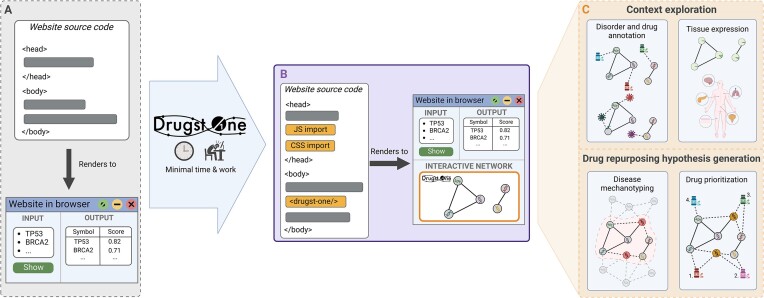
Abstract
In recent decades, the development of new drugs has become increasingly expensive and inefficient, and the molecular mechanisms of most pharmaceuticals remain poorly understood. In response, computational systems and network medicine tools have emerged to identify potential drug repurposing candidates. However, these tools often require complex installation and lack intuitive visual network mining capabilities. To tackle these challenges, we introduce Drugst.One, a platform that assists specialized computational medicine tools in becoming user-friendly, web-based utilities for drug repurposing. With just three lines of code, Drugst.One turns any systems biology software into an interactive web tool for modeling and analyzing complex protein-drug-disease networks. Demonstrating its broad adaptability, Drugst.One has been successfully integrated with 21 computational systems medicine tools. Available at https://drugst.one, Drugst.One has significant potential for streamlining the drug discovery process, allowing researchers to focus on essential aspects of pharmaceutical treatment research.
Graphical Abstract
Graphical Abstract.
Introduction
In recent years, rapid technological advancements and unmet medical needs have fueled the development of computational tools that leverage systems biology methodologies to decipher complex biomedical data (1). These tools frequently target the identification of specific proteins or genes in a given disease context, such as marker genes indicative of disease progression (2,3). The visualization of these results in a biomedical network context can greatly improve their interpretability, allowing us to better understand the underlying disease mechanisms and the interrelationships among the identified entities (4). This principle applies to a variety of biomedical fields, including oncology (5,6), virology (7) and disease subtype identification and patient stratification through differential gene expression analysis (8,9). Rendering these intricate cellular processes as graphs aids researchers in tailoring more precise pharmaceutical treatments, minimizing side effects (10,11) and opening prospects for novel therapeutic and diagnostic strategies, such as mechanistic drug repurposing (12).
Key challenges in the development of systems biology platforms include the integration of comprehensive biomedical data and the creation of flexible graphical user interfaces for data analysis, prioritization, and visualization. Stand-alone software such as Cytoscape (13) visualizes biological networks but necessitates local installation for each user. To circumvent this, developers often provide online solutions dependent solely on browser compatibility. However, this presents additional hurdles for researchers who may lack sufficient web-development skills and need to establish and maintain an infrastructure, including a server hosting a database and a website. Beyond network visualization, the collection, harmonization, integration, and incorporation of diverse biomedical data demand a significant time investment (14). Moreover, the database should be maintained and regularly updated, a chore that is often not addressed by bioinformatics tools that primarily provide a result overview with a limited set of features. Thus, if network exploration is not neglected due to the additional workload, unique solutions are being developed from scratch, resulting in network visualizers and explorers of varying quality (7,8,15).
We developed Drugst.One to reduce software engineering overhead, bundle development capacities, and to standardize and simplify network analysis and visual network exploration for biomedical web tools. With minimal programming effort, Drugst.One can turn any gene or protein-based systems biology tool into a powerful online toolkit for network integration and visualization, as well as mechanistic drug repurposing. Drugst.One is a customizable plug-and-play solution for web-application developers in need of a feature-rich network explorer coupled with a biomedical protein-drug-disease network data warehouse. With as little as three lines of code, Drugst.One can be added to any biomedical web tool, highlighting opportunities for drug repurposing and elucidating disease mechanisms. Incorporating multiple state-of-the-art databases (Supplementary Table S2) to complement visualized data, Drugst.One provides an intuitive interface for applying algorithms for exploratory network analyses, drug target, and drug repurposing candidate identification and prioritization. Weekly updates guarantee the relevance of its database for frequently changing data. Currently, Drugst.One is integrated into 21 systems medicine software resources (Supplementary Table S1), including mirDIP (Supplementary Figure S5) (16), pathDIP (17) and WikiPathways (18). In this article, we describe the functionality of Drugst.One and demonstrate its utility on the basis of two studies – on drug repurposing for inflammatory bowel disease (IBD) and on exploring the smooth muscle cell (SMC) proliferation pathway.
Materials and methods
Database
Drugst.One integrates 14 data sources, forming a large, heterogeneous database (Supplementary Table S2). Basic entities in Drugst.One are proteins/genes, drugs, and diseases. The available ID spaces for gene or protein entities are HGNC (19), UniProt (20), Ensemble (21), and Entrez (22). For drugs Drugst.One uses DrugBank and for disorders MONDO (23) identifiers. To describe links between the different entities Drugst.One integrates four different relational layer types, namely protein-protein, protein-drug, protein-disorder, and drug-disorder data.
