doi:16 may 2021, preprint: not peer reviewedhttps://arxiv.org/abs/2009.01411.
- 15. Correia B.E., Bates J.T., Loomis R.J., Baneyx G., Carrico C., Jardine J.G., Rupert P., Correnti C., Kalyuzhniy O., Vittal V. et al. Proof of principle for epitope-focused vaccine design. Nature. 2014; 507:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sesterhenn F., Yang C., Bonet J., Cramer J.T., Wen X., Wang Y., Chiang C.-I., Abriata L.A., Kucharska I., Castoro G. et al. De novo protein design enables the precise induction of RSV-neutralizing antibodies. Science. 2020; 368:eaay5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The models are available via web interface https://sema.airi.net. The source code, including code for the dataset generation and models training, is available at GitHub (https://github.com/AIRI-Institute/SEMAi) and Zenodo (https://doi.org/10.5281/zenodo.11076971).