Abstract
The repetitive ETn (early transposon) family of sequences represents an active “mobile mutagen” in the mouse genome. The presence of long terminal repeats (LTRs) and other diagnostic features indicate that ETns are retrotransposons but they contain no long open reading frames or documented similarity to the genes of known retroviruses or other retroelements. Thus, the mechanisms responsible for the mobility of this family have been unknown. In this study, we used computer searches to detect a small region of previously unrecognized type D retroviral pol homology within ETn elements. This small region was used to isolate two mouse endogenous proviral elements with gag, pro, and pol genes similar to simian type D viruses. This new family of mouse endogenous proviruses, termed MusD, is present in several hundred copies in the genome. Interestingly, the MusD LTRs, 3′ internal region, and the 5′ region expected to contain the packaging signal are very closely related to members of the ETn subfamily that have recently transposed. Analysis of different mouse strains indicates that MusD elements predate the existence of the mobile subfamily of ETns. These findings indicate that the ETn family was likely created via recombination events resulting in a near complete substitution of MusD coding sequences with unrelated DNA. Furthermore, these results suggest that ETn transcripts retrotranspose using proteins provided by MusD proviruses.
ETn (early transposon) elements were first described in 1983 as a family of middle repetitive sequences transcribed during early mouse embryogenesis (4). ETn expression peaks between 3.5 and 7.5 days and is found primarily in undifferentiated cells of the inner cell mass and embryonic ectoderm (3). These elements were initially classified as retrotransposon-like because they contain long terminal repeats (LTRs) and retrovirus-like primer binding sites and are flanked by target site duplications (11, 26). However, sequence analysis of full-length copies revealed no long open reading frames (ORFs) and no significant homology to known retroviral genes (26). Although this enigmatic structure might indicate that these elements are old and extensively mutated, this is not the case. Copies cloned at random can be closely related to each other, which suggests a relatively recent dispersion in the genome. Furthermore, it is evident that some ETn elements remain active as retrotransposons. At least eight mouse mutations at different loci are due to ETn insertions (1, 9, 10, 18, 24, 27, 28) and several somatic insertions have also been reported (17, 22, 29). Despite the bona fide mutagenic activity of ETns, very little has been done to investigate their mode of retrotransposition. ETn transcripts are presumably recognized by reverse transcriptase and other proteins encoded by another type of endogenous retrovirus or retrotransposon, but the identity of these putative coding-competent elements is unknown. Interestingly, it was noted several years ago (23) that new ETn insertions into the immunoglobulin (Ig) region in cell lines are members of a subfamily which differ completely from the first randomly isolated elements in the 3′ part of the LTR and approximately 300 bp of sequence just internal to the 5′ LTR—a region which typically contains the retroviral packaging signal (8). It was therefore suggested that this sequence difference allows members of the “active” ETn subfamily to be preferentially packaged and to retrotranspose (23).
Here we report that ETn elements contain a small region of similarity to the 3′ end of pol genes from simian type D retroviruses. This finding led us to characterize full-length mouse endogenous retroviral genomes, termed MusD elements, with extensive similarity to the gag, pro, and pol genes of primate type D viruses. Interestingly, another group has recently detected type D mouse endogenous sequences associated with particles budding from a cell line established from a thymic lymphoma (21). Only short regions (∼500 bp) of pol sequence were reported in that study, but they are closely related to the sequences identified here, indicating that the elements belong to the MusD family. The LTRs, 5′ internal segment, and 3′ internal region of these type D sequences are very similar to the analogous regions in the active ETn subfamily. The origin of ETn elements and their ability to retrotranspose are discussed in light of these findings.
MATERIALS AND METHODS
PCR, library screening, and DNA sequencing.
To amplify the 3.4-kb deleted region, a gag primer based on the sequence of mouse EST AA142642 (gag3, aaaggatccgcGGTTGCAAGCAGGCCGTGCC, nucleotides 374 to 395) and a pol primer based on the MusD sequence from the mouse T-cell receptor locus (accession no. AE000665) (pol2, tccccgcgGATCCGCTGCAGCTGCCCT) were used in an Elongase (Life Technologies) PCR with C57BL/6 DNA as template. BamHI and SstII sites were incorporated at the 5′ end of each primer (lowercase letters). PCR conditions were as follows: 200 μM concentrations of each deoxynucleoside triphosphate, 200 nM concentrations of each primer, 60 mM Tris-SO4 (pH 9.1), 18 mM (NH4)2SO4, 2 mM MgSO4, and 2 μl of Elongase enzyme mix with 100 ng of C57BL/6 DNA in a 50-μl volume; 25 cycles of 30 s at 94°C and 3 min 30 s at 68°C. PCR products of the expected size were obtained and subcloned.