Using the secondary database NeDRexDB (6), which is updated on a weekly basis, all datasets from NeDRexDB are automatically updated weekly. While all protein-protein and drug-target interaction datasets included in NeDRex are available individually, the NeDRex datasets represent a combination of all individual data sources. Some data sources have restrictive reuse licenses attached, e.g., for use in a commercial scenario. In Drugst.One, we provide both, licensed and openly available datasets, but the access to licensed data has to be unlocked with a configuration parameter.
Input and output
The required input is a list of proteins or genes in HGNC, UniProt, Ensembl or Entrez ID space. On demand, these entities are integrated into the interactome and automatically annotated with clinically relevant information, e.g. targeting drugs or known disease associations. Exploratory functions allow the visualization of known drug indications and disease associations as well as an overlay for tissue-specific expression information. For most information-enriching functions, Drugst.One provides several data sources to choose from (Supplement 3.1). Additionally, network algorithms allow to find and rank directly and indirectly associated drugs.
Algorithms
Drugst.One originates from the network-based drug repurposing platforms CoVex (7) and CADDIE (5), developed for the application in SARS-CoV-2 and cancer, respectively. While they provide disease-specific information, both tools share underlying principles and algorithms (Supplementary Table S3). These tested and published methods form the Drugst.One algorithmic toolkit (Supplement 3.2). Module identification algorithms provide means to identify additional potential drug targets from the interactome to enrich the mechanistic context. In a second step, drugs that are directly or indirectly linked can be ranked. This allows the assessment of the compound’s potential to be repurposed using network-based algorithms. Although both steps work automatically, users can infuse their expert knowledge by adjusting input gene sets. Users can choose among seven drug prioritization and drug target identification algorithms to rank small molecules directly or indirectly targeting disease proteins, thus serving as potential drug repurposing candidates (Supplement 3.2). Information on the execution time scaling behavior of module identification and drug prioritization algorithms can be found in supplementary material (Supplementary Figure S3 and S4, respectively). Overrepresentation analysis using g:Profiler (24) or functional coherence validation using DIGEST (25) on all loaded proteins in the network can be run with one click.
Results
Integration and customization
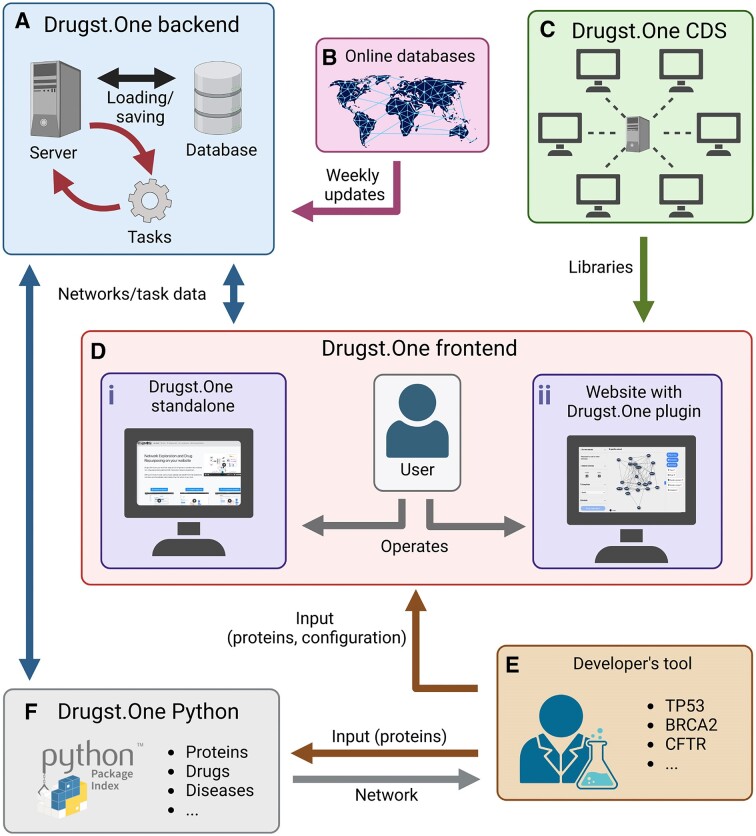
The Drugst.One ecosystem is a multi-component platform consisting of a website, the web plugin, a server, a content delivery system (CDS) and a Python package (Figure 1, Supplement 1). Drugst.One is free and open to all users and no login is required.
Figure 1.
The Drugst.One ecosystem: the Drugst.One server (A) updates weekly from online databases (B), executes computationally demanding tasks, and provides data to the Drugst.One plugin (D i and D ii). The frontend is loaded from the content delivery system (CDS), (C), receives the network data from the developer integrating Drugst.One (E) and presents it to the user. Drugst.One can also be accessed programmatically through a Python package (F).
