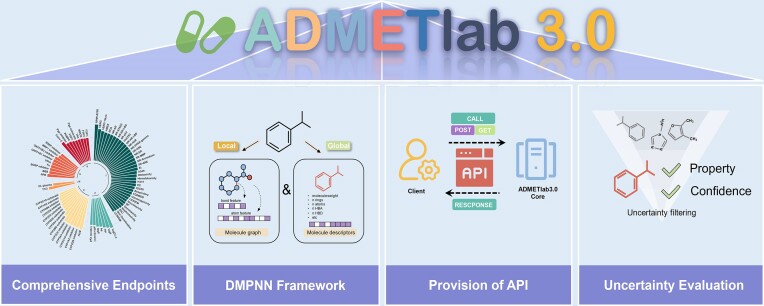
Abstract
ADMETlab 3.0 is the second updated version of the web server that provides a comprehensive and efficient platform for evaluating ADMET-related parameters as well as physicochemical properties and medicinal chemistry characteristics involved in the drug discovery process. This new release addresses the limitations of the previous version and offers broader coverage, improved performance, API functionality, and decision support. For supporting data and endpoints, this version includes 119 features, an increase of 31 compared to the previous version. The updated number of entries is 1.5 times larger than the previous version with over 400 000 entries. ADMETlab 3.0 incorporates a multi-task DMPNN architecture coupled with molecular descriptors, a method that not only guaranteed calculation speed for each endpoint simultaneously, but also achieved a superior performance in terms of accuracy and robustness. In addition, an API has been introduced to meet the growing demand for programmatic access to large amounts of data in ADMETlab 3.0. Moreover, this version includes uncertainty estimates in the prediction results, aiding in the confident selection of candidate compounds for further studies and experiments. ADMETlab 3.0 is publicly for access without the need for registration at: https://admetlab3.scbdd.com.
Graphical Abstract
Graphical Abstract.
Introduction
Drug development is a long journey full of high risk, high investments and extremely high attrition rates. The preclinical stage lays the crucial groundwork and, as a demanding phase in drug development, confronts a considerable attrition rate of approximately 93% (1). Even if certain drug candidates progress to clinical studies, over 75% of them are likely to fail in phases I, II and III clinical trials, and during the subsequent drug approval process (2,3). In this process, undesirable pharmacokinetic properties, including absorption, distribution, metabolism, and excretion (ADME) properties, play a crucial role, leading to the failure of approximately 40% of candidate molecules. Addition to them, toxicity, another important evaluation indicator in the drug development process, also accounts for up to 30% of drug development failures (2,4). This underscores the substantial impact of ADME and toxicity (ADMET) properties on the overall success or failure of drug development efforts, and the importance of early assessment of ADMET properties for drug development.
To evaluate the ADMET properties of new molecular entities (NMEs) as early as possible, various in vitro and in vivo methods including medium- and high-throughput screening have been developed, which also facilitate the rapid accumulation of experimental data. However, as the number of NMEs continues to increase, these experimental approaches have shown several inherent shortcomings: time-consuming, costly, and animal welfare issues involved, which have greatly limited their application and spurred the emergence of in silico methods for predicting ADMET properties. In recent decades, with the rapid development of computer science and the accumulation of ADMET experimental data, in silico predictive models and derived web tools aimed at facilitating the efficient evaluation of ADMET properties have been greatly developed. Representative tools include admetSAR (5), SwissADME (6) and ProTox-II (7), as well as several web-based platforms developed after the release of ADMETlab 2.0 (8), such as ADMETboost (9) and Interpretable-ADMET (10). Despite their evident utility, current platforms still have certain limitations including narrower coverage of endpoints and lower calculation efficiency, etc. Understanding and addressing these limitations is crucial for improving the effectiveness and reliability of computer tools in drug development.
Since its first release in 2018 and the updated version in 2021, we have been dedicated to enhancing the performance and user experience of the ADMETlab webserver. As one of the most popular ADMET prediction platforms, our platform has not only garnered numerous appreciative feedback from ordinary users but has also earned recognition in the field of AI-driven drug discovery thanks to its superior performance and wider ADMET predictions coverage (11–13). To date, the article publishing ADMETlab 2.0 has been cited 955 times according to Google Scholar, and the web server has been visited more than 1.7 million times. Inspired by increasingly frequent access and user expectations for enhancements, including the development of Application Programming Interface (API) capabilities to facilitate batch evaluation, we have upgraded ADMETlab to version 3.0 to address existing issues and further optimize its user experience. ADMET 3.0 currently covers a more comprehensive set of ADMET endpoints, including 400 000 high-quality entries and 119 endpoints, marking an enhancement of 31 additional endpoints in comparison to its predecessor. The multi-task deep message passing neural networks (DMPNN) framework combined with molecular descriptors was applied to construct predictive models for various endpoints, which significantly improved the performance and robustness of these models (14,15). More than that, in response to the escalating demand for enhanced in-batch evaluation and analysis experiences, we implemented API capabilities in ADMETlab 3.0, specifically designed for those dealing with substantial volumes of data. Additionally, an uncertainty estimation module using evidential deep learning techniques is deployed in ADMETlab 3.0, which can evaluate the reliability of prediction results by providing precise measurement of uncertainties in the outcomes of ADMET predictions, thereby facilitating informed decision-making processes regarding candidate prioritization during virtual screening. In summary, we believe that the updated ADMET 3.0 is expected to provide drug developers and chemists with a more comprehensive, reliable, and accurate service based on its broader coverage, improved performance, API functionality, and decision support. ADMETlab 3.0 is freely available for all users without login at: https://admetlab3.scbdd.com.
