Abstract
Absorption, distribution, metabolism, excretion and toxicity (ADMET) properties play a crucial role in drug discovery and chemical safety assessment. Built on the achievements of admetSAR and its successor, admetSAR2.0, this paper introduced the new version of the series, admetSAR3.0, as a comprehensive platform for chemical ADMET assessment, including search, prediction and optimization modules. In the search module, admetSAR3.0 hosted over 370 000 high-quality experimental ADMET data for 104 652 unique compounds, and supplemented chemical structure similarity search function to facilitate read-across. In the prediction module, we introduced comprehensive ADMET endpoints and two new sections for environmental and cosmetic risk assessments, empowering admetSAR3.0 to provide prediction for 119 endpoints, more than double numbers compared to the previous version. Furthermore, the advanced multi-task graph neural network framework offered robust and reliable support for ADMET prediction. In particular, a module named ADMETopt was added to automatically optimize the ADMET properties of query molecules through transformation rules or scaffold hopping. Finally, admetSAR3.0 provides user-friendly interfaces for multiple types of input data, such as SMILES string, chemical structure and batch molecule file, and supports various output types, including digital, chart displays and file downloads. In summary, admetSAR3.0 is anticipated to be a valuable and powerful tool in drug discovery and chemical safety assessment at http://lmmd.ecust.edu.cn/admetsar3/.
Graphical Abstract
Graphical Abstract.
Introduction
Absorption, distribution, metabolism, excretion and toxicity (ADMET) properties play a crucial role in drug discovery and chemical safety assessment. Undesirable pharmacokinetic properties and unacceptable toxicity pose potential risks to human health and the environment, and constitute principal causes of drug development failure (1,2). Research suggested that integration of ADMET into the evaluation of new chemical entities could significantly reduce attrition rates (3). Consequently, the early examination and optimization of ADMET properties in chemicals represent a critical scientific endeavor.
Computational methods have significantly advanced the study of chemical ADMET properties, offering both an economical and a time-efficient approach (4–7). Recently, a vast array of free online resources and tools for ADMET assessment has been available. These resources include databases such as DrugBank (8), ChEMBL (9) and PKKB (10), as well as web servers including SwissADME (11), ADMETlab2.0 (12), ProTox-II (13), and admetSAR (14,15). Especially, admetSAR was launched in 2012 by our group, the first one among them, to facilitate ADMET property assessment. admetSAR initially provided free access to over 210 000 high-quality experimental data points for nearly 96 000 unique compounds, and 27 models for assessment of chemical ADMET properties. It was upgraded to admetSAR2.0 in 2019, thereby enhancing the quality of predictions for 47 ADMET endpoints. As of 7 January 2024, admetSAR and admetSAR2.0 have been extensively utilized, receiving 1291 and 584 citations from the Web of Science, respectively, while admetSAR2.0 has completed over 711 400 user tasks. Despite its success, admetSAR2.0 has several drawbacks to be improved, such as ADMET endpoint coverage, computational speed and user interface.
In this study, we introduced admetSAR3.0, an enhanced version of our comprehensive platform for chemical ADMET assessment, including search, prediction, and optimization. Notably, admetSAR3.0 features an expanded range of ADMET endpoints, complemented by two new sections dedicated to environmental and cosmetic risk assessments. This expansion allows admetSAR3.0 to offer predictions for 119 endpoints, more than double of its predecessor's capacity. The integration of an advanced multi-task graph neural network framework ensures robust and efficient ADMET prediction. admetSAR3.0 further boasts a user-friendly interface, supports multiple input and output formats, and delivers clear result display and practical guidance. Overall, admetSAR3.0 is positioned to emerge as an even more valuable and powerful tool in drug discovery and chemical safety assessment.
Materials and methods
Data collection
The ADMET Search module, a feature underpinned by the core database of admetSAR3.0, enables the retrieval of chemical and biological information of compounds based on standard SMILES notation. These experimental data were collected from peer-reviewed scientific literature and open-source databases such as ChEMBL (9), DrugBank (8), CPDB (16), ECOTOX (https://cfpub.epa.gov/ecotox/) and OpenFoodTox (https://www.efsa.europa.eu/en/data-report/chemical-hazards-database-openfoodtox). Compounds were characterized as standard SMILES through Open Babel (Version 2.4.1) (17), ensuring the elimination of duplicates. Furthermore, leveraging RDKit (https://www.rdkit.org/), we calculated ten fundamental properties for each compound in the database, such as molecular weight, log P and the quantitative estimate of drug-likeness (QED) value. These properties are crucial for drug-likeness screening and application domain assessment. Details of the application domain check are available in Text S2.
