Abstract
Enrichment analysis, crucial for interpreting genomic, transcriptomic, and proteomic data, is expanding into metabolomics. Furthermore, there is a rising demand for integrated enrichment analysis that combines data from different studies and omics platforms, as seen in meta-analysis and multi-omics research. To address these growing needs, we have updated WebGestalt to include enrichment analysis capabilities for both metabolites and multiple input lists of analytes. We have also significantly increased analysis speed, revamped the user interface, and introduced new pathway visualizations to accommodate these updates. Notably, the adoption of a Rust backend reduced gene set enrichment analysis time by 95% from 270.64 to 12.41 s and network topology-based analysis by 89% from 159.59 to 17.31 s in our evaluation. This performance improvement is also accessible in both the R package and a newly introduced Python package. Additionally, we have updated the data in the WebGestalt database to reflect the current status of each source and have expanded our collection of pathways, networks, and gene signatures. The 2024 WebGestalt update represents a significant leap forward, offering new support for metabolomics, streamlined multi-omics analysis capabilities, and remarkable performance enhancements. Discover these updates and more at https://www.webgestalt.org.
Graphical Abstract
Graphical Abstract.
Introduction
The rapid advancement of omics technologies has provided unprecedented opportunities for understanding complex biological systems. However, the analysis and integration of diverse omics data remain challenging due to differences in data types, standards, and analytical requirements. WebGestalt (WEB-based Gene SeT AnaLysis Toolkit) has been a widely used tool in functional enrichment analysis, enabling researchers to interpret omics data through over-representation analysis (ORA), gene set enrichment analysis (GSEA), and network topology-based analysis (NTA) (1–4). For users who are new to these methods, we recommend referring to previous publications from our team and other researchers (3,5). These publications detail the unique utilities, advantages, and disadvantages of the three complementary enrichment analysis methods. In response to the evolving demands of the research community, the WebGestalt 2024 update not only revitalizes the underlying database and enhances the platform's existing capabilities but also introduces essential new features, including support for metabolomics and a new multi-list analysis functionality.
Metabolomics, the comprehensive analysis of small molecule metabolites in biological systems, offers insights into the metabolic status and biochemical activities (6). Pathway analysis has become a critical tool for the functional interpretation of metabolomics datasets (7–9). Yet, this analysis has faced significant challenges, such as the lack of standardization in metabolite identification, the incomplete representation of metabolites in individual pathway databases, and the complexities of selecting an appropriate background set for the widely adopted ORA method (10). To address these challenges, WebGestalt 2024 introduces support for a wide array of metabolite ID types, taps into multiple pathway databases, and employs both ORA and GSEA methods to enhance the pathway analysis of human metabolomics data.
Furthermore, the integration of omics data—whether across independent studies for meta-analysis or within a single study across multiple omics platforms for multi-omics analysis—offers a distinct opportunity to achieve a more robust and in-depth understanding of biological systems (11,12). The introduction of multi-list analysis in WebGestalt 2024 caters to both meta-analysis and multi-omics analysis, addressing the challenges related to increased data complexity, computational requirements, and the interpretation of results. To navigate these intricacies, we have developed user-friendly input and output interfaces, implemented advanced pathway visualizations, and leveraged the Rust programming language for the enrichment analysis algorithms.
This manuscript details the significant updates introduced in WebGestalt 2024, highlighting our efforts to address key challenges in metabolomics analysis, enabling multi-omics integration and delivering significant performance improvements. These and other updates can be accessed at https://www.webgestalt.org.
Data update and support for metabolomics
WebGestalt integrates ID mapping information, gene sets, and networks from various data sources into its database. This integration enables users to perform enrichment analyses across a broad spectrum of knowledge, utilizing different types of IDs as input. In the 2024 update, the existing data in the database have been updated to reflect the current state of each source. Moreover, new data has been incorporated to expand the variety of analyte/ID types, pathways, networks, and gene signatures available in the database, leading to a total of 663 247 analyte sets for enrichment analysis (Supplementary Table S1).