Methods and webserver description
Data collection
To provide a broader and deeper insight into the ADMET profiles, molecular physicochemical properties, and medicinal chemistry rules for query compounds, our team performed an extensive re-collection and reorganization based on the existing ADMET dataset. We incorporated a wide spectrum of open-access bioactivity databases including ChEMBL (16), PubChem (17) and OCHEM (18), as well as rigorously peer-reviewed literature (19–24). To ensure the quality, consistency and reliability of the data, and to build more accurate and generalizable models based on it, a series of preprocessing was performed: organometallic compounds, isomeric mixtures and chemical mixtures were selectively removed, salts were neutralized, counterions were eliminated, and canonical SMILES strings were adopted as model input format. After these pretreatments, we finally obtained over 400 000 molecules covering 77 ADMET-related endpoints for model building. The detailed information of 77 datasets and their respective data splitting information can be seen in Supplementary Table S1.
Directed message passing neural network (DMPNN)
The ADMETlab 2.0 employed the multi-task graph attention (MGA) model, a multi-task framework based on relation graph convolutional networks (RGCN). Although version 2.0 has shown significant improvement compared to its predecessor, we are continually seeking further enhancements to achieve superior performance without compromising computational speed. In ADMETlab 3.0, we leveraged directed message passing neural network (DMPNN), a subclass of graph convolutional neural networks (GCNN), by utilizing Chemprop (25,26), a DMPNN-based package developed by a team from the Massachusetts Institute of Technology. DMPNN considers directed edges and learns molecular encodings through bond-centered convolutions instead of atom-centered convolutions, thus avoiding unnecessary loops during the message passing phase. ADMETlab 2.0′s superior prediction ability over its predecessor and other webservers primarily owed to its multi-task graph neural network structure. To inherit the multi-tasking benefits from the previous version—simultaneously improving model performance and computational speed—we implemented Chemprop's multi-task training approach in ADMETlab 3.0.
Recent studies have demonstrated that incorporating external features, rather than relying solely on SMILES, can further enhance the performance of DMPNN (14,27). The advantage of this combination likely stems from the complementary relationship between the global information of molecules from descriptors and the local information extracted by DMPNN. RDKit 2D descriptors offer a comprehensive overview of molecular characteristics, including molecular size and shape, topological information, and functional groups. Our approach adopted the combination of DMPNN with RDKit 2D descriptors (hereinafter referred to as DMPNN-Des), an optional combination provided in Chemprop, to significantly enhance predictive performance. Presently, DMPNN methods have been successfully applied in various fields of drug discovery, including antibiotic discovery (28–30), reaction property prediction (31), infrared spectra prediction (32), solute parameters prediction (14), and molecular optical peak prediction (15).
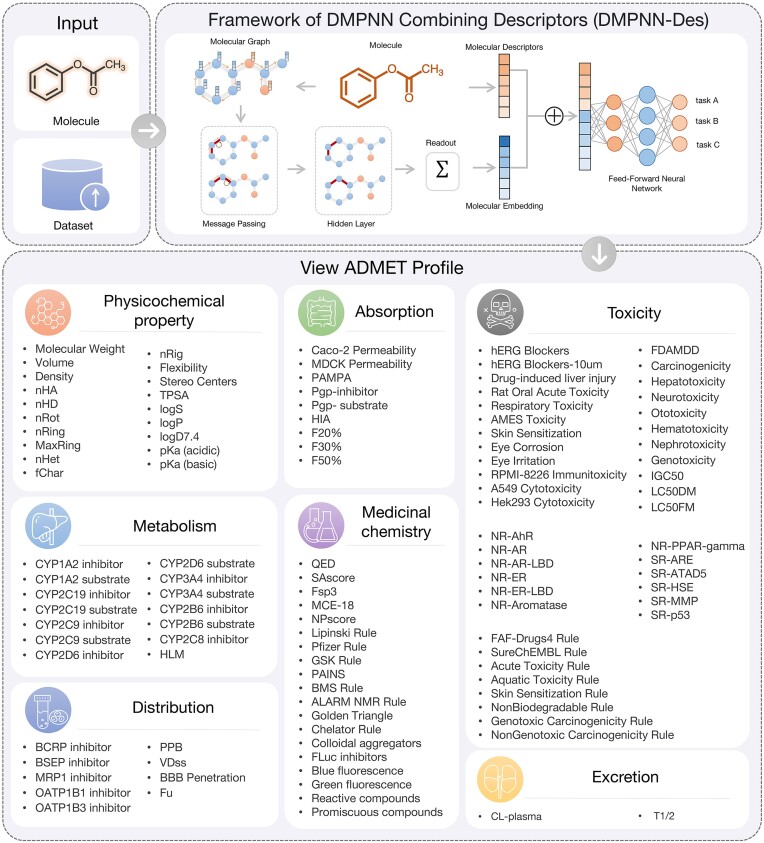
The multi-task DMPNN-Des framework in this work consists of four key modules. Firstly, there is a feature-extraction function that transforms an input molecule into both a molecular graph and a RDKit 2D descriptor vector. Secondly, a DMPNN structure is employed to learn atomic and bond embeddings from molecular graph features. Subsequently, an aggregation function is utilized to concatenate the graph readout feature and RDKit 2D descriptors, creating a more comprehensive molecular embedding. Lastly, a standard feed-forward neural network is employed to transform these molecular embeddings into target property values. The overview of ADMET profiles and the framework of the DMPNN-Des model is summarized in Figure 1.
Figure 1.
Overview scheme of ADMETlab 3.0 data and DMPNN-Des model framework.