Model building
The ADMET Prediction module was supported by a contrastive learning based multi-task graph neural network framework (CLMGraph), and this unsupervised pre-training strategy has been validated through a study on drug sensitivity (18). During the pre-training phase, molecular pairs for contrastive learning strategies were constructed using the QED values of 10 million small molecules, thereby enhancing the overall representational capability of the models. In the multi-task property prediction phase, we employed a fine-tuning strategy to fully leverage the strengths of the pre-trained model for completing ADMET prediction tasks. The ADMET models were optimized using specific task-oriented loss functions. Specifically, MSELoss was utilized for the ADMET regression tasks, while BCELoss was employed for the classification tasks. The models underwent comprehensive evaluations, including five-fold cross-validation and external validation.
Optimization strategy
The ADMET Optimization module facilitates the guidance for optimizing query molecules through scaffold hopping and a transformation rule library. Specifically, the tools ADMETopt (19) and newly introduced ADMETopt2 support property optimization within the admetSAR3.0 framework. While ADMETopt focuses on scaffold hopping optimization, ADMETopt2 employs matched molecular pair analysis (MMPA) (20) technique to extract transformation rules for guiding the optimization of chemical properties. This strategy was proven effective and showed potential in applications, particularly in the context of chemical mutagenicity (21).
Implementation
The front-end of admetSAR3.0 was developed using HTML, CSS and JavaScript, ensuring a robust and interactive user experience. jQuery (version 2.1.1) facilitated page control and event handling, while Bootstrap (version 3.3.7) enhanced the client-side interface. A JavaScript-based molecular editor, JSME (updated on 10 June 2022) (22), enabled easy input of chemical structures. For data visualization, Apache ECharts (version 5.4.0) was incorporated. Server-side interactivity was managed via PHP scripts (version 5.4.16), with MariaDB (version 5.5.64) serving as the database management system. The predictive models utilized Python (version 3.9.18), supported by chemical informatics tools including PyTorch (version 1.11.0) (https://pytorch.org/), DGL (version 0.9.1) (https://www.dgl.ai/), DGL-LifeSci (version 0.3.2) (23) and the RDKit package (version 2022.03.2). admetSAR3.0 is freely available and does not require login for accessing its features.
Results and discussion
Overview of admetSAR3.0
Functional modules
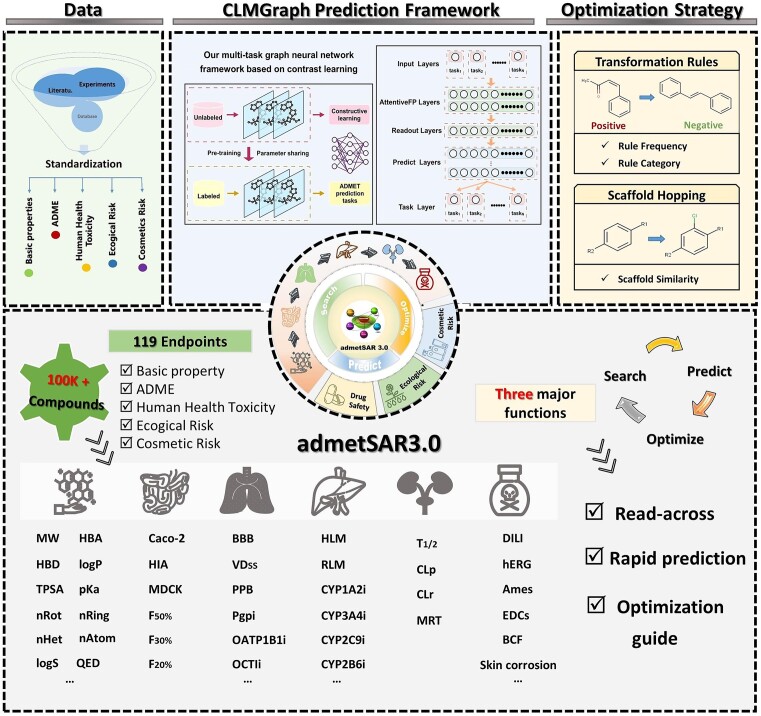
With comprehensive enhancements that include functional modules, endpoint data, predictive models, and a user-friendly web interface, admetSAR3.0 is poised to emerge as a more useful and powerful ADMET assessment tool (Figure 1). First, admetSAR3.0 now features a comprehensive assessment module. It offers property search, prediction and optimization capabilities, thus providing a one-stop convenience for the research of ADMET properties. The application of this tool primarily focuses on drug development, environmental and cosmetic risk assessment.