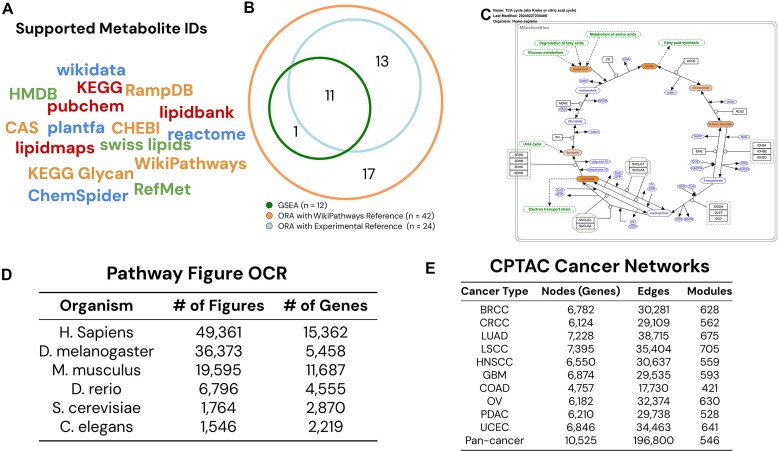
This update places a particular emphasis on introducing support for metabolomics. A major challenge in metabolomic analysis is the lack of standardization. The ID types used in metabolomic studies often vary, sometimes even within the same study, posing a significant barrier to analysis. To address this challenge and facilitate flexible ID input for metabolites, WebGestalt has adopted support for 16 metabolite ID types from the publicly available RaMP-DB (7) (Figure 1A). These IDs are mapped to RaMP-DB IDs, which are further linked to a standardized set of metabolite names using RefMet (13) to allow easy comprehension. For metabolites not in the RefMet database, the name given by RaMP-DB is used.
Figure 1.
New data and support for metabolomics. (A) WebGestalt 2024 supports 16 ID types for metabolites with automatic ID detection. (B) Choosing all metabolites in WikiPathways as the reference/background set in ORA results in many enriched pathways not found in GSEA. Limiting the reference set to all experimentally quantified metabolites increases the concordance with GSEA. (C) WebGestalt provides colored pathway maps for WikiPathways and KEGG. For GSEA analysis, identified metabolites are shaded by their input values, as shown in the down-regulated (orange-shade) metabolites in the TCA cycle pathway. (D) WebGestalt 2024 adds tens of thousands of pathways from Pathway Figure OCR for six organisms. (E) WebGestalt 2024 adds 11 new cancer networks generated from CPTAC data and a total of 6488 modules.
For the pathway analysis of metabolomics data, RaMP-DB is used as our primary source of pathway data. This database aggregates human metabolic pathways from several well-established repositories, including KEGG, Reactome, HMDB and WikiPathways (7). Of note, each database covers different metabolites, with many being exclusive to a single database. Specifically, the HMDB contains 49 968 metabolites, of which 48 933 (98%) are unique to this database. Although the other databases feature fewer metabolites, >44% of the metabolites in Reactome and WikiPathways are unique to each respective database. Overall, fewer than 1% of the metabolites listed in RaMP-DB are present in all four databases. Users have the flexibility to choose either an individual pathway collection or opt for the full RaMP-DB pathway collection for enrichment analysis. Because RaMP-DB simply concatenates pathways from individual databases, there is a significant amount of pathway redundancy. Thus, when conducting enrichment analysis with the full RaMP-DB pathway collection, it is helpful to utilize the redundancy reduction feature in WebGestalt (1,14). This allows the removal of redundant pathways from the analysis results using different computational algorithms, as described in detail in the Performance Improvements and New Software packages section, thereby facilitating clearer interpretation.