Model validation methods
In this latest update, a comprehensive set of 77 prediction models including 59 classification models and 18 regression models, has been implemented. Additionally, the platform includes 34 other directly computable endpoints and 8 drug-related rules, which cover the computation of physicochemical properties utilizing the RDKit package, the generation of medicinal chemistry and toxicophore rules using Scopy (33), and the incorporation of six frequent hitter detection methods developed by our team (34–37). For each ADMET endpoint, the dataset was randomly split into training, validation, and test sets by a ratio of 8:1:1. The larger part was used for training, and the validation and test sets were used to optimize hyperparameters and test the predictive capacity of each model, respectively. We built and compared models based on sole DMPNN and DMPNN-Des for all the 77 endpoints with a view to selecting the best prediction model to deploy on the website. The Adam optimizer method (38) was applied for training the models, with Bayesian optimization was adopted for hyperparameter optimization. The detailed information of the optimal hyperparameters of DMPNN and DMPNN-Des models is listed in Supplementary Tables S2 and S3, respectively. To evaluate the performance of the models, R-square (R2), root mean squared error (RMSE), and mean absolute error (MAE) were applied for regression tasks, while the area under the ROC curve (AUC), accuracy (ACC), and Matthews correlation coefficient (MCC) (39) were applied for classification tasks. To obtain robust and accurate prediction models, each training process was repeated five times with randomly partitioned data, and the best-performing models were incorporated into the online platform.
Webserver implementation
Similar to its predecessor, the ADMETlab 3.0 application has been crafted using Django as its backbone. The front end, using frameworks such as Bootstrap and JQuery, is responsible for obtaining user queries, communicating with the back end, and selecting visual components in a user-friendly interface. Better than ADMET1.0 and 2.0, ADMETlab 3.0 introduces an API that can be easily integrated into various research pipelines. This API, implemented using Django Ninja, is responsible for crucial tasks such as storing computational data, overseeing the execution and management of predictive analyses for ADMET-related compounds, and enabling interoperability with external applications through a public API. Furthermore, a noteworthy addition to the latest version is the incorporation of a caching mechanism. This module temporarily stores user calculation results on the server through database storage, which aims to improve the overall speed and efficiency of the web service. Chemprop was utilized for the implementation of prediction models. The web server has been successfully tested on the latest versions of popular browsers, including Mozilla Firefox, Google Chrome, Microsoft Edge and Apple Safari.
New features and updates
Comprehensive coverage of ADMET endpoints
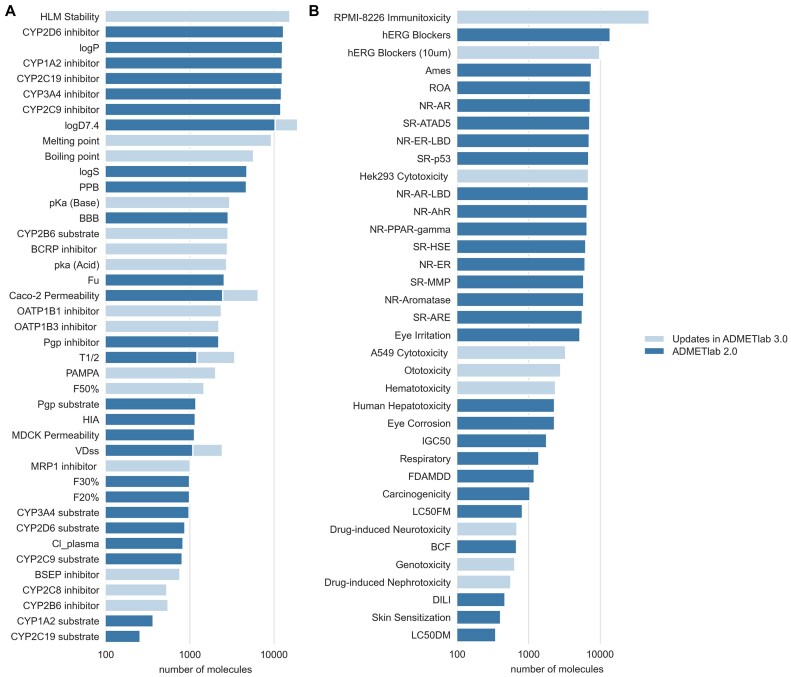
To obtain as much ADMET-related data as possible and further improve the practicability of the ADMET webserver, we collected new data from two aspects: new data for new endpoints and new data for existing endpoints. Up to now, we extended the number of predictable endpoints from 88 in ADMETlab 2.0 to 119 in ADMET 3.0, that is, we have incorporated 31 new endpoints. In addition, we also collected new molecules for four previously existing endpoints. As a result, the number of updated entries is 1.5 times larger than the previous version, which is over 400 000. Herein, we presented a comparison between the training datasets available in ADMETlab versions 2.0 and 3.0 and the details can be shown in Figure 2.
Figure 2.
(A) The dataset sizes for absorption, distribution, metabolism, and physicochemical properties. (B) The dataset sizes for toxicity properties. The wathet blue (lighter) bars represent the volume of renewed data, while the Aegean blue (darker) bars indicate the existing data volume.
The 119 ADMET endpoints in ADMET 3.0 are composed of 21 physicochemical, 20 medical chemistry, 9 absorption, 9 distribution, 14 metabolism, 2 excretion, 36 toxicity properties and 8 toxicophore rules. The detailed information of the 119 endpoints including their data description, results interpretation, and empirical decision can be seen in the ‘Help’ section of the website. Compared with ADMET 2.0, the 31 newly added endpoints are as follows: 4 physicochemical, 7 medical chemistry, 2 absorption, 5 distribution, 4 metabolism properties and 9 toxicity endpoints. Among these, 7 medicinal chemistry endpoints are directly computable, requiring no additional data support. The data for the remaining 24 endpoints are utilized to expand the ADMET database, constructing more comprehensive and precise models.
The update includes new physicochemical properties such as pKa(acid), pKa(base), melting point and boiling point. For absorption, two endpoints—human oral bioavailability (50%) and Parallel Artificial Membrane Permeability Assay (PAMPA)—were integrated. Distribution data covers trans-membrane transporter inhibitors. Metabolism updates feature CYP inhibition and human liver microsomal stability data. The toxicity section now includes five types of toxicity (nephrotoxicity, neurotoxicity, ototoxicity, hematotoxicity, genotoxicity) and in vitro toxicity assessments (hERG blockers, RPMI-8226 immunotoxicity, A549 and Hek293 cytotoxicity). Notably, hematotoxicity data stems from recent research on chemical structure and hematotoxicity relationships.