The web plugin can be added to any webpage by importing one JavaScript and one stylesheet file from the https://cdn.drugst.one distribution server, and by adding the ‘drugst-one’ HTML tag to the source code of any system medicine tool’s website (Supplementary Figure S2). Features can be customized to a high degree through JSON configuration strings that are passed as attributes. This includes default states of on/off toggles, the network, and the node and edge groups that define the network style. The plugin is responsive to changes during runtime, allowing developers to add buttons or other controls to the host page, for example switching between networks. For seamless integration of the rendered plugin into any website, styling and coloring are controllable by adding specific CSS variables to the website stylesheet (Supplementary Figure S1). To assist developers in the integration process, the Drugst.One website provides conclusive documentation of available parameters, features, and styles. It further offers an interactive configuration page at https://drugst.one/playground where configuration options are categorized, and the replication of a configured Drugst.One instance is achieved by simple copy-pasting of the generated code snippets to the developers’ websites. This low-code approach allows bioinformatics researchers to provide the community with an interactive mechanism mining web tool within hours or even minutes instead of days. The lightweight Drugst.One JavaScript library connects to the Drugst.One data warehouse server, which handles all the computationally expensive work like data annotation, mapping, and asynchronous algorithm execution.
Alternatively, a standalone integration of Drugst.One is provided at https://drugst.one/standalone, which can be accessed and customized using URLs or POST-based requests. This way, results from any website or even a command line tool can be redirected to Drugst.One through a simple web service request (Supplement 2). Detailed documentation about all Drugst.One integration options can be found at https://drugst.one/doc.
Integration examples
Drugst.One plugin integration with BiCoN
BiCoN (8) is a systems medicine tool for simultaneous patient stratification and disease mechanism identification, i.e. network-based endotyping. BiCoN uses a molecular interaction network as input and identifies two subgroups of patients along with a subnetwork that is enriched for differentially expressed genes between the two groups. These subnetworks can serve as composite biomarkers but may also be enriched for putative drug targets. Since BiCoN also features a web version (https://exbio.wzw.tum.de/bicon), we integrated the Drugst.One plugin for enhancing the result presentation by interactively visualizing the identified subnetworks. This allows users to explore possible drug repurposing candidates targeting the newly identified disease mechanisms, which can subsequently be experimentally validated.
Drugst.One link-out from WikiPathways
WikiPathways (26) is a widely used, community-driven platform for exploring molecular pathways. It allows users to upload, edit, browse, and download a constantly growing pool of pathway datasets. Pathway data can be used to identify and understand key players in metabolism, which is critical for understanding rare or common diseases such as COVID-19 (26). Thus, pathways allow for the prediction of drug target and drug repurposing candidates and are commonly used in the development of new disease treatments (27). When inspecting individual pathways on the WikiPathways platform, users now have the option to forward the pathway genes to the Drugst.One standalone version by clicking a ‘Query Drugst.One’ link now provided by WikiPathways, located in the search menu of the ‘Participants’ table. The link redirects the user to the Drugst.One website, visualizing pathway genes, drugs directly targeting them, and offering the complete toolset of Drugst.One. In the following, we give an example of the Drugst.One usage for exploration of the smooth muscle differentiation and proliferation pathway (WP1991).
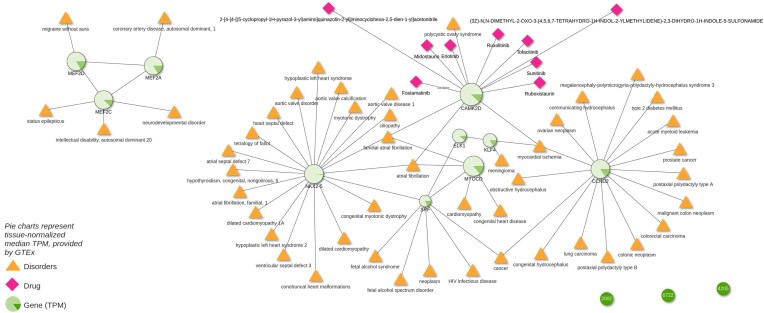
WikiPathway WP1991 describes the mechanism behind smooth muscle cell (SMC) differentiation and proliferation. The WikiPathways web interface now incorporates a button to export the pathway genes into Drugst.One (using the magnifier glass in the table showing the proteins participating in the pathway, state 29.01.24) and visualizing their interactions with drugs in Drugst.One directly. To gain a general overview of the complications (symptoms, comorbidities, etc.) associated with dysfunctional SMC development and their drivers, Drugst.One allows for extending the WikiPathways-exported network by the corresponding disorders and their associated pathway genes. Several disease nodes appear in the network, mainly representing various cardiovascular disorders (CVDs), e.g. cardiomyopathy, coronary artery disease, and aortic valve disease (Figure 2). Further, genes with a strong link to vascular function are present: NKX2-5 is associated with heart and cardiac muscle development (28); KLF4 is involved in cytoskeletal stabilization of vascular SMCs (29); MYOCD is critical for cardiovascular development and vascular SMC autophagy regulation (30). This leads to the assumption that the mechanism vascular SMC proliferation at least partially overlaps with WP1991, the pathway of SMC. The importance of SMCs for proper vascular functionality (31,32) and thus to atherosclerosis, hypertension, myocardial infarction, and other cardiovascular diseases was reported before (33–35). An isolated subnetwork community of proteins is formed by the three myocyte enhancer factors MEF2A, MEF2C, and MEF2D. Besides their obvious cardiovascular implications, these factors play a role in neurological processes (36). An impact of SMCs on status epilepticus was shown in mouse models (37), and a connection between migraine and SMC dysfunction was suggested as well (38,39).
Figure 2.
