Abstract
Neurodevelopmental disorders are major indications for genetic referral and have been linked to more than 1500 loci including genes encoding transcriptional regulators. The dysfunction of transcription factors often results in characteristic syndromic presentations; however, at least half of these patients lack a genetic diagnosis. The implementation of machine learning approaches has the potential to aid in the identification of new disease genes and delineate associated phenotypes.
Next generation sequencing was performed in seven affected individuals with neurodevelopmental delay and dysmorphic features. Clinical characterization included reanalysis of available neuroimaging datasets and 2D portrait image analysis with GestaltMatcher. The functional consequences of ZSCAN10 loss were modelled in mouse embryonic stem cells (mESCs), including a knockout and a representative ZSCAN10 protein truncating variant. These models were characterized by gene expression and western blot analyses, chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR) and immunofluorescence staining. Zscan10 knockout mouse embryos were generated and phenotyped.
We prioritized bi-allelic ZSCAN10 loss-of-function variants in seven affected individuals from five unrelated families as the underlying molecular cause. RNA-sequencing analyses in Zscan10−/− mESCs indicated dysregulation of genes related to stem cell pluripotency. In addition, we established in mESCs the loss-of-function mechanism for a representative human ZSCAN10 protein truncating variant by showing alteration of its expression levels and subcellular localization, interfering with its binding to DNA enhancer targets. Deep phenotyping revealed global developmental delay, facial asymmetry and malformations of the outer ear as consistent clinical features. Cerebral MRI showed dysplasia of the semicircular canals as an anatomical correlate of sensorineural hearing loss. Facial asymmetry was confirmed as a clinical feature by GestaltMatcher and was recapitulated in the Zscan10 mouse model along with inner and outer ear malformations.
Our findings provide evidence of a novel syndromic neurodevelopmental disorder caused by bi-allelic loss-of-function variants in ZSCAN10.
Keywords: neurodevelopmental disorders, zinc finger transcription factor, oto-facial syndrome, semicircular canal dysplasia
Laugwitz et al. describe a novel syndromic neurodevelopmental disorder with characteristic malformations of the face and of the inner and outer ear. The autosomal recessive disorder is caused by biallelic loss-of-function variants in the ZSCAN10 gene.
Introduction
Major congenital anomalies are observed in 2%–5% of live births and are often accompanied by developmental delay (DD) and intellectual disability (ID).1 These syndromic forms of neurodevelopmental disorders are among the most common indications for genetic referral and represent a heterogenous group of conditions that can severely affect a child’s development. To date, more than 1500 loci have been linked to syndromic and non-syndromic forms of DD/ID, which collectively have a global prevalence of approximately 1%.2,3 While diverse mechanisms can cause neurodevelopmental disorders, pathogenic genetic variation in developmentally important genes has a major contribution.4,5 Some of these genes encode transcriptional regulators, and individuals affected with these disease entities often present with numerous clinical features in addition to DD/ID such as organ malformations and facial abnormalities.6
The largest human transcription factor family, the zinc finger proteins, contains >800 members, several of which are associated with syndromic neurodevelopmental disorders, e.g. GLI3: OMIM #175700; ZIC1: OMIM #618736; and ZNF462: OMIM #618619.7-11ZSCAN10 (also known as ZFP206: OMIM *618365) encodes a human zinc finger-transcription factor of the C2H2 zinc finger subfamily with an additional N-terminal SCAN domain (Fig. 1).8,12 It is expressed in human embryonic stem cells where it is believed to function in the maintenance of pluripotency within the transcriptional network of other transcription factors like OCT4, SOX2 and NANOG.13-16 Compatible with a predominant role in early embryonic development ZSCAN10 mRNA expression decreases with age but remains postnatally detectable in the brain, testis and pituitary gland.17,18
Figure 1.
Pedigrees of investigated families and structure of ZSCAN10. (A) Pedigrees of five families with pathogenic variants in ZSCAN10, illustrating affected (filled symbol), healthy (open symbol) family members. Heterozygous carriers are indicated (open symbols containing central dot). Unaffected siblings and fetuses were not tested unless indicated. Families F5a and F5b were remotely related. Individual F5b:II.1 exhibited thalassaemia minor as an additional phenotype. Individual F5b.II.2 was intentionally aborted due to genetically confirmed thalassaemia major (asterisk); the fetus was a carrier of the c.1456C>T variant in ZSCAN10. (B) Structure of ZSCAN10 and the encoded protein with known domains and position of identified variants. aa = amino acid; CDS = coding sequence.