Increased diversity and quantity of data contribute to a more general understanding of the relationships between molecular features and the corresponding properties to be learned, leading to more robust and accurate predictions. Therefore, in addition to the above new endpoints and their datasets, we also expanded the volume of data for four important datasets for drug absorption, distribution, and excretion, namely Caco-2, logD7.4, VDss and half-life, with increasing rates of 20%, 20%, 125% and 20%, respectively. We made a thorough analysis on data distribution and diversity for each endpoint. Users can access these information in the ‘Diversity & Distribution’ item under ‘Help’ section of the website to gain a deeper understanding of the data composition for the model.
Overall, the high-quality updates to the data have made ADMETlab 3.0 the most comprehensive ADMET online prediction platform known to date. These processed datasets laid a solid foundation for subsequent model construction, accurate prediction, and in vitro or in vivo in-depth study of ADMET properties of new compound molecules.
Robust and accurate multi-task DMPNN models
There are a total of 77 predictive models based on DMPNN-Des or DMPNN, of which 18 are regression models and 59 are classification models. The performance scores for the classification and regression models for both MPNN and MPNN-Des were summarized in Supplementary Tables S4 and S5, respectively. For these regression models, R2 values of the existing 13 endpoints mainly ranged from 0.75 to 0.95. Even for the endpoint with the lowest performance, the R2 value for LC50FM still reaches a satisfactory 0.68. Regarding the newly added five regression endpoints, the R2 values for four of them fell between 0.8 and 0.9. The remaining one, T1/2 (a classification endpoint in ADMET2.0), although hindered by its complex pharmacological mechanism and a small dataset, still achieved an R2 of near 0.7. For these classification models, the AUC values of existing 40 endpoints were in the range of 0.72 to 0.99, and the AUC values of the newly added 20 classification endpoints were between 0.73 and 0.96, which have also been proven to be practical and reliable to a certain extent. In conclusion, the overall performance of the ADMET predictive models based on DMPNN was excellent for both regression and classification endpoints and we believe that the online predictive webserver developed based on these models can provide extensive, accurate and detailed ADMET information for drug developers and medicinal chemists.
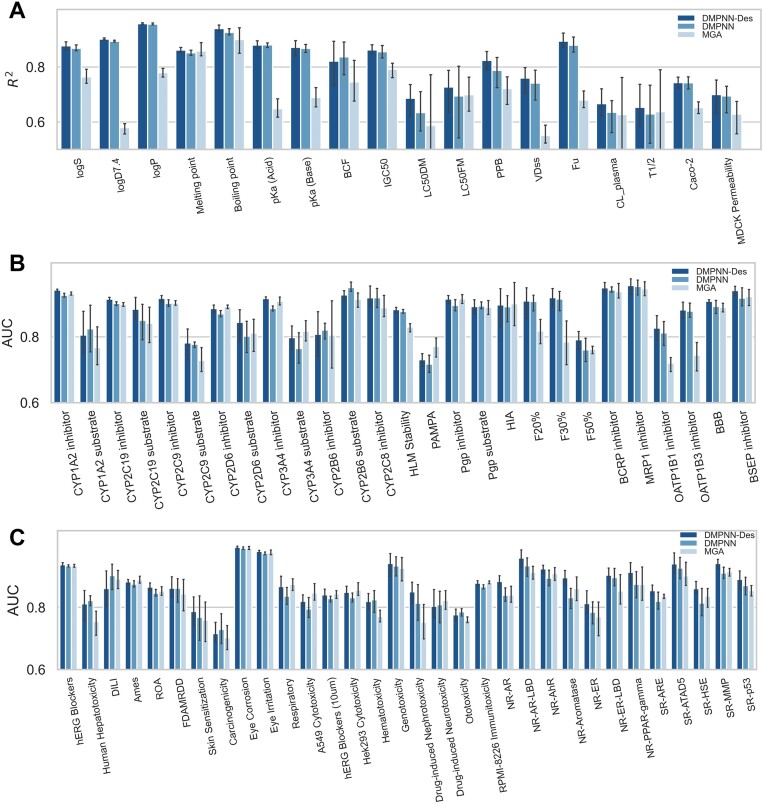
To further assess the performance and efficacy of our models, we conducted a comprehensive comparison of the two types of models embedded in ADMETlab 3.0, namely DMPNN-Des and DMPNN, against MGA models. All these models were trained and tested on the same dataset and split method used in this study. The outcomes of this comparative analysis were visually depicted in Figure 3. As illustrated in Figure 3A, for most tasks in regression tasks, both DMPNN-Des and DMPNN models exhibited significantly superior R2 performance compared to MGA.
Figure 3.
Comparison among DMPNN-Des, DMPNN and MGA in regression and classification tasks. (A) R2 of regression tasks. (B) AUC of pharmacokinetics parameters (ADME) in classification tasks. (C) AUC of toxicity parameters in classification tasks.
However, it's noteworthy that for certain specific tasks, such as melting point, LC50FM and half-life, MGA demonstrated slightly superior performance compared to the naïve MPNN in the past. Nevertheless, its performance remained lower than that achieved by MPNN-Des. In classification tasks, both DMPNN-Des and DMPNN consistently showed significantly higher AUC scores when compared with MGA. Notably, these methods outperformed MGA in 47 out of 59 tasks, underscoring their superior performance in the classification domain. In terms of the AUC of ADME (Figure 3B), MGA demonstrated superior AUC scores for only four of the 27 tasks, including two cytochrome enzyme endpoints and two absorption endpoints. For the performance of toxicity tasks (Figure 3C), MGA only achieved higher AUC scores for seven out of 32 endpoints, including AMES, respiratory toxicity, nephrotoxicity, hERG 10 μm, and three others. The above results suggested that DMPNN-Des or DMPNN generally exhibited a more dominant and superior classification performance over the MGA method. The overall performance of DMPNN-Des was slightly better than DMPNN; however, the addition of descriptors can slightly reduce processing speed (see in the following runtime analysis). To provide users with flexibility, both DMPNN-Des, and DMPNN are available as options in the webserver API, allowing users to select their preferred model for compound evaluation.