Participants of WikiPathway WP1991 displayed in Drugst.One. Adjacent diseases and drugs are enabled, as well as diseases linked to drugs targeting this smooth muscle cell (SMC) and differentiation pathway. To investigate if there is the chance that WP1991 also represents vascular SMC proliferation, normalized median expression values for ‘Artery - Aorta’ are overlaid as pie charts, where 360○ represent the maximum observed transcripts per million (TPM) in the selected tissue and all other TPMs are exponentially scaled.
Drugst.One allows for the projection of (gene) expression data from GTEx on the proteins in the network. The relative expression of these genes appears to be quite high in arteries, indicating again a relevancy of these genes in vascular SMC. Further, the expression also seems to be elevated in organs that have to perform physical motion, like heart, lung, bladder and skeletal muscles, but with observable fluctuations between the tissues in the relative expression of genes like CCND2.
With one mouse click, we import drug target information for drug repurposing candidate prediction. Despite SMCs relation to cardiovascular diseases, no corresponding CVD drugs have been identified. Mainly anti-cancer drugs (e.g. sunitinib, erlotinib, midostaurin and ruxolitinib) targeting calcium/calmodulin-dependent protein kinase II delta (CAMK2D), which is associated with cancer growth (40), are found. Notably, however, CAMK2D also plays a role in calcium signaling, which is essential for the upkeep of SMC function (41,42). Hence, this may explain the observed cardiovascular side effects of CAMK2D-targeting drugs. According to SIDER (43), sunitinib may cause hypertensive symptoms and corresponding studies suggest that midostaurin has cardiotoxic effects (44). Algorithms integrated in Drugst.One can extend the search space by looking for indirectly connected drugs. The selection menu offers a function to automatically add all displayed proteins to the selection, serving as the starting point (seeds) of subsequent searches. The harmonic centrality algorithm (Supplement 3.2.3) was used to extend the network by the ten drugs with the highest score, including indirectly (transitively) connected drugs from the NeDRex database. Through this search, the tyrosine kinase inhibitor nintedanib, which has shown promising effects in pulmonary arterial smooth muscle cells and intestinal smooth muscle cells (45,46), can be identified.
This shows the identification potential of mechanism-associated drugs through the network-based drug repurposing functions Drugst.One incorporates. Whereas before only drugs primarily used in cancer were present through direct association with SMC pathway participants, Drugst.One suggested more relevant options for this context.
Due to the weekly automatic updates of the Drugst.One data warehouse, analyses performed on the Drugst.One website may not always be fully reproducible. For instances where long-term reproducibility is prioritized over the most current data, we offer an alternative at https://stable.drugst.one/, which receives updates only on the first day of each year. Similarly, a Drugst.One plugin is available that connects to this stable database.
More Drugst.One use cases
Other exmplary case studies using Drugst.One on more diseases can be found in the supplementary material section 4. We recreated a drug repurposing example case proposed by Sadegh et al. 2021 on inflammatory bowel disease (6) using the same seed genes (Supplementary Table S4) and matching algorithms (Supplementary Table S5) and through Drugst.One we were able to identify even more promising treatment options than the original study did (Supplementary Table S6). Using the integrated version of Drugst.One in the web tool mirDIP, we explored the IBD case from the angle of microRNA targets. A short report about ROBUST-Web, a web application for module detection, that uses Drugst.One as primary tool for visualization can be found in section 5 of the supplementary material.
Discussion
Biomedical research generates a wealth of data that could inform the development of novel therapies or treatments. However, despite this potential, a significant portion of the analyses conducted in this field fail to translate into clinical trials, leading to major issues in the effectiveness of public health research (47). To this end, Drugst.One has the potential to help transform specifically omics-based research results into actionable hypotheses with potential clinical impact by closing the gap between disease mechanism mining and hypothesis generation for drug repurposing. Drugst.One offers a community-driven solution to streamline the knowledge distributed over many online resources for multi-omics analyses and other biomedical tools (48) to turn the results of biomedical analyses into concrete candidate drug targets and drug repurposing hypotheses. Still, we emphasize that the drug target and drug repurposing predictions are merely candidates and supervision with expert knowledge is still required before experimental validation. Drugst.One delivers explainable indications based on established biological data like expression and known disease associations or drug indications, however, the interpretation of their application in the case-specific context is up to the user. Therefore, we designed Drugst.One to be operated with maximal transparency and allow optional user input for every step of the analysis.
With the infrastructure and the resources being provided, Drugst.One helps to find a community-wide solution for standardization and streamlining the visualization of explainable disease modules and their pharmacological implications. Drugst.One provides various interfaces to be highly accessible and customizable by all members of the community while maintaining up-to-date database information and network analysis algorithms. Smooth integration into most biomedical websites and tools is confirmed by 21 resources already integrating Drugst.One. For future developers who wish to customize Drugst.One before its integration, an interactive web interface provides copy-paste-able code for customized plugin integration with their own website. An endpoint for developers who want to link out from any of their websites, apps, or command line tools is provided by Drugst.One as well. By default, all endpoints connect to the database that is updated weekly. However, we provide adjusted versions or configuration options to use the stable database, which is updated only once a year, to facilitate long-term reproducibility of results.