Here, we report seven affected individuals from five families with a clinically recognizable form of syndromic DD/ID due to bi-allelic loss-of-function variants in ZSCAN10 (Fig. 1).
Materials and methods
Clinical assessment
Clinical data were provided by the primary physicians. Consent to publish images was obtained from legal guardians. All procedures were performed in accordance with the Declaration of Helsinki (Supplementary material).
Genetic studies
After obtaining written informed consent, exome or genome sequencing was performed as previously described (Supplementary material).19,20 Assembly of this international collaborative study cohort resulted from personal communication between collaborators (Families F4 and F5) and was facilitated by GeneMatcher (Family F3).21 Runs of homozygosity were evaluated using GSvar (https://github.com/imgag/ngs-bits/blob/master/doc/GSvar/roh_analysis.md). The possibility of shared ancestry was evaluated based on the available sequencing datasets by manual inspection of genotypes in the genomic region surrounding the ZSCAN10 locus.
Zscan10 knockout in mouse embryonic stem cells
Feeder-free HM1 mouse embryonic stem cells (mESCs) used in this study were previously described.22 HM1 mESCs were grown in standard medium [Dulbecco’s modified Eagle medium supplemented with 15% fetal bovine serum (FBS), ES-qualified, (ThermoFisher Scientific, 16141-079), 1000 U/ml leukemia inhibitory factor (Millipore), 1× Minimum Essential Medium Non-Essential Amino Acids solution, 1 mM of sodium pyruvate, 1× GlutaMAX™ supplement, 1× penicillin and streptomycin and 0.1 mM of 2-mercaptoethanol] supplemented with 1 µM MEK1/2 inhibitor (PD0325901, Sigma) and 3 µM GSK3 inhibitor (CHIR99021, Sigma). Cells were grown on 0.2% gelatinized (Sigma, G1890) tissue culture plates or dishes.
The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system was used to knock out exon 2 of Zscan10 to generate Zscan10−/− mESCs as previously described.23 The guide RNAs (gRNAs) were designed using CRISpick (https://portals.broadinstitute.org/gppx/crispick/public) and synthesized by biomers (www.biomers.net). Two guide RNAs targeting the upstream and downstream of Zscan10 exon2 were picked up and sub-cloned into the pX459 vector (upstream gRNA1: ATACTGCGTTAAGATCTGAC; downstream gRNA2: AGAGGTGAGTGGAAAGAAAC). PCR and Sanger sequencing were used to select the positive cell clones (forward primer: ACTCCTGGTCCTCTAGACTC; reverse primer: CTAGCTATGCTCACTGCCTT) (Supplementary Fig. 1).
RNA-sequencing and expression analysis
RNA-sequencing (RNA-seq) and data analysis were performed as previously described.24 A total of 100 ng total RNA extracted from mESCs was subjected to polyA enrichment and cDNA sequencing libraries were prepared using a NEBNext Ultra II Directional RNA Library Preparation Kit for Illumina (NEB, E7760L). Libraries were sequenced as paired-end reads on a NovaSeq6000 (Illumina) with at least 20 million reads each. After quality control, RNA-seq data were aligned using HISAT2 (version 2.2.1) to Ensemble Mouse GRCm38 (mm10). Normalized read counts for all genes were obtained using featureCount (v3.18.1). Differentially expressed genes [false discovery rate (FDR) < 0.05] were determined in comparisons between experimental and control groups using Differential Expression Analysis for Sequence Count Data (DESeq, v1.22.1).
Characterization of the impact of the human ZSCAN10 c.1456C>T variant
The human wild-type ZSCAN10 cDNA was purchased from Genscript (OHu73812D) and PCR amplified and subcloned into pcDNA3/C-SF vector using HindIII and EcoRI.23 The ZSCAN10 c.1456C>T variant cDNA was obtained using PCR amplification of the first 1455 bp coding region of the full-length ZSCAN10 cDNA, encoding the first 485 amino acids of the ZSCAN10 protein. The mutant ZSCAN10 cDNA was subcloned into the pcDNA3/C-SF vector using HindIII and EcoRI. The full-length ZSCAN10 (ZSCAN10 full) and c.1456C>T ZSCAN10 (ZSCAN10 485) expression vectors were transfected into mESCs using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the cells were harvested for western blot analysis, immunofluorescence staining and chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR) analysis (Supplementary material).
