Abstract
Cells function as dynamic platforms for executing exogenous genetic programs in synthetic biology, resulting in highly context-dependent circuit performance. Recent years have seen an increasing interest in comprehending the intricacies of circuit-host relationships, their influence on the synthetic bioengineering workflow, and the development of strategies to alleviate undesired effects. We provide an overview of how emerging circuit-host interactions, such as growth feedback and resource competition, impact both deterministic and stochastic circuit behaviors. Additionally, we emphasize control strategies for mitigating these unwanted effects. This review summarizes the latest advancements and the current state of host-aware and resource-aware design of synthetic gene circuits.
Keywords: metabolic burden, circuit-host interactions, feedback contextual factors, control-embedded circuit design
Why context-dependent redesign of synthetic gene circuits?
The goal of synthetic biology is to increase understanding of the design principles behind biological parts, devices, and systems, and to extend these principles to the development of deployable biological constructs in a wide range of applications in health, agriculture, sustainability, and smart/programmable material science [1–5]. However, despite significant progress over the past two decades, the field is not without its maladies. Currently, the predictability of the design process of synthetic biological constructs is hampered by “circuit-host interactions” that have gained significant attention over recent years (reviewed in [6–16]). These interactions fall within the realm of context-dependent phenomena, where the behavior and effectiveness of synthetic gene circuits are influenced by the specific genetic characteristics and host cell environment. While previous review articles have underscored the challenges posed by host cell context in the predictive engineering of gene circuits, this review highlights the underlying mechanism driving emergent dynamics resulting from these non-regulatory interactions, followed by the context-dependent redesign of gene circuits incorporating various embedded control strategies. Special emphasis is placed on the essential understanding of new noise sources arising from these interactions and the implementation of multi-module control strategies. It is consensus that gene circuits do not exist in a vacuum, rather their function is intricately linked to various contexts, such as intragenetic context (Box 1), intergenetic context (Box 2), and host context, showing a strong degree of context dependence that results in lengthy design-build-test-learn (DBTL) cycles (Figure 1, Key Figure) and causes constructs to display a limited propensity for out-of-lab deployment [17]. The non-modularity resulting from inefficiencies in interfacing synthetic constructs and the host system contravenes the principles of predictability and independent behavior typically foundational in other engineering disciplines [6,9,15].
Outstanding Questions.
Can the emergent phenomena and control strategies identified in simple circuits be successfully extended to more complex circuit architectures and diverse environmental conditions?
What are the limitations in predicting and controlling the context-dependent behavior of multi-module circuits, and what strategies can be developed to overcome these challenges?
To what extent can the new redesign principles for circuit-host interactions be generalized and reliably applied across different types of circuits and host organisms?
To what extent does the context dependence of synthetic biological constructs differ between mammalian and bacterial systems, and how can the distinct contextual factors be leveraged for achieving predictable and stable outcomes in both cellular environments?
What are the key factors influencing the scalability and applicability of these control strategies, and how can they be optimized to ensure their effectiveness in various contexts?
Box 2. Single Intergenic Context Factors.
Intergenic context refers to the potential interactions between genes or genetic parts that affect the regulation and expression of the gene and its neighbors. Intergenic factors can introduce context dependence to circuit activity via retroactivity, syntax dependence, and supercoiling.
Retroactivity refers to the phenomenon where downstream nodes in a network can adversely affect or interfere with upstream nodes in an unintended manner. This interference occurs when downstream nodes sequester or modify the signals that are used by the upstream nodes, leading to unexpected changes in the network dynamics or behavior (Figure IA) [31]. This phenomenon effectively illustrated how a module downstream from a reporter module can reduce the reported circuit output by sequestering the input signal to the reporter module [32]. A “load driver” device has been developed to mitigate the undesirable impact of retroactivity [33].
Circuit syntax involves the relative order and orientation of the genes in a construct. Three possible basic syntaxes exist between two operons: convergent, divergent, and tandem orientations (Figure IB). The transcriptional interference in divergent and tandem-oriented genes is primarily mediated by DNA supercoiling [94], a phenomenon mediated by the twisting of the DNA helix upon itself which can cause regions of DNA to become under/over-wound. Positive supercoiling occurs ahead of the transcription bubble where the DNA double helix is coiled right-handedly in its natural turn direction resulting in more tight coiling, which slows down transcription initiation and halts elongation [31]. Negative supercoiling occurs behind the transcription bubble where the DNA helix is coiled left-handedly resulting in fewer twists than the relaxed state, generally facilitating transcription initiation [32]. Expression of genes can be actively altered by supercoiling caused by the expression of upstream genes[33]. Whether these supercoiling effects are activatory/inhibitory/neutral is highly dependent on the circuit syntax and boundary conditions (circular plasmid boundaries or linear chromosomal boundaries).
Under certain gene syntaxes, supercoiling effects from multiple operons can influence each other to form supercoiling-mediated feedback (Figure IC). Consequently, two adjacent genes in one synthetic gene circuit can affect the transcription activity and dynamics of each other, leading to a bidirectional feedback phenomenon that adds a layer of complexity to the synthetic gene circuit [94,95]. The accumulation of positive/negative supercoiling in the intergenic region has been demonstrated to enhance/diminish mutual inhibition between toggle switch genes getting expressed in convergent/divergent manners respectively [94,95].
Figure I:

Single intergenic context factors include (A) retroactivity, (B) circuit syntax and supercoiling, and (C) supercoiling-mediated feedback.
Figure 1. Feedback context factors context-dependent redesign of synthetic gene circuits.

(A) the canonical design-build-test-learn (DBTL) cycle in engineering synthetic gene circuits through fine-tuning individual contextual factors. (B-E) the engineering cycle by understanding the emergent dynamics mediated by growth feedback contextual factors (including growth feedback and resource competition), predicting circuit behavior with host- and resource-aware modeling, and redesigning circuits with a control strategy embedded.
Here, we categorize contextual factors into two primary groups: individual contextual factors capable of independently influencing circuit gene expression alone, such as gene part and orientation choice (Box 1–2), and feedback contextual factors that are not simply contingent on the specific individual component but emerge as systemic properties resulting from complex interplays between circuit and host. The design of gene circuits necessitates consideration of individual contextual factors and requires optimization of individual factors to circumvent circuit constraints (Box 1–2). However, sometimes exhaustive fine-tuning of individual contextual factors doesn’t guarantee the optimal functionality of the circuits as intended (Figure 1A). Feedback rather than single contextual factors pose more intractable bottlenecks to design because a circuit’s stability and robustness require not just component-wise consideration but a comprehensive understanding of emergent dynamics from these feedback interactions (Figure 1B), employing mathematical modeling frameworks with dynamic consideration of host (host-aware) and resource (resource-aware) contributions to the system (Figure 1C), and implementing control-embedded design (Figure 1D).
Growth feedback is a multiscale feedback loop characterized by reciprocal interactions between a synthetic circuit and the host cell’s growth rate, wherein the cellular burden resulting from the utilization of limited transcriptional/translational resources in the host cell during the circuit gene expression leads to a reduction in the host growth rate, and the growth rate, in turn, alters the behavior of the circuit (Figure 1B, left) [18–24]. The state of cellular growth considerably impacts the functionality of synthetic constructs due to its cell-wide scope. Growth has the ubiquitous effect of elevating dilution rates of circuit products, as mRNA/proteins are diluted across a growing population. Additionally, changes in the growth rate or genetic context of the host cell are accompanied by changes in the physiological state of the cell, resource supply, and cell-wide parameters such as ribosome abundance and transcription/translation rates [25,26].
Resource competition represents a form of context dependence arising from the competition among multiple modules in a synthetic biological system for a finite pool of shared resources [27–30]. When this occurs, modules indirectly repress each other by diminishing the pool of resources available for other modules (Figure 1B, right). A related yet distinct phenomenon is retroactivity, wherein the downstream module either sequesters or modifies the signal employed by the upstream module [31–33] (Box 2). A prevalent form of resource competition that operates at a cell-wide scope is competition over transcriptional (RNAP) and translational (ribosome) resources. It is noteworthy that in bacterial cells the primary source of competition lies in translational resources [29,27,34,35], competition for transcriptional resources is more dominant in mammalian cells [36–38]. As these expression resources globally promote gene expression throughout the cell, competition over RNAPs and ribosomes are significant sources of bottlenecks [27,36,37]. Aside from these two primary constraints, competition can also occur at the level of transcriptional families, between parallel-placed downstream modules, and between mRNA and protein moieties over sigma factors, shared transcription factors, dCas9, and nuclease/protease degradation machinery respectively [39–44].
Intertwining multiple feedback contextual factors convolutes behavior further. Modeling frameworks have been developed with a focus on either resource competition [27–30] or growth feedback [19,20,23]. Attempts have been made to consider both, aiming to explore the complexity of intricate interactions among feedback factors [21,45,46]. Figure 1C shows how each of the discussed interactions can be put together to form a comprehensive framework for the relationships between these three nodes. The operation of the circuit causes cellular burden by reducing the level of free resources within the cell, while the resource pools themselves stimulate the production of circuit proteins and host growth, and stimulate themselves via autosynthesis. Host growth on the other hand upregulates the cellular transcriptional/translational resource pools, while typically reducing the circuit output concentration due to enhanced dilution. This framework provides a solid foundation for predictive modeling of synthetic circuits.
Emergent dynamics resulting from global growth feedback
Emergence and Loss of Qualitative States
Bi-/Multistability (see Glossary) is a special function that has been achieved by several different synthetic gene circuits. However, the growth feedback could alter the number of steady states achieved by a synthetic genetic system. The rate-balance plot in Figure 2A (top) shows how growth feedback results in the loss of the high-expression (“ON”) state in a bistable self-activation switch by increasing the protein dilution rate to a point where the production and degradation rate curves no longer intersect above their low-expression state [23]. While the loss of bistability here is attributed to dilution, the cellular burden imposed by the circuits could reduce the growth rate and dilution, potentially leading to the emergence of a high-expression state. For example, the significant burden on host cells from a self-activation circuit with a noncooperative promoter resulted in emergent bistability in this circuit that would otherwise display only monostable behavior (Figure 2A, middle) [22]. Cellular burden stymies the dilution rate, resulting in a low-expression high-growth state and a high-expression low-growth state. Lastly, ultrasensitive growth feedback can shift the degradation curve non-monotonically to result in emergent tristability in a self-activation circuit (Figure 2A, bottom) [47].
Figure 2. Emergent phenomena resulting from growth feedback.
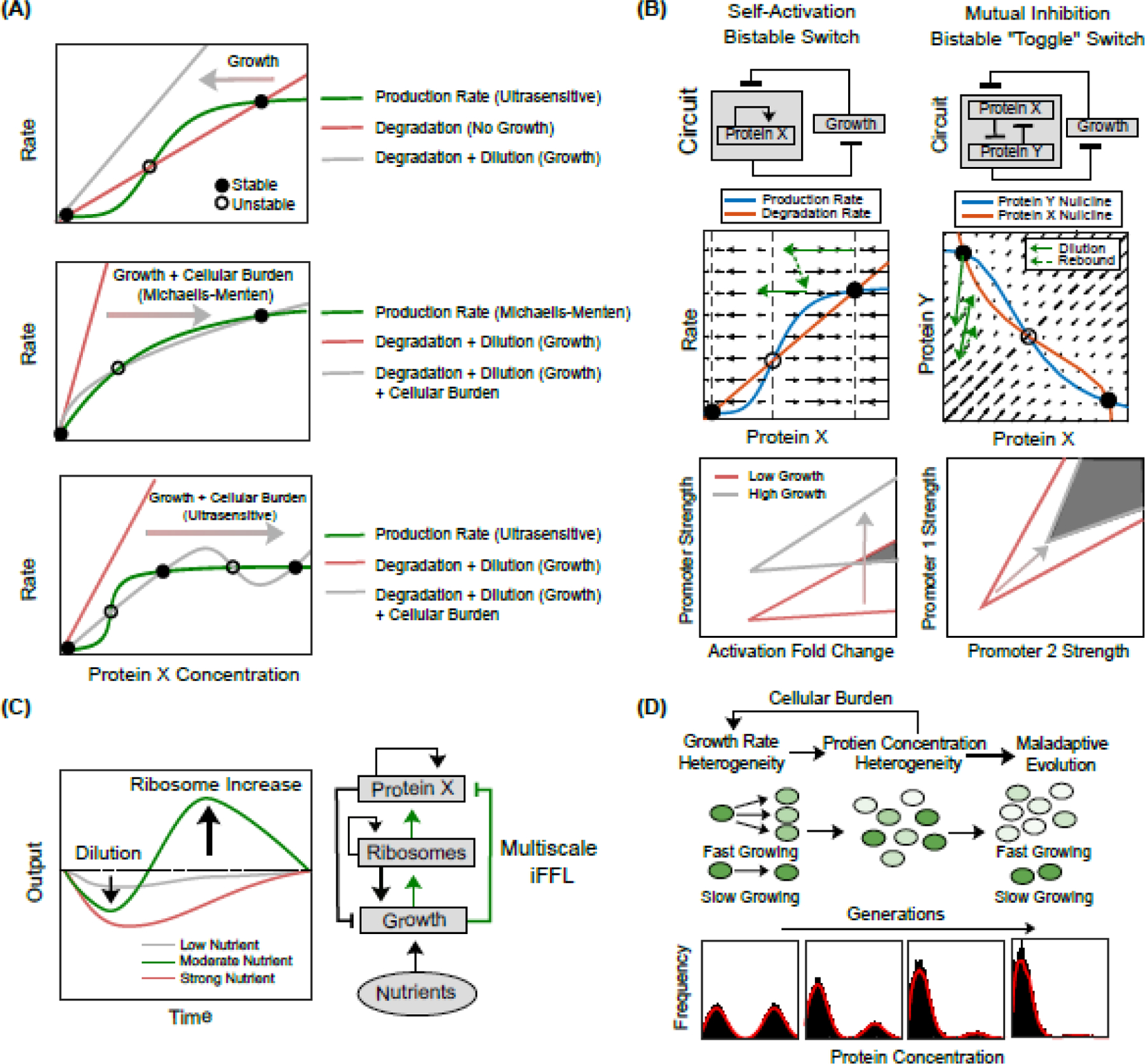
(A) (Top) Loss of qualitative state due to growth feedback. A bistable system with a self-activation ultrasensitive promoter loses its high-expression state under fast growth conditions, where the enhanced dilution rate of proteins only intersects with the production rate curve at a low-expression level in the rate-balance plot [23]. (Middle) Emergence of a qualitative state in a self-activation system with a noncooperative promoter due to the cellular burden placed on host cells by output production, which enables a diminished degradation curve to intersect the production curve three times [22]. (Bottom) Emergent tristability in a self-activation system driven by ultrasensitive growth feedback [47]. (B) Loss of circuit memory due to growth feedback depends on the circuit topology. A bistable switch with self-activation topology is sensitive to growth because the dilution direction is perpendicular to the system separatrix, while a toggle switch is robust given that the dilution direction is more parallel to the separatrix [23]. (C) The bistable region across a range of growth rates for the self-activation switch (left) and the toggle switch (right) shows that the toggle switch is more robust than the self-activation switch level in responding to host growth [18]. (D) Emergent damped oscillation in a self-activation circuit induced by nutrient-modulating growth feedback [52]. (E) The heterogeneity in cell-to-cell growth rates and protein concentrations are coupled through growth feedback. Over time, the fast-growing cell population with low circuit gene expression outcompetes the cell population with high gene expression, leading to the degeneration of qualitative states.
Interestingly, the memory loss in response to growth strongly depends on the circuit topology [23]. Bistability can be attained by a self-activation switch or a toggle switch. However, only the self-activation switch loses bistability under transient high-growth conditions. The difference lies in how growth causes the system to change in the direction field as shown in the rate-balance plot or nullcline analysis in Figure 2B. For the self-activation circuit (Figure 2B, left), growth causes an increased dilution rate for the circuit protein, moving the system away from the ON state. Notably, this direction is perpendicular to the separatrix (middle dashed line), resulting in the system arriving and crossing the separatrix very quickly. On the other hand, growth accelerates the dilution of both proteins in the toggle switch (Figure 2B, right), forcing the system to take a path that is much more parallel to the separatrix thereby increasing the difficulty of crossing it. This topology-dependent effect can also be seen via two-parameter bifurcation (Figure 2C), where an increased growth rate causes a shifting of the activation thresholds in a way that results in a quickly shrinking overlap bistable region across a range of growth rates in the self-activation circuit and a much slower bistable collapse in the toggle switch [18]. In addition, the bistable region of the toggle switch diminishes or may even vanish upon nutrient variation [48]. Growth variations can lead to an unbalanced response in gene expression, potentially reshaping cell fate decisions within the toggle switch [49].
Dynamic ribosomal composition and growth-dependent damped oscillation
Ribosomal upregulation during growth can lead to interesting emergent phenomena [50,51]. When the cell begins to exit the exponential growth phase, the return of ribosomal levels to baseline lags under some growth conditions which results in a temporary burgeoning of translational resources [52]. This may result in a slight overshoot in trivial circuits due to the increased resource pool, however, self-activation switches can capitalize highly on this burgeoning effect, resulting in a significant spike in this time frame (Figure 2D). As this peak typically follows a trough in circuit output caused by the growth-dependent dilution, circuits containing positive feedback topologies could yield an emergent damped oscillation.
Effect of host cell growth on the noise level of synthetic gene expression
Host cell growth encompasses several unique sources of noise. A significant level of cell-to-cell variability is introduced at the moment of cell splitting, whereby plasmids, transcription factors, ribosomal/RNAP fractions, etc., can be unevenly distributed between the emerging daughter cells. Differential attributes conferred to these cells during their split can persist for several generations before disappearing under other forms of noise [53]. The growth rate can also vary from cell to cell due to the variability in metabolic burden caused by synthetic gene circuits (Figure 2E, top), which in turn also contributes to the spread of cell-to-cell protein distribution via growth feedback [54]. Bimodal gene expression was observed, with one population exhibiting a high expression but a slow growth state, and the other population displaying a low expression but a fast growth state [22]. The emerging heterogeneity in cell-to-cell protein distribution simultaneously also feeds into growth rate heterogeneity by introducing varying levels of cellular burden. Over time, growth rate heterogeneity will cause the population distribution to change as slow-growing cells with a high metabolic burden are outcompeted by faster-growing cells (Figure 2E, bottom) [55].
Emergent phenomena resulting from resource competition
Resource competition between circuit modules can exert a substantial impact on gene circuits. The sequestration of resources by a “strong module” results in diminished expression of the ‘weak module.’ Various outcomes are reported depending on the circuit complexity.
Linear interdependence in a simple two-gene system
The expression levels of two genes in the presence of a constrained resource (Figure 3A) were found negatively correlated (Figure 3B) in E. coli, interpreted as an isocost line [27]. The horizontal dashed line (Figure 3B, left) shows an ideal system with no resource constraint present where the expression of either gene is completely independent of the other. Application of a resource constraint forces the conserved resource to be split between the involved modules resulting in a negative correlation, with the ratio of the RBS activity between the two genes and the plasmid copy number determining the slope and y-intercept of the curve respectively (Figure 3B) [27].
Figure 3. Emergent phenomena resulting from resource competition.

(A) Simple two-gene system for studying resource competition. The two genes are independent of one another other than the shared RNAP and ribosomes for transcription and translation. (B) Linear Isocost dependence was observed between the expression levels of two competing genes. The RBS activity ratio and the circuit copy number determine the isocost line’s slope and y-intercept respectively [27]. (C) Nonlinear resource competition shapes the dose-response curve non-monotonically rather than monotonically in an activation cascade circuit because the increasing resource consumption from the upstream module limits the expression of the downstream at high inducer levels [29]. (D) “Winner-Take-All” resource competition excludes the coactivation of two self-activation modules in a cascading bistable switches circuit because the activation of one switch precludes the activation of the other one, leading to the solitary module’s activation (bottom), in contrast to the coactivation in the ideal scenario with no resource constraint (top) [30]. (E) Analytic expression of the protein noise level in a two-gene system under a resource-constrained context includes noise from 1) the stochastic birth/death process of its mRNA, 2) the stochastic birth/death process of protein, and 3) the stochastic birth/death process of the other mRNA. The last term is “resource competitive noise”, which completely results from resource competition [56]. (F) The dependences of total noise and its three components on the translational capacity show double-edged effects of resource competition on circuit gene expression noise [56].
Nonmonotonic rather than expected monotonic dose-response curve in activation cascade circuit
Although a single resource-constrained interaction between two genes can be easily predicted via linear interdependence curves, multiple interactions in more complex circuits and the influence of intrinsically present nonlinearity in the circuit can lead to unintuitive behavior. For example, the presence of resource competition in an activation cascade circuit can result in a nonmonotonic dose-response curve in E. Coli, where output actually falls despite increasing inducer levels [29]. As shown in Figure 3C, two activation modules can on their own demonstrate monotonically increasing output with inducer levels, but when placed together, the resource consumption from upstream nodes can sequester resources from the final output module.
Winner-takes-all rather than expected coactivation behavior in cascading bistable switch (CBS)
The presence of resource competition can also result in higher-order emergent properties in more complex circuits, such as changing the number/nature of qualitative states in a system. For example, a CBS circuit with two positively connected self-activation modules was found to display winner-takes-all (WTA) behavior instead of expected coactivation in E. coli (Figure 3D) [30]. The theoretical simulation predicted that the system could achieve two successive cell fate transitions going from both modules in the OFF state to single module activation and then to coactivation of both modules with increasing inducer (Figure 3D, top). However, experimental data supported the existence of the first two states (Figure 3D, bottom), but due to resource competition, the coactivation state proved experimentally elusive. Even when disconnected, subtle imbalances between the switches would result in a runaway behavior where the more active switch could take increasingly more resources and prevent the activation of the other.
Effect of resource competition on gene expression noise
The role of resource constraints on intracellular noise was investigated using a simple model system involving the independent expression of two proteins, GFP and RFP, under a constrained ribosomal pool (Figure 3A) [56]. Interestingly, resource competition was found to have unintuitive double-edged effects on circuit noise, introducing a novel noise term coined “resource competitive noise” while also providing a global repressive effect that places a soft limit on the range of values mRNA and protein concentrations can achieve (Figure 3E). Which of these effects is more dominant depends on the circuit’s transcriptional/translational capacity (Figure 3F). Large shared pools of resources increase gene transcription via increased burst size, not burst frequency, resulting in larger variance than sums of small discrete pools [57]. It is suggested that resource competition diminishes the robustness of the toggle switch to noise [58–60]. Further investigation can enhance our understanding of how resource competition influences noise behavior in more intricate circuits, guiding efforts to explore how to control and employ this noise effectively
Embedding control mechanisms within circuit design to mitigate the impacts of feedback contextual factors
Control strategies targeting resource availability
Orthogonal resources.
Several groups have attempted to disentangle resource-coupled synthetic circuits by employing orthogonal transcriptional/translational resources. “O-ribosomes” are constructed by expressing an orthogonal 16S rRNA (o-16S) that can bind to ribosomal proteins and recognize an altered Shine-Dalgarno (SD) over the wild-type [61]. The Ribo-T system tethers 50S ribosomal units via linker sequences to the mutated 30S sequences to prevent competition between mutated and wild-type 30S units over the shared pool of 50S units (Figure 4A) [62]. On the other hand, orthogonal transcriptional machinery adapts naturally occurring RNAP moieties, such as the T7 bacteriophage RNAP that operates independently from endogenous E. coli RNAP, or engineering RNAP targeting specificity via sigma factors [63,64].
Figure 4. Redesign synthetic gene circuits to reduce context dependence.

(A) Ribo-T system for fully orthogonal translation by tethering two subunits (the core 16S and 23S rRNAs) of the engineered ribosome [62]. (B) The OSYRIS system allows for the high-efficiency production of synthetic proteins by swapping the roles of the wild type and Ribo-T ribosomes [65]. (C) Ribosomal resource allocator utilizing NFL to control the expression of orthogonal ribosomes (oR) [70]. (D) Combatting cellular burden by upregulating ribosomes and growth rate using a modified SpoT (SpoTH) that is expressed alongside the reporter gene to lower ppGpp level for more ribosomes [71]. (E) Reducing host resource consumption to reallocate resources to the circuit by using the nuclease MazF that only splices host mRNAs to reduce the host’s ribosomal usage. (F) NFL at the transcriptional level to decouple modules accessing shared resource pools [73]. (G) A burden-driven feedback using a burden-driven promoter to express sgRNA which works with dCas9 to shut off protein production and alleviate the burden [75]. (H) NFL mediated via covalent modification cycles where the phosphatase is expressed alongside the circuit to inactivate the transcription factor [38]. (I) Quasi-integral controller created by expressing a transcription factor ECF32 alongside the circuit reporter to produce an antisense sRNA that binds to the GOI mRNA and degrades it [35]. (J) Antithetic integral negative feedback in which output transcription factor leads to an antisense RNA and an RNA-binding protein NES-L7Ae that degrades output mRNA or inhibits its translation [78]. (K) Endoribonuclease (ERN) feedforward loop where the shared resources produce both the circuit output and an ERN that binds to the output mRNA and degrades it [40]. (L-M) Multi-module control topologies utilizing NFL (L) and iFFL (M) [83,84]. Local controllers contain a control loop for each module, global controllers contain one control loop for the entire circuit, and competitive controllers design additional competition between two control loops for dCas9 before inhibiting individual modules.
Orthogonal resource systems suffer from their own set of issues, particularly the reduced expression rate resulting from the limited binding affinity of the orthogonal SD sequence. The OSYRIS system (Figure 4B) attempts to solve this by trading host growth rate for increased translation of the target gene sequences by swapping the roles of the Ribo-T and wild-type ribosomes [65]. Furthermore, although orthogonal transcriptional/translational systems exist, modeling has demonstrated that creating a fully orthogonal expression system with both may be challenging as merely combining existing systems can lead to a plethora of emergent instability [66].
Resource reallocation.
Transcriptional and translational resources can be selectively allocated to genes of interest [67–69]. For example, UBER system serves as a portable platform, offering an insulated resource environment for heterologous infrastructure [67]. An RNAP resource allocator system has been constructed to effectively coordinate expression across multiple modules within a synthetic system depending on their transcriptional budget [68]. Several groups have attempted to integrate host resource nodes into synthetic negative feedback loops (NFL) and incoherent feedforward loops (iFFL) [69–72]. An NFL was designed to control a pool of o-ribosomes dedicated to circuit protein production (Figure 4C) [69,70]. Here, repression of o-ribosome production was mediated by a repressor that was transcribed by the o-ribosomes as well; increased circuit expression would result in sequestration of the o-ribosomes, reduction in repressor production, and upregulation of o-ribosome production. An iFFL architecture was achieved by simultaneously activating a modified SpoT enzyme to upregulate intracellular ribosome concentrations alongside increasing GOI activity (Figure 4D) [71]. The modified SpoT enzyme reduces the concentration of ppGpp, a global regulator of growth rate and rRNA/ribosomal synthesis in E. coli, resulting in a derepression of rRNA, enhanced ribosome production rate, and replenishing of translational resources upon GOI induction. Another strategy was devised by altering the resource allocation between the host and the circuit in E. Coli (Figure 4E) [72]. The authors achieved this by expressing a global RNase mazF to downregulate host translation and free up resources for GOI translation while protecting circuit proteins by engineering mazF binding sequences out of the circuit. In these systems, higher resource capacity is expected to reduce gene expression noise as it diminishes with increasing protein steady-state concentration.
Control strategies targeting one single module of interest
Negative feedback control has also been utilized as a module-specific control architecture to mitigate the effects of resource competition. For example, including an NFL in the circuit could decouple resource-coupled gene expression and mitigate the growth burden in E. Coli [73,74] (Figure 4F).
It is noted that resource allocation and growth feedback are intricately linked as both either indicate or emerge under conditions that impose heightened stress or cellular burden. Ceroni and coworkers developed a “resource capacity monitor” by inserting a constitutively expressed GFP reporter into the host genome in bacteria and demonstrated the close correlation of ribosome consumption, heterologous gene expression, and growth rate [34]. As the GFP was constitutively expressed, any changes in GFP fluorescence were indicative of fluctuating levels of intracellular resources and growth rate. Sensors like these can in turn be utilized to drive NFL control for mitigating burden-inducing behavior. In a following study, a burden-based feedback system was constructed by placing a CRISPR-dCas9 transcriptional silencing motif under the expression of a burden-activated promoter to insulate GOIs from fluctuations in global cellular resource pools (Figure 4G) [75]. Under stressful conditions, the CRISPR-dCas9 silencing motif would upregulate and reduce transcription of GOIs, alleviating cellular burden.
A form of NFL control mediated via covalent modification cycles was utilized to reduce context effects in mammalian cells (Figure 4H) [38]. Here, the transcription factor required to activate the GOI production was chosen to only exhibit promoter-binding capabilities when phosphorylated, the activation of which was dependent on a dynamic equilibrium between the kinase and phosphatase specific to the transcription factor. NFL was mediated by co-expressing phosphatase along with the GOI.
Antithetic feedback control is a form of control that realizes integral feedback using two mutually annihilating signals [76,77]. A quasi-integral controller was utilized to stabilize gene expression whereby GOI expression led to the expression of an sRNA which was complementary to the GOI’s mRNA, leading to its degradation in E. Coli (Figure 4I) [35]. Under this type of control, a sudden increase/decrease in resource capacity would result in an upregulation/downregulation of the inhibitory sRNA, buffering any significant change in GOI mRNA. In addition, antithetic feedback can be combined with NFL to create a more sophisticated controller displaying both proportional and integral feedback control by coexpressing the NES-L7Ae RNA-binding protein (proportional) and an antisense RNA (antithetic integral) alongside the output mRNA in mammalian cells (Figure 4J) [78].
Feedforward control has also been incorporated in synthetic circuits for module-specific control. iFFL was utilized to stabilize expression levels against fluctuating resource environments by coexpressing an endoribonuclease alongside the GOI which accelerated the GOI’s mRNA degradation rate in mammalian cells (Figure 4K) [40]. A relative increase/decrease in the global resource pool would result in a relative increase/decrease in the concentration of the endoribonuclease, stifling any changes in the GOI’s expression rate. A similar architecture was established through the coexpression of a miRNA with GOI to inhibit the GOI transcript in mammalian cells [36]. Incorporating a supplementary repressive link in a growth-sensitive self-activation circuit converts the overall system into a multiscale iFFL that enhances the circuit robustness in response to growth feedback in E. Coli [79].
Control strategies targeting multiple modules
While the control strategies detailed above have been proven to be successful in controlling one single module of interest in systems, proper control of more elaborate, multi-module systems may require more extensive control mechanisms. Division of labor and higher-order topological control strategies provide methods for controlling multi-module gene systems.
Division of labor.
Division of labor has been utilized for metabolic engineering, whereby reaction steps in desired biochemical pathways requiring similar machinery are segmented into their own substrains/microenvironments, improving reaction rates and titers while simultaneously reducing the host burden [80–82]. WTA resource competition could be attenuated via a two-strain consortium by splitting two switch modules into different subpopulation strains and linking them together with a quorum-sensing system in E. coli [30]. In this fashion, each switch was limited to its own intracellular resource pool and could not steal resources from the other, nullifying the WTA behavior seen in the single-strain system. This strategy faces challenges as quorum-sensing signaling links circuit behavior to population density and intermodule effects may arise between consortium strains. For example, competitive effects between different consortium strains in nutrient-limited media could result in a WTA resource competition at the population level rather than the gene expression level.
Global, local, and competitive control.
The NFL and iFFL control strategies discussed above can be extrapolated to control all the circuit modules. These multi-module control strategies can generally take three different architectural forms (Figure 4L–M), defined by the mechanism through which regulation across modules is mediated [9,11,83,84]. Local controllers act via module-specific control loops, i.e., each module has its own NFL or iFFL (Figure 4L–M). Global controller instead uses one circuit-wide control loop on the entire circuit. The resource reallocation strategies discussed above are considered one form of global controller, operating globally to regulate the resource distribution between the circuit and the host [9,11]. Here, we highlight an alternative form of global controller that does not control the resource allocation but uses a circuit-wide NFL or iFFL to repress all the circuit modules. In this context, any increase in activity in a particular module leads to a simultaneous reduction in activity levels across all circuit modules (Figure 4L–M) [83,84]. The local controller exhibits greater flexibility given that the activity of each module can be independently modulated and the global controller could enhance the stability against fluctuation in available resources for the whole circuit. To alleviate the WTA resource competition, another controller type was introduced, referred to as negatively competitive regulation (NCR) that introduces additional synthetic competition between the control nodes to counterbalance resource competition intrinsic to the cell (Figure 4L–M) [83,84]. In this way, combined repressive and cross-activatory effects could be achieved simultaneously, giving an advantage to the low-activity module while inhibiting the high-activity module. Modeling and simulation results have indicated that NCR controllers could have an advantage over local and global control architectures in combating WTA resource competition and circuit noise [56,83,84].
Concluding Remarks and Future Perspectives
In this review, we have discussed emergent phenomena caused by circuit-host interactions as well as corresponding strategies to control these behaviors. However, it is important to note that the phenomena and control strategies discussed thus far have primarily focused on relatively simple gene circuits. Currently, these strategies have neither seen widespread adoption by others nor found practical applications beyond laboratory settings. To fully understand the potential of these new design principles, it is essential to explore their scalability and applicability to more complex circuits in diverse conditions (see Outstanding Questions). By assessing their function in broader conditions, we can gain insights into the generality of the discussed design principles and their reliability across various circuits and hosts. This knowledge will be crucial for advancing the field of synthetic biology and enabling the development of sophisticated multi-agent systems that are deployable outside of the lab. The investigation of circuit-host interactions and the development of tailored control strategies for synthetic consortia systems represent exciting avenues for future research in synthetic biology. These avenues encompass the refinement of regulatory mechanisms for precise gene expression control, the orchestration of multiple modules within consortia for seamless coordination, and the development of systems capable of adaptable responses to environmental cues. In addition, a holistic approach integrating synthetic and systems biology would provide us with a comprehensive understanding of complex biological processes [85,86]. By expanding our knowledge and capabilities in this area, we can unlock the full potential of engineered microbial communities and pave the way for innovative biotechnological solutions.
Highlights.
Unavoidable circuit-host interactions confound the modularity in engineering synthetic gene circuits.
Unintuitive emergent dynamics can be better comprehended by incorporating the influence of contextual feedback factors into the system.
Mechanistic understanding of the intricate relationships between circuits and hosts enhances our ability to predict and control them.
Complementary control strategies can be incorporated into circuit design to mitigate unwanted effects.
Identification of contextually dependent modes of failure early can accelerate circuit development.
Box 1. Single Intragenic Context Factors.
Intragenic contextual factors are the elements within the coding and non-coding regions of one gene, such as promoter, ribosome binding sites (RBS), and terminators (Figure IA), that determine the dynamic range of that gene expression.
Maintaining the essential properties of single genetic parts when they are placed in a larger system is one of the primary challenges faced in the DBTL cycle [6]. The flanking sequences of individual parts, such as promoters, RBS, and terminator sequences, can alter the behavior of the part through various structural interactions [17]. Transcriptional initiation and promoter activity are influenced specifically by the genetic sequences located directly upstream of the – 35 element and downstream of the −10 element, while RBS and terminator strength are strongly affected by interactions with the 5’ UTR region and the sequences directly upstream the terminator respectively [87,88].
This context-dependency makes it difficult to establish accurate predictions of protein expression rates for different promoter-RBS sets. To avoid this sequence-dependent behavior, the activity of promoters can be insulated from the genetic context by flanking promoters with adjoining insulator sequences (Figure IB), which place physical distance between adjacent parts to limit their interaction and themselves display a neutral effect on the parts they flank [89]. To eliminate flanking sequence dependence at the translational level, the activity of RBSs can be isolated from any interaction with the 5’ UTR region by utilizing CRISPR or ribozymes to cleave off the 5’ UTR from the precursor mRNA, nullifying the possibility of 5’UTR-RBS interactions [90–92]. High-dimensional biokinetic models have also been developed to harness computational power to predict genetic context effects, design promoters with desired transcription rates, and identify undesirable promoters before construction [93].
Figure I: Single intragenic context factors include.

(A) a wide array of cis and trans-acting parts for tuning gene expression levels, including promoters, RBS, terminators, untranslated sequences, transcription factors/binding sites, etc. (B) Unintended structural interactions can arise between adjacent transcriptional regions (ATRs).
Acknowledgments
This project was supported by the National Science Foundation, USA grant (2143229) and the National Institutes of Health, United States grant (R35GM142896).
Glossary
- Bi-/multistability
the phenomenon where a system has two or more stable steady states for a given input value.
- Circuit topology
the designed structure and connectivity of genetic elements within an engineered genetic network.
- Division of labor
a family of strategies that attempt to address the limitations of resource-constrained systems by dividing tasks between multiple agents to prevent indirect resource-econstrained crosstalk between modules.
- Isocost lines
a concept originally derived from microeconomics, are employed in this context to illustrate a linear cellular economy of gene expression when faced with limited resources.
- Nullcline analysis
the stability analysis method for a two-dimensional biochemical system. There are two nullcline curves, each of them denoting the zero rate of change in concentrations of one variable. The intersections of the two nullcline curves determine the steady states of the system.
- Orthogonal transcriptional/translational resources
molecular components that operate independently of the endogenous cellular machinery for the expression of genes within synthetic constructs.
- Rate-balance plot
the production and degradation rates of a circuit protein on its concentration for one-dimensional biochemical systems. The values of protein concentration for which production and degradation curves intersect represent the steady states of the system.
- Separatrix
the boundary separating two stable steady states in a multistable system. One side of the separatrix leads to one stable state, and the other side leads to a different stable state.
- Winner-takes-all (WTA)
a phenomenon characterized by a resource-dependent discontinuous change in the system, wherein a single module acquires the majority of resources and emerges as the exclusive activated entity.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no competing interests.
References
- 1.Wurtzel ET et al. (2019) Revolutionizing agriculture with synthetic biology. Nat. Plants 5, 1207–1210 [DOI] [PubMed] [Google Scholar]
- 2.Moe-Behrens GHG et al. (2013) Preparing synthetic biology for the world. Front. Microbiol 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang T-C et al. (2020) Materials design by synthetic biology. Nat. Rev. Mater 6, 332–350 [Google Scholar]
- 4.Nguyen PQ et al. (2023) Harnessing synthetic biology to enhance ocean health. Trends Biotechnol. 41, 860–874 [DOI] [PubMed] [Google Scholar]
- 5.Meng F and Ellis T (2020) The second decade of synthetic biology: 2010–2020. Nat Commun 11, 5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Del Vecchio D (2015) Modularity, context-dependence, and insulation in engineered biological circuits. Trends Biotechnol. 33, 111–119 [DOI] [PubMed] [Google Scholar]
- 7.Borkowski O et al. (2016) Overloaded and stressed: whole-cell considerations for bacterial synthetic biology. Curr. Opin. Microbiol 33, 123–130 [DOI] [PubMed] [Google Scholar]
- 8.Boo A et al. (2019) Host-aware synthetic biology. Curr. Opin. Syst. Biol 14, 66–72 [Google Scholar]
- 9.Grunberg TW and Del Vecchio D (2020) Modular Analysis and Design of Biological Circuits. Curr Opin Biotechnol 63, 41–47 [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Osorio I et al. (2020) Quantitative modeling of the interplay between synthetic gene circuits and host physiology: experiments, results, and prospects. Curr Opin Microbiol 55, 48–56 [DOI] [PubMed] [Google Scholar]
- 11.McBride C et al. (2021) Design of genetic circuits that are robust to resource competition. Curr. Opin. Syst. Biol 28, 100357 [Google Scholar]
- 12.Shakiba N et al. (2021) Context-aware synthetic biology by controller design: Engineering the mammalian cell. Cell Syst. 12, 561–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Şimşek E et al. (2022) Toward predictive engineering of gene circuits. Trends Biotechnol. 41, 760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barajas C and Del Vecchio D (2022) Synthetic biology by controller design. Curr. Opin. Biotechnol 78, 102837. [DOI] [PubMed] [Google Scholar]
- 15.Ilia Katherine and Domitilla Del Vecchio (2022) Squaring a Circle: To What Extent Are Traditional Circuit Analogies Impeding Synthetic Biology? GEN Biotechnol. 1, 150–155 [Google Scholar]
- 16.Kumar S and Hasty J (2023) Stability, robustness, and containment: preparing synthetic biology for real-world deployment. Curr. Opin. Biotechnol 79, 102880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardinale S and Arkin AP (2012) Contextualizing context for synthetic biology – identifying causes of failure of synthetic biological systems. Biotechnol. J 7, 856–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klumpp S et al. (2009) Growth Rate-Dependent Global Effects on Gene Expression in Bacteria. Cell 139, 1366–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiße AY et al. (2015) Mechanistic links between cellular trade-offs, gene expression, and growth. Proc. Natl. Acad. Sci. U. S. A 112, E1038–E1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao C et al. (2017) An integrative circuit–host modelling framework for predicting synthetic gene network behaviours. Nat. Microbiol 2, 1658–1666 [DOI] [PubMed] [Google Scholar]
- 21.Nikolados E-M et al. (2019) Growth Defects and Loss-of-Function in Synthetic Gene Circuits. ACS Synth. Biol 8, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 22.Tan C et al. (2009) Emergent bistability by a growth-modulating positive feedback circuit. Nat. Chem. Biol 5, 842–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang R et al. (2020) Topology-dependent interference of synthetic gene circuit function by growth feedback. Nat. Chem. Biol 16, 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong L-W et al. (2023) Effects of growth feedback on gene circuits: A dynamical understanding. eLife 12 [Google Scholar]
- 25.Klumpp S and Hwa T (2014) Bacterial growth: global effects on gene expression, growth feedback and proteome partition. Curr. Opin. Biotechnol 28, 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardinale S et al. (2013) Effects of Genetic Variation on the E. coli Host-Circuit Interface. Cell Rep. 4, 231–237 [DOI] [PubMed] [Google Scholar]
- 27.Gyorgy A et al. (2015) Isocost Lines Describe the Cellular Economy of Genetic Circuits. Biophys. J 109, 639–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbonell-Ballestero M et al. (2016) Dealing with the genetic load in bacterial synthetic biology circuits: convergences with the Ohm’s law. Nucleic Acids Res. 44, 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian Y et al. (2017) Resource Competition Shapes the Response of Genetic Circuits. ACS Synth Biol 6, 1263–1272 [DOI] [PubMed] [Google Scholar]
- 30.Zhang R et al. (2021) Winner-takes-all resource competition redirects cascading cell fate transitions. Nat. Commun 12, 853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Del Vecchio D et al. (2008) Modular cell biology: retroactivity and insulation. Mol. Syst. Biol 4, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jayanthi S et al. (2013) Retroactivity Controls the Temporal Dynamics of Gene Transcription. ACS Synth. Biol 2, 431–441 [DOI] [PubMed] [Google Scholar]
- 33.Mishra D et al. (2014) A load driver device for engineering modularity in biological networks. Nat. Biotechnol 32, 1268–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceroni F et al. (2015) Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat. Methods 12, 415–418 [DOI] [PubMed] [Google Scholar]
- 35.Huang H-H et al. (2018) A quasi-integral controller for adaptation of genetic modules to variable ribosome demand. Nat. Commun 9, 5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frei T et al. (2020) Characterization and mitigation of gene expression burden in mammalian cells. Nat Commun 11, 4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberto Di Blasi et al. (2023) Resource-aware construct design in mammalian cells. Nat Commun 14, 3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones RD et al. (2022) Robust and tunable signal processing in mammalian cells via engineered covalent modification cycles. Nat. Commun 13, 1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mauri M and Klumpp S (2014) A Model for Sigma Factor Competition in Bacterial Cells. PLOS Comput. Biol 10, e1003845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones RD et al. (2020) An endoribonuclease-based feedforward controller for decoupling resource-limited genetic modules in mammalian cells. Nat Commun 11, 5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butzin NC and Mather WH (2018) Crosstalk between Diverse Synthetic Protein Degradation Tags in Escherichia coli. ACS Synth. Biol 7, 54–62 [DOI] [PubMed] [Google Scholar]
- 42.Huang H-H et al. (2021) dCas9 regulator to neutralize competition in CRISPRi circuits. Nat. Commun 12, 1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butzin NC et al. (2017) Entrainment of a Bacterial Synthetic Gene Oscillator through Proteolytic Queueing. ACS Synth. Biol 6, 455–462 [DOI] [PubMed] [Google Scholar]
- 44.Cookson NA et al. (2011) Queueing up for enzymatic processing: correlated signaling through coupled degradation. Mol. Syst. Biol 7, 561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Melendez-Alvarez JR et al. (2023) Growth feedback confers cooperativity in resource-competing synthetic gene circuits. Chaos Solitons Fractals 173, 113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sechkar K et al. (2023) A coarse-grained bacterial cell model for resource-aware analysis and design of synthetic gene circuits. bioRxiv, 2023.04.08.536106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melendez-Alvarez JR and Tian X-J (2022) Emergence of qualitative states in synthetic circuits driven by ultrasensitive growth feedback. PLOS Comput. Biol 18, e1010518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sickle JJ et al. (2020) Integrative Circuit-Host Modeling of a Genetic Switch in Varying Environments. Sci Rep 10, 8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu J et al. (2023) Unbalanced response to growth variations reshapes the cell fate decision landscape. Nat. Chem. Biol 19, 1097–1104 [DOI] [PubMed] [Google Scholar]
- 50.Dai X and Zhu M (2020) Coupling of Ribosome Synthesis and Translational Capacity with Cell Growth. Trends Biochem Sci 45, 681–692 [DOI] [PubMed] [Google Scholar]
- 51.Scott M et al. (2010) Interdependence of Cell Growth and Gene Expression: Origins and Consequences. Science 330, 1099–1102 [DOI] [PubMed] [Google Scholar]
- 52.Melendez-Alvarez J et al. (2021) Emergent Damped Oscillation Induced by Nutrient-Modulating Growth Feedback. ACS Synth. Biol 10, 1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas P et al. (2018) Sources, propagation and consequences of stochasticity in cellular growth. Nat. Commun 9, 4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuru S et al. (2009) Noisy cell growth rate leads to fluctuating protein concentration in bacteria. Phys. Biol 6, 036015. [DOI] [PubMed] [Google Scholar]
- 55.Blanchard AE et al. (2018) Circuit-Host Coupling Induces Multifaceted Behavioral Modulations of a Gene Switch. Biophys. J 114, 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goetz H et al. (2022) Double‐ Edged Role of Resource Competition in Gene Expression Noise and Control. Adv. Genet 3, 2100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caveney PM et al. (2017) Resource Sharing Controls Gene Expression Bursting. ACS Synth Biol 6, 334–343 [DOI] [PubMed] [Google Scholar]
- 58.Yong C et al. (2021) Stability and Robustness of Unbalanced Genetic Toggle Switches in the Presence of Scarce Resources. Life 11, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gyorgy A (2020) Scarcity of Cellular Resources Decreases the Robustness of Toggle Switches to Noise. in 2020 American Control Conference (ACC), pp. 4264–4269 [Google Scholar]
- 60.Gyorgy A (2019) How Cell-to-Cell Heterogeneity and Scarce Resources Shape the Population-Level Stability Profile of Toggle Switches. in 2019 IEEE 58th Conference on Decision and Control (CDC), pp. 6622–6627 [Google Scholar]
- 61.Rackham O and Chin JW (2005) A network of orthogonal ribosome·mRNA pairs. Nat. Chem. Biol 1, 159–166 [DOI] [PubMed] [Google Scholar]
- 62.Orelle C et al. (2015) Protein synthesis by ribosomes with tethered subunits. Nature 524, 119–124 [DOI] [PubMed] [Google Scholar]
- 63.Qin C et al. (2023) Precise programming of multigene expression stoichiometry in mammalian cells by a modular and programmable transcriptional system. Nat. Commun 14, 1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Temme K et al. (2012) Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 40, 8773–8781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aleksashin NA et al. (2020) A fully orthogonal system for protein synthesis in bacterial cells. Nat. Commun 11, 1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Darlington APS and Bates DG (2020) Architectures for Combined Transcriptional and Translational Resource Allocation Controllers. Cell Syst 11, 382–392 e9 [DOI] [PubMed] [Google Scholar]
- 67.Kushwaha M and Salis HM (2015) A portable expression resource for engineering cross-species genetic circuits and pathways. Nat. Commun 6, 7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Segall-Shapiro TH et al. (2014) A “resource allocator” for transcription based on a highly fragmented T7 RNA polymerase. Mol Syst Biol 10, 742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darlington APS et al. (2018) Dynamic allocation of orthogonal ribosomes facilitates uncoupling of co-expressed genes. Nat. Commun 9, 695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Darlington APS et al. (2018) Engineering Translational Resource Allocation Controllers: Mechanistic Models, Design Guidelines, and Potential Biological Implementations. ACS Synth Biol 7, 2485–2496 [DOI] [PubMed] [Google Scholar]
- 71.Barajas C et al. (2022) Feedforward growth rate control mitigates gene activation burden. Nat. Commun 13, 7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Venturelli OS et al. (2017) Programming mRNA decay to modulate synthetic circuit resource allocation. Nat Commun 8, 15128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shopera T et al. (2017) Decoupling Resource-Coupled Gene Expression in Living Cells. ACS Synth Biol 6, 1596–1604 [DOI] [PubMed] [Google Scholar]
- 74.Guan Y et al. (2022) Mitigating Host Burden of Genetic Circuits by Engineering Autonegatively Regulated Parts and Improving Functional Prediction. ACS Synth. Biol 11, 2361–2371 [DOI] [PubMed] [Google Scholar]
- 75.Ceroni F et al. (2018) Burden-driven feedback control of gene expression. Nat. Methods 15, 387–393 [DOI] [PubMed] [Google Scholar]
- 76.Briat C et al. (2016) Antithetic Integral Feedback Ensures Robust Perfect Adaptation in Noisy Biomolecular Networks. Cell Syst. 2, 15–26 [DOI] [PubMed] [Google Scholar]
- 77.Aoki SK et al. (2019) A universal biomolecular integral feedback controller for robust perfect adaptation. Nature 570, 533–537 [DOI] [PubMed] [Google Scholar]
- 78.Frei T et al. (2022) A genetic mammalian proportional–integral feedback control circuit for robust and precise gene regulation. Proc. Natl. Acad. Sci 119, e2122132119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stone A et al. (2024) Enhancing circuit stability under growth feedback with supplementary repressive regulation. Nucleic Acids Res. gkad1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lindemann SR (2020) A piece of the pie: engineering microbiomes by exploiting division of labor in complex polysaccharide consumption. Curr. Opin. Chem. Eng 30, 96–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thommes M et al. (2019) Designing Metabolic Division of Labor in Microbial Communities. mSystems 4, e00263–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsoi R et al. (2018) Metabolic division of labor in microbial systems. Proc. Natl. Acad. Sci. U. S. A 115, 2526–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stone A et al. (2021) Coupling Shared and Tunable Negative Competition Against Winner-Take-All Resource Competition Via CRISPRi Moieties. in 2021 American Control Conference (ACC), New Orleans, LA, USA, pp. 1–6 [Google Scholar]
- 84.Stone A et al. (2022) Negatively Competitive Incoherent Feedforward Loops Mitigate Winner-Take-All Resource Competition. ACS Synth. Biol 11, 3986–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Contreras-Llano LE et al. (2020) Holistic engineering of cell-free systems through proteome-reprogramming synthetic circuits. Nat. Commun 11, 3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marucci L et al. (2020) Computer-Aided Whole-Cell Design: Taking a Holistic Approach by Integrating Synthetic With Systems Biology. Front. Bioeng. Biotechnol 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rhodius VA et al. (2012) Predicting the strength of UP-elements and full-length E. coli σE promoters. Nucleic Acids Res. 40, 2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kosuri S et al. (2013) Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci 110, 14024–14029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Davis JH et al. (2011) Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 39, 1131–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bashor CJ and Collins JJ (2012) Insulating gene circuits from context by RNA processing. Nat. Biotechnol 30, 1061–1062 [DOI] [PubMed] [Google Scholar]
- 91.Qi L et al. (2012) RNA processing enables predictable programming of gene expression. Nat. Biotechnol 30, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 92.Lou C et al. (2012) Ribozyme-based insulator parts buffer synthetic circuits from genetic context. Nat. Biotechnol 30, 1137–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.LaFleur TL et al. (2022) Automated model-predictive design of synthetic promoters to control transcriptional profiles in bacteria. Nat. Commun 13, 5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yeung E et al. (2017) Biophysical Constraints Arising from Compositional Context in Synthetic Gene Networks. Cell Syst. 5, 11–24.e12 [DOI] [PubMed] [Google Scholar]
- 95.Johnstone CP and Galloway KE (2022) Supercoiling-mediated feedback rapidly couples and tunes transcription. Cell Rep. 41, 111492. [DOI] [PMC free article] [PubMed] [Google Scholar]