Pathway enrichment analysis of metabolomics data is conducted with either the ORA or GSEA algorithms implemented in WebGestalt. GSEA is a computationally intensive approach that utilizes the statistical outcomes from all metabolites, eliminating the need for selecting specific metabolites based on differential abundance and a background set. This contrasts with ORA, which is more time-efficient but necessitates the pre-selection of differentially abundant metabolites along with a suitable background set for analysis. WebGestalt addresses potential annotation bias by restricting the background set to metabolites documented in the utilized pathway databases for enrichment analysis. To further mitigate experimental bias, it is recommended that users refine the background set by providing the list of metabolites quantified in their study. The importance of selecting an appropriate background set for ORA is exemplified using an untargeted metabolomics dataset (15). This dataset compares HeLa cells treated with ionophore carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), which disrupts mitochondrial membrane potential, to control cells treated with DMSO, with each condition having eight replicates. Analysis of this dataset through GSEA in WebGestalt, utilizing the signed negative logarithm of P-values calculated by Limma (16) and employing the WikiPathways database, revealed 12 significantly downregulated pathways (FDR < 0.01). Analysis of the same dataset through ORA, using Limma-identified significantly downregulated metabolites (adjusted P < 0.01) and a background set limited to quantified metabolites, identified 24 significantly downregulated pathways (FDR < 0.01), and 11 (46%) overlapped with the pathways identified by GSEA (Figure 1B). Expanding the background set to include all metabolites annotated in WikiPathways led to the identification of an additional 18 downregulated pathways (FDR < 0.01), and only 1 of these (5.6%) overlapped with the pathways identified by GSEA (Figure 1B). These results suggest possible false positives introduced by experimental bias. Following the enrichment analysis, users can access color-coded pathway maps for the enriched pathways. These maps effectively highlight the involvement of different metabolites within the pathways, providing an intuitive understanding of the metabolic interactions and functions implicated in the input metabolomics data (Figure 1C). These examples are accessible from the front webpage of WebGestalt 2024.
For gene-based pathway analysis, gene sets from existing pathway databases are well-curated but do not completely encompass the wealth of pathway information available in the published literature. Using a combination of machine learning, optical character recognition (OCR), and manual curation, the Pathway Figure OCR (PFOCR) project has successfully identified tens of thousands of pathway figures within the published literature and extracted genes from these figures, providing a valuable resource for pathway analysis (17,18). We have incorporated these pathway figure-derived gene sets into the WebGestalt gene set database, including 49 361 for humans, 36 373 for fruit flies, 19 595 for mice, 6796 for zebrafish, 1764 for yeast and 1546 for roundworms (Figure 1D). This collection includes thousands of genes absent from conventional pathway databases, thus presenting new avenues for discovery and research. One distinct advantage of using the pathways in PFOCR for enrichment analysis lies in their comprehensive coverage of pathways documented in the published literature, especially those from recent publications absent in conventional pathway databases. For example, PFOCR features hundreds of pathways related to SARS-CoV-2, compared to only one such pathway in KEGG. Indeed, it has been shown that the disease coverage by PFOCR significantly exceeds that of KEGG, Reactome, and WikiPathways in both breadth and depth, leading to new biological discoveries (18). Moreover, PFOCR pathways are linked to the original articles, providing unique contextual insights and specific experimental information. However, a significant challenge arises when multiple publications describe the same pathway, leading to an accumulation of duplicated pathways in PFOCR. The redundancy reduction feature in WebGestalt (1,14) is particularly valuable for addressing this issue.
For network-based analysis, we have generated protein co-expression networks for 10 cancer types based on the recently harmonized pan-cancer proteogenomics data from the National Cancer Institute's Clinical Proteomic Tumor Analysis Consortium (CPTAC) (19), using a previously described method (20). From these networks, hierarchical network modules were identified using the NetSAM algorithm (21). These networks and modules have been incorporated into WebGestalt to enhance network-based analysis through ORA, GSEA or NTA approaches (Figure 1E). In addition, we have constructed an integrated pan-cancer functional association network that combines all protein and RNA expression data from the CPTAC pan-cancer dataset. This comprehensive network, along with its derived dense modules and hierarchical modules, has also been incorporated into WebGestalt. Moreover, we have pre-computed the enriched GO terms for each module, similar to those derived from other networks. Users can easily explore these GO terms to gain insights into the function of each module by clicking on the module name within the HTML report. Together, these new additions significantly enhance the capabilities of WebGestalt for network-based analysis, particularly in the field of human cancer research.