The P1 bacteriophage genomic library filters of C57BL/6 DNA (obtained from the Resource Center/Primary Database, German Human Genome Project) were hybridized to a combination of three 32P-labeled oligonucleotides with the 5′-to-3′ sequences TGCGCTGGTCACTGTATAAACTC, ATGAAAAAGGACAAAATAACTCTGAC, and TTGATTCTTGATGGAAAAGGCTTTG. Hybridization conditions were 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.5% sodium dodecyl sulfate (SDS), 0.1% Ficoll, 0.1% bovine serum albumin, and 0.1% polyvinylpyrrolidone at 63°C (5°C below the melting temperature [Tm]). Each of the labeled oligonucleotides was added to a level of 1.5 × 105 dpm per ml. After overnight hybridization, washing was performed with 3× SSC and 1% SDS at room temperature for 15 min. DNA from positive clones was isolated using the Nucleobond AX500 kit (Clontech) and characterized by restriction mapping.
Sequencing was performed on plasmids using the Prism Big Dye Cycle Sequence Ready Reaction Kit (PE Biosystems) in an ABI 310 sequencing machine. Analysis was done using Genetics Computer Group software, DNA Strider for the Macintosh, and internet resources.
Genomic Southern analysis.
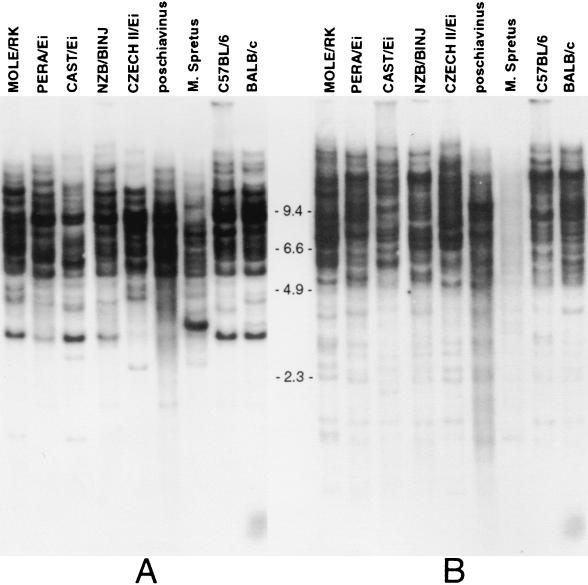
Genomic DNA from various mouse strains was obtained from Jackson Laboratories or isolated using standard protocols. Four micrograms of each DNA was digested with EcoRI, electrophoresed overnight in a 0.8% agarose gel, and transferred onto zeta-probe nylon membrane (Bio-Rad) in 20× SSC. For Fig. 7a, a 32P-labeled 250-bp Eco0109I-PstI fragment from the protease region of MusD1 was used as a probe with hybridization conditions as described previously (14) at a temperature of 65°C. The final posthybridization wash was at 65°C in 0.1× SSC. This probe has no similarity to ETns and was not homologous to any mouse proviral sequence in GenBank. The blot decayed for 6 months before being rehybridized with a 32P end-labeled oligonucleotide of the sequence 5′ ACCTAGCAAGTTAATTAAAGAGCA 3′ for Fig. 7b. The hybridization was performed at 50°C (14°C below the Tm) in 5× SSPE (0.9 M NaCl, 50 mM NaH2PO4, 5 mM EDTA), 0.5% SDS, 0.1% Ficoll, 0.1% bovine serum albumin, 0.1% polyvinylpyrrolidone, 60 μg of boiled, sheared salmon sperm DNA/ml, and 2 × 106 dpm/ml of probe. The blot was washed twice for 10 min in 5× SSC–0.1% SDS at room temperature and then for 15 min in 3× SSC–0.1% SDS prewarmed to 50°C.
FIG. 7.
(A) Genomic Southern analysis of EcoRI-digested DNAs from different mouse strains by using a 250-bp MusD-specific probe (25-h exposure). (B) Rehybridization of the same blot with a 24-mer oligonucleotide probe specific for the type 2 ETn subfamily (10-day exposure).
EST database screening.
The National Center for Biotechnology Information version of BLAST was used to screen the mouse EST database (on 14 December 1999). Query sequences were the 2,945-bp ETn segment not found in MusD elements and the 4,885-bp sequence of MusD2 not present in ETn elements. The results were examined, and redundant entries due to multiple matches to the same clone were eliminated.
Nucleotide sequence accession numbers.
The sequences of MusD1 and MusD2 have been submitted to GenBank with the accession no. AF246632 and AF246633.
RESULTS AND DISCUSSION
ETn elements have a small segment of type D retroviral pol homology.
As mentioned above, no similarity to known retroviral genes has been reported for ETn elements. However, by conducting BLAST searches using ETn sequences translated into all possible reading frames, we detected a short but significant region of strong similarity to the 3′ end of pol genes from simian type D retroviruses. Figure 1 shows the region of translated ETn sequence compared to Mason-Pfizer monkey virus (MPMV) (25). There is 65% amino acid identity in a 47-amino-acid region which corresponds to the very end of the pol gene. This segment is found in recently inserted ETn elements (Fig. 1) and in randomly isolated elements (e.g., accession no. M16478) (26). No other regions of similarity to retroviral genes were detected in ETn elements using this approach. Interestingly, in the original report describing the ETn sequence, it was mentioned that the closest similarity of the ETn LTR was to the LTR of MPMV (26). The two LTRs were reported to be 67% homologous but a sequence alignment was not shown. It was also reported that ETn and MPMV have the same primer binding site and a similar polypurine tract. Indeed, ETn no. M16478 and MPMV do have an identical 19-bp primer binding site but our computer comparisons detected an overall level of LTR identity of only 40 to 45%. If only portions of the ETn and MPMV LTRs are compared, the highest level of identity we could detect was 63% over a 135-bp region if two large gaps are allowed (data not shown). We are therefore uncertain as to how the figure of 67% was derived. No similarity was detected 3′ to the primer binding site.