Figure 1.
An overview of the new developments in admetSAR3.0.
In the search module, it encompasses over 370 000 high-quality experimental ADMET data entries for 104 652 unique compounds and features a chemical structure similarity search function to facilitate read-across.
In the prediction module, admetSAR3.0 now has a broader endpoint coverage, boasting 119 ADMET-related endpoints, divided into five categories, namely basic properties, ADME properties, human health toxicities, environmental risk assessment, and cosmetic risk assessment. Compared to its predecessor, admetSAR3.0 demonstrates a 9.01% increase in compound numbers, a 78.08% increase in data records, and a 108.77% increase in endpoint numbers. Additionally, the advanced CLMGraph framework enhances the speed and accuracy of ADMET property predictions.
In the optimization module, it facilitates ADMET property optimization through scaffold hopping and transformation rules. In ADMETopt, over 50 000 unique scaffolds from ChEMBL and Enamine databases aid in similar scaffold matching. The newly introduced ADMETopt2 (not published yet), a transformation rule-based optimization tool, utilizes MMPA and a transformation rule library for 21 ADMET endpoints. The transformation rules are freely accessible on figshare at https://figshare.com/articles/dataset/ADMETopt2Transformation_Rules/25472317. More details about ADMETopt2 are available in Supplementary Text S1 and Supplementary Table S1. ADMETopt2 introduces innovative strategies for the structural modification of compounds with undesirable ADMET characteristics, thereby enhancing the scope of ADMETopt and providing a more comprehensive and efficient approach to optimize chemical properties.
At last, the updated interface of admetSAR3.0 offers users clear and concise result displays and practical guidance.
Web interface
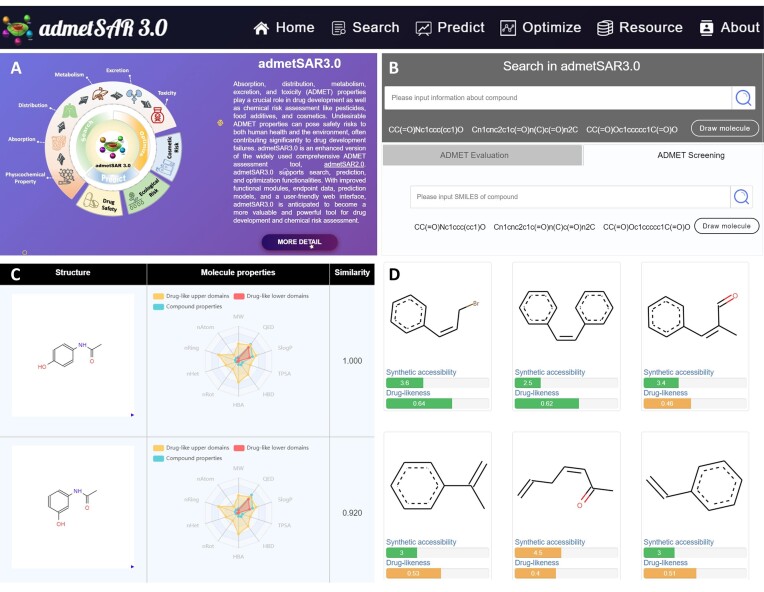
As depicted in Figure 2, it presents an overview of the admetSAR3.0 interface. Figure 2A displays the homepage, which provides user-friendly access to detailed information and functional modules via a navigation bar. Figure 2A illustrates the search and prediction interface. In search module, users can obtain ADMET property information of compounds by entering their SMILES strings and chemical structures. Meanwhile, admetSAR3.0 provides two prediction services: single-molecule evaluation and batch screening. Users have the option to select the evaluation mode according to their needs, and are able to assess up to 1000 compounds per batch.