To facilitate the identification of cell types involved in various biological states and processes, we have curated gene signatures for 63 human and 52 mouse cell types, derived from two landmark cell landscape studies (22,23). These signatures have been integrated into the WebGestalt gene set database within a newly established ‘cell type’ category. This integration facilitates enrichment analysis against cell type signatures, offering a deeper understanding of cellular landscape dynamics associated with the input data. One limitation of our enrichment analysis methods is that they do not apply weighting to the genes in the signatures. This is particularly significant when the user's input gene list is small, as gene-weighting can be crucial for accurately estimating cell type enrichment in this scenario. Therefore, for small input gene lists, we recommend users to verify WebGestalt's outputs with machine learning-based approaches such as scPred (24) and SingleCellNet (25).
Multi-list analysis
This update introduces new support for enrichment analysis with multi-list inputs in both the WebGestalt web application and its software packages. This development aims to facilitate both meta-analysis and multi-omics analysis. Meta-analysis combines results from independent studies, typically within the same omics layer, to increase the statistical power. In contrast, multi-omics analysis integrates datasets across various omics layers, such as genomics, transcriptomics, proteomics and metabolomics, to offer a comprehensive understanding of molecular mechanisms.
This new feature requires the input of multiple analyte lists for both ORA and GSEA. It supports four types of analytes: genes, proteins, post-translational modifications (PTMs), and metabolites. To facilitate the use of this new feature, we have implemented a tab-based interface, which allows users to input, organize, and label multiple lists for one analysis. The format for input data remains consistent with that used for single list analysis. Specifically, for ORA, users can input a list of analytes in a single column and may also include an optional reference list in the same format. For GSEA, users must provide a single column list of all analytes from the study along with a corresponding numerical value for each analyte. These values, which can represent fold changes, signed minus log p-values, correlation coefficients, or other statistical measurements, are used to rank the analytes for enrichment analysis. To further simplify data entry, we have also introduced a new ID detection and database filtering system. This system automatically identifies the type of IDs entered into the web interface and filters out incompatible databases based on the selected analyte type, thereby streamlining the user experience.
For multi-list analysis, we implemented a late-stage integration approach (26). Specifically, WebGestalt runs the enrichment analysis for each list separately, then the P values from each individual input dataset for every pathway are integrated using the Stouffer's Z-score method, as implemented in the metaP R-package (https://cran.rproject.org/web/packages/metap/index.html). The resulting meta-P values and corresponding multiple test adjusted P values (meta-FDRs) are reported.
On the HTML output page, users can review the outcomes of the integrated analysis, with interactive bar graphs and tables. Users can also use tabs to view individual results for each dataset. For KEGG and WikiPathways, multi-colored pathway maps are created, providing a visual representation of individual input datasets for easy comparison.
