Abstract
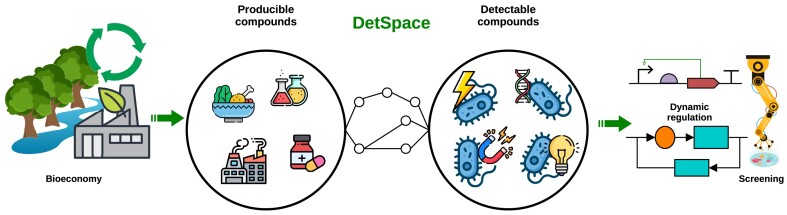
Tackling climate change challenges requires replacing current chemical industrial processes through the rational and sustainable use of biodiversity resources. To that end, production routes to key bio-based chemicals for the bioeconomy have been identified. However, their production still remains inefficient in terms of titers, rates, and yields; because of the hurdles found when scaling up. In order to make production more efficient, strategies like automated screening and dynamic pathway regulation through biosensors have been applied as part of strain optimization. However, to date, no systematic way exists to design a genetic circuit that is responsive to concentrations of a given target compound. Here, the DetSpace web server provides a set of integrated tools that allows a user to select and design a biological circuit that performs the sensing of a molecule of interest by its enzymatic conversion to a detectable molecule through a transcription factor. In that way, the DetSpace web server allows synthetic biologists to easily design biosensing routes for the dynamic regulation of metabolic pathways in applications ranging from genetic circuits design, screening, production, and bioremediation of bio-based chemicals, to diagnostics and drug delivery.
Graphical Abstract
Graphical Abstract.
Introduction
Biomanufacturing initiatives are currently thriving worldwide to address the societal challenges and threats posed by climate change (1). Policymakers and global stakeholders have established bioeconomy goals to achieve carbon neutrality in the next decade (2). A well-established objective towards that end is to replace current production processes, for a substantial amount of key compounds in the consumer value chain (which are primarily carried out by the chemical industry based on fossil resources), by greener biomanufacturing processes (3). In the last years, metabolic engineering efforts combined with prospective studies based on retrobiosynthetic tools (4) have identified biosynthetic routes for production of chemicals of industrial interest (5). However, most of the known routes have only been validated at small volumes. Therefore, in order to achieve industrial levels, it would be necessary to ensure that their bioproduction processes are still efficient and reproducible in terms of titer, yield, and rate (6,7).
Achieving desirable strains with optimal production, at fermentation levels, requires both an efficient way for high-throughput screening and a strategy for pathway robustness against context-dependency uncertainty, notably by engineering dynamic regulation in the production pathway (8). Transcription factor-based biosensors, which are genetic circuits that can detect the concentration of the target compound and elicit a response, provide a promising approach for achieving such goal (9,10). However, the number of known molecules that can act as effectors for allosteric transcription factors is limited (11). To alleviate this limitation, a strategy has been proposed based on the use of extended biosensors. The strategy consists of a pathway where through enzymatic conversions the target compound is transformed into a detectable compound (12,13); defining in that way an extended metabolic detectable space for the target compounds. The extent of such space is yet to be charted, and its determination is nowadays crucial and instrumental for the bioeconomy. This is due to the fact that this specific knowledge will substantially improve our understanding of the biomanufacturing potential capabilities for bio-based chemicals.
Here, we present DetSpace: a web server that allows a user to navigate through a comprehensive map of bio-based products of industrial interest, and to select (in a desired microbial chassis) their associated genetic circuits consisting of detectable pathways; thus leading to known effectors, as well as their associated transcription factors. Integrating the DetSpace extended detectable pathway design as part of the Design-Build-Test-Learn pipeline of biomanufacturing and biofoundries contributes to more robust and efficient bio-based production; bringing closer the fulfillment of the goals fostered by the future bioeconomy.
Materials and methods
Datasets
Chassis selection
A selection of genome-scale models from conventional and alternative chassis at lab and industrial scale (14) were compiled and their metabolomes were obtained from the BiGG database (15) or the BioCyc collection (16).
Producible compounds
The set of target compounds of industrial interest was obtained from a curated map of bio-based chemical compounds (5).
Detectable compounds
The list of detectable molecules and their associated transcription factors was obtained from Sensbio, a comprehensive database of biosensors based on allosteric transcription factors (11).