Figure 2.
Homepage, search, prediction and optimization interface of admetSAR3.0. (A) The homepage includes a description of the platform and ‘More Detail’ access. (B) The search and prediction input interface. (C) The search result preview interface, including compound structure, properties and similarity. (D) Optimization result interface based on transformation rule.
Results presentation
admetSAR3.0 is designed to be user-friendly, catering to a diverse spectrum of users, including those lacking expertise in the field. The platform includes a ‘Tutorial’ page that provides comprehensive guidance on its usage. Reflecting our commitment to user convenience, a focus has been placed on simplifying both data input and output interpretation.
admetSAR3.0 accepts various types of input data, such as SMILES strings, chemical structures and batch molecular files, and offers diverse output formats, including numerical data, graphical displays and downloadable files. For specific project details, users can access contextual assistance by hovering over question marks or boxes on the page. The search module supports structure-based similarity searching, evaluating the similarity between query molecules and compounds in the database using ECFP fingerprints. Compounds with a similarity score >0.9 are displayed, enhancing read-across (Figure 2C).
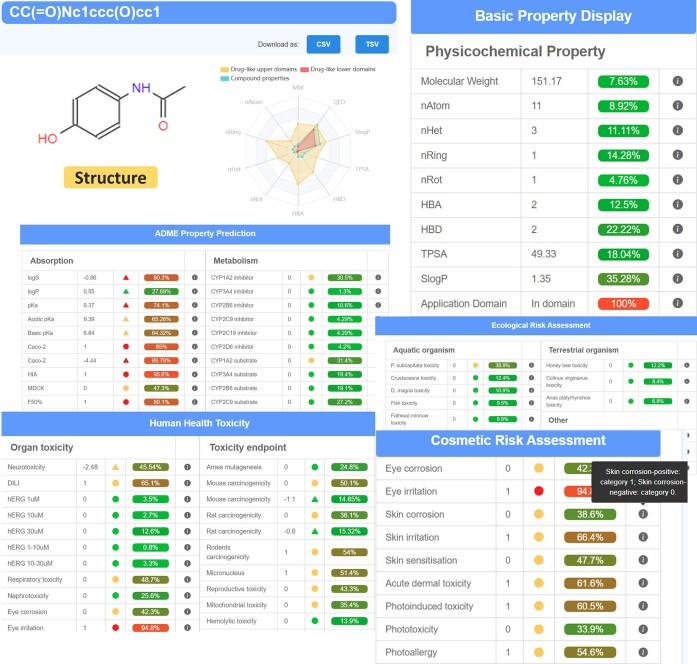
Figure 3 demonstrates the user-friendly prediction interface, featuring basic information about the query molecule, including SMILES and structural details. The prediction results are presented using a traffic light format and brief descriptions of each endpoint are immediately available for quick reference. In the prediction module, an optimal model developed by the CLMGraph framework is integrated for rapid and accurate prediction of 119 ADMET endpoints. For classification models, the average AUC (area under the curve) value for 90 endpoints stands at 0.870, with most models demonstrating satisfactory predictive performance, with the exception of a few endpoints such as renal clearance. For regression models, >82% of endpoints have PCC (pearson correlation coefficient) values >0.70, accompanied by low MAE (mean absolute error) and RMSE (root mean square error) values. However, some endpoints, including half-life and mean residence time, do not exhibit satisfactory predictive performance. In addition to internal validation, external validation sets were employed to further assess model performance, demonstrating the models' generalization ability. Meanwhile, the provision of an applicability domain assessment serves to guide the prediction models.
Figure 3.
ADMET property prediction results display.
Figure 2D shows the optimization results of ADMETopt2, displaying the transformation results for query molecule. Users are able to download the structures of optimized compounds, and can view synthetic accessibility (SA) and QED values. Detailed optimization information, such as transformation rules and rule occurrence frequencies, can be accessed by clicking on a compound's structure.