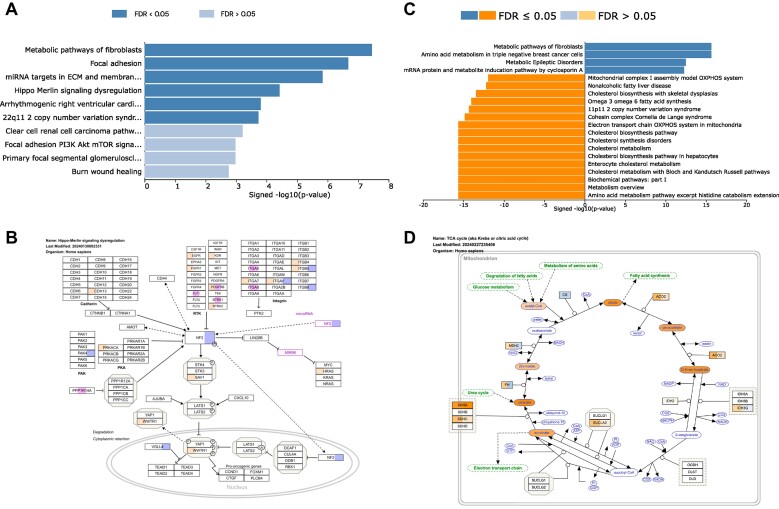
On the homepage of WebGestalt 2024, we have provided two examples for users to explore the newly introduced multi-list analysis feature. The first example demonstrates an ORA-based meta-analysis aimed at identifying pathways associated with resistance against pembrolizumab treatment in melanoma. This analysis incorporates data from three independent clinical trials sourced from ClinicalOmicsDB (27), using the top 500 genes with increased abundance in resistant tumors from each study as input. The analysis against WikiPathways identifies enriched pathways for each dataset individually (Supplemental Figure S1A–C) as well as collectively (Figure 2A). For pathways identified in the meta-analysis, corresponding multi-colored pathway maps highlight genes from individual input gene lists (Figure 2B). The second example involves a GSEA-based multi-omics analysis against the WikiPathways database, and the input includes the differential metabolomics data used for Figure 1B, alongside RNASeq and proteomics data from the same study. This analysis identifies enriched pathways for each type of omics data individually (Supplemental Figure S1D–F) and for all three combined (Figure 2C). For pathways identified in the multi-omics analysis, corresponding multi-colored pathway maps visualize the GSEA rank metrics from different data types for all leading-edge genes identified in the analysis (Figure 2D).
Figure 2.
Examples of multi-list analysis output. (A) An ORA-based meta-analysis of three input gene lists reports enriched pathways from the integrated analysis. (B) A multi-colored pathway map for a pathway enriched in the meta-analysis, highlighting genes from the individual input gene lists. (C) A GSEA-based multi-omics analysis reports enriched pathways from the integrated analysis of RNA, protein, and metabolite data. (D) A multi-colored pathway map visualizing input values from different data types for all leading-edge genes identified in the analysis.
Performance improvements and new software packages
Enrichment analysis, especially GSEA, is computationally expensive. In WebGestalt 2019, running GSEA on a typical dataset could take up to three minutes. With the addition of multi-list analysis, the amount of computation is further increased. To reduce computational time, we have reengineered the core of the computational backend using Rust. Rust is a high-performance language that allows finer control over computational resources and can be integrated as a library with other programming languages. We have recoded the ORA, GSEA and NTA algorithms in Rust, and used this library as the foundation for the existing R package. This new architecture allows users of both the web application and the R package to experience improved performance without changing their existing workflows. As Rust can be compiled for use across multiple programming languages, we further developed a new Python package, WebGestaltPy, to expand the access to WebGestalt. WebGestaltPy provides an API that supports ORA and GSEA analysis for both single list and multi-list inputs. Additionally, the Rust library has been made publicly available, allowing developers to integrate its capabilities into their own tools.
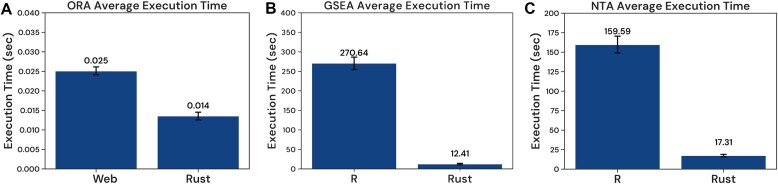
To evaluate the improvements in enrichment analysis speed within WebGestalt 2024, we conducted a comparative assessment of computational efficiency relative to WebGestalt 2019. Differential gene expression data from resistant versus sensitive breast tumors in the BrighTNess trial's veliparib treatment arm (28), downloaded from ClinicalOmicsDB (27), were used for enrichment against the KEGG database. The ORA analysis used the top 500 most significantly altered genes. The computational time for the ORA analysis was reduced from 0.025 s to 0.014 s, a decrease of 0.011 s or 44% (Figure 3A). For the GSEA analysis, the computational time was dramatically shortened from 270.64 s to 12.41 s, equating to a time saving of 258.23 s or 95% (Figure 3B). For the NTA analysis, the top 500 most significantly altered genes were used as input to identify the top 50 genes in the BioGrid protein-protein interaction network using the random walk with restart algorithm. The new R package reduced the computation time from 160 s to 17 s, an 89% reduction (Figure 3C). Notably, the enhanced speed has facilitated swift generation of results for ORA and GSEA analyses when querying large databases of analyte sets. As a demonstration, we reanalyzed the FCCP-responsive metabolites, used as input for Figure 1B, against the entire RaMP-DB pathway collection, which comprises 52301 pathways. Through the new web interface, the ORA analysis delivered results in just 6 s, and the GSEA analysis completed in 17 s, both processes including the generation of HTML reports with visualizations.