Bioretrosynthesis
The set of reaction rules RetroRules (17) was used to connect detectable to producible compounds using bioretrosynthesis. Such set can be tuned in terms of specificity by selecting the diameter, which is a parameter that determines the description of the reaction based on atom vicinity around the reaction center.
Calculation of detectable routes
Detectable routes connecting bio-based compounds were determined in the extended metabolic space through a four-step bioretrosynthesis procedure as detailed in the following.
Metabolic expansion
The RetroRules database allows the application of a generative algorithm for expanding the metabolic space by an iterative backward prediction of compounds that can be used as reactants, starting from the detectable compounds.
Monte Carlo tree search
Since the metabolic expansion is highly combinatorial, an artificial intelligence approach based on Monte Carlo tree search was used to limit the search (18). The Monte Carlo was launched for each pair of detectable and producible compounds. At each step, the new nodes of the tree are scored using Tanimoto similarity with the target molecule in order to guide the search. See Supplementary Note 1 for a detailed explanation of the algorithm.
Pathway enumeration
Once a producible compounds is reached, the set of detectable pathways is determined by combining all the shortest pathways for each branched node within the scope of biochemical transformations linking to the detectable compound. See Supplementary Note 2 for a detailed explanation of the algorithm.
Pathway ranking
Reaction rules in the RetroRules database have a penalty score, which was used to rank the pathways by selecting the higher individual score as the global score. Moreover the estimated steady-state flux of the production pathway precursors was included in the ranking calculation.
Calculation of producible routes
Starting from a comprehensive metabolic map of bio-based compounds of industrial interest (19), major natural and engineered production pathways were determined, linking the compounds to the central metabolism of the chassis. The map contains a total of 532 producible compounds and 438 biological reactions (enzyme-catalyzed reactions), which were further processed for detailed annotation and cross-link database references.
Genetic design
Each chemical reaction included in the pathways generated by DetSpace contains a link to query the Selenzyme web server (20), a free online enzyme selection tool for metabolic pathway design. This search provides the user with additional information about the enzymes that catalyze specific reactions in different chassis organisms, in order to choose the best candidates for a given pathway step; thus allowing a more accurate and rational enzyme selection. Furthermore, the resulting pathway can be exported into JSON Cytoscape.js (21) format and as SBML, allowing further processing for the generation of the genetic circuit associated with the pathway (see Supplementary Figure S1) using genetic design tools, such as the Galaxy SynBioCAD workflow (22).
Implementation
DetSpace web development
The DetSpace web server runs on the Django platform in Python 3.11, using the JQuery framework for the front end interface and the Cytoscape.js library (21) for the visualization of the metabolic networks and pathways. The code is freely available at https://github.com/dbtl-synbio/detspace.
API features
DetSpace provides an API based on the REST framework with the following functionalities: retrieval of the list of chassis organisms; producible and detectable compounds and its corresponding paired combinations; information regarding the transcription factors associated with each detectable compound; and the list of detectable pathways in both SBML and JSON format.
Description of the web server
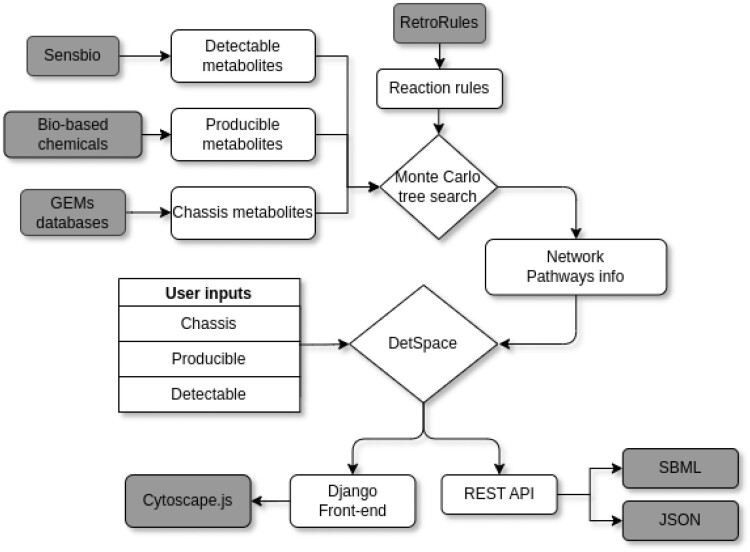
Figure 1 shows the DetSpace workflow and data sources. The entry point of the web server is a target compound selected from a list of bio-based chemicals of major interest for metabolic engineering (19) and the desired microbial host as industrial chassis. The server calculates the different alternative steps needed to convert the target chemical into one of the effector molecules reported in the literature that can be detected through a transcription factor, as well as the required precursors in the pathway. The design process consists of the following steps:
Figure 1.