FIG. 1.
Similarity between ETn elements and primate type D retroviruses. The amino acid translation of nucleotides 3618 to 3764 of the ETn element at the tyrosinase locus (10) is compared to the 3′ end of the pol protein of MPMV. Identical residues are in bold.
Identification of type D-related mouse provirus-like elements.
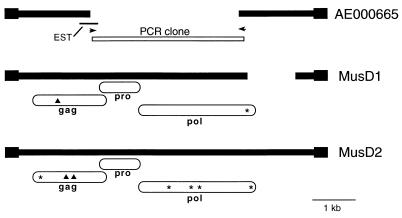
The discovery of remnants of retroviral type D-related sequences in ETn elements led us to conduct a search of the mouse genomic databases for type D-related sequences. This search revealed a region from the mouse T-cell receptor (TCR) locus (accession no. AE000665, positions 95366 to 99319) containing a retrovirus-like sequence with a largely intact gag gene but a mostly deleted pol gene. This sequence was used to search the mouse EST database, and several matches were found, including an EST from mouse heart (accession no. AA142642, clone ID 604576) that extended ∼130 bp into the deleted gag region. Using primers designed to amplify the deleted segment, we conducted PCR on C57BL/6 mouse genomic DNA and obtained a major product of 3.4 kb. One PCR product was cloned and sequenced, and the 3,398-bp clone revealed intact ORFs for the pro and pol genes throughout the extent of the clone. This sequence was compared to the four short pol region sequences published by Ristevski et al. (21). Two of those sequences (AF093700 and AF093701) were over 90% identical to our PCR clone, but one had a termination codon and one had a 24-bp deletion. This comparison was then used to design oligonucleotide probes which were used to screen a C57BL/6 P1 genomic library. Because our PCR product had ORFs but the related published segments had mutations, three oligonucleotides where chosen which matched the sequence of our clone but had mismatches with the published sequences to maximize the chances of isolating full-length functional genomic elements. Twelve positive clones were obtained but several rearranged during growth so only five clones were characterized further.
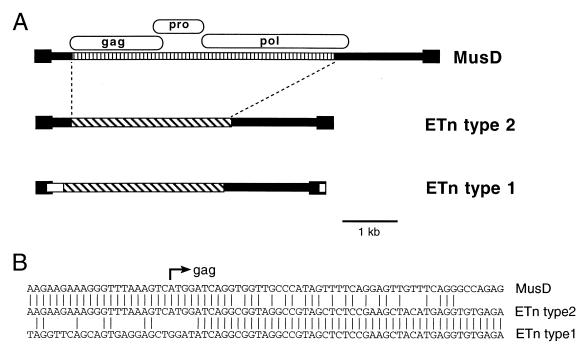
Results from limited sequencing in short regions led to the selection of two clones for full-scale sequencing because we encountered ORF-destroying mutations in the other three clones. The two elements, termed MusD1 and MusD2 (for mouse type D elements 1 and 2), are 6,286 and 7,398 bp in length, respectively. Overall, they are 98% identical except that MusD1 has a 1.1-kb deletion that deletes the 3′ terminus of the pol gene. MusD1 is identical in sequence to our 3.4-kb PCR clone. Figure 2 shows MusD1 and MusD2 and their regions of similarity to the gag, pro, and pol genes of type D viruses. The extent of the TCR sequence derived from GenBank is also shown. Both MusD1 and MusD2 have an intact ORF for pro, which is in the −1 frame with respect to gag as seen for other type D viruses. The gag genes are both mutated, with MusD1 having a 14-bp deletion compared to MusD2 and AE000665. MusD2 has three ORF-disrupting mutations with respect to the other two clones, a 1-bp insertion, a 14-bp deletion (different from the gag deletion in MusD1), and a 4-bp deletion. The pol ORF in MusD1 is intact except for one stop codon but is missing the last 45 amino acids due to the large 1.1-kb deletion. The MusD2 pol gene has four mutations which destroy the ORF, two 1-bp deletions, and two nucleotide substitutions creating stop codons.
FIG. 2.
Representation of three MusD elements. Thick lines are MusD sequences, and the filled boxes indicate the LTRs. The location of the EST AA142642 is shown as a thin line, and locations of the PCR primers used to amplify the region deleted in AE000665 are shown as small arrows. The gag, pro, and pol genes are represented as ovals, with the ORF-destroying mutations shown as asterisks (for single-nucleotide-length differences or substitutions) and triangles (for the 4- and 14-bp deletions described in the text).
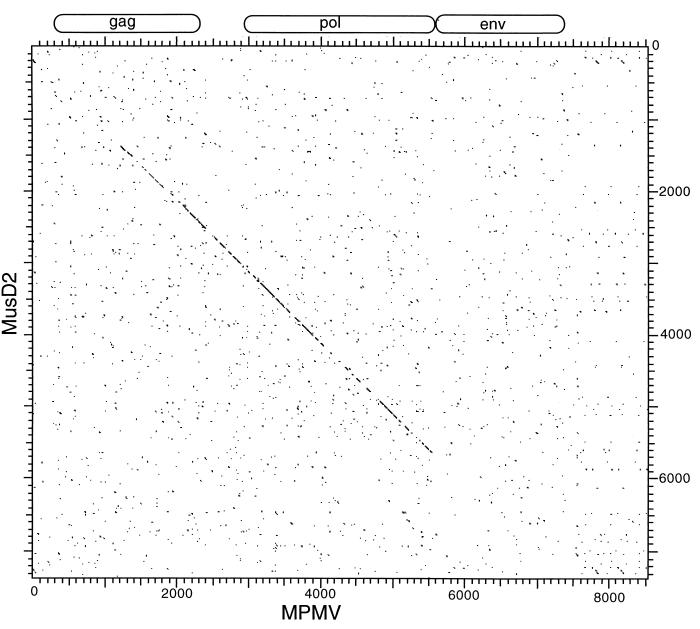
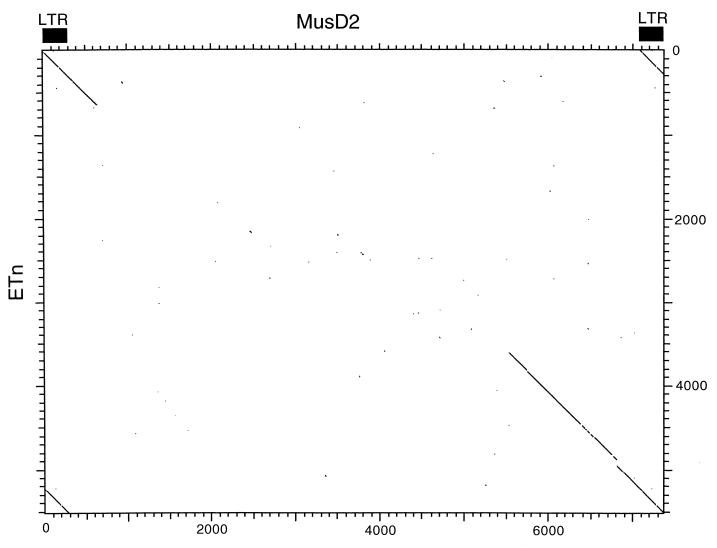
Interestingly, neither these clones nor the element in the TCR locus shows evidence of an env gene. The sequence between the end of pol and the 3′ LTR lacks any vestige of an ORF, and translations of all six frames revealed no similarity to env proteins or any other known genes. This lack of an env-like region is also illustrated in Fig. 3, which is a dot plot DNA comparison of MusD2 and MPMV. Similarity at the DNA level is evident through parts of gag, pro, and pol but ceases at the end of pol. Risteveski et al. used PCR with pol consensus primers to amplify segments of MusD-related elements from type D-like retroviral particles budding from a cell line established from a thymic lymphoma (21). It was suggested that a novel type D-related endogenous virus exists in the mouse and may be associated with a high incidence of thymomas in SCID mice. Apparently mature particles were observed in that study, suggesting that they are encoded by complete retroviral genomes. In addition, 8.5-kb RNAs, the approximate expected size of full-length retroviral genomes, were detected using Northern blots of RNA from a cell line producing the type D-like particles. Therefore, although the elements described in this study lack an env gene, it is possible that related proviruses with an intact env gene also exist. Alternatively, it is possible that unrelated elements provide the env function in trans.
FIG. 3.
Dot matrix nucleotide comparison of MusD2 and MPMV. Extents of the MPMV genes are indicated. The stringency of comparison was 15 out of 23.
The 5′ LTR and 5′ internal region of the MusD1 element is shown in Fig. 4a, with various features highlighted. The LTRs of the three sequenced elements are all highly related. The 5′ and 3′ LTRs of MusD1, MusD2, and the TCR sequence AE000665 are 98.5, 98.1, and 98.1% identical, respectively, and the LTRs between different elements differ by less than 5%. The elements are flanked by 6-bp target site duplications. Just downstream of the 5′ LTR in all three elements is an 18-bp sequence with 16 out of 18 matches to the 3′ end of Lys3-tRNA, which presumably serves as the primer binding site (Fig. 4a). A 16-bp polypurine rich stretch occurs just inside the 3′ LTR (not shown).
FIG. 4.
(A) DNA sequence of the 5′ LTR and 5′ internal region of MusD1. The LTR is outlined, and potential TATAA and polyadenylation signals are boxed. The putative Lys-tRNA primer binding site is underlined, and the region forming the stem-loop structure shown in panel B is overlined. The gag initiation codon is shown with a double underline. (B) Potential stem-loop structure with the ACC motif present in the loop.
The packaging signal of retroviruses remains poorly defined in many cases but is thought to involve secondary structures of the viral RNA. Harrison et al. (8) conducted a study of potential secondary structures in the 5′ leader region of MPMV known to encompass the packaging signal. They identified a stable stem-loop structure upstream of the gag initiation codon with the triplet ACC in the loop of 7 nucleotides. Similar secondary structures were identified in the leader region of nine other retroviruses with the ACC motif, or a slight modification, being conserved (8). It was suggested that this is a common structural motif which may be involved in genomic packaging. Figure 4b shows that the 5′ leader region of MusD elements also contains a potential stem-loop structure with an ACC motif within the loop. This stem loop was present in the four most stable secondary structure configurations predicted by the Mfold program of Mathews et al. (15) for the region between the 5′ LTR and the start of gag.
Protein similarities to MPMV.
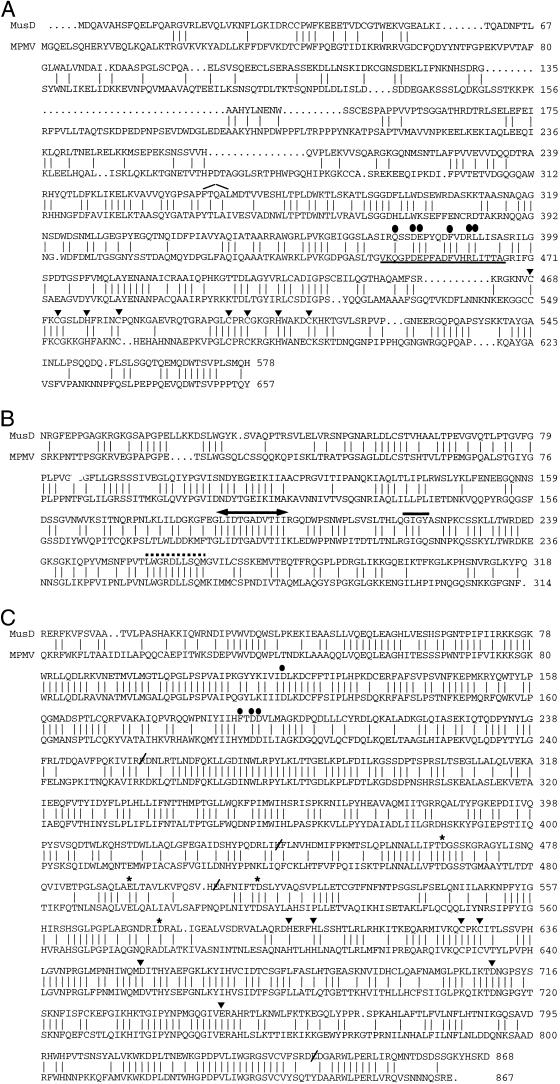
Amino acid alignments of the gag, pro, and pol genes compared to proteins of the type D retrovirus MPMV are shown in Fig. 5. For the purposes of these comparisons, the ORF-destroying mutations in the MusD1 gag gene and the MusD2 pol gene were “corrected” by comparing them to the other sequences to keep the reading frame intact. Overall, the translated gag gene of MusD1 is 34% identical to MPMV gag (Fig. 5a). The degree of similarity in the 5′ part of gag corresponding to MPMV core proteins p10 (matrix), pp24, and p12 is quite limited, but the degree of relatedness increases in the p27 (capsid), p14 (nucleocapsid), and p4 regions. This 3′ half of the MusD1 gag gene is 50% identical at the amino acid level to MPMV. One of the most characteristic conserved sequences in retroviral gag genes is the Cys-His zinc finger motif in the nucleocapsid, which has the structure CX2CX4HX4C (5). Type D retroviruses have two such motifs and the MusD1 gag gene has both, as shown in Fig. 5a. Another conserved region in gag genes is the major homology region, a 20-amino-acid stretch in the 3′ part of the capsid protein (30). This region is highlighted in Fig. 5a. MusD1 has all of the most conserved residues except for the glycine residue at position 4 within the motif. All three MusD elements shown in Fig. 2 have a serine at that position.
FIG. 5.
Amino acid comparisons of MusD to MPMV. (A) Comparison of the translated MusD1 sequence to the MPMV gag products. MPMV p10, pp24, and p12 span residues 1 to 299 and p27, p14, and p4 span residues 300 to 657. The location of the 14-bp insertion added to maintain the MusD1 reading frame is indicated (residues 266 to 270 of MusD1). The major homology region is underlined, and the highly conserved residues are shown with filled circles. The conserved cysteine and histidine residues in the two zinc finger motifs are indicated with a filled triangle. (B) Comparison of the translated MusD2 sequence to the MPMV pro product. The enzymatic active site, the “flap” region, and the GRDLL conserved domains are shown by an arrowed line, a solid line, and a dashed line, respectively. (C) Comparison of the translated MusD2 sequence to the MPMV pol product. The four positions which were corrected based on other MusD sequences to maintain the ORF are indicated with a slanted line through the sequence. The highly conserved residues in the reverse transcriptase, the RNase H, and the integrase domains discussed in the text are indicated by filled circles, asterisks, and triangles, respectively.
The predicted pro gene of MusD2 is 51% identical to MPMV pro (Fig. 5b). In MPMV, the protease PR is encoded by the 3′ half of the pro gene, with the 5′ part encoding the dUTPase (16). The most highly conserved parts of retroviral proteases are the active site, the “flap” region, and the GRDLL domain (20), all of which are intact in the MusD2 predicted protein. This indicates that some MusD elements may encode functional proteases.
There is 59% overall amino acid identity between the “ORF-corrected” 868-amino-acid pol gene of MusD2 and the 867-amino-acid pol gene of MPMV (Fig. 5c). In the reverse transcriptase region, the MusD element has the absolutely conserved F/YXDD motif (positions 191 to 194) and the D at position 118 which are required for reverse transcriptase activity (13, 19). Four residues of catalytic importance are absolutely conserved among RNaseH domains (6) and are also found in the MusD2 sequence. Within the integrase, the most conserved features are the HHCC zinc finger motif found in the amino-terminal part of the protein and the universal DD35E motif, which forms the catalytic core of the enzyme (7, 12). The MusD element is intact for all these residues as shown in Fig. 5c.
ETns and MusD elements share LTRs and 5′ and 3′ internal segments.
Because we had detected a small region of MusD pol sequence within ETn elements, we compared the two types of elements throughout their length. Figure 6 is a dot matrix DNA comparison of MusD2 versus the ETn recently inserted into the tyrosinase locus (10). The two sequences are highly related in the LTRs (5′ LTRs are 94% identical) and in the 5′ and 3′ internal regions. The 5′ internal stretch of homology extends to include the first 13 bp of the gag ORF, and the 3′ region of homology includes the last 166 bp of the pol gene, which is the region originally detected in our BLAST searches. These 5′ and 3′ internal regions are 94 to 95% identical between the two element types. No other regions of similarity were detected, even at a reduced stringency of comparison.
FIG. 6.
Dot matrix nucleotide comparison of MusD2 and the ETn element at the tyrosinase locus (10). The stringency of comparison was 17 out of 23.
Genomic complexity of MusD and ETn elements.
To examine the copy number and distribution of MusD elements in the genome, Southern blot analysis was performed on DNAs from different mouse strains cut with EcoRI. All DNAs were of Mus musculus origin except for one DNA sample from Mus spretus. The probe used was derived from the protease region so it will not detect ETn elements. The results, shown in Fig. 7a, indicate that MusD elements are highly repetitive in the genomes of both M. musculus and M. spretus. The banding pattern is too complex to accurately determine copy number, but we estimate that it is several hundred, given the strength of the hybridization signal. Variations in banding patterns also indicate that these elements are polymorphic between strains but the extent of this polymorphism is masked by the high copy number.
Previous estimates of ETn copy numbers using Southern hybridizations could have been complicated by the fact that MusD and ETn elements share sequences. To analyze copy numbers of only the active subfamily of ETn elements (see below), we exploited a small (28-bp) deletion which occurs in the two fully sequenced recently inserted ETn elements (10) with respect to other ETn elements in GenBank. The region surrounding this deletion is not shared with MusD elements. An oligonucleotide probe spanning this deletion was used to rehybridize the same genomic blot as shown in Fig. 7a. Figure 7b shows that this probe detects a large number of ETn elements with different banding patterns in different M. musculus strains. Interestingly, hybridization of this specific probe to M. spretus DNA is very weak, suggesting that this particular ETn subfamily is not present. Since M. spretus has similar numbers of MusD elements compared to M. musculus (Fig. 7a), this suggests that the ETn active subfamily is younger, being amplified in M. musculus after divergence from M. spretus approximately 1 to 2 million years ago (2).
Possible confusion between ETns and MusD sequences.
As mentioned in the Introduction, it was noted in 1990 that ETn elements newly inserted into Ig regions in myeloma cell lines differed in the 3′ part of the LTR and the 5′ internal region with respect to the original, randomly isolated ETns (23). Thus, two subfamilies of ETn elements were defined, which we will call type 1 (original) and type 2 (Ig insertions). It was previously suggested that type 2 may be more active or mobile in the genome (23). Indeed, since 1990, several ETn insertions have been described and, in all cases for which DNA sequence is available, they appear to be type 2. However, the discovery of MusD elements complicates the matter, since the internal sequence was not determined in most reports of ETn insertions. Figure 8a illustrates how the ETn types differ from each other and from the MusD elements described here. The only two recently inserted ETn elements that have been completely sequenced (10) are 98% identical and serve as the prototype for type 2. As is clear from the figure, it would be difficult to distinguish between ETn type 2 and MusD elements without sufficient DNA sequencing in the interior of the element. Notably, the LTR sequences between the two element types are 94 to 96% identical. It is therefore possible that some of the recently inserted elements described as ETns may actually be MusD sequences. Figure 8b shows the 5′ point of divergence between ETns and MusD elements. It is intriguing that this point occurs so close to the gag initiation codon, but the possible relevance of this is unknown.
FIG. 8.
(A) Representation of the relationship between MusD elements and ETns. The vertically striped region shows MusD-specific sequences. Diagonally striped boxes show ETn-specific sequences, and white boxes show regions specific to the type 1 ETn subfamily. (B) Nucleotide sequence of the three element types at the start of the gag gene. MusD is the MusD1 sequence, ETn type 2 is the tyrosinase locus element (10), and ETn type 1 is GenBank accession no. M16478 (26).
Expression patterns of ETn and MusD elements.
It has been previously shown that transcription of ETn elements peaks between days 3.5 and 7.5 of embryogenesis (3). To compare the expression level of ETns and MusD sequences, we conducted BLAST searches of the mouse EST database by using the entire segments of the elements, which are specific for each. For ETn elements, this segment is ∼3 kb (Fig. 8a), and the MusD-specific region is ∼4.9 kb. The number of independent EST clones identified, using a cutoff probability value of e−10, was determined and compared. This analysis suggests that ETn elements are expressed at a higher level during embryogenesis. A total of 32 ETn ESTs but only 6 MusD ESTs were found from several independent libraries representing different stages of embryonic development. Nine additional ETn ESTs but no MusD ESTs were identified in libraries from embryonic stem cells or embryonic carcinoma cells. From all tissue sources, a total of 88 ETn ESTs and 23 MusD ESTs were identified. Thus, it appears that the level of ETn transcripts is generally higher.
Summary and conclusions.
We have shown that ETns share sequences with the novel family of MusD elements described here. Specifically, the LTRs and the 5′ and 3′ internal regions are essentially indistinguishable between the MusD elements and the type 2, or active, ETn subfamily. Southern blot analysis has also shown that type 2 ETn elements are younger than MusD sequences. It is therefore probable that ETn elements arose via recombination events resulting in a near total replacement of the MusD gene-coding sequences with sequences of unknown origin. Other recombination events affecting the LTRs and 5′ internal region could have generated the type 1 ETn elements. However, more extensive phylogenetic analyses will be needed to determine the evolutionary history and relationships of these different types of sequences.
The similarity of ETn elements, particularly the type 2 subfamily, to MusD sequences strongly suggests that ETn transcripts retrotranspose by utilizing MusD gene-encoded reverse transcriptase and other proteins. Such a pseudotyping mechanism would be analogous to the highly defective VL30 proviral elements which are efficiently packaged by Moloney leukemia viral proteins. The MusD clones analyzed here have a few mutations which would prevent protein production, but their high copy number makes it likely that some coding competent elements are present in the genome. Results of screening the EST database indicate that ETn transcripts are present at a higher level than MusD transcripts in the embryo. This suggests that the frequency of ETn retrotransposition would also be higher. The fact that no new MusD insertions have been documented supports this suggestion. However, as discussed above, some of the less-well-characterized new inserts reported to be of ETn origin solely on the basis of LTR sequence could potentially be MusD elements. Reasons for the higher level of transcription of ETn elements are not known, but there are at least three possibilities. First, slight sequence differences between the closely related MusD and ETn LTRs, which contain the transcriptional regulatory elements, could be the explanation. However, sequence comparisons have not revealed an obvious difference in transcriptional control motifs likely to result in the observed expression differences. Second, it is possible that MusD elements have transcriptional suppressors in the internal region which constrain their expression. Finally, the noncoding DNA found in ETn-specific internal regions could contain transcriptional enhancer elements. If either of the last two possibilities is true, it is tempting to speculate that the recombination event which replaced MusD coding sequences with unrelated DNA to create the ETn family may have contributed to the amplification and continued retrotransposition of these elements. In conclusion, the findings reported here provide insight into the potential basis for the ongoing retrotranspositional activity of ETn elements, a family that has essentially remained a mystery since it was first described.
ACKNOWLEDGMENTS
We thank Diana Juriloff and Muriel Harris for discussions which led to the initiation of this work. We also thank Patrik Medstrand for helpful comments on the manuscript and during the course of this study.
This work was supported by a grant from the Medical Research Council of Canada with core support provided by the British Columbia Cancer Agency.
REFERENCES
- 1.Adachi M, Watanabe-Fukunaga R, Nagata S. Aberrant transcription caused by the insertion of an early transposable element in an intron of the Fas antigen gene of lpr mice. Proc Natl Acad Sci USA. 1993;90:1756–1760. doi: 10.1073/pnas.90.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck J A, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig J T, Festing M F, Fisher E M. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 3.Brulet P, Condamine H, Jacob F. Spatial distribution of transcripts of the long repeated ETn sequence during early mouse embryogenesis. Proc Natl Acad Sci USA. 1985;82:2054–2058. doi: 10.1073/pnas.82.7.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brulet P, Kaghad M, Xu Y S, Croissant O, Jacob F. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc Natl Acad Sci USA. 1983;80:5641–5645. doi: 10.1073/pnas.80.18.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covey S N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986;14:623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doolittle R F, Feng D F, Johnson M S, McClure M A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989;64:1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- 7.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison G P, Hunter E, Lever A M. Secondary structure model of the Mason-Pfizer monkey virus 5′ leader sequence: identification of a structural motif common to a variety of retroviruses. J Virol. 1995;69:2175–2186. doi: 10.1128/jvi.69.4.2175-2186.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann B G, Labeit S, Poustka A, King T R, Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann M, Harris M, Juriloff D, Boehm T. Spontaneous mutations in SELH/Bc mice due to insertions of early transposons: molecular characterization of null alleles at the nude and albino loci. Genomics. 1998;52:107–109. doi: 10.1006/geno.1998.5409. [DOI] [PubMed] [Google Scholar]
- 11.Kaghad M, Maillet L, Brulet P. Retroviral characteristics of the long terminal repeat of murine E.Tn sequences. EMBO J. 1985;4:2911–2915. doi: 10.1002/j.1460-2075.1985.tb04022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larder B A, Purifoy D J, Powell K L, Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. Nature. 1987;327:716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- 14.Mager D L, Goodchild N L. Homologous recombination between the LTRs of a human retrovirus-like element causes a 5-kb deletion in two siblings. Am J Hum Genet. 1989;45:848–854. [PMC free article] [PubMed] [Google Scholar]
- 15.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 16.McGeoch D J. Protein sequence comparisons show that the ‘pseudoproteases’ encoded by poxviruses and certain retroviruses belong to the deoxyuridine triphosphatase family. Nucleic Acids Res. 1990;18:4105–4110. doi: 10.1093/nar/18.14.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitreiter K, Schmidt J, Luz A, Atkinson M J, Hofler H, Erfle V, Strauss P G. Disruption of the murine p53 gene by insertion of an endogenous retrovirus-like element (ETn) in a cell line from radiation-induced osteosarcoma. Virology. 1994;200:837–841. doi: 10.1006/viro.1994.1253. [DOI] [PubMed] [Google Scholar]
- 18.Moon B C, Friedman J M. The molecular basis of the obese mutation in ob2J mice. Genomics. 1997;42:152–156. doi: 10.1006/geno.1997.4701. [DOI] [PubMed] [Google Scholar]
- 19.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao J K, Erickson J W, Wlodawer A. Structural and evolutionary relationships between retroviral and eucaryotic aspartic proteinases. Biochemistry. 1991;30:4663–4671. doi: 10.1021/bi00233a005. [DOI] [PubMed] [Google Scholar]
- 21.Ristevski S, Purcell D F, Marshall J, Campagna D, Nouri S, Fenton S P, McPhee D A, Kannourakis G. Novel endogenous type D retroviral particles expressed at high levels in a SCID mouse thymic lymphoma. J Virol. 1999;73:4662–4669. doi: 10.1128/jvi.73.6.4662-4669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shell B, Szurek P, Dunnick W. Interruption of two immunoglobulin heavy-chain switch regions in murine plasmacytoma P3.26Bu4 by insertion of retroviruslike element ETn. Mol Cell Biol. 1987;7:1364–1370. doi: 10.1128/mcb.7.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shell B E, Collins J T, Elenich L A, Szurek P F, Dunnick W A. Two subfamilies of murine retrotransposon ETn sequences. Gene. 1990;86:269–274. doi: 10.1016/0378-1119(90)90289-4. [DOI] [PubMed] [Google Scholar]
- 24.Shiels A, Bassnett S. Mutations in the founder of the MIP gene family underlie cataract development in the mouse. Nat Genet. 1996;12:212–215. doi: 10.1038/ng0296-212. [DOI] [PubMed] [Google Scholar]
- 25.Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 26.Sonigo P, Wain-Hobson S, Bougueleret L, Tiollais P, Jacob F, Brulet P. Nucleotide sequence and evolution of ETn elements. Proc Natl Acad Sci USA. 1987;84:3768–3771. doi: 10.1073/pnas.84.11.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinmeyer K, Klocke R, Ortland C, Gronemeier M, Jockusch H, Grunder S, Jentsch T J. Inactivation of muscle chloride channel by transposon insertion in myotonic mice. Nature. 1991;354:304–308. doi: 10.1038/354304a0. [DOI] [PubMed] [Google Scholar]
- 28.Thien H, Ruther U. The mouse mutation Pdn (Polydactyly Nagoya) is caused by the integration of a retrotransposon into the Gli3 gene. Mamm Genome. 1999;10:205–209. doi: 10.1007/s003359900973. [DOI] [PubMed] [Google Scholar]
- 29.Weiss S, Johansson B. Integration of the transposon-like element ETn upstream of V lambda 2 in the cell line P3X63Ag8. J Immunol. 1989;143:2384–2388. [PubMed] [Google Scholar]
- 30.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]