API integration and architecture upgrades
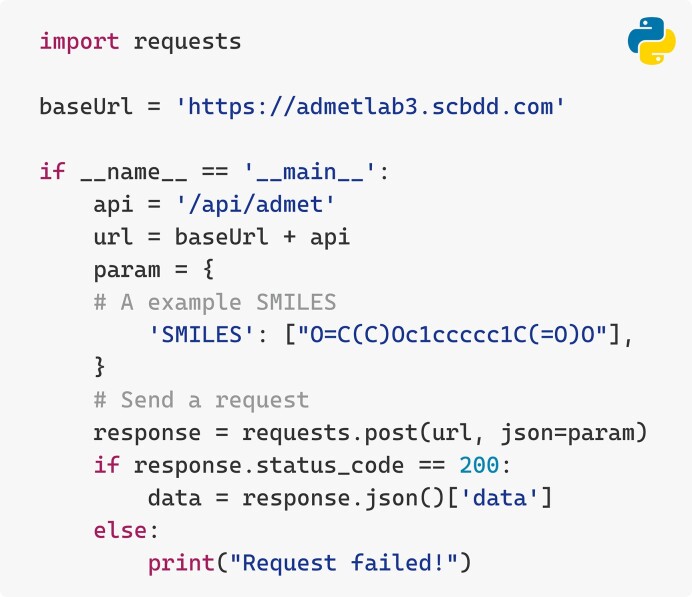
The Application Programming Interface (API) in ADMETlab 3.0 provides the option for researchers to use the command line for efficient access. This is particularly beneficial for tasks involving large volume of data. The API achieves accessibility by employing well-established protocols that are compatible with widely used programming languages, thus simplifying interactions with the core functionalities of the web server. Users can retrieve comprehensive calculation results conveniently via a simple script (Figure 4). A tutorial introducing the utility and providing detailed code examples can be found in the ‘API Tutorial’ section on the website.
Figure 4.
A Python code example for making predictions by calling an API.
ADMETlab 3.0′s API provides two key functions: (i) Molecule Wash. This includes practical functions crucial for generating a reliable outcome, including standardizing molecules, addressing fragments, assigning charges, handling tautomerism, isotope correction, and managing stereochemistry, which are crucial for generating a reliable outcome. (ii) Off-website Batch Prediction. the API returns all the 119 ADMET-related properties for a request molecule. Users can follow the tutorial or use the example scripts to obtain the full prediction results in CSV format.
Additionally, the API can return the ‘structure’ as an SVG string representing the molecular structure and a unique identifier ‘taskid’ for result retrieval. Note that uncertainty estimates for each prediction result are exclusively available through the API. To maintain server stability, we recommend no more than five requests per second. For optimal user experience, web interface queries take precedence over API requests. Notably, users can choose between DMPNN or DMPNN-Des as the prediction model. The API’s flexibility also encourages developers to leverage its functionality for broader applications or integrations, including the development of repositories, graphic user interfaces, and web applications for ADMET evaluation. By making full use of API functionality, the efficiency of high throughout ADMET evaluation by medicinal chemists is expected to be greatly improved.
Uncertainty evaluation for predicted results
In predictive modelling, offering uncertainty estimates for predictive results is a crucial metric to evaluate the accuracy and reliability of predictions. This metric reflects the model's confidence, with lower uncertainty indicating increased confidence and higher uncertainty signalling an unreliable prediction for a given molecule.
For the regression model, an evidence-based deep learning approach is utilized to predict target properties and deduce the parameters of the underlying evidential distribution (40). The method integrates an evidential layer after feature extraction in a strategic position to capture nuanced support for each prediction. Implemented in Chemprop, we adopted the ‘evidential_total’ estimation method for the regression model, which goes beyond conventional uncertainty assessments by calculating both evidential epistemic and aleatoric uncertainty. For the classification model, Monte Carlo dropout is employed to assess uncertainty across different properties (41). Monte Carlo dropout assesses uncertainty across different properties by simulating an ensemble of sub-models within a single neural network. It generates a distribution of outcomes, offering a prediction mean as the predicted value and a variance as an uncertainty score for each property.
We presented the model's RMSE range within distinct uncertainty intervals for regression tasks and provided uncertainty thresholds with their corresponding maximum Youden's index for classification tasks. In classification, the uncertainty score exceeding maximum Youden's index designates the model's prediction as ‘low confidence’, while prediction uncertainty below this threshold indicates ‘high confidence’ in the model's prediction. Users can retrieve prediction results with these confidence labels through the API, and additional details can be found in Supplementary Tables S6 and S7 for reference on confidence values.