Workflow of the DetSpace functionality and data sources. The set of detectable and producible metabolites go through a Monte Carlo tree search algorithm in order to generate all pathways. This information is combined on the webserver with the user-selected chassis, producible and detectable compounds.
Chassis selection
The user can select the chassis organism where the pathway will be engineered from two lists: one containing a taxonomic selection of main industrial and prototyping chassis used by the synthetic biology and metabolic engineering community classified; and the other one grouped by application. The selected chassis will determine the number of endogenous and heterologous metabolites present in the pathway. Typically, a DetSpace pathway should contain a high ratio of heterologous metabolites in order to avoid cross-talk and flux diversion and, therefore, this consideration might determine in some cases the selection of the appropriate chassis.
Target selection
The user selects the target compound of interest from a list that contains a comprehensive metabolic map for production of bio-based chemicals. DetSpace provides a curated list of bio-based chemicals that are essential building blocks and compounds for the bioeconomy, based on the metabolic map collected by Lee et al. (5). Only those compounds associated with detectable pathways are available for selection in the list.
Effector selection
The next step is the selection of the effector compound of a transcription factor that can be connected to the target product. A curated list of detectable molecules that can act as allosteric effectors for transcription factors is provided based on the molecule-transcription factor pairs database collected by Carbonell and colleagues in 2023 (11). For each selected target compound, only those effectors that can be associated through a DetSpace pathway to the compound are shown in the list.
Retrosynthetic analysis
Retrosynthetic routes for production of the precursors in the chassis are graphically displayed, as well as required supplements to add to the growth media. The routes are calculated based on a Monte Carlo tree search algorithm that prioritizes pathways involving less complex biochemical transformations from the RetroRules database of reaction rules. Each given solution pair of target product and effectors can lead to multiple alternative pathways. A prioritization list is provided based on the reaction rules scores. The pathways are displayed graphically on an interactive map where the user can select individual pathways and explore in more detail the intermediates in the pathway, as well as the information about each enzymatic transformation.
Production routes design
The main backbone upstream routes for bio-based production of the target compound can be accessed. Main production routes of bio-based compounds are provided based on their metabolic map, which can be further analyzed through the use of bioretrosynthesis tools such as RetroPath 2.0 (23).
Detectable routes design
DetSpace provides the list of enzyme sequences, selected through the Selenzyme algorithm (20) and the combinatorial library of the genetic circuits for building the detectable pathway. The selected genes and pathways can be exported into SBML format so that they can be transferred to the Galaxy SynBioCAD genetic design workflow (22), in order to build and assembly a combinatorial library for further engineering and optimization of the DetSpace detectable pathway for bio-based production.
Results
Overall results
Overall results are shown on Table 1. In total, 10 231 producible-detectable pairs were identified that can be connected through 914 192 pathways. The metabolic scope contains 2227 intermediate compounds, 1042 precursors (molecules that are in the chassis or at least a production pathway exists to produce it from the chassis according to the literature (5) or the retrosynthetic analysis (23), 1959 supplement compounds that need to be added to the media in order to make viable some of the pathways. These results were calculated for the Escherichia coli chassis. Table 2 shows chassis-independent specific numbers for each type of compound. For the producible set, there are 198 products with available DetSpace pathway, with and average number of pathway per product of 4617 and an average number of pairs close to 52. For the detectable set, there are 303 effectors with available DetSpace pathway, with and average number of pathway per effector of 3017 and an average number of pairs close to 34.
Table 1.
Overall results (for Escherichia coli)
| Pairs | Pathways | Intermediates | Precursors | Supplements | |
|---|---|---|---|---|---|
| Total | 10 231 | 914 192 | 2227 | 1042 | 1959 |
Table 2.