Drugst.One complies with community standards regarding data management as defined by the FAIRness principles (49). Download links for any data shown in Drugst.One are provided at any step, whether it is a table with drug target and drug candidates or the visualized network with all activated extensions like expression information. Export to current community standards is supported via exporting compatible .graphml files, which can be loaded directly into, e.g. Cytoscape (13). To further increase reproducibility and interoperability, concrete plans to implement community standards, such as the export of Drugst.One networks to NDEx (50,51) are made (Supplementary Table S7). In summary, Drugst.One offers an important service to the systems medicine research community to tackle the widely recurring problem of web-based disease mechanism mining and drug repurposing candidate prediction by capturing the results of biomedical assays.
Supplementary Material
Acknowledgements
The Graphical abstract and Figure 1 and Supplementary Figures S1, S2 and S5 were created with BioRender.com.
Author contributions: A.M. and M.H. developed and implemented the Drugst.One ecosystem, and wrote the manuscript. J.B. and O.Z. supervised the project and provided continuous feedback and ideas during the development. The rest of the Drugst.One Initiative integrated the Drugst.One ecosystem into their platforms, tested, provided helpful feedback, and revised the manuscript. All authors agreed to the final version of the manuscript.
Contributor Information
Andreas Maier, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany.
Michael Hartung, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany.
Mark Abovsky, Division of Orthopaedic Surgery, Schroeder Arthritis Institute, Toronto, Canada; Data Science Discovery Centre for Chronic Diseases, Krembil Research Institute, Toronto, ON M5T 0S8, Canada.
Klaudia Adamowicz, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany.
Gary D Bader, Department of Molecular Genetics, University of Toronto, Toronto, ON, Canada; The Donnelly Centre, University of Toronto, Toronto, ON, Canada; Department of Computer Science, University of Toronto, Toronto, ON, Canada; Princess Margaret Cancer Centre, University Health Network, Toronto, ON, Canada; The Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, ON, Canada.
Sylvie Baier, Data Science in Systems Biology, TUM School of Life Sciences, Technical University of Munich, Munich, Germany.
David B Blumenthal, Department Artificial Intelligence in Biomedical Engineering (AIBE), Friedrich-Alexander University Erlangen-Nürnberg (FAU), 91052 Erlangen, Germany.
Jing Chen, Department of Medicine, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA.
Maria L Elkjaer, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany; Department of Neurology, Odense University Hospital, Odense, Denmark; Institute of Clinical Research, University of Southern Denmark, Odense, Denmark; Institute of Molecular Medicine, University of Southern Denmark, Odense, Denmark.
Carlos Garcia-Hernandez, Barcelona Supercomputing Center (BSC), 08034 Barcelona, Spain.
Mohamed Helmy, Vaccine and Infectious Disease Organization (VIDO), University of Saskatchewan, Canada; School of Public Health, University of Saskatchewan, Canada; Department of Computer Science, University of Saskatchewan, Canada; Department of Computer Science, Lakehead University, Canada; Department of Computer Science, Idaho State University, USA; Bioinformatics Institute (BII), A*STAR, Singapore.
Markus Hoffmann, Data Science in Systems Biology, TUM School of Life Sciences, Technical University of Munich, Munich, Germany; Institute for Advanced Study, Technical University of Munich, Germany; National Institute of Diabetes, Digestive, and Kidney Diseases, Bethesda, MD 20892, USA.
Igor Jurisica, Division of Orthopaedic Surgery, Schroeder Arthritis Institute, Toronto, Canada; Data Science Discovery Centre for Chronic Diseases, Krembil Research Institute, Toronto, ON M5T 0S8, Canada; Departments of Medical Biophysics and Computer Science, University of Toronto, Toronto, Canada; Institute of Neuroimmunology, Slovak Academy of Sciences, Bratislava, Slovakia.
Max Kotlyar, Division of Orthopaedic Surgery, Schroeder Arthritis Institute, Toronto, Canada; Data Science Discovery Centre for Chronic Diseases, Krembil Research Institute, Toronto, ON M5T 0S8, Canada.
Olga Lazareva, Division of Computational Genomics and Systems Genetics, German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany; Junior Clinical Cooperation Unit Multiparametric methods for early detection of prostate cancer, German Cancer Research Center (DKFZ), Heidelberg, Germany; European Molecular Biology Laboratory, Genome Biology Unit, 69117 Heidelberg, Germany.
Hagai Levi, Blavatnik School of Computer Science, Tel-Aviv University, Tel-Aviv, Israel.
Markus List, Data Science in Systems Biology, TUM School of Life Sciences, Technical University of Munich, Munich, Germany.
Sebastian Lobentanzer, Heidelberg University, Faculty of Medicine, and Heidelberg University Hospital, Institute for Computational Biomedicine, Bioquant, Heidelberg, Germany.
Joseph Loscalzo, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA.
Noel Malod-Dognin, Barcelona Supercomputing Center (BSC), 08034 Barcelona, Spain.
Quirin Manz, Data Science in Systems Biology, TUM School of Life Sciences, Technical University of Munich, Munich, Germany.
Julian Matschinske, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany; Data Science in Systems Biology, TUM School of Life Sciences, Technical University of Munich, Munich, Germany.
Miles Mee, Department of Molecular Genetics, University of Toronto, Toronto, ON, Canada; The Donnelly Centre, University of Toronto, Toronto, ON, Canada; Department of Computer Science, University of Toronto, Toronto, ON, Canada.
Mhaned Oubounyt, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany.
Chiara Pastrello, Division of Orthopaedic Surgery, Schroeder Arthritis Institute, Toronto, Canada; Data Science Discovery Centre for Chronic Diseases, Krembil Research Institute, Toronto, ON M5T 0S8, Canada.
Alexander R Pico, Institute of Data Science and Biotechnology, Gladstone Institutes, 1650 Owens Street, San Francisco, 94158 California, USA.
Rudolf T Pillich, Department of Medicine, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA.
Julian M Poschenrieder, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany; Data Science in Systems Biology, TUM School of Life Sciences, Technical University of Munich, Munich, Germany.
Dexter Pratt, Department of Medicine, University of California San Diego, 9500 Gilman Drive, La Jolla, CA 92093, USA.
Nataša Pržulj, Barcelona Supercomputing Center (BSC), 08034 Barcelona, Spain; Department of Computer Science, University College London, London WC1E 6BT, UK; ICREA, Pg. Lluís Companys 23, 08010 Barcelona, Spain.
Sepideh Sadegh, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany; Data Science in Systems Biology, TUM School of Life Sciences, Technical University of Munich, Munich, Germany; Department of Clinical Genetics, Odense University Hospital, Odense, Denmark; Clinical Genome Center, Department of Clinical Research, University of Southern Denmark, Odense, Denmark.
Julio Saez-Rodriguez, Heidelberg University, Faculty of Medicine, and Heidelberg University Hospital, Institute for Computational Biomedicine, Bioquant, Heidelberg, Germany.
Suryadipto Sarkar, Department Artificial Intelligence in Biomedical Engineering (AIBE), Friedrich-Alexander University Erlangen-Nürnberg (FAU), 91052 Erlangen, Germany.
Gideon Shaked, Blavatnik School of Computer Science, Tel-Aviv University, Tel-Aviv, Israel.
Ron Shamir, Blavatnik School of Computer Science, Tel-Aviv University, Tel-Aviv, Israel.
Nico Trummer, Data Science in Systems Biology, TUM School of Life Sciences, Technical University of Munich, Munich, Germany.
Ugur Turhan, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany.
Rui-Sheng Wang, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA.
Olga Zolotareva, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany; Data Science in Systems Biology, TUM School of Life Sciences, Technical University of Munich, Munich, Germany.
Jan Baumbach, Institute for Computational Systems Biology, University of Hamburg, Hamburg, Germany; Computational Biomedicine Lab, Department of Mathematics and Computer Science, University of Southern Denmark, Odense, Denmark.
Data availability
The authors declare that all data supporting the findings of this study are available publicly and their integration is described accordingly within the paper and its supplementary information file. All code of the Drugst.One platform is available on GitHub (https://github.com/drugst-one) and on Zenodo (plugin: https://doi.org/10.5281/zenodo.11073055, backend: https://doi.org/10.5281/zenodo.11073096, Python package: https://doi.org/10.5281/zenodo.11073116, integration examples: https://doi.org/10.5281/zenodo.11073112, Django template: https://doi.org/10.5281/zenodo.11073104, runtime evaluation: https://doi.org/10.5281/zenodo.11073073).
Supplementary data
Supplementary Data are available at NAR Online.
Funding
REPO-TRIAL: this project has received funding from the European Union’s Horizon 2020 research and innovation programme [777111]; this publication reflects only the authors’ view and the European Commission is not responsible for any use that may be made of the information it contains; RePo4EU: this project is funded by the European Union [101057619]; views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or European Health and Digital Executive Agency (HADEA). Neither the European Union nor the granting authority can be held responsible for them; Swiss State Secretariat for Education, Research and Innovation (SERI) [22.00115]; German Federal Ministry of Education and Research (BMBF) within the framework of ‘CLINSPECT-M/-2’ [F031L0214A, 161L0214A, 16LW0243K]; Technical University Munich – Institute for Advanced Study, funded by the German Excellence Initiative; Intramural Research Programs (IRPs) of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) [422216132]; J.B. was partially funded by his VILLUM Young Investigator Grant [13154]; European Research Council (ERC) Consolidator Grant [770827]; Spanish State Research Agency AEI 10.13039/501100011033 [PID2019-105500GB-I00]; I.J. was supported in part by funding from Natural Sciences Research Council [NSERC #203475], Canada Foundation for Innovation [CFI #225404, #30865]; Ontario Research Fund [RDI #34876, RE010-020]; IBM and Ian Lawson van Toch Fund; S.L. has received funding from the European Union’s Horizon 2020 research and innovation programme [965193] for DECIDER. Funding for open access charge: Horizon Europe project Repo4EU.
Conflict of interest statement. J.S.R. reports funding from GSK, Pfizer and Sanofi and fees from Travere Therapeutics and Astex Pharmaceuticals.
References
- 1. Hufsky F., Lamkiewicz K., Almeida A., Aouacheria A., Arighi C., Bateman A., Baumbach J., Beerenwinkel N., Brandt C., Cacciabue M. et al. Computational strategies to combat COVID-19: useful tools to accelerate SARS-CoV-2 and coronavirus research. Brief. Bioinform. 2021; 22:642–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., O’Meara M.J., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R. et al. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. Nature. 2020; 583:459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zolotareva O., Kleine M. A survey of gene prioritization tools for mendelian and complex human diseases. J. Integr. Bioinform. 2019; 16:20180069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hütter C.V.R., Sin C., Müller F., Menche J. Network cartographs for interpretable visualizations. Nat. Comput. Sci. 2022; 2:84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hartung M., Anastasi E., Mamdouh Z.M., Nogales C., Schmidt H.H.H.W., Baumbach J., Zolotareva O., List M. Cancer driver drug interaction explorer. Nucleic Acids Res. 2022; 50:W138–W144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sadegh S., Skelton J., Anastasi E., Bernett J., Blumenthal D.B., Galindez G., Salgado-Albarrán M., Lazareva O., Flanagan K., Cockell S. et al. Network medicine for disease module identification and drug repurposing with the NeDRex platform. Nat. Commun. 2021; 12:6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sadegh S., Matschinske J., Blumenthal D.B., Galindez G., Kacprowski T., List M., Nasirigerdeh R., Oubounyt M., Pichlmair A., Rose T.D. et al. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat. Commun. 2020; 11:3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lazareva O., Canzar S., Yuan K., Baumbach J., Blumenthal D.B., Tieri P., Kacprowski T., List M. BiCoN: Network-constrained biclustering of patients and omics data. Bioinformatics. 2020; 37:2398–2404. [DOI] [PubMed] [Google Scholar]
- 9. Zolotareva O., Khakabimamaghani S., Isaeva O.I., Chervontseva Z., Savchik A., Ester M. Identification of differentially expressed gene modules in heterogeneous diseases. Bioinformatics. 2021; 37:1691–1698. [DOI] [PubMed] [Google Scholar]
- 10. Nelissen E., van Hagen B.T.J., Argyrousi E.K., van Goethem N.P., Heckman P.R.A., Paes D., Mulder-Jongen D.A.J., Ramaekers J.G., Blokland A., Schmidt H.H.H.W. et al. Soluble guanylate cyclase stimulator riociguat improves spatial memory in mice via peripheral mechanisms. Neurosci. Lett. 2022; 788:136840. [DOI] [PubMed] [Google Scholar]
- 11. Casas A.I., Hassan A.A., Larsen S.J., Gomez-Rangel V., Elbatreek M., Kleikers P.W.M., Guney E., Egea J., López M.G., Baumbach J. et al. From single drug targets to synergistic network pharmacology in ischemic stroke. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:7129–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goh K.-I., Cusick M.E., Valle D., Childs B., Vidal M., Barabási A.-L. The human disease network. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lobentanzer S., Aloy P., Baumbach J., Bohar B., Carey V.J., Charoentong P., Danhauser K., Doğan T., Dreo J., Dunham I. et al. Democratizing knowledge representation with BioCypher. Nat. Biotechnol. 2023; 41:1056–1059. [DOI] [PubMed] [Google Scholar]
- 15. Levi H., Elkon R., Shamir R. DOMINO: a network-based active module identification algorithm with reduced rate of false calls. Mol. Syst. Biol. 2021; 17:e9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hauschild A.-C., Pastrello C., Ekaputeri G. K.A., Bethune-Waddell D., Abovsky M., Ahmed Z., Kotlyar M., Lu R., Jurisica I. MirDIP 5.2: tissue context annotation and novel microRNA curation. Nucleic Acids Res. 2023; 51:D217–D225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pastrello C., Kotlyar M., Abovsky M., Lu R., Jurisica I. PathDIP 5: improving coverage and making enrichment analysis more biologically meaningful. Nucleic Acids Res. 2024; 52:D663–D671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pico A.R., Kelder T., van Iersel M.P., Hanspers K., Conklin B.R., Evelo C. WikiPathways: pathway editing for the people. PLoS Biol. 2008; 6:e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shows T.B., McAlpine P.J. The catalog of human genes and chromosome assignments. A report on human genetic nomenclature and genes that have been mapped in man. Cytogenet. Cell Genet. 1978; 22:132–145. [DOI] [PubMed] [Google Scholar]
- 20. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017; 45:D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R. et al. Ensembl 2022. Nucleic Acids Res. 2022; 50:D988–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maglott D., Ostell J., Pruitt K.D., Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007; 35:D26–D31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mungall C.J., McMurry J.A., Köhler S., Balhoff J.P., Borromeo C., Brush M., Carbon S., Conlin T., Dunn N., Engelstad M. et al. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2017; 45:D712–D722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019; 47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Adamowicz K., Maier A., Baumbach J., Blumenthal D.B. Online in silico validation of disease and gene sets, clusterings or subnetworks with DIGEST. Brief. Bioinform. 2022; 23:bbac247. [DOI] [PubMed] [Google Scholar]
- 26. Martens M., Ammar A., Riutta A., Waagmeester A., Slenter D.N., Hanspers K., A Miller R., Digles D., Lopes E.N., Ehrhart F. et al. WikiPathways: connecting communities. Nucleic Acids Res. 2021; 49:D613–D621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akhurst R.J., Hata A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 2012; 11:790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lints T., Parsons L., Hartley L., Lyons I., Harvey R. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993; 119 2:419–431. [DOI] [PubMed] [Google Scholar]
- 29. Liu Y., Zheng B., Zhang X., juan Nie C., hui Li Y., Wen J. Localization and function of KLF4 in cytoplasm of vascular smooth muscle cell. Biochem. Biophys. Res. Commun. 2013; 436:162–168. [DOI] [PubMed] [Google Scholar]
- 30. Shi D., Ding J., Xie S., Huang L., Zhang H., Chen X., Ren X., Zhou S., He H., Ma W., Zhang T. et al. Myocardin/microRNA-30a/Beclin1 signaling controls the phenotypic modulation of vascular smooth muscle cells by regulating autophagy. Cell Death Dis. 2022; 13:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Steucke K.E., Tracy P.V., Hald E.S., Hall J.L., Alford P.W. Vascular smooth muscle cell functional contractility depends on extracellular mechanical properties. J. Biomech. 2015; 48:3044–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jaminon A., Reesink K., Kroon A., Schurgers L. The role of vascular smooth muscle cells in arterial remodeling: focus on calcification-related processes. Int. J. Mol. Sci. 2019; 20:5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhuge Y., Zhang J., Qian F., Wen Z., Niu C., Xu K., Ji H., Rong X., Chu M., Jia C. Role of smooth muscle cells in cardiovascular disease. Int. J. Biol. Sci. 2020; 16:2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu M., Gomez D. Smooth muscle cell phenotypic diversity. Arterioscler. Thromb. Vasc. Biol. 2019; 39:1715–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allahverdian S., Chaabane C., Boukais K., Francis G.A., Bochaton-Piallat M.-L. Smooth muscle cell fate and plasticity in atherosclerosis. Cardiovasc. Res. 2018; 114:540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chaudhary R., Agarwal V., Kaushik A.S., Rehman M. Involvement of myocyte enhancer factor 2c in the pathogenesis of autism spectrum disorder. Heliyon. 2021; 7:e06854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cozart M.A., Phelan K.D., Wu H., Mu S., Birnbaumer L., Rusch N.J., Zheng F. Vascular smooth muscle TRPC3 channels facilitate the inverse hemodynamic response during status epilepticus. Sci. Rep. 2020; 10:812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Napoli R., Guardasole V., Zarra E., Matarazzo M., D’Anna C., Saccà F., Affuso F., Cittadini A., Carrieri P.B., Saccà L. Vascular smooth muscle cell dysfunction in patients with migraine. Neurology. 2009; 72:2111–2114. [DOI] [PubMed] [Google Scholar]
- 39. Napoli R., Guardasole V., Zarra E., De Sena A., Saccà F., Ruvolo A., Grassi S., Giugliano S., De Michele G., Cittadini A., Carrieri P.B., Saccà L. Migraine attack restores the response of vascular smooth muscle cells to nitric oxide but not to norepinephrine. World J Cardiol. 2013; 5:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He Q., Li Z. The dysregulated expression and functional effect of CaMK2 in cancer. Cancer Cell Int. 2021; 21:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hill-Eubanks D.C., Werner M.E., Heppner T.J., Nelson M.T. Calcium signaling in smooth muscle. Cold Spring Harb. Perspect. Biol. 2011; 3:a004549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adelstein R.S., Sellers J.R. Effects of calcium on vascular smooth muscle contraction. Am. J. Cardiol. 1987; 59:4B–10B. [DOI] [PubMed] [Google Scholar]
- 43. Kuhn M., Letunic I., Jensen L.J., Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016; 44:D1075–D1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giudice V., Vecchione C., Selleri C. Cardiotoxicity of novel targeted hematological therapies. Life. 2020; 10:344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tsutsumi T., Nagaoka T., Yoshida T., Wang L., Kuriyama S., Suzuki Y., Nagata Y., Harada N., Kodama Y., Takahashi F. et al. Nintedanib ameliorates experimental pulmonary arterial hypertension via inhibition of endothelial mesenchymal transition and smooth muscle cell proliferation. PLoS One. 2019; 14:e0214697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kataria J., Kerr J., Lourenssen S.R., Blennerhassett M.G. Nintedanib regulates intestinal smooth muscle hyperplasia and phenotype in vitro and in TNBS colitis in vivo. Sci. Rep. 2022; 12:10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Drolet B.C., Lorenzi N.M. Translational research: understanding the continuum from bench to bedside. Transl. Res. 2011; 157:1–5. [DOI] [PubMed] [Google Scholar]
- 48. Luo J., Wu M., Gopukumar D., Zhao Y. Big Data application in biomedical research and health care: a literature review. Biomed. Inform. Insights. 2016; 8:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wilkinson M.D., Dumontier M., Aalbersberg I. J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016; 3:160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pratt D., Chen J., Pillich R., Rynkov V., Gary A., Demchak B., Ideker T. NDEx 2.0: a clearinghouse for research on cancer pathways. Cancer Res. 2017; 77:e58–e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pillich R.T., Chen J., Churas C., Liu S., Ono K., Otasek D., Pratt D. NDEx: accessing network models and streamlining network biology workflows. Curr. Protoc. 2021; 1:e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available publicly and their integration is described accordingly within the paper and its supplementary information file. All code of the Drugst.One platform is available on GitHub (https://github.com/drugst-one) and on Zenodo (plugin: https://doi.org/10.5281/zenodo.11073055, backend: https://doi.org/10.5281/zenodo.11073096, Python package: https://doi.org/10.5281/zenodo.11073116, integration examples: https://doi.org/10.5281/zenodo.11073112, Django template: https://doi.org/10.5281/zenodo.11073104, runtime evaluation: https://doi.org/10.5281/zenodo.11073073).