Figure 3.
Execution time comparison between the 2024 and 2019 implementations. (A) Execution times for the ORA implementations. (B) Execution times for the GSEA implementations. (C) Execution times for the NTA implementations. (A), (B) and (C) are based on data from 10 runs.
To mitigate the issue of functionally similar enriched sets cluttering the output and reducing interpretability, we introduced a redundancy removal feature in WebGestalt 2019 (1,14). This feature programmatically filters enrichment results to display the most representative gene sets, using either the weighted set cover or affinity propagation methods. Building on this, WebGestalt 2024 incorporates a new k-Medoid method, which selects up to k gene sets that best represent the full spectrum of enriched gene sets, based on Jaccard similarity and employing the Partitioning Around Medoids (PAM) approach. While both affinity propagation and k-Medoid are based on assessing similarity between gene sets, k-Medoid offers a more straightforward approach for interpretation. To conserve computational resources, users have the flexibility to choose which redundancy removal methods to execute. The results page features tabs for viewing the output of each redundancy removal method, along with an option to view results without redundancy removal.
Discussion
WebGestalt 2024 marks a significant upgrade to the functional enrichment analysis platform. This update not only brings the database up to date but also enhances the tool's capabilities by incorporating support for metabolomics and introducing new pathways, networks, and gene signatures. WebGestalt 2024 provides a unique tool for enrichment analysis, with support for multiple features within a single tool (Supplemental Table S2). The introduction of multi-list analysis now supports essential functions for both meta-analysis and multi-omics analysis. Moreover, enhancements in the implementation and algorithms have enabled the web server to accommodate a higher volume of requests, process large datasets with greater efficiency, deliver rapid results for user queries, and generate concise, non-redundant outputs that facilitate scientific discovery. Additionally, we also launched new software packages, providing advanced users with the means to integrate WebGestalt into their own workflows and tools. For future developments, we aim to enhance the multi-list HTML output to more effectively visualize the impact of individual lists. Additionally, we plan to expand the functionality of our multi-list pathway maps by including legends and introducing animations.
A major challenge encountered during this update was the analysis of metabolomic data, primarily due to the lack of standardized ID formats across studies. To mitigate this, we have implemented support for 16 metabolite ID types, along with automatic detection of these IDs upon entry. Despite this advancement, challenges persist with metabolites that lack standardized IDs. This issue is particularly problematic in metabolomic studies, which typically identify fewer analytes of interest compared to other omics studies. As a result, the loss of analytes due to identification issues can significantly affect the outcomes of an enrichment analysis. Moreover, WebGestalt currently supports only human metabolic pathways. In future developments, we plan to expand our metabolomics support to include other organisms.
WebGestalt 2024 supports meta-analysis and multi-omics analysis by enabling users to aggregate individual p-values into a meta-P value for each pathway, exemplifying a late-stage integration approach (26). On the other hand, early-stage integration can be applied at the gene level, where ranks of a gene across multiple lists are consolidated into a singular ranked list, serving as the input for a single enrichment analysis. We plan to evaluate the efficacy of early-stage integration versus late-stage integration. Based on this assessment, we will consider incorporating early-stage integration as an option in our future updates, should it prove beneficial.
The adoption of Rust in WebGestalt has led to significant performance improvements. In particular, Rust's efficiency and speed, combined with its memory safety features, have contributed to a markedly expedited GSEA and NTA process. The broader integration of Rust reflects our ongoing commitment to adopting cutting-edge technologies, emphasizing our dedication to the continuous enhancement of WebGestalt and our aim to provide users with superior tools and services.
Supplementary Material
Acknowledgements
B.Z. is a McNair Scholar.
Author contributions: J.M.E. led the development of new features, with Y.L. and Z.S. contributing support. Q.Z. curated the cell type signatures. A.R.P. provided expert advice on developing the Pathway Figure OCR and WikiPathways features. B.Z. oversaw the entire study. J.M.E. and B.Z. wrote the manuscript, while all authors participated in the review and approval of the final manuscript.
Contributor Information
John M Elizarraras, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA.
Yuxing Liao, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA.
Zhiao Shi, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA.
Qian Zhu, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Alexander R Pico, Institute of Data Science and Biotechnology, Gladstone Institutes, San Francisco, CA 94158, USA.
Bing Zhang, Lester and Sue Smith Breast Center, Baylor College of Medicine, Houston, TX 77030, USA; Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
Data availability
WebGestalt can be accessed at https://www.webgestalt.org. This website is free and open to all users and there is no login requirement.
WebGestaltR is available online (bzhanglab.github.io/webgestaltr) and its source code is hosted on Github (https://github.com/bzhanglab/WebGestaltR) and Zenodo (https://doi.org/10.5281/zenodo.11186329).
WebGestaltPy is available on PyPI (https://pypi.org/project/webgestaltpy/) with source code on GitHub (https://github.com/bzhanglab/webgestaltpy/) and Zenodo (https://doi.org/10.5281/zenodo.11186400).
webgestalt-lib is available on crates.io (https://crates.io/crates/webgestalt_lib) with source code on GiHub (https://github.com/bzhanglab/webgestalt_rust/) and Zenodo (https://doi.org/10.5281/zenodo.11186377).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health (NIH) grants from the National Cancer Institute (NCI) [U24 CA271076]; McNair Medical Institute at the Robert and Janice McNair Foundation. Funding for open access charge: NCI [U24 CA271076].
Conflict of interest statement. B.Z. received research funding from AstraZeneca and consulting fee from Inotiv.
References
- 1. Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019; 47:W199–W205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang J., Duncan D., Shi Z., Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013; 41:W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang J., Vasaikar S., Shi Z., Greer M., Zhang B. WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017; 45:W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang B., Kirov S., Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005; 33:W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khatri P., Sirota M., Butte A.J. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput. Biol. 2012; 8:e1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu X., Locasale J.W. Metabolomics: a primer. Trends Biochem. Sci. 2017; 42:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braisted J., Patt A., Tindall C., Sheils T., Neyra J., Spencer K., Eicher T., Mathe E.A. RaMP-DB 2.0: a renovated knowledgebase for deriving biological and chemical insight from metabolites, proteins, and genes. Bioinformatics. 2023; 39:btac726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu Y., Pang Z., Xia J. Comprehensive investigation of pathway enrichment methods for functional interpretation of LC-MS global metabolomics data. Brief. Bioinform. 2023; 24:bbac553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pang Z., Chong J., Zhou G., de Lima Morais D.A., Chang L., Barrette M., Gauthier C., Jacques P.E., Li S., Xia J. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021; 49:W388–W396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wieder C., Frainay C., Poupin N., Rodriguez-Mier P., Vinson F., Cooke J., Lai R.P., Bundy J.G., Jourdan F., Ebbels T. Pathway analysis in metabolomics: recommendations for the use of over-representation analysis. PLoS Comput. Biol. 2021; 17:e1009105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017; 18:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toro-Dominguez D., Villatoro-Garcia J.A., Martorell-Marugan J., Roman-Montoya Y., Alarcon-Riquelme M.E., Carmona-Saez P. A survey of gene expression meta-analysis: methods and applications. Brief. Bioinform. 2021; 22:1694–1705. [DOI] [PubMed] [Google Scholar]
- 13. Fahy E., Subramaniam S. RefMet: a reference nomenclature for metabolomics. Nat. Methods. 2020; 17:1173–1174. [DOI] [PubMed] [Google Scholar]
- 14. Savage S.R., Shi Z., Liao Y., Zhang B. Graph algorithms for condensing and consolidating gene set analysis results. Mol. Cell. Proteomics. 2019; 18:S141–S152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Quiros P.M., Prado M.A., Zamboni N., D’Amico D., Williams R.W., Finley D., Gygi S.P., Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017; 216:2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hanspers K., Riutta A., Summer-Kutmon M., Pico A.R. Pathway information extracted from 25 years of pathway figures. Genome Biol. 2020; 21:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shin M.G., Pico A.R. Using published pathway figures in enrichment analysis and machine learning. BMC Genomics [Electronic Resource]. 2023; 24:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liao Y., Savage S.R., Dou Y., Shi Z., Yi X., Jiang W., Lei J.T., Zhang B. A proteogenomics data-driven knowledge base of human cancer. Cell Syst. 2023; 14:777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J., Ma Z., Carr S.A., Mertins P., Zhang H., Zhang Z., Chan D.W., Ellis M.J., Townsend R.R., Smith R.D. et al. Proteome profiling outperforms transcriptome profiling for coexpression based gene function prediction. Mol. Cell. Proteomics. 2017; 16:121–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shi Z., Wang J., Zhang B. NetGestalt: integrating multidimensional omics data over biological networks. Nat. Methods. 2013; 10:597–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F. et al. Mapping the mouse cell atlas by Microwell-Seq. 2018; 172:1091–1107. [DOI] [PubMed] [Google Scholar]
- 23. Han X., Zhou Z., Fei L., Sun H., Wang R., Chen Y., Chen H., Wang J., Tang H., Ge W. et al. Construction of a human cell landscape at single-cell level. Nature. 2020; 581:303–309. [DOI] [PubMed] [Google Scholar]
- 24. Alquicira-Hernandez J., Sathe A., Ji H.P., Nguyen Q., Powell J.E. scPred: accurate supervised method for cell-type classification from single-cell RNA-seq data. Genome Biol. 2019; 20:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tan Y., Cahan P. SingleCellNet: a computational tool to classify single cell RNA-seq data across platforms and across species. Cell Syst. 2019; 9:207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cai Z., Poulos R.C., Liu J., Zhong Q. Machine learning for multi-omics data integration in cancer. iScience. 2022; 25:103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moon C.I., Elizarraras J.M., Lei J.T., Jia B., Zhang B. ClinicalOmicsDB: exploring molecular associations of oncology drug responses in clinical trials. Nucleic Acids Res. 2024; 52:D1201–D1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loibl S., O'Shaughnessy J., Untch M., Sikov W.M., Rugo H.S., McKee M.D., Huober J., Golshan M., von Minckwitz G., Maag D. et al. Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): a randomised, phase 3 trial. Lancet Oncol. 2018; 19:497–509. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WebGestalt can be accessed at https://www.webgestalt.org. This website is free and open to all users and there is no login requirement.
WebGestaltR is available online (bzhanglab.github.io/webgestaltr) and its source code is hosted on Github (https://github.com/bzhanglab/WebGestaltR) and Zenodo (https://doi.org/10.5281/zenodo.11186329).
WebGestaltPy is available on PyPI (https://pypi.org/project/webgestaltpy/) with source code on GitHub (https://github.com/bzhanglab/webgestaltpy/) and Zenodo (https://doi.org/10.5281/zenodo.11186400).
webgestalt-lib is available on crates.io (https://crates.io/crates/webgestalt_lib) with source code on GiHub (https://github.com/bzhanglab/webgestalt_rust/) and Zenodo (https://doi.org/10.5281/zenodo.11186377).