GestaltMatcher analysis
We analysed facial similarities among affected subjects with confirmed genetic disorders harbouring pathogenic variants in string interacting genes including ZSCAN10, CHD7, PBX1, SALL4 and SMAD4 using GestaltMatcher.25,26 The dataset consisted of 90 frontal images in total. 2D images of five subjects with ZSCAN10 deficiency (Individuals F1:II.6, F2:II.2, F2:II.3, F3:II.1 and F5B:II.1) were available for facial analysis (Supplementary Table 2). GestaltMatcher was first trained on 3438 images with 139 different disorders from the GestaltMatcher database to learn the facial dysmorphic features and later encoded each image into a 320-dimensional facial phenotype descriptor (FPD). The FPDs formed a Clinical Face Phenotype Space, and the facial syndromic similarity was quantified by the cosine distance between two FPDs in the space. We performed t-distributed stochastic neighbour embedding to visualize the 90 FPDs in 2D space and expanded the model to recognize facial asymmetry (Supplementary Table 3).27
Dysmorphism analyses in mouse embryos
We used mice homozygous for a gene targeting Zscan10 allele generated by homologous recombination in ESCs using the knockout-first allele method,28 adopting a strategy identifying exon 6 common to all transcripts of Zscan10. This strategy perturbs the expression of the targeted gene by inclusion of a large LacZ cassette in the upstream region of exon 6 that interferes with transcription producing the Zscan10tm2a(EUCOMM)Wtsi constitutive knockout allele. The mouse model was validated according to standardized quality control procedures (https://www.mousephenotype.org/data/alleles/qc_data/mouse/MDNC/).
Episcopic image stacks were obtained from the Deciphering the Mechanisms of Developmental Disorders (DMDD) consortium for morphometric analysis as described previously.29,30 Ten paired landmarks and six unpaired landmarks were digitized three times on the 3D surface of each head surface using the R package digit3DLand (https://github.com/morphOptics/digit3DLand). Morphometric analysis with a focus on facial asymmetry was performed blinded for the genotype using 26 well-defined landmarks on mouse 3D surface meshes including 10 midline structures and 16 lateral/medial structures. For inner ear reconstructions, 22 points were digitized on the manually segmented models. A full generalized Procrustes superimposition with object symmetry was performed using the R package Morpho.31 A Procrustes ANOVA for object symmetry was computed to estimate fluctuating asymmetry (FA) and the among-individuals variance.32,33 The sum of squares of these variance components was decomposed according to the genotypes. FA was defined according to the following equation: FA = 2(mean squareindividual×side − mean squareerror) (Supplementary material).
Results
Genetic characterization
Genetic diagnostic testing failed to identify pathogenic or likely pathogenic variants in established disease genes that have been associated with the affected individuals’ clinical phenotypes. However, a subsequent search for genes carrying ultra-rare, predicted protein-truncating variants (PTVs) prioritized compound heterozygous or homozygous variants in ZSCAN10 in all seven affected individuals (Fig. 1 and Table 1). No homozygous PTVs in ZSCAN10 were listed in gnomAD (v2.1.1) or in-house databases. Carrier testing on available family members showed full co-segregation of the ZSCNA10 variants with the clinical phenotype (Fig. 1).
Table 1.
Clinical and genetic findings in individuals with bi-allelic ZSCAN10 variants
| Individual | F1:II.6 | F2:II.2 | F2:II.3 | F3.II.1 | F4:II.2 | F5a:II.3 | F5b:II.1 |
|---|---|---|---|---|---|---|---|
| Gender | male | female | male | male | female | female | male |
| Country of origin | Turkey | Iran | Iran | India | Pakistan | Iran | Iran |
| cDNA change(s)a | c.1112del | c.1456C>T | c.1456C>T | c.1250C>A; c.2050del | c.1456C>T | c.1456C>T | c.1456C>T |
| Protein change(s)b | p.Pro371Argfs*49 | p.Gln486* | p.Gln486* | p.Ser417*; p.His684Thrfs*153 | p.Gln486* | p.Gln486* | p.Gln486* |
| Allelic status | Homozygous | Homozygous | Homozygous | Cmpd. heterozygous | Homozygous | Homozygous | Homozygous |
| Age at onset/last examination | Congenital/9 y 6 m | Congenital/15 y | Congenital/11 y | Congenital/1 y 8 m | Congenital/2 y | Neonatal period/3 y 7 m | Congenital/10 y 3 m |
| Cognitive impairment | Moderate | Moderate, minimal speech development | Mild | Moderate | