Abstract
The major immediate-early proteins of human cytomegalovirus (HCMV) play a pivotal role in controlling viral and cellular gene expression during productive infection. As well as negatively autoregulating its own promoter, the HCMV 86-kDa major immediate early protein (IE86) activates viral early gene expression and is known to be a promiscuous transcriptional regulator of cellular genes. IE86 appears to act as a multimodal transcription factor. It is able to bind directly to target promoters to activate transcription but is also able to bridge between upstream binding factors such as CREB/ATF and the basal transcription complex as well as interacting directly with general transcription factors such as TATA-binding protein and TFIIB. We now show that IE86 is also able to interact directly with histone acetyltransferases during infection. At least one of these factors is the histone acetyltransferase CBP-associated factor (P/CAF). Furthermore, we show that this interaction results in synergistic transactivation by IE86 of IE86-responsive promoters. Recruitment of such chromatin-remodeling factors to target promoters by IE86 may help explain the ability of this viral protein to act as a promiscuous transactivator of cellular genes.
Human cytomegalovirus (HCMV) is a ubiquitous herpesvirus which rarely causes disease upon primary infection and remains latent in the infected host throughout its life (54). Infection usually becomes clinically apparent only in the newborn or immunocompromised upon primary infection or reactivation (67). As with all herpesviruses, productive infection with HCMV results in a regulated cascade of viral gene expression which has been operationally defined as immediate-early (IE), early (E), and late (L) (17, 53, 78, 88, 89).
IE proteins are expressed in the absence of de novo protein synthesis, and the major IE proteins (IE72 and IE86) result from differential splicing of the major IE region (29, 32, 77, 79, 87). IE72 and IE86 are known to transactivate E and L genes (1, 38, 50, 70, 71) and either positively or negatively autoregulate their own expression (10, 11, 28, 45, 62, 76). While IE72 is able to weakly activate some cellular promoters, IE86 is a strong transcriptional activator of cellular gene expression (6, 15, 25, 26, 31, 40, 48, 51, 55, 85) and appears to be able to act at the promoter level by a number of mechanisms. It is able to bind DNA directly so repressing its own promoter (12, 28, 30, 33, 35, 44, 45, 49, 60, 95) and has recently been shown to bind directly to at least one cellular promoter region, resulting in transcriptional activation (5). Similarly, IE86 has been shown to bind to a number of cellular factors (6, 19, 24, 36, 42, 47, 48, 69, 73, 74, 97), suggesting that IE86 may also act by bridging between transcription factors bound upstream of the basal promoter and the transcription preinitiation complex. IE86 is also able to directly activate basal promoter elements containing only a TATA motif, and this may result from the ability of IE86 to interact directly with general transcription factors such as TFIIB and TFIID (7, 34, 73). However, it is not understood how these interactions are able to effect increases in transcription, as IE86 appears not to be able to increase recruitment of general transcription factors to the preinitiation complex (6, 36, 73).
Chromatin configuration has been known for some time to play an important role in transcriptional activation (81, 90, 93). Transcription must occur through DNA, which is tightly associated with histone proteins in nucleosome arrays (18, 57). Packaging of DNA in this form generally represses transcription. Consequently, activated transcription requires remodeling of such chromatin structure and can be considered, to some extent, a derepression mechanism (20, 37, 91). Such remodeling of chromatin to a transcriptionally active form has been correlated to the levels of acetylation of nucleosomal histones (21, 66, 86, 92, 94). Histone acetylation neutralizes the basic residues of core histones, weakening the histone-DNA interactions and causing nucleosome repositioning and loosening of the higher-order structure of the chromatin (46). In vivo, histone hyperacetylation is known to be associated with activation of gene expression (27, 58, 82, 83). Recently, it has become clear that transcriptional activation by numerous sequence-specific transcription factors requires coactivators such as CREB-binding protein (CBP) and p300 (3, 4, 14, 16, 22, 59, 80). It has been believed that such coactivators bridge between DNA sequence-specific transcription factors and the basal transcription complex, resulting in stabilization of the preinitiation complex and recruitment of additional activation domains to promoters (41, 98). However, transcriptional coactivator proteins such as CBP and p300 are also known to have intrinsic histone acetyltransferase (HAT) activity (2, 52, 56, 68), and they can also complex with other cellular factors such as CBP-associated factor P/CAF and steroid receptor coactivator 1, which are also known to contain intrinsic HAT activity (9, 75, 96). Consequently, the mechanism by which such activation complexes stimulate transcription is likely to include some aspect of chromatin remodeling mediated by histone acetylation (20, 84).
We have therefore examined whether the ability of IE86 to promiscuously activate cellular gene expression may be due to the ability of this viral protein to bind cellular proteins with HAT activity. Here, we show that IE86 is also able to interact directly with P/CAF in vivo and in vitro and that activation of promoters by IE86 in transient and stable transfection assays is synergized by the presence of P/CAF, suggesting a role for histone acetylation in transcriptional regulation of cellular promoters by IE86.
MATERIALS AND METHODS
Cell culture and virus.
Human U373 (glioblastoma) and human U2-OS (osteogenic bone sarcoma) cells were maintained in Eagle's minimal essential medium–10% fetal calf serum at 37°C and in 5% CO2. U373 cells are fully permissive for HCMV infection, whereas U2-OS cells undergo abortive infection, expressing high levels of viral IE proteins. Infection with HCMV (AD169) was carried out as described previously (64).
Plasmids.
TGFβ77CAT, a chloramphenicol acetyltransferase (CAT) reporter vector containing the transforming growth factor β2 (TGF-β2) promoter, and TGFβ77MCAT, a CAT reporter vector containing the TGF-β2 promoter with a mutated CREB/ATF binding site, were gifts from Michael Green and have been described elsewhere (39). pcDNA3IE72 was made by cloning the EcoRI/XhoI fragment from pBSIE1 (7) containing the HCMV major IE72 cDNA into pcDNA3 (Invitrogen). Similarly, pcDNA3IE86 was constructed by cloning the HCMV IE86 cDNA from pGex3XIE2 (7) into pcDNA3, using BamHI/EcoRI. pCX-Flag-P/CAF, a full-length P/CAF cDNA also encoding a Flag Tag epitope (Invitrogen), also under the control of the HCMV major IE promoter, was a kind gift from Xiang-Jiao Yang (Bethesda, Md.) and has been described elsewhere (96).
All plasmids for in vitro transcription and translation have been described previously (7, 12) except pBSP/CAF, which was made by inserting a HindIII/EcoRI P/CAF fragment from pCX-Flag-P/CAF into pBluescript KS+.
pGexIE72, pGexIE86, and pGEXTFIID have also been described previously (7). pGexIE86[290–390] was made by PCR amplification of the 300-bp coding region for amino acids 290 to 390 of IE86 from pBSIE86 with EcoRI/SalI linkers and cloning into pGex3XP. Similarly, pGexIE86[290–542] was constructed by PCR amplification of a 756-bp region encoding amino acids 290 to 542 of IE86 from pBSIE86 with EcoRI/SalI linkers into pGex3XP. pGexCBP, a gift from Xiang-Jiao Yang, has been described elsewhere (96). pGexP/CAF was constructed by inserting an EcoRI/HindIII fragment from pCX-Flag-P/CAF containing the P/CAF cDNA into pGex3XP. pQE10IE2, a bacterial expression vector to generate His-tagged IE86 protein, was a gift from Thomas Stamminger (Erlangen, Germany) and has been described elsewhere (43).
Transient and stable transfection assays.
For transient transfection, approximately 5 × 106 U373 cells were transfected with 2.5 μg of reporter gene together with 5 μg of cotransfected plasmid by calcium phosphate precipitation. Cells were assayed 48 h posttransfection for CAT activity. CAT activity, expressed as percentage conversion of chloramphenicol, was quantified from thin-layer chromatography sheets using a Hewlett-Packard Instant Imager. The results are an average of three independent experiments.
For stable transfections, approximately 5 × 106 U373 cells were transfected with 5 μg of pcDNA3 and 15 μg of TGFβ77CAT or TGFβ77MCAT. Cells were selected for 3 weeks in G418 (500 μg/ml). For transient supertransfection of these stable cell lines, 10 μg of effector DNA was introduced into cell lines by calcium phosphate coprecipitation.
Immunoprecipitation assays.
For immunoprecipitation of HAT activity (HAT-IP), approximately 5 × 107 U373 cells were transfected with 20 μg of plasmid DNA, in total, using Fugene (Boehringer Mannheim) as described by the manufacturer. Alternatively, cells were infected with HCMV (AD169) at a multiplicity of infection of 5. HAT activity of the immunoprecipitated complexes was assayed as previously described (2). Briefly, cells were harvested and resuspended in 1 ml HAT-IP buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 0.5 mM phenylmethylsulfonyl fluoride with aprotinin, leupeptin, and pepstatin), rotated for 1 h at 4°C, and cleared by centrifugation (13,000 rpm, 15 min). For infected or mock-infected cells, HCMV gB-specific monoclonal antibody as a control or an HCMV IE86-specific monoclonal antibody (SMX) (63) was added, and samples were rotated for 1 h at 4°C. Alternatively, for transfected cells, a monoclonal antibody to a true late 55-kDa HCMV structural protein (Chemicon) as a control, antibody M2 (Amersham), against the Flag epitope of P/CAF, or antibody E13 (Biosys), which recognizes an epitope common to IE72 and IE86, was used. Following addition of 30 μl of a 1:1 mixture of protein A-Sepharose beads and protein G-Sepharose beads, samples were rotated overnight. The immune complexes were pelleted by centrifugation (6,000 rpm, 2 min), washed three times in HAT-IP buffer, and resuspended in 30 μl. Following addition of 14−C-labeled acetyl coenzyme A, samples were incubated at 37°C for 45 min; 20 μl of the reaction was then spotted onto Whatman P81 filter paper. Filters were washed three times with 50 mM sodium carbonate (pH 9.2), soaked in acetone, and air dried for 1 min prior to counting in a liquid scintillation counter.
For double immunoprecipitation assays, approximately 5 × 107 U2-OS cells were transfected with 30 μg of plasmid DNA in total. At 24 h posttransfection, cells were labeled with 250 μCi of [35S]methionine in methionine-free minimal essential medium–1% fetal calf serum for a further 24 h. Cells were harvested, lysed in EBC buffer (23), and analyzed by double immunoprecipitation (23) using anti-murine cyclin D1 (HD11; Santa Cruz) as a negative control, anti-Flag antibody M2; and antibody E13.
GST fusion assays.
Glutathione S-transferase (GST) fusion proteins were prepared essentially as described by Smith and Johnson (72) except that bacteria were cultured for 3 h and then induced with isopropyl-β-d-thiogalactopyranoside for a further 3 h. GST pull-down assays were carried out as described previously (7). In vitro transcription-translation or coupled transcriptions-translations (Promega) were used to generate [35S]methionine-labeled proteins as described by the manufacturer. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% polyacrylamide gels and analyzed by autoradiography.
For assays using 32P-labeled IE86, radiolabeled IE86 protein was prepared from induction of a culture of pQE10IE2 and incubation with Ni-agarose beads. The protein was renatured on the beads using decreasing concentrations of urea (43) and labeled with [32P]dATP using casein kinase II. 32P-labeled IE86 was eluted from the Ni-agarose beads with imidazole buffer and dialyzed in NET buffer (7) overnight. GST pull-down assays were carried out as described above except that 32P-labeled IE86 protein was used as the probe, and reactions were scaled up 10-fold.
Yeast two-hybrid assays.
Yeast two-hybrid analysis was carried out using the Matchmaker two-hybrid system (Clontech) as described by the manufacturer. pGBT10:P/CAF was constructed by insertion of an EcoRI/HindIII fragment containing full-length P/CAF, which had been blunt ended with Klenow enzyme, into pGBT10 (6) which had been cut with EcoRI and SalI and then also blunt ended. To make pGAD425:IE86, pGAD425 (6) was digested with BamHI and EcoRI and ligated with a BamHI-EcoRI IE86 cDNA fragment from pGex3X-IE2. pGBT10:IE72 and pGBT10:IE86 have been described elsewhere (6). Liquid β-galactosidase assays were carried out as described by the manufacturer.
RESULTS
HAT activity coprecipitates with IE86.
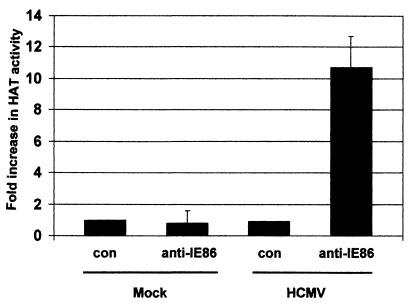
Infection of U373 cells with HCMV results in a full productive infection of this cell line. Figure 1 shows that after virus infection for 24 h, immunoprecipitation of IE86 coprecipitates HAT activity. HAT activity is specific for virally infected cells and coprecipitates only with antibodies specific for IE86, not a control antibody that recognizes HCMV gB, a late structural protein.
FIG. 1.
HAT activity coimmunoprecipitates with IE86 in infected cells. U373 cells were either mock infected (Mock) or infected with HCMV (HCMV) at approximately 5 PFU/cell; 24 h postinfection, cells were lysed and extracts were immunoprecipitated with a control monoclonal antibody to HCMV gB (con) or a monoclonal antibody to IE86 (anti-IE86). Immunocomplexes were then assayed for HAT activity. Data represent average fold increases in HAT activity over the corresponding control antibody immunoprecipitations from three independent experiments.
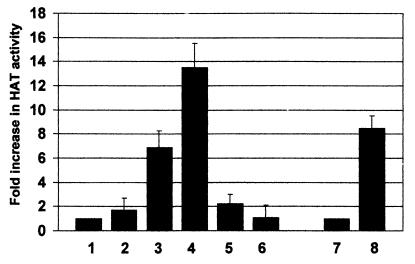
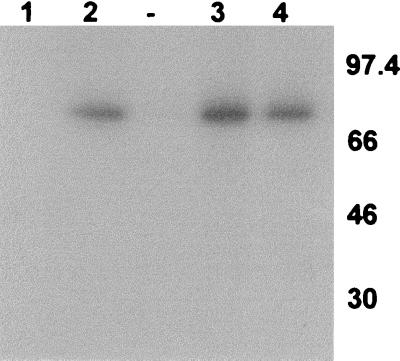
It has already been shown that IE86 is able to bind directly to CBP in vitro (42, 69) and that CBP has intrinsic HAT activity. Consequently, it was possible that the HAT activity coprecipitating with IE86 may have been due to CBP. However, transient cotransfections of cells with IE86 and P/CAF expression vectors (Fig. 2) showed clearly that immunoprecipitations from cells cotransfected with P/CAF and IE86 (lane 4), using IE86-specific antibodies, resulted in complexes with significantly higher HAT activity than when cells were transfected with IE86 alone (lane 3). This increase in HAT activity that coimmunoprecipitates with IE86 in IE86-P/CAF-transfected cells does not result from an increase in levels of expression of IE86 or P/CAF in cotransfected cells, as Western blot analysis shows no such increase in expression of transfected IE86 or P/CAF compared to cells transfected with IE86 or P/CAF alone (data not shown). Similarly IE86 expression does not lead to any increase in endogenous CBP expression (data not shown).
FIG. 2.
HAT activity coimmunoprecipitates with IE86 in transfected cells. U373 cells were transfected with pcDNA3 (bar 1), pcDNA3IE72 (bar 2), pcDNA3IE86 (bar 3), pcDNA3IE86 plus pCX-Flag-P/CAF (P/CAF) (bar 4), pcDNA3IE72P/CAF (bar 5), or P/CAF alone (bar 8). At 48 h posttransfection, cells were lysed and extracts were immunoprecipitated with E13 antibody, specific for HCMV IE72/IE86 (bars 1 to 5). An equivalent amount of extract from pcDNA3IE86+P/CAF-transfected cells was also immunoprecipitated with a monoclonal antibody to an HCMV late structural protein (Chemicon) as a control (bar 6). Data represent average fold increases in HAT activity over the HAT activity detected for pcDNA3 transfections from three independent experiments. Extracts from P/CAF-transfected cells were also immunoprecipitated with the control late HCMV antibody (bar 7) or an anti-Flag antibody, which detects the Flag epitope on P/CAF (lane 8), to act as a positive control for HAT detection. Immunocomplexes were then assayed for HAT activity. Data represent average fold increases in HAT activity over the HAT activity detected in the corresponding control antibody immunoprecipitations from three independent experiments.
IE2 and P/CAF interact directly in vitro via domains important for transcriptional activation.
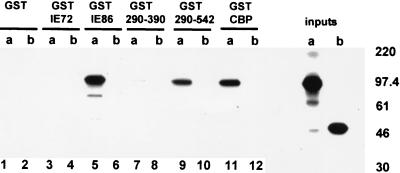
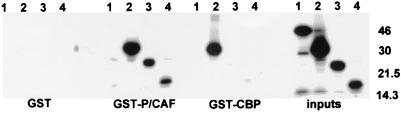
Since cotransfection of IE86 and P/CAF clearly resulted in increased HAT activity in immunocomplexes after immunoprecipitation with anti-IE86 antibodies, we asked whether IE86 and P/CAF were able to physically interact with each other. We first tested this interaction in vitro using GST pull-down assays. Figure 3 shows that [35S]methionine-labeled P/CAF binds to GST-CBP beads as expected (lane 11) and also to GST-IE86 beads (lane 5) but not to GST (lane 1) or GST-IE72 (lane 3) beads. We also analyzed the ability of P/CAF to bind to specific domains of IE86. Figure 3, lane 7, shows that P/CAF was unable to bind a known retinoblastoma protein (RB) binding domain (amino acids 290 to 390) of IE86 (23). However, GST-IE86 beads bearing amino acids 290 to 542 of IE86 (lane 9) were able to interact with P/CAF. In contrast, a control for nonspecific binding, gelsolin, failed to interact with either GST-IE86 or GST-CBP (lanes 6 and 12, respectively).
FIG. 3.
P/CAF interacts with IE86 in vitro. In vitro-transcribed and -translated P/CAF (lanes a) or gelsolin (lanes b) was analyzed by GST fusion pull-down assays for the ability to bind GST, GST-IE72, GST-IE86, or GST-CBP beads. GST fusions to the 290–390 (GST 290–390) or 290–542 (GST 290–542) amino acid domain of IE86 were also analyzed. Inputs were 25% of the amount of IE86 or gelsolin used in each assay. Here and in subsequent figures, positions of marker proteins are indicated in kilodaltons.
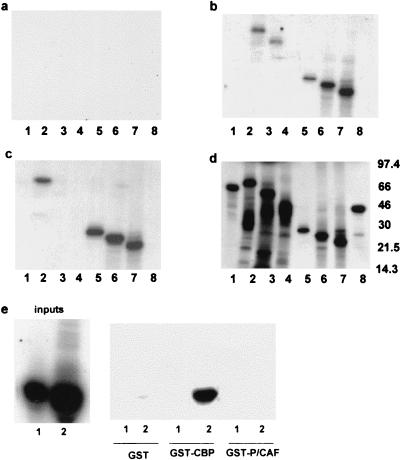
We repeated these assays using GST-P/CAF beads as targets for [35S]methionine domains of IE86 (Fig. 4a to d). Full-length IE86, but not IE72 or gelsolin, bound specifically to GST-P/CAF (Fig. 4c, lanes 2, 1, and 8, respectively) but not GST control beads (Fig. 4a, lane 2). However, as we have observed before for binding to a number of cellular proteins (7), full-length IE86 appears to bind less efficiently than short domains of IE86, possibly because N-terminal regions of IE86 interfere with interaction domains at the C terminus of IE86 (7, 73). Consistent with the analysis shown in Fig. 3, residues 290 to 579, 290 to 542, and 290 to 504 (Fig. 4c, lanes 5, 6, and 7, respectively) but not residues 1 to 290 (lane 4) of IE86 all appeared to interact with P/CAF, and these domains did not interact with GST control beads (Fig. 4a). In most cases, these same regions of IE86 also bound equally well to GST-CBP (Fig. 4b); interestingly, they have all been suggested as being important for autoregulation and transactivation (61). However, residues 1 to 390 of IE86 showed no binding to P/CAF (Fig. 4c, lane 3) but did bind to CBP (Fig. 4b, lane 3). Further deletion analysis (Fig. 4e) showed that a minimal 290–390 domain of IE86 was able to bind to CBP but not P/CAF, suggesting that CBP and P/CAF require different domains of IE86 for interaction. Similarly, Fig. 5 confirms that CBP binding absolutely requires the 290–390 domain of IE86 and no additional binding site for CBP is present in amino acids 388 to 579 of IE86.
FIG. 4.
Minimal domains of IE86 interact with P/CAF in vitro. GST (a), GST-CBP (b), and GST-P/CAF (c) beads were used as targets for GST pull-down assays using [35S]methionine-labeled IE72 (lanes 1), full-length IE86 (lanes 2), amino acids 1 to 390 (lanes 3), 1 to 290 (lanes 4), 290 to 579 (lanes 5), 290 to 542 (lanes 6), and 290 to 504 (lanes 7) of IE86, as well as gelsolin (lane 8). Input protein (25% of the amount of protein used in each assay) is shown in panel d. (e) GST, GST-CBP, and GST-P/CAF beads were used as targets in GST pull-down assays using [35S]methionine-labeled amino acids 1 to 85 (1) or 290 to 390 (2) of IE86; 25% of the amount of protein used in each assay is shown as inputs.
FIG. 5.
P/CAF and CBP require different domains of IE86 for interaction. GST, GST-P/CAF, and GST-CBP beads were used as targets for GST pull-down assays using [35S]methionine-labeled amino acids 290 to 579 (lane 2), 388 to 579 (lane 3), or 428 to 579 (lane 4) of IE86 and gelsolin (lane 1). Inputs represent 25% of the amount of protein used in each assay.
It has been suggested that reticulocyte lysates may contain high levels of eukaryotic nuclear proteins. To ensure that the interaction between IE86 and P/CAF could not be due to a protein present in the reticulocyte lysate bridging between them, we carried out GST fusion protein interaction assays using proteins that had been expressed in and purified from bacteria. Figure 6 shows that bacterially expressed IE86 protein bound specifically to GST-P/CAF beads (lane 2) but not to GST beads alone (lane 1). IE86 also bound to GST–TATA-binding protein (TBP) and GST-CBP (lanes 3 and 4, respectively), as expected. We have observed no such binding of IE72 to TBP, CBP, or P/CAF in these types of assays (data not shown). Consequently, the direct interaction between IE86 and P/CAF is not mediated by any other eukaryotic protein.
FIG. 6.
Interaction between P/CAF and IE86 requires no other eukaryotic protein. Bacterially expressed IE86 protein was labeled in vitro with 32P and used in GST pull-down assays to analyze binding to GST (lane 1), GST-P/CAF (lane 2), GST-TBP (lane 3), and GST-CBP (lane 4). A lane between lanes 2 and 3 was left unloaded to ensure no accidental overspill of the GST-TBP positive control sample.
IE2 interacts with P/CAF in vivo.
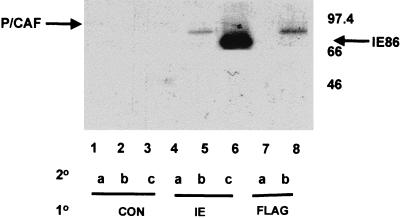
We next confirmed the ability of IE86 and P/CAF to interact in vivo. To demonstrate this interaction, we performed double immunoprecipitation assays in cells transfected with IE86 and P/CAF expression vectors (Fig. 7). As expected, cells cotransfected with pcDNA3IE86 plus pCX-Flag-P/CAF and immunoprecipitated with an anti-IE antibody followed by reprecipitation with the same antibody showed high levels of IE86 protein in the immunoprecipitated complex (lane 6). Similarly, double immunoprecipitation using an anti-Flag tag antibody for both primary and secondary immunoprecipitations showed P/CAF (lane 8) present in the immunocomplex, also as expected. Interestingly, immunoprecipitation using an anti-IE antibody as the primary antibody followed by an anti-Flag tag antibody as the secondary antibody (lane 5) clearly showed the presence of P/CAF in IE86 immunocomplexes.
FIG. 7.
IE86 and P/CAF interact in vivo. U2-OS cells were cotransfected with pcDNA3IE86 and pCX-Flag-P/CAF and labeled with [35S]methionine; 48 h posttransfection, cells were lysed in EBC buffer and extracts were subjected to double immunoprecipitation assays using a control anti-murine cyclin D1 monoclonal antibody (CON), an anti-IE antibody (IE), and an anti-Flag monoclonal antibody (FLAG) for primary immunoprecipitations (10). Complexes were washed and reimmunoprecipitated using the control (lanes a), anti-Flag (lanes b), and anti-IE (lanes c) antibodies for secondary (20) immunoprecipitations. Complexes were separated by SDS-PAGE and autoradiographed.
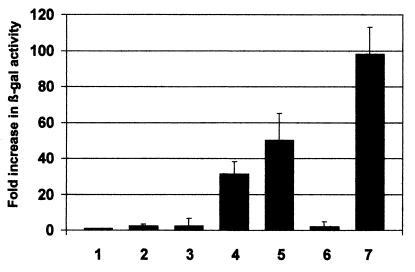
We also confirmed the interaction between IE86 and P/CAF in vivo using the yeast two-hybrid system (Fig. 8). P/CAF was expressed as a fusion protein to the DNA binding domain of GAL4 (pGBT10:P/CAF), and IE86 was expressed as a fusion protein to the GAL4 activation domain (pGAD:IE86). Combinations of plasmids were transformed into yeast and tested for the ability to up-regulate expression of a β-galactosidase gene under the control of a yeast promoter bearing GAL4 binding sites. Coexpression of pGBT10:P/CAF and pGAD:IE2 resulted in much higher levels of β-galactosidase activity (lane 4) compared to a panel of negative controls (i.e., pGBT10 plus pGAD425 [lane 1], pGBT10 plus pGAD:IE2 [lane 2], pGBT10:P/CAF plus pGAD425 [lane 3], and pGBT10:P/CAF plus pGADIE72 [lane 6]). This confirmed the direct interaction between IE86 and P/CAF in vivo.
FIG. 8.