Results by compound
| Number of | Average no. | Average no. | |
|---|---|---|---|
| compounds | of pathways | of pairs | |
| Producible | 198 | 4617 | 51.67 |
| Detectable | 303 | 3017 | 33.76 |
Case study 1
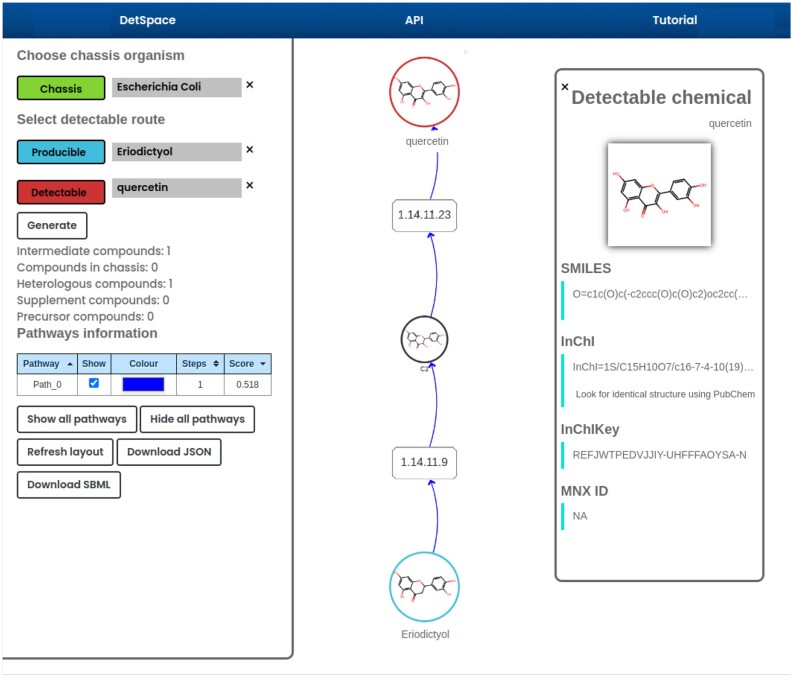
(2S)-Eriodyctiol is a flavonoid belonging to the flavanones subclass that is widespread in citrus fruits, vegetables, and medicinally important plants, and with important antioxidant and anticancer properties (24). Eriodyctiol is an hydroxylated product of (2S)-naringenin and therefore its production is of interest in the production of high-value molecules from agricultural waste. Detecting eriodyctiol through a biosensor circuit will therefore open also the possibility for indirect sensing of naringenin, with potential applications for screening and dynamic regulation (12), including directed evolution for efficient production (25). According to DetSpace, eriodictyol can be downstream transformed into two molecules with associated allosteric transcription factor, i.e., into quercetin, which can be detected by the transcription factor LmrA from Bacillus subtilis (26) by a two-step transformation involving taxifolin (see Figure 2), and into catechin by a three-step transformation (see Supplementary Figure S2), which can be detected by the transcription factor QdoR from Bacillus subtilis (26), belonging both to the repressor TetR family.
Figure 2.
Detectable pathway for the (2S)-eriodyctiol to quercetin.
Case study 2
Campesterol is a plant sterol with cholesterol lowering and anticarcinogenic effects (27). Interestingly, multiple pathways were found by DetSpace converting campesterol into acyl-CoA thioesters, like feruloyl-CoA (4 pathways), caffeoyl-CoA (12 pathways), shown in Supplementary Figure S3, oleoyl-CoA and palmitoyl-CoA (24 pathways), which are fatty esters involved in the biosynthesis of flavonoids and various alkaloids. Several species have developed allosteric transcription factors related to those compounds to regulate fatty acid metabolism, such as FadR (28) and FerC (29), in the case of feruloyl-CoA. Detection pathways linking campesterol to those effectors were found containing between five and seven steps.
Conclusions and future perspectives
Determining the extended detectable biochemical space of key bioeconomy bio-based compounds through DetSpace has a major utility, as it allows to quickly visualize and explore biosensing pathways for screening and dynamic regulation that are expected to become instrumental in order to scale up production for a substantial amount of bio-sourced compounds into industrial levels. In the near future we plan to incorporate to the DetSpace server a predictive machine learning model of biochemical properties of compounds with industrial interest (pharmacological, aromatic, toxic potential...), and to automate the design of the detectable pathways under desired specifications for dynamic and operating ranges.
Supplementary Material
Acknowledgements
H.M. and P.C. acknowledge financial support from Generalitat Valenciana through grant CIAICO/2021/159 (SmartBioFab). R.M. and P.C. acknowledge MCIN/AEI /10.13039/501100011033 and European Union NextGenerationEU/ PRTR funding through grant TED2021-131049B-I00 (BioEcoDBTL).
Contributor Information
Hèctor Martín Lázaro, Institute of Industrial Control Systems and Computing (AI2), Universitat Politècnica de València (UPV), Camí de Vera s/n, 46022 València, Spain.