Comparison with other web-based tools
Numerous outstanding online servers, such as ADMETlab2.0 (12), SwissADME (11), vNN-ADMET (24), pkCSM (25), ADMETboost (26), ProTOX-II (13), ADMET-AI (https://doi.org/10.1101/2023.12.28.573531) and Danish QSAR Models (https://qsarmodels.food.dtu.dk/), efficiently facilitate the assessment of ADMET properties in an economical way. This study presents a thorough comparison of these readily available online tools with admetSAR3.0, detailed in Table 1. admetSAR3.0 along with its counterparts, including ADMETlab2.0, pkCSM, ADMETboost, and vNN-ADMET, emphasizes the comprehensive evaluation of ADMET properties. SwissADME specializes in assisting researchers to assess the physicochemical properties and drug-likeness of chemicals, particularly with detailed log P prediction. ADMET-AI provides unique context for ADMET predictions by comparing the query molecule properties with the approved drugs. The Danish QSAR Models and ProTOX-II both cover various toxicity endpoints, with ProTOX-II additionally providing insights into potential adverse outcome pathways (AOPs) and toxicity target analysis for a deeper understanding of toxicological mechanisms. In contrast, admetSAR3.0 excels by providing the widest range of predicted endpoints, including essential ones vital for medicinal chemists. Furthermore, our platform is distinguished by its thorough and robust nature, offering functionalities for ADMET property search, prediction and optimization, while boasting a faster computational speed.
Table 1.
Statistics and comparison between admetSAR3.0 and existing tools
| admetSAR3.0 | admetSAR2.0 | ADMETlab2.0 | SwissADME | vNN-ADMET | pkCSM | ADMET-AI | ADMETboost | ProTOX-II | Danish QSAR Models | |
|---|---|---|---|---|---|---|---|---|---|---|
| Endpoints | ||||||||||
| Basic property | 18 | 6 | 30 | 28 | 0 | 7 | 8 | 9 | 9 | 0 |
| ADME | 39 | 24 | 23 | 9 | 9 | 19 | 22 | 16 | 0 | 2 |
| Human health toxicity | 43 | 21 | 23 | 0 | 6 | 8 | 18 | 4 | 17 | 48 |
| Ecological risk | 16 | 5 | 4 | 0 | 0 | 2 | 0 | 0 | 0 | 4 |
| Cosmetic risk | 9 | 3 | 3 | 1 | 0 | 1 | 1 | 0 | 0 | 7 |
| Total | +++++ | +++ | ++++ | ++ | ++ | ++ | +++ | ++ | ++ | +++ |
| Functions | ||||||||||
| Search | √ | √ | √ | |||||||
| Optimization | √ | √ | ||||||||
| Prediction | +++++ | +++ | ++++ | +++ | +++ | ++ | ++++ | + | +++ | +++ |
| Batch evaluation | +++ | + | ++ | +++ | + | ++ | +++ | - | - | - |
| Application domain | √ | √ | √ | √ | √ | |||||
| Speed (1000 molecules) | 12s | 315s | 96s | 1830s | 2411s | 1910s | 101s | - | - | - |
Case studies
Property prediction
Ogsiveo tablets, representing the first FDA-approved medication for treating hard fibromas, are an orally-administered γ-secretase small molecule inhibitor (27). Detailed information about its ADMET-related properties is readily available. Employing admetSAR3.0, we performed a detailed ADMET property analysis, and the specific results are presented in Supplementary Figure S1. Given its oral administration, Ogsiveo demonstrates good absorption in the small intestine and high oral bioavailability. It displays a serum protein binding rate of 99.6%, aligning with admetSAR3.0 prediction. The drug is robustly predicted as both a P-glycoprotein (P-gp) substrate and inhibitor, which corroborates in vitro experimental findings. Metabolically, Ogsiveo is mainly processed by CYP3A4 (85%), while also involving CYP3A4, 2C19, 2C9 and 2D6. Although predicted as substrates for CYP3A4, 2C19 and 2D6, primarily for CYP3A4, the potential of Ogsiveo as a CYP2C9 substrate warrants additional investigation. In non-clinical toxicology studies, no statistically significant tumors emerged in mouse carcinogenicity studies, indicating a lack of carcinogenic risk in mice. This finding aligns with our platform's prediction of no carcinogenicity in both mice and rats, supporting ongoing rat carcinogenicity evaluation. Furthermore, the platform accurately predicted the outcomes of Ames test mutagenicity. Finally, the prediction results highlight potential adverse reactions linked to Ogsiveo, including hepatotoxicity, ovarian toxicity and respiratory toxicity.