IE86 and P/CAF interact in yeast two-hybrid assays. The parental plasmid for expression in yeast of the DNA binding domain of GAL4 (pGBT10) was fused to P/CAF (pGBT:P/CAF) or IE86 (pGBT:IE86). The parental plasmid for expression in yeast of the GAL4 activation domain (pGAD425) was fused to IE86 (pGAD:IE86) or IE72 (pGAD:IE72). pGBT10 plus pGAD425 (bar 1), pGBT10 plus pGAD:IE86 (bar 2), pGBT:P/CAF plus pGAD425 (bar 3), pGBT:P/CAF plus pGAD:IE86 (bar 4), pGBT:IE86 plus pGAD:IE86 (bar 5), pGBT:P/CAF plus pGAD:IE72 (bar 6), and pVA3-1 plus pTD1-1 containing a GAL4 DNA binding domain murine p53 fusion protein and a GAL4 activation domain simian virus 40 T-antigen fusion, respectively (bar 7), were transformed into yeast, and β-galactosidase expression was measured in liquid culture. Data represent average fold increases in β-galactosidase activity over activity obtained from cotransformation with pGBT10 plus pGAD425 from three independent experiments.
IE86 and P/CAF act synergistically in transient transfection assays.
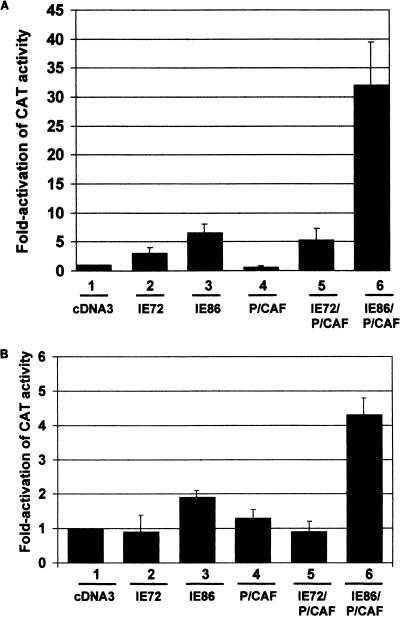
To determine whether the physical interaction between IE86 and P/CAF was of functional importance, we examined whether IE86 and P/CAF could affect promoter activity in transient cotransfection assays (Fig. 9A). U373 cells were transfected with a CAT reporter construct based on the TGF-β2 promoter (TGFβ77MCAT) which contains a mutation in the known single ATF site (39). Consequently, this promoter does not respond to activation by CREB/ATF (39), but we have previously shown that it is responsive to IE86 (8). TGFβ77MCAT was transfected into cells together with IE expression vectors in the presence or absence of P/CAF. Consistent with previous observations (8), IE86 is able to mildly activate the TGFβ77MCAT reporter (lane 3). In contrast, P/CAF alone appeared to have little effect on this promoter (lane 4). Cotransfection of both IE86 and P/CAF, however, resulted in high levels of activation, much higher than that observed for IE86 alone (compare lanes 3 and 6). We have ruled out that this effect may be mediated via IE86 activating transfected P/CAF expression or P/CAF activating transfected IE86 expression directly, as Western blot analysis of IE86-P/CAF-cotransfected cells shows no increase in levels of transfected P/CAF or IE86 compared to cells transfected with P/CAF or IE86 alone (data not shown). Similarly IE86 expression does not increase levels of endogenous CBP (data not shown). In contrast and as expected, IE72 showed no such synergistic activation with P/CAF (lane 5). However, coexpression of IE72 and P/CAF did show levels of activation slightly higher than with IE72 alone. How P/CAF elicits such an effect is unclear at present but appears not to be due to any direct interaction between these proteins.
FIG. 9.
IE86 and P/CAF synergistically activate transiently or stably transfected target promoters in transfection assays. (A) U373 cells were transiently transfected with TGFβ77MCAT together with pcDNA3 (bar 1), pcDNA3IE72 (bar 2), pcDNA3IE86 (bar 3), pCX-Flag-P/CAF (bar 4), pcDNA3IE72 and pCX-Flag-P/CAF (bar 5), or pcDNA3IE86 and pCX-Flag-P/CAF (bar 6). Data represent the average of three independent experiments. (B) U373 cells that had been stably transfected with TGFβ77CAT were transiently transfected with pcDNA3 (bar 1), pcDNA3IE72 (bar 2), pcDNA3IE86 (bar 3), pCX-Flag-P/CAF (bar 4), pcDNA3IE72 and pCX-Flag-P/CAF (bar 5), or pcDNA3IE86 and pCX-Flag-P/CAF (bar 6). Data represent the average of three independent experiments.
While it is likely that transiently transfected DNA will chromatinize to some extent, it is unlikely that full chromatinization of reporter templates will occur under transient transfection conditions. Consequently, it was important to determine if the ability of IE86 and P/CAF to activate transiently transfected reporter constructs also occurred with reporter constructs that were integrated into host genomic DNA and, hence, fully chromatinized. We therefore also analyzed the effects of IE86 and P/CAF on cells that had been stably transfected with CAT reporter constructs. Figure 9B shows that cells stably transfected with TGFβ77CAT also show modest activation by IE86 (lane 3) or P/CAF (lane 4) alone. In contrast, cotransfection with IE86 and P/CAF results in activation of the stably integrated TGFβ77CAT (lane 6). As expected, no such synergistic activation is observed with IE72 and P/CAF (lane 5). The differences in fold activation by IE86 and P/CAF observed in transient transfections compared to supertransfection of stable cells expressing the TGFβ–CAT reporter is likely to be due to transient transfection efficiencies. The transient transfection efficiency of U373 cells is about 5 to 10% (T. H. Sinclair, unpublished observations). In the transient transfections, all cells expressing the TGFβ–CAT reporter DNA will also have taken up IE86 and P/CAF plasmids. In stable cells expressing the TGFβ–CAT reporter, however, 90% of these cells will not be affected by IE86 and P/CAF supertransfection.
DISCUSSION
The mechanism by which IE86 activates cellular and viral promoters is still unclear. However, the apparent promiscuity of promoter activation by IE86 reflects its ability to act multimodally. IE86 can interact directly with sequence-specific transcription factors, presumably bridging between these DNA bound factors and the basal transcription complex, components of which IE86 is also able to bind directly (7, 19, 24, 36, 42, 47, 48, 69, 73, 74, 97). The interaction between IE86 and cellular factors such as RB, which is known to sequester cellular transcription factors such as E2F, also results in the release of functional E2F from RB, allowing activation of E2F-dependent promoters (13, 23, 73). However, IE86 is also able to activate minimal basal promoters that have little or no upstream DNA sequences (24, 25). Consequently, IE86 appears to be able to act on the transcription preinitiation complex directly in the absence of any specific recruitment to the basal promoter. In the case of some promoters, activation by IE86 may be mediated by inhibition of cellular transcriptional repressors that inhibit basal transcription (6, 40).
Here, we have shown that IE86 interacts directly with the chromatin acetylation factor P/CAF. HAT activity is coprecipitated with IE86 in HCMV-infected cells, and this HAT activity was substantially increased when P/CAF was specifically coexpressed with IE86 in cotransfection assays. Clearly, the HAT activity associated with IE86 that was detected during infection may also have been due, at least in part, to CBP. However, the increase in IE86-associated HAT activity observed upon cotransfection of cells with P/CAF and IE86 argues for a direct interaction between P/CAF and IE86, independently of CBP. Also, importantly, the interaction between IE86 and these HATs appears not to inhibit HAT activity.