Ricardo Marín Bautista, Institute of Industrial Control Systems and Computing (AI2), Universitat Politècnica de València (UPV), Camí de Vera s/n, 46022 València, Spain.
Pablo Carbonell, Institute of Industrial Control Systems and Computing (AI2), Universitat Politècnica de València (UPV), Camí de Vera s/n, 46022 València, Spain; Institute for Integrative Systems Biology I2SysBio, Universitat de València-CSIC, Escardino Street 9, Paterna, 46980 València, Spain.
Data availability
DetSpace is an open-source application, available at GitHub (https://github.com/dbtl-synbio/detspace) and FigShare at https://doi.org/10.6084/m9.figshare.25103978. The DetSpace website is free and open to all users, there is no login requirement, available at https://detspace.carbonelllab.org/.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Conselleria d'Educació, Investigació, Cultura i Esport [CIAICO/2021/159 (SmartBioFab)]; MCIN/AEI/10.13039/501100011033; European Union NextGenerationEU/PRTR [TED2021-131049B-I00 (BioEcoDBTL)]. Funding for open access charge: CIAICO/2021/159; MCIN/AEI /10.13039/501100011033, European Union NextGenerationEU/PRTR and Universitat Politècnica de València.
Conflict of interest statement. None declared.
References
- 1. Philp J. Bioeconomy and net-zero carbon: lessons from Trends in Biotechnology, volume 1, issue 1. Trends Biotechnol. 2023; 41:307–322. [DOI] [PubMed] [Google Scholar]
- 2. Akinsemolu A., Onyeaka H., Fagunwa O., Adenuga A.H. Toward a resilient future: the promise of microbial bioeconomy. Sustainability. 2023; 15:7251. [Google Scholar]
- 3. Attal-Juncqua A., Dods G., Crain N., Diggans J., Dodds D., Evans S., Fackler N., Flyangolts K., Gibson K., Kosal M.E. et al. Shaping the future US bioeconomy through safety, security, sustainability, and social responsibility. Trends Biotechnol. 2023; 10.1016/j.tibtech.2023.11.015. [DOI] [PubMed] [Google Scholar]
- 4. Otero-Muras I., Carbonell P. Automated engineering of synthetic metabolic pathways for efficient biomanufacturing. Metab. Eng. 2020; 63:61–80. [DOI] [PubMed] [Google Scholar]
- 5. Jang W.D., Kim G.B., Lee S.Y. An interactive metabolic map of bio-based chemicals. Trends Biotechnol. 2023; 41:10–14. [DOI] [PubMed] [Google Scholar]
- 6. Cordell W.T., Avolio G., Takors R., Pfleger B.F. Milligrams to kilograms: making microbes work at scale. Trends Biotechnol. 2023; 41:1442–1457. [DOI] [PubMed] [Google Scholar]
- 7. Lux M.W., Strychalski E.A., Vora G.J. Advancing reproducibility can ease the ‘hard truths’ of synthetic biology. Synth. Biol. 2023; 8:ysad014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ni C., Dinh C.V., Prather K.L. Dynamic control of metabolism. Annu. Rev. Chem. Biomol. Eng. 2021; 12:519–541. [DOI] [PubMed] [Google Scholar]
- 9. Pham C., Stogios P.J., Savchenko A., Mahadevan R. Advances in engineering and optimization of transcription factor-based biosensors for plug-and-play small molecule detection. Curr. Opin. Biotechnol. 2022; 76:102753. [DOI] [PubMed] [Google Scholar]
- 10. Tellechea-Luzardo J., Stiebritz M.T., Carbonell P. Transcription factor-based biosensors for screening and dynamic regulation. Front. Bioeng. Biotechnol. 2023; 11:1118702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tellechea-Luzardo J., Martín Lázaro H., Moreno López R., Carbonell P. Sensbio: an online server for biosensor design. BMC Bioinformatics. 2023; 24:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Boada Y., Vignoni A., Picó J., Carbonell P. Extended metabolic biosensor design for dynamic pathway regulation of cell factories. iScience. 2020; 23:101305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delépine B., Libis V., Carbonell P., Faulon J.-L. SensiPath: computer-aided design of sensing-enabling metabolic pathways. Nucleic Acids Res. 2016; 44:W226–W231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim J., Salvador M., Saunders E., González J., Avignone-Rossa C., Jiménez J. Properties of alternative microbial hosts used in synthetic biology: towards the design of a modular chassis. Essays Biochem. 