Property optimization
In the study by Ellis et al. (28), lead compound 42 was identified as a highly selective inhibitor of spleen tyrosine kinase, crucial in the treatment of autoimmune diseases such as rheumatoid arthritis. However, it exhibited mutagenic potential in Ames tests and prompted Salmonella colonization. This necessitated the need for structural modification and optimization. To address this, lead compound 42 underwent processing via the ADMETopt2 optimization module, utilizing the mutagenicity transformation rule base for enhancement. The ADMETopt2 platform eventually proposed 12 structural modification strategies, resulting in the generation of corresponding novel molecules. These findings are detailed in Supplementary Figure S2 and Supplementary Table S1. Notably, optimization strategy 9 mirrored techniques commonly used by medicinal chemists. Experimental validation demonstrated that the revised compound 9 was devoid of mutagenic properties while maintaining its high pharmacological efficacy (IC50 = 1 nM), validating the effectiveness of our proposed structural modification approach. Additionally, we expect that additional optimization strategies outlined by ADMETopt2 will provide valuable insights into lead optimization.
Conclusions
We are pleased to introduce admetSAR3.0, a significantly enhanced and comprehensive online platform dedicated to the search, prediction and optimization of chemical ADMET properties. This updated platform is efficient, state-of-the-art and user-friendly. admetSAR3.0 provides predictive services for 119 ADMET property endpoints, making it the most comprehensive among open-source online services currently available. The integration of the advanced CLMGraph framework enhances the platform's ability to deliver robust and rapid predictions, facilitating batch processing of ADMET property information for compounds. Additionally, the optimization module provides significant guidance in the structural modification and optimization of compounds. To improve usability and enhance user experience, admetSAR3.0 now features a completely redesigned interface, offering clear results and practical guidance, thereby ensuring accessibility for both expert and novice users. We believe that admetSAR3.0 will become an invaluable and powerful tool in the fields of drug development and chemical risk assessment.
Supplementary Material
Acknowledgements
In this work, all three co-authors contributed equally. Y.G. collected and processed data for 119 ADMET endpoints, prepared relevant images and textual materials for the website pages, and finalized the article manuscript. Z.Y. completed the construction of the website and implemented the relevant functions of the service. Y.W. established the Contrastive Learning based Multi-Task Graph Neural Network framework (CLMGraph) for ADMET property prediction. The corresponding author, Y.T., received funding support and provided guidance for the entire project.
Contributor Information
Yaxin Gu, Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China.
Zhuohang Yu, Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China.
Yimeng Wang, Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China.
Long Chen, Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China.
Chaofeng Lou, Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China.
Chen Yang, Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China.
Weihua Li, Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China.
Guixia Liu, Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China.
Yun Tang, Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, Shanghai Key Laboratory of New Drug Design, School of Pharmacy, East China University of Science and Technology, Shanghai 200237, China.
Data availability
The data is provided via admetSAR3.0 website (http://lmmd.ecust.edu.cn/admetsar3/).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Key Research and Development Program of China [2023YFF1204904]; National Natural Science Foundation of China [U23A20530 and 82173746]; Shanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism (Shanghai Municipal Education Commission). Funding for open access charge: National Key Research and Development Program of China [2023YFF1204904].
Conflict of interest statement. None declared.