We confirmed the putative direct interaction between P/CAF and IE86 in vitro. In vitro-translated and radiolabeled P/CAF protein bound specifically to GST-IE86. In contrast to its binding to CBP, which required only amino acids 290 to 390 of IE86, the minimum region of IE86 required for binding to P/CAF encompassed amino acids 290 to 504; the 290–390 region alone was unable to bind P/CAF. GST fusion interaction assays using 32P-labeled bacterially expressed IE86 also confirmed the direct interaction between IE86 and P/CAF in the absence of any other eukaryotic protein. These analyses would be consistent with IE86 being able to contact both P/CAF and CBP simultaneously. As yet, we do not know the domains in P/CAF or CBP responsible for interaction with IE86. Consequently, we do not know if interaction of IE86 with P/CAF prevents interaction of CBP with P/CAF as has been shown for E1A (65). The question as to why IE86 should need to interact independently with two HATs is a valid one. However, in vitro, CBP and P/CAF acetylate different subsets of histones (2, 56, 96), and it has been suggested that promoter recruitment of these two HATs is selective and that different promoters may require different HAT activities for activation (65). Consequently, it is possible that activation of different target promoters by IE86 may be mediated by different HATs.
We also analyzed the ability of IE86 to interact with P/CAF in vivo. Immunoprecipitation of cells transfected with IE86 and P/CAF expression vectors showed that IE86 immunocomplexes also contained P/CAF. Similarly, yeast two hybrid analysis confirmed this in vivo interaction.
The physical interaction between IE86 and P/CAF proteins was also reflected functionally. In the case of E1A, interaction with P/CAF has been shown to abrogate P/CAF's ability to activate the Rous sarcoma virus long terminal repeat in transient transfection assays (65). In contrast, IE86 appears to act in concert with P/CAF to synergistically activate target promoters. In the case of the TGF-β2 promoter devoid of a CREB binding site, P/CAF appears to have a slightly repressive effect, probably due to squelching. However, transient cotransfection with IE86 results in levels of TGF-β2 promoter activation, from TGFβ277MCAT, much higher than that observed with IE86 alone. Similar results were observed when stably transfected TGF-β2 reporter constructs were used. Clearly, while we and others have emphasized the many functional similarities between HCMV IE86 and adenovirus E1A, the effect of IE86 on P/CAF is distinctly different from that seen with E1A. The ability of HCMV to activate cellular gene expression and the role of HCMV IE86 in the promiscuous activation of cellular promoters is well established. It is also likely that IE86-mediated promoter activation occurs by a number of mechanisms involving direct interaction with the basal transcription complex. Here, we have shown that IE86 is able to physically and functionally interact with the HAT P/CAF.
We propose that IE86, by means of its direct interaction with general transcription factors, is able to recruit P/CAF directly to target promoters, and we believe that virus-induced chromatin remodeling plays a pivotal role in HCMV-mediated gene activation.
ACKNOWLEDGMENTS
We are grateful to M. Green, Y. Nakatani, T. Stamminger, and X.-J. Yang for plasmids.
This work was supported by the British Medical Research Council and The Wellcome Trust.
REFERENCES
- 1.Arlt H, Lang D, Gebert S, Stamminger T. Identification of binding sites for the 86-kilodalton IE2 protein of human cytomegalovirus within an IE2-responsive viral early promoter. J Virol. 1994;68:4117–4125. doi: 10.1128/jvi.68.7.4117-4125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 3.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 5.Bresnahan W A, Albrecht T, Thompson E A. The cyclin E promoter is activated by human cytomegalovirus 86-kDa immediate early protein. J Biol Chem. 1998;273:22075–22082. doi: 10.1074/jbc.273.34.22075. [DOI] [PubMed] [Google Scholar]
- 6.Caswell R, Bryant L, Sinclair J. Human cytomegalovirus immediate-early 2 (IE2) protein can transactivate the human hsp70 promoter by alleviation of Dr1-mediated repression. J Virol. 1996;70:4028–4037. doi: 10.1128/jvi.70.6.4028-4037.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caswell R, Hagemeier C, Chiou C J, Hayward G, Kouzarides T, Sinclair J. The human cytomegalovirus 86K immediate early (IE) 2 protein requires the basic region of the TATA-box binding protein (TBP) for binding, and interacts with TBP and transcription factor TFIIB via regions of IE2 required for transcriptional regulation. J Gen Virol. 1993;74:2691–2698. doi: 10.1099/0022-1317-74-12-2691. [DOI] [PubMed] [Google Scholar]
- 8.Caswell R, Hagemeier C, Hayhurst G, Kouzarides T, Sinclair J. Mechanisms of transactivation of cellular promoters by the HCMV major immediate early proteins involve direct interactions with cellular transcription factors. Amsterdam, The Netherlands: Elsevier Science; 1993. [Google Scholar]
- 9.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 10.Cherrington J M, Khoury E L, Mocarski E S. Human cytomegalovirus ie2 negatively regulates α gene expression via a short target sequence near the transcription start site. J Virol. 1991;65:887–896. doi: 10.1128/jvi.65.2.887-896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the α promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiou C J, Zong J, Waheed I, Hayward G S. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J Virol. 1993;67:6201–6214. doi: 10.1128/jvi.67.10.6201-6214.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi K S, Kim S J, Kim S. The retinoblastoma gene product negatively regulates transcriptional activation mediated by the human cytomegalovirus IE2 protein. Virology. 1995;208:450–456. doi: 10.1006/viro.1995.1175. [DOI] [PubMed] [Google Scholar]
- 14.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 15.Colberg-Poley A M, Santomenna L D, Harlow P P, Benfield P A, Tenney D J. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai P, Akimaru H, Tanaka Y, Hou D X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. CBP as a transcriptional coactivator of c-Myb. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 17.DeMarchi J M, Schmidt C A, Kaplan A S. Patterns of transcription of human cytomegalovirus in permissively infected cells. J Virol. 1980;35:277–286. doi: 10.1128/jvi.35.2.277-286.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenfeld G, Emerson B M, Jackson P D, Lewis C D, Nickol J M. Chromatin structure near transcriptionally active genes. Prog Clin Biol Res. 1986;218:63–74. [PubMed] [Google Scholar]
- 19.Furnari B A, Poma E, Kowalik T F, Huong S M, Huang E S. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J Virol. 1993;67:4981–4991. doi: 10.1128/jvi.67.8.4981-4991.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory P D, Horz W. Chromatin and transcription—how transcription factors battle with a repressive chromatin environment. Eur J Biochem. 1998;251:9–18. doi: 10.1046/j.1432-1327.1998.2510009.x. [DOI] [PubMed] [Google Scholar]