Comparison with other web-based tools
We compared endpoint information and processing efficiency among ADMETlab 3.0, ADMETlab 2.0, and several popular ADMET prediction platforms, including SwissADME, admetSAR2.0, FAF-Drugs4, pkCSM, vNN-ADMET, ADMET-boost and Interpret-ADMET. Details are summarized in Table 1. As indicated by the results, ADMETlab 3.0 and 2.0 version unquestionably exhibits superior data support and evaluation performance on SwissADME, admetSAR2.0, FAF-Drugs4, pkCSM and vNN-ADMET. In comparison with the two latest platforms, ADMET-boost and Interpret-ADMET, ADMETlab 3.0 also showed overall better coverage and utility performance. ADMET-boost covered 22 ADMET related parameters plus 7 physiochemical properties. Its optimal model turned out to be extreme gradient boosting, a method that has proven powerful on moderate or smaller training datasets (42). Interpret-ADMET, on the other hand, can predict 59 ADMET properties and physiochemical properties. Specifically, Interpretable-ADMET not only made predictions but also offered an interpretation module and an optimization module to identify key substructures for specific properties and optimize the structure afterwards. However, due to its limitations on user access and the lack of an interface to perform batch calculations, its practicality is largely limited. ADMETlab 3.0, on the other hand, demonstrates interpretability by providing uncertainty estimation scores for prediction results, utilizing coloured dots to represent empirical decision state of each output, and highlighting alert substructures. Additionally, we have significantly improved the user experience by introducing elaborate documentation, better guidance, and enhanced web design compared to 2.0 version. The detailed information for endpoint explanation, data distribution and other aspects are summarized in Supplementary Document and the ‘help’ section on the website. In runtime analysis, ADMETlab 3.0 exhibited a slightly longer runtime compared to ADMETlab 2.0 in the web portal. This is considered a commendable performance, given that ADMETlab 3.0 has 31 more endpoints to calculate. Detailed runtime assessment results of ADMETlab 3.0 with different modelling options can be found in Supplementary Table S8 and Supplementary Figure S1. For users who prioritize uncertainty reference, the combination of DMPNN-Des and uncertainty demonstrated optimal performance on runtime. However, for those primarily concerned with runtime efficiency, DMPNN-Des can be the preferred choice, as it exhibited superior computational efficiency across varying dataset sizes while maintained a better performance over sole DMPNN.
Table 1.
Comparison of the main features of ADMETlab (2.0 and 3.0 version) with other web-based platforms
| Features | ADMETlab 3.0 | ADMETlab 2.0 | SwissADME | admetSAR 2.0 | FAF-Drugs4 | pkCSM | vNN-ADME | ADMETboost | Interpretable-ADMET |
|---|---|---|---|---|---|---|---|---|---|
| Physicochemical property | 21 | 17 | 12 | 5 | 20 | 6 | 0 | 7 | 10 |
| Medicinal chemistry | 20 | 13 | 10 | 0 | 16 | 0 | 0 | 0 | 0 |
| ADME | 34 | 23 | 9 | 35 | 0 | 20 | 9 | 18 | 20 |
| Toxicity | 36 | 27 | 0 | 12 | 0 | 10 | 6 | 4 | 29 |
| Toxicophoric rule | 8 | 8 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| PAINS included | Yes | Yes | Yes | No | Yes | No | No | No | Yes |
| Batch evaluation/API support | +++ | ++ | + | + | ++ | ++ | ++ | + | + |
| Explanation | +++ | ++ | ++ | + | ++ | ++ | + | + | +++ |
| Uncertainty estimation | Yes | No | No | No | No | No | No | No | No |
| Availability | Free | Free | Free | Free | Free | Free | Registration required | Free | Restricted visit |
| Computation time (1000 molecules) | 87 | 84 | 1560 | 267 | 967 | 1845 | 2400 | 5 (per molecule) | Restricted evaluation |
*Medicinal chemistry contains drug-likeness rules, chemical friendly measures, and substructural rules of frequent hitters; ADME contains absorption, distribution, metabolism, and excretion related endpoints; Toxicity contains human toxicity, animal toxicity, environmental toxicity, and toxic pathways. A higher number of ‘+’ symbols indicates better support in the respective item. Runtime assessment for each platform was conducted ten times, and the average runtime value in seconds was demonstrated.
URL links:
ADMETlab 2.0: https://admetmesh.scbdd.com/
SwissADME: http://www.swissadme.ch/
admetSAR 2.0: http://lmmd.ecust.edu.cn/admetsar2/
FAF-Drugs4: https://fafdrugs4.rpbs.univ-paris-diderot.fr/
FAF-Drugs4: https://fafdrugs4.rpbs.univ-paris-diderot.fr/
ADMETboost: https://ai-druglab.smu.edu/admet
Interpretable-ADMET: http://cadd.pharmacy.nankai.edu.cn/interpretableadmet/
Conclusions and future plans
ADMETlab 3.0 marks a significant advance in the field of in silico tools for drug development. Building upon the successes of its predecessor, ADMETlab 3.0 not only rectifies limitations related to narrower coverage, uncertainty estimation deficiencies, and integration capabilities but also introduces innovative features to align with the dynamic requirements of the field. By incorporating the multi-task DMPNN modeling method to aggregate local molecular information, and further enhancing it by combining RDKit 2D descriptors with global molecular information, the updated model structure ensures the precision, efficiency, and reliability of ADMET predictions. The augmentation of the dataset, encompassing over 400 000 entries and 31 additional endpoints firmly establish ADMETlab 3.0 as a comprehensive and potent platform. The integration of API capabilities streamlines in-batch evaluation, especially beneficial for users handling substantial data volumes. Furthermore, the implementation of an uncertainty estimation module addresses challenges associated with interpretation, out-of-domain regimes, and robustness guarantees. This enhancement provides a valuable tool for informed decision-making in candidate prioritization during virtual screening. With these advancements, ADMETlab 3.0 is poised to play a vital role as an indispensable resource for researchers and practitioners in accelerating drug research and development.
Supplementary Material
Acknowledgements
We acknowledge Haikun Xu, and the High-Performance Computing Center of Central South University for support. The study was approved by the university's review board.
Contributor Information
Li Fu, Xiangya School of Pharmaceutical Sciences, Central South University, Changsha, Hunan 410013, P.R. China.
Shaohua Shi, School of Chinese Medicine, Hong Kong Baptist University, Kowloon, Hong Kong SAR, 999077, P.R. China.
Jiacai Yi, School of Computer Science, National University of Defense Technology, Changsha, Hunan 410073, P.R. China.
Ningning Wang, Xiangya Hospital of Central South University, Changsha, Hunan 410008, P.R. China.