2016; 60:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. King Z.A., Lu J., Dräger A., Miller P., Federowicz S., Lerman J.A., Ebrahim A., Palsson B.O., Lewis N.E. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016; 44:D515–D522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karp P.D., Billington R., Caspi R., Fulcher C.A., Latendresse M., Kothari A., Keseler I.M., Krummenacker M., Midford P.E., Ong Q. et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019; 20:1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duigou T., Du Lac M., Carbonell P., Faulon J.-L. Retrorules: a database of reaction rules for engineering biology. Nucleic Acids Res. 2019; 47:D1229–D1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koch M., Duigou T., Faulon J.-L. Reinforcement learning for bioretrosynthesis. ACS Synth. Biol. 2019; 9:157–168. [DOI] [PubMed] [Google Scholar]
- 19. Jang W. D. e.a. An interactive metabolic map of bio-based chemicals. Trends Biotechnol. 2022; 41:10–14. [DOI] [PubMed] [Google Scholar]
- 20. Carbonell P., Wong J., Swainston N., Takano E., Turner N.J., Scrutton N.S., Kell D.B., Breitling R., Faulon J.-L. Selenzyme: enzyme selection tool for pathway design. Bioinformatics. 2018; 34:2153–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Franz M., Lopes C.T., Fong D., Kucera M., Cheung M., Siper M.C., Huck G., Dong Y., Sumer O., Bader G.D. Cytoscape.js 2023 update: a graph theory library for visualization and analysis. Bioinformatics. 2023; 39:btad031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hérisson J., Duigou T., du Lac M., Bazi-Kabbaj K., Sabeti Azad M., Buldum G., Telle O., El Moubayed Y., Carbonell P., Swainston N. et al. The automated Galaxy-SynBioCAD pipeline for synthetic biology design and engineering. Nat. Commun. 2022; 13:5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Delépine B., Duigou T., Carbonell P., Faulon J.-L. RetroPath2.0: A retrosynthesis workflow for metabolic engineers. Metab. Eng. 2018; 45:158–170. [DOI] [PubMed] [Google Scholar]
- 24. Zhang X., Feng Y., Hua Y., Zhang C., Fang B., Long X., Pan Y., Gao B., Zhang J.Z.H., Li L. et al. Biosynthesis of eriodictyol in citrus waster by endowing P450BM3 activity of naringenin hydroxylation. Appl. Microbiol. Biotechnol. 2024; 108:84. [DOI] [PubMed] [Google Scholar]
- 25. Gao S., Xu X., Zeng W., Xu S., Lyv Y., Feng Y., Kai G., Zhou J., Chen J. Efficient Biosynthesis of (2S)-Eriodictyol from (2S)-Naringenin in Saccharomyces cerevisiae through a combination of promoter adjustment and directed evolution. ACS Synth. Biol. 2020; 9:3288–3297. [DOI] [PubMed] [Google Scholar]
- 26. Hirooka K. Transcriptional response machineries of Bacillus subtilis conducive to plant growth promotion. Biosci. Biotechnol. Biochem. 2014; 78:1471–1484. [DOI] [PubMed] [Google Scholar]
- 27. Salehi B., Quispe C., Sharifi-Rad J., Cruz-Martins N., Nigam M., Mishra A.P., Konovalov D.A., Orobinskaya V., Abu-Reidah I.M., Zam W. et al. Phytosterols: from preclinical evidence to potential clinical applications. Front. Pharmacol. 2021; 11:599959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gao R., Li D., Lin Y., Lin J., Xia X., Wang H., Bi L., Zhu J., Hassan B., Wang S. et al. Structural and functional characterization of the fadr regulatory protein from Vibrio alginolyticus. Front. Cell. Infect. Microbiol. 2017; 7:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kasai D., Kamimura N., Tani K., Umeda S., Abe T., Fukuda M., Masai E. Characterization of FerC, a MarR-type transcriptional regulator, involved in transcriptional regulation of the ferulate catabolic operon in Sphingobium sp. strain SYK-6. FEMS Microbiol. Lett. 2012; 332:68–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DetSpace is an open-source application, available at GitHub (https://github.com/dbtl-synbio/detspace) and FigShare at https://doi.org/10.6084/m9.figshare.25103978. The DetSpace website is free and open to all users, there is no login requirement, available at https://detspace.carbonelllab.org/.