- 21.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 22.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 23.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 1994;13:2897–2903. doi: 10.1002/j.1460-2075.1994.tb06584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagemeier C, Walker S, Caswell R, Kouzarides T, Sinclair J. The human cytomegalovirus 80-kilodalton but not the 72-kilodalton immediate-early protein transactivates heterologous promoters in a TATA box-dependent mechanism and interacts directly with TFIID. J Virol. 1992;66:4452–4456. doi: 10.1128/jvi.66.7.4452-4456.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagemeier C, Walker S M, Sissons P J, Sinclair J H. The 72K IE1 and 80K IE2 proteins of human cytomegalovirus independently trans-activate the c-fos, c-myc and hsp70 promoters via basal promoter elements. J Gen Virol. 1992;73:2385–2393. doi: 10.1099/0022-1317-73-9-2385. [DOI] [PubMed] [Google Scholar]
- 26.Hayhurst G P, Bryant L A, Caswell R C, Walker S M, Sinclair J H. CCAAT box-dependent activation of the TATA-less human DNA polymerase alpha promoter by the human cytomegalovirus 72-kilodalton major immediate-early protein. J Virol. 1995;69:182–188. doi: 10.1128/jvi.69.1.182-188.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hebbes T R, Thorne A W, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hermiston T W, Malone C L, Stinski M F. Human cytomegalovirus immediate-early two protein region involved in negative regulation of the major immediate-early promoter. J Virol. 1990;64:3532–3536. doi: 10.1128/jvi.64.7.3532-3536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hermiston T W, Malone C L, Witte P R, Stinski M F. Identification and characterization of the human cytomegalovirus immediate-early region 2 gene that stimulates gene expression from an inducible promoter. J Virol. 1987;61:3214–3221. doi: 10.1128/jvi.61.10.3214-3221.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L, Stinski M F. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J Virol. 1995;69:7612–7621. doi: 10.1128/jvi.69.12.7612-7621.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwamoto G K, Monick M M, Clark B D, Auron P E, Stinski M F, Hunninghake G W. Modulation of interleukin 1β gene expression by the immediate early genes of human cytomegalovirus. J Clin Investig. 1990;85:1853–1857. doi: 10.1172/JCI114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jahn G, Knust E, Schmolla H, Sarre T, Nelson J A, McDougall J K, Fleckenstein B. Predominant immediate-early transcripts of human cytomegalovirus AD169. J Virol. 1984;49:363–370. doi: 10.1128/jvi.49.2.363-370.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins D E, Martens C L, Mocarski E S. Human cytomegalovirus late protein encoded by ie2: a trans-activator as well as a repressor of gene expression. J Gen Virol. 1994;75:2337–2348. doi: 10.1099/0022-1317-75-9-2337. [DOI] [PubMed] [Google Scholar]
- 34.Jupp R, Flores O, Nelson J A, Ghazal P. The DNA-binding subunit of human transcription factor IID can interact with the TATA box as a multimer. J Biol Chem. 1993;268:16105–16108. [PubMed] [Google Scholar]
- 35.Jupp R, Hoffmann S, Depto A, Stenberg R M, Ghazal P, Nelson J A. Direct interaction of the human cytomegalovirus IE86 protein with the cis repression signal does not preclude TBP from binding to the TATA box. J Virol. 1993;67:5595–5604. doi: 10.1128/jvi.67.9.5595-5604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jupp R, Hoffmann S, Stenberg R M, Nelson J A, Ghazal P. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J Virol. 1993;67:7539–7546. doi: 10.1128/jvi.67.12.7539-7546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 38.Kerry J A, Priddy M A, Stenberg R M. Identification of sequence elements in the human cytomegalovirus DNA polymerase gene promoter required for activation by viral gene products. J Virol. 1994;68:4167–4176. doi: 10.1128/jvi.68.7.4167-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim S J, Wagner S, Liu F, O'Reilly M A, Robbins P D, Green M R. Retinoblastoma gene product activates expression of the human TGF-beta 2 gene through transcription factor ATF-2. Nature. 1992;358:331–334. doi: 10.1038/358331a0. [DOI] [PubMed] [Google Scholar]
- 40.Klucher K M, Sommer M, Kadonaga J T, Spector D H. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol Cell Biol. 1993;13:1238–1250. doi: 10.1128/mcb.13.2.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 42.Lang D, Gebert S, Arlt H, Stamminger T. Functional interaction between the human cytomegalovirus 86-kilodalton IE2 protein and the cellular transcription factor CREB. J Virol. 1995;69:6030–6037. doi: 10.1128/jvi.69.10.6030-6037.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang D, Stamminger T. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J Virol. 1993;67:323–331. doi: 10.1128/jvi.67.1.323-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lang D, Stamminger T. Minor groove contacts are essential for an interaction of the human cytomegalovirus IE2 protein with its DNA target. Nucleic Acids Res. 1994;22:3331–3338. doi: 10.1093/nar/22.16.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B, Hermiston T W, Stinski M F. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J Virol. 1991;65:897–903. doi: 10.1128/jvi.65.2.897-903.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loidl P. Histone acetylation: facts and questions. Chromosoma. 1994;103:441–449. doi: 10.1007/BF00337382. [DOI] [PubMed] [Google Scholar]
- 47.Lukac D M, Harel N Y, Tanese N, Alwine J C. TAF-like functions of human cytomegalovirus immediate-early proteins. J Virol. 1997;71:7227–7239. doi: 10.1128/jvi.71.10.7227-7239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukac D M, Mannupello J R, Alwine J C. Transcriptional activation by the human cytomegalovirus immediate-early proteins: requirements for simple promoter structures and interactions with multiple components of the transcription complex. J Virol. 1994;68:5184–5193. doi: 10.1128/jvi.68.8.5184-5193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macias M P, Stinski M F. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc Natl Acad Sci USA. 1993;90:707–711. doi: 10.1073/pnas.90.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malone C L, Vesole D H, Stinski M F. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J Virol. 1990;64:1498–1506. doi: 10.1128/jvi.64.4.1498-1506.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Margolis M J, Pajovic S, Wong E L, Wade M, Jupp R, Nelson J A, Azizkhan J C. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J Virol. 1995;69:7759–7767. doi: 10.1128/jvi.69.12.7759-7767.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonough S H, Spector D H. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology. 1983;125:31–46. doi: 10.1016/0042-6822(83)90061-2. [DOI] [PubMed] [Google Scholar]
- 54.Merigan T C, Resta S. Cytomegalovirus: where have we been and where are we going? Rev Infect Dis. 1990;12(Suppl. 7):693–700. doi: 10.1093/clinids/12.supplement_7.s693. [DOI] [PubMed] [Google Scholar]
- 55.Monick M M, Geist L J, Stinski M F, Hunninghake G W. The immediate early genes of human cytomegalovirus upregulate expression of the cellular genes myc and fos. Am J Respir Cell Mol Biol. 1992;7:251–256. doi: 10.1165/ajrcmb/7.3.251. [DOI] [PubMed] [Google Scholar]