References
- 1. Fan J., de Lannoy I.A.M. Pharmacokinetics. Biochem. Pharmacol. 2014; 87:93–120. [DOI] [PubMed] [Google Scholar]
- 2. Wu P., Lin S., Cao G., Wu J., Jin H., Wang C., Wong M.H., Yang Z., Cai Z. Absorption, distribution, metabolism, excretion and toxicity of microplastics in the human body and health implications. J. Hazard. Mater. 2022; 437:129361. [DOI] [PubMed] [Google Scholar]
- 3. Waring M.J., Arrowsmith J., Leach A.R., Leeson P.D., Mandrell S., Owen R.M., Pairaudeau G., Pennie W.D., Pickett S.D., Wang J. et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015; 14:475–486. [DOI] [PubMed] [Google Scholar]
- 4. McGibbon M., Shave S., Dong J., Gao Y., Houston D.R., Xie J., Yang Y., Schwaller P., Blay V. From intuition to AI: evolution of small molecule representations in drug discovery. Brief. Bioinform. 2024; 25:bbad422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferreira L.L.G., Andricopulo A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today. 2019; 24:1157–1165. [DOI] [PubMed] [Google Scholar]
- 6. Jia C., Li J., Hao G., Yang G. A drug-likeness toolbox facilitates ADMET study in drug discovery. Drug Discov. Today. 2020; 25:248–258. [DOI] [PubMed] [Google Scholar]
- 7. Chan H.C.S., Shan H., Dahoun T., Vogel H., Yuan S. Advancing drug discovery via artificial intelligence. Trends Pharmacol. Sci. 2019; 40:592–604. [DOI] [PubMed] [Google Scholar]
- 8. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z. DrugBank 5.0: a major update to the DrugBank database for. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cao D., Wang J., Zhou R., Li Y., Yu H., Hou T. ADMET evaluation in drug discovery. 11. PharmacoKinetics Knowledge Base (PKKB): a comprehensive database of pharmacokinetic and toxic properties for drugs. J. Chem. Inf. Model. 2012; 52:1132–1137. [DOI] [PubMed] [Google Scholar]
- 11. Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017; 7:42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A. et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021; 49:W5–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banerjee P., Eckert A., Schrey A., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018; 46:W257–W263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng F., Li W., Zhou Y., Shen J., Wu Z., Liu G., Lee P.W., Tang Y. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012; 52:3099–3105. [DOI] [PubMed] [Google Scholar]
- 15. Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019; 35:1067–1069. [DOI] [PubMed] [Google Scholar]
- 16. Fitzpatrick R.B. CPDB: carcinogenic potency database. Med. Ref. Serv. Q. 2008; 27:303–311. [DOI] [PubMed] [Google Scholar]
- 17. O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminform. 2011; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y., Yu X., Gu Y., Li W., Zhu K., Chen L., Tang Y., Liu G. XGraphCDS: an explainable deep learning model for predicting drug sensitivity from gene pathways and chemical structures. Comput. Biol. Med. 2023; 168:107746. [DOI] [PubMed] [Google Scholar]
- 19. Yang H., Sun L., Wang Z., Li W., Liu G., Tang Y. ADMETopt: a Web Server for ADMET Optimization in Drug Design via Scaffold Hopping. J. Chem. Inf. Model. 2018; 58:2051–2056. [DOI] [PubMed] [Google Scholar]
- 20. Dossetter A.G., Griffen E.J., Leach A.G. Matched molecular pair analysis in drug discovery. Drug Discov. Today. 2013; 18:724–731. [DOI] [PubMed] [Google Scholar]
- 21. Lou C., Yang H., Deng H., Huang M., Li W., Liu G., Lee P.W., Tang Y. Chemical rules for optimization of chemical mutagenicity via matched molecular pairs analysis and machine learning methods. J. Cheminform. 2023; 15:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bienfait B., Ebert P. JSME: a free molecule editor in JavaScript. J. Cheminform. 2013; 5:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li M., Zhou J., Hu J., Fan W., Zhang Y., Gu Y., Karypis G. DGL-LifeSci: an Open-Source Toolkit for Deep Learning on Graphs in Life Science. ACS Omega. 2021; 6:27233–27238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schyman P., Liu R., Desai V., Wallqvist A. vNN web server for ADMET predictions. Front. Pharmacol. 2017; 8:889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pires D.E.V., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015; 58:4066–4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tian H., Ketkar R., Tao P. ADMETboost: a web server for accurate ADMET prediction. J. Mol. Model. 2022; 28:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gounder M., Ratan R., Alcindor T., Schöffski P., van der Graaf W.T., Wilky B.A., Riedel R.F., Lim A., Smith L.M., Moody S. et al. Nirogacestat, a γ-secretase inhibitor for desmoid tumors. N. Engl. J. Med. 2023; 388:898–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ellis J.M., Altman M.D., Bass A., Butcher J.W., Byford A.J., Donofrio A., Galloway S., Haidle A.M., Jewell J., Kelly N. et al. Overcoming mutagenicity and ion channel activity: optimization of selective spleen tyrosine kinase inhibitors. J. Med. Chem. 2015; 58:1929–1939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is provided via admetSAR3.0 website (http://lmmd.ecust.edu.cn/admetsar3/).