Yuanhang He, Xiangya School of Pharmaceutical Sciences, Central South University, Changsha, Hunan 410013, P.R. China.
Zhenxing Wu, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, Zhejiang 310058, P.R. China.
Jinfu Peng, Xiangya School of Pharmaceutical Sciences, Central South University, Changsha, Hunan 410013, P.R. China.
Youchao Deng, Xiangya School of Pharmaceutical Sciences, Central South University, Changsha, Hunan 410013, P.R. China.
Wenxuan Wang, Xiangya School of Pharmaceutical Sciences, Central South University, Changsha, Hunan 410013, P.R. China.
Chengkun Wu, School of Computer Science, National University of Defense Technology, Changsha, Hunan 410073, P.R. China.
Aiping Lyu, School of Chinese Medicine, Hong Kong Baptist University, Kowloon, Hong Kong SAR, 999077, P.R. China.
Xiangxiang Zeng, Department of Computer Science, Hunan University, Changsha, Hunan 410082, P.R. China.
Wentao Zhao, School of Computer Science, National University of Defense Technology, Changsha, Hunan 410073, P.R. China.
Tingjun Hou, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, Zhejiang 310058, P.R. China.
Dongsheng Cao, Xiangya School of Pharmaceutical Sciences, Central South University, Changsha, Hunan 410013, P.R. China.
Data availability
ADMETlab 3.0 is publicly accessible without registration at https://admetlab3.scbdd.com or via API. Results are promptly displayed on the website and available for download in optional formats.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Key Research and Development Program of China [2021YFF1201400]; National Natural Science Foundation of China [22173118, 22220102001]; Hunan Provincial Science Fund for Distinguished Young Scholars [2021JJ10068]; Science and Technology Innovation Program of Hunan Province [2021RC4011]; Natural Science Foundation of Hunan Province [2022JJ80104]; 2020 Guangdong Provincial Science and Technology Innovation Strategy Special Fund [2020B1212030006, Guangdong-Hong Kong-Macau Joint Lab]. Funding for open access charge: HKBU Strategic Development Fund project [SDF19-0402-P02].
Conflict of interest statement. None declared.
References
- 1. Pammolli F., Magazzini L., Riccaboni M. The productivity crisis in pharmaceutical R&D. Nat. Rev. Drug Discov. 2011; 10:428–438. [DOI] [PubMed] [Google Scholar]
- 2. Dowden H., Munro J. Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 2019; 18:495–496. [DOI] [PubMed] [Google Scholar]
- 3. Takebe T., Imai R., Ono S. The current status of drug discovery and development as originated in United States academia: the influence of industrial and academic collaboration on drug discovery and development. Clin. Transl. Sci. 2018; 11:597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harrison R.K. Phase II and phase III failures: 2013-2015. Nat. Rev. Drug Discov. 2016; 15:817–818. [DOI] [PubMed] [Google Scholar]
- 5. Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019; 35:1067–1069. [DOI] [PubMed] [Google Scholar]
- 6. Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017; 7:42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banerjee P., Eckert A.O., Schrey A.K., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018; 46:W257–W263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A. et al. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021; 49:W5–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tian H., Ketkar R., Tao P. ADMETboost: a web server for accurate ADMET prediction. J. Mol. Model. 2022; 28:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei Y., Li S., Li Z., Wan Z., Lin J. Interpretable-ADMET: a web service for ADMET prediction and optimization based on deep neural representation. Bioinformatics. 2022; 38:2863–2871. [DOI] [PubMed] [Google Scholar]
- 11. Dulsat J., Lopez-Nieto B., Estrada-Tejedor R., Borrell J.I. Evaluation of free online ADMET tools for academic or small biotech environments. Molecules. 2023; 28:776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hasselgren C., Oprea T.I. Artificial intelligence for drug discovery: are we there yet. Annu. Rev. Pharmacol. Toxicol. 2023; 64:527–550. [DOI] [PubMed] [Google Scholar]
- 13. Tran T.T.V., Surya Wibowo A., Tayara H., Chong K.T. Artificial intelligence in drug toxicity prediction: recent advances, challenges, and future perspectives. J. Chem. Inf. Model. 2023; 63:2628–2643. [DOI] [PubMed] [Google Scholar]
- 14. Chung Y., Vermeire F.H., Wu H., Walker P.J., Abraham M.H., Green W.H. Group contribution and machine learning approaches to predict Abraham Solute parameters, solvation free energy, and Solvation enthalpy. J. Chem. Inf. Model. 2022; 62:433–446. [DOI] [PubMed] [Google Scholar]
- 15. Greenman K.P., Green W.H., Gomez-Bombarelli R. Multi-fidelity prediction of molecular optical peaks with deep learning. Chem. Sci. 2022; 13:1152–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Felix E., Magarinos M.P., Mosquera J.F., Mutowo P., Nowotka M. et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B. et al. PubChem 2023 update. Nucleic Acids Res. 2023; 51:D1373–D1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sushko I., Novotarskyi S., Korner R., Pandey A.K., Rupp M., Teetz W., Brandmaier S., Abdelaziz A., Prokopenko V.V., Tanchuk V.Y. et al. Online chemical modeling environment (OCHEM): web platform for data storage, model development and publishing of chemical information. J Comput. Aided Mol. Des. 2011; 25:533–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Arnot J.A., Brown T.N., Wania F. Estimating screening-level organic chemical half-lives in humans. Environ. Sci. Technol. 2014; 48:723–730. [DOI] [PubMed] [Google Scholar]