- 56.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 57.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 58.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 59.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 60.Pizzorno M C, Hayward G S. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target located near the cap site. J Virol. 1990;64:6154–6165. doi: 10.1128/jvi.64.12.6154-6165.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pizzorno M C, Mullen M A, Chang Y N, Hayward G S. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J Virol. 1991;65:3839–3852. doi: 10.1128/jvi.65.7.3839-3852.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pizzorno M C, O'Hare P, Sha L, LaFemina R L, Hayward G S. Transactivation and autoregulation of gene expression by the immediate-early region 2 gene products of human cytomegalovirus. J Virol. 1988;62:1167–1179. doi: 10.1128/jvi.62.4.1167-1179.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plachter B, Britt W, Vornhagen R, Stamminger T, Jahn G. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology. 1993;193:642–652. doi: 10.1006/viro.1993.1172. [DOI] [PubMed] [Google Scholar]
- 64.Poma E E, Kowalik T F, Zhu L, Sinclair J H, Huang E S. The human cytomegalovirus IE1-72 protein interacts with the cellular p107 protein and relieves p107-mediated transcriptional repression of an E2F-responsive promoter. J Virol. 1996;70:7867–7877. doi: 10.1128/jvi.70.11.7867-7877.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reid J L, Bannister A J, Zegerman P, Martinez-Balbas M A, Kouzarides T. E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J. 1998;17:4469–4477. doi: 10.1093/emboj/17.15.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roth S Y. Something about silencing. Nat Genet. 1996;14:3–4. doi: 10.1038/ng0996-3. [DOI] [PubMed] [Google Scholar]
- 67.Rubin R H, Cosimi A B, Tolkoff-Rubin N E, Russell P S, Hirsch M S. Infectious disease syndromes attributable to cytomegalovirus and their significance among renal transplant recipients. Transplantation. 1977;24:458–464. doi: 10.1097/00007890-197712000-00010. [DOI] [PubMed] [Google Scholar]
- 68.Schiltz R L, Mizzen C A, Vassilev A, Cook R G, Allis C D, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and P/CAF within nucleosomal substrates. J Biol Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 69.Schwartz R, Helmich B, Spector D H. CREB and CREB-binding proteins play an important role in the IE2 86-kilodalton protein-mediated transactivation of the human cytomegalovirus 2.2-kilobase RNA promoter. J Virol. 1996;70:6955–6966. doi: 10.1128/jvi.70.10.6955-6966.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schwartz R, Sommer M H, Scully A, Spector D H. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J Virol. 1994;68:5613–5622. doi: 10.1128/jvi.68.9.5613-5622.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scully A L, Sommer M H, Schwartz R, Spector D H. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J Virol. 1995;69:6533–6540. doi: 10.1128/jvi.69.10.6533-6540.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 73.Sommer M H, Scully A L, Spector D H. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J Virol. 1994;68:6223–6231. doi: 10.1128/jvi.68.10.6223-6231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Speir E, Modali R, Huang E-S, Leon M B, Shawl F, Finkel T, Epstein S E. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–394. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 75.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 76.Stenberg R M, Fortney J, Barlow S W, Magrane B P, Nelson J A, Ghazal P. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J Virol. 1990;64:1556–1565. doi: 10.1128/jvi.64.4.1556-1565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stenberg R M, Witte P R, Stinski M F. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J Virol. 1985;56:665–675. doi: 10.1128/jvi.56.3.665-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stinski M F. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J Virol. 1978;26:686–701. doi: 10.1128/jvi.26.3.686-701.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stinski M F, Thomsen D R, Stenberg R M, Goldstein L C. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1983;46:1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Trouche D, Cook A, Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsukiyama T, Wu C. Chromatin remodeling and transcription. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 82.Turner B M. Histone acetylation and control of gene expression. J Cell Sci. 1991;99:13–20. doi: 10.1242/jcs.99.1.13. [DOI] [PubMed] [Google Scholar]
- 83.Turner B M, O'Neill L P. Histone acetylation in chromatin and chromosomes. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 84.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 85.Wade M, Kowalik T F, Mudryj M, Huang E S, Azizkhan J C. E2F mediates dihydrofolate reductase promoter activation and multiprotein complex formation in human cytomegalovirus infection. Mol Cell Biol. 1992;12:4364–4374. doi: 10.1128/mcb.12.10.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wade P A, Pruss D, Wolffe A P. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 87.Wathen M W, Stinski M F. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol. 1982;41:462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wathen M W, Thomsen D R, Stinski M F. Temporal regulation of human cytomegalovirus transcription at immediate early and early times after infection. J Virol. 1981;38:446–459. doi: 10.1128/jvi.38.2.446-459.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilkinson G W, Akrigg A, Greenaway P J. Transcription of the immediate early genes of human cytomegalovirus strain AD169. Virus Res. 1984;1:101–106. doi: 10.1016/0168-1702(84)90067-4. [DOI] [PubMed] [Google Scholar]
- 90.Wolffe A P. Histones, nucleosomes and the roles of chromatin structure in transcriptional control. Biochem Soc Trans. 1997;25:354–358. doi: 10.1042/bst0250354. [DOI] [PubMed] [Google Scholar]
- 91.Wolffe A P, Kurumizaka H. The nucleosome: a powerful regulator of transcription. Prog Nucleic Acid Res Mol Biol. 1998;61:379–422. doi: 10.1016/s0079-6603(08)60832-6. [DOI] [PubMed] [Google Scholar]
- 92.Wolffe A P, Pruss D. Targeting chromatin disruption: transcription regulators that acetylate histones. Cell. 1996;84:817–819. doi: 10.1016/s0092-8674(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 93.Workman J L, Buchman A R. Multiple functions of nucleosomes and regulatory factors in transcription. Trends Biochem Sci. 1993;18:90–95. doi: 10.1016/0968-0004(93)90160-o. [DOI] [PubMed] [Google Scholar]
- 94.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 95.Wu J, Jupp R, Stenberg R M, Nelson J A, Ghazal P. Site-specific inhibition of RNA polymerase II preinitiation complex assembly by human cytomegalovirus IE86 protein. J Virol. 1993;67:7547–7555. doi: 10.1128/jvi.67.12.7547-7555.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 97.Young Do Y, Chiou C J, Kyeong Sook C, Yi Y, Michelson S, Kim S, Hayward G S, Kim S J. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor β1 gene through an Egr-1 binding site. J Virol. 1996;70:7062–7070. doi: 10.1128/jvi.70.10.7062-7070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. Human p300 protein is a coactivator for the transcription factor MyoD. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]