- 20. Wang Y., Liu H., Fan Y., Chen X., Yang Y., Zhu L., Zhao J., Chen Y., Zhang Y. In Silico prediction of Human intravenous pharmacokinetic parameters with improved accuracy. J. Chem. Inf. Model. 2019; 59:3968–3980. [DOI] [PubMed] [Google Scholar]
- 21. Duan Y.J., Fu L., Zhang X.C., Long T.Z., He Y.H., Liu Z.Q., Lu A.P., Deng Y.F., Hsieh C.Y., Hou T.J. et al. Improved GNNs for log D(7.4) prediction by transferring knowledge from low-fidelity data. J. Chem. Inf. Model. 2023; 63:2345–2359. [DOI] [PubMed] [Google Scholar]
- 22. Dong J., Wang N.-N., Liu K.-Y., Zhu M.-F., Yun Y.-H., Zeng W.-B., Chen A.F., Cao D.-S. ChemBCPP: a freely available web server for calculating commonly used physicochemical properties. Chemom. Intell. Lab. Syst. 2017; 171:65–73. [Google Scholar]
- 23. Wu J., Wan Y., Wu Z., Zhang S., Cao D., Hsieh C.Y., Hou T. MF-SuP-pK(a): multi-fidelity modeling with subgraph pooling mechanism for pK(a) prediction. Acta Pharm. Sin. B. 2023; 13:2572–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu J., Wang J., Zhao H., Gao J., Kang Y., Cao D., Wang Z., Hou T. Organic compound synthetic accessibility prediction based on the graph attention mechanism. J. Chem. Inf. Model. 2022; 62:2973–2986. [DOI] [PubMed] [Google Scholar]
- 25. Yang K., Swanson K., Jin W., Coley C., Eiden P., Gao H., Guzman-Perez A., Hopper T., Kelley B., Mathea M. et al. Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 2019; 59:3370–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heid E., Greenman K.P., Chung Y., Li S.C., Graff D.E., Vermeire F.H., Wu H., Green W.H., McGill C.J. Chemprop: a machine learning package for chemical property prediction. J. Chem. Inf. Model. 2024; 64:9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cai H., Zhang H., Zhao D., Wu J., Wang L. FP-GNN: a versatile deep learning architecture for enhanced molecular property prediction. Brief Bioinform. 2022; 23:bbac408. [DOI] [PubMed] [Google Scholar]
- 28. Stokes J.M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N.M., MacNair C.R., French S., Carfrae L.A., Bloom-Ackermann Z. et al. A deep learning approach to antibiotic discovery. Cell. 2020; 180:688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu G., Catacutan D.B., Rathod K., Swanson K., Jin W., Mohammed J.C., Chiappino-Pepe A., Syed S.A., Fragis M., Rachwalski K. et al. Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii. Nat. Chem. Biol. 2023; 19:1342–1350. [DOI] [PubMed] [Google Scholar]
- 30. Wong F., Zheng E.J., Valeri J.A., Donghia N.M., Anahtar M.N., Omori S., Li A., Cubillos-Ruiz A., Krishnan A., Jin W. et al. Discovery of a structural class of antibiotics with explainable deep learning. Nature. 2023; 626:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heid E., Green W.H. Machine learning of reaction properties via learned representations of the condensed graph of reaction. J. Chem. Inf. Model. 2022; 62:2101–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McGill C., Forsuelo M., Guan Y., Green W.H. Predicting infrared spectra with message passing neural networks. J. Chem. Inf. Model. 2021; 61:2594–2609. [DOI] [PubMed] [Google Scholar]
- 33. Yang Z.Y., Yang Z.J., Lu A.P., Hou T.J., Cao D.S. Scopy: an integrated negative design python library for desirable HTS/VS database design. Brief. Bioinform. 2021; 22:bbaa194. [DOI] [PubMed] [Google Scholar]
- 34. Yang Z.Y., Yang Z.J., Dong J., Wang L.L., Zhang L.X., Ding J.J., Ding X.Q., Lu A.P., Hou T.J., Cao D.S. Structural analysis and identification of colloidal aggregators in drug discovery. J. Chem. Inf. Model. 2019; 59:3714–3726. [DOI] [PubMed] [Google Scholar]
- 35. Yang Z.Y., Dong J., Yang Z.J., Yin M., Jiang H.L., Lu A.P., Chen X., Hou T.J., Cao D.S. ChemFLuo: a web-server for structure analysis and identification of fluorescent compounds. Brief. Bioinform. 2021; 22:bbaa282. [DOI] [PubMed] [Google Scholar]
- 36. Yang Z.Y., Dong J., Yang Z.J., Lu A.P., Hou T.J., Cao D.S. Structural analysis and identification of false positive hits in luciferase-based assays. J. Chem. Inf. Model. 2020; 60:2031–2043. [DOI] [PubMed] [Google Scholar]
- 37. Yang Z.Y., He J.H., Lu A.P., Hou T.J., Cao D.S. Frequent hitters: nuisance artifacts in high-throughput screening. Drug Discov. Today. 2020; 25:657–667. [DOI] [PubMed] [Google Scholar]
- 38. Kingma D.P., Ba J. Adam: a method for stochastic optimization. 2014; arXiv doi:22 December 2014, preprint: not peer reviewed 10.48550/arXiv.1412.6980. [DOI]
- 39. Chicco D., Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. Bmc Genomics [Electronic Resource]. 2020; 21:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Soleimany A.P., Amini A., Goldman S., Rus D., Bhatia S.N., Coley C.W. Evidential deep learning for guided molecular property prediction and discovery. ACS Cent. Sci. 2021; 7:1356–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gal Y., Ghahramani Z.. Maria Florina B., Kilian Q.W. Proceedings of the 33rd International Conference on Machine Learning. 2016; 48:PMLR, Proceedings of Machine Learning Research; 1050–1059. [Google Scholar]
- 42. Chen C., Zhang Q., Yu B., Yu Z., Lawrence P.J., Ma Q., Zhang Y. Improving protein-protein interactions prediction accuracy using XGBoost feature selection and stacked ensemble classifier. Comput. Biol. Med. 2020; 123:103899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ADMETlab 3.0 is publicly accessible without registration at https://admetlab3.scbdd.com or via API. Results are promptly displayed on the website and available for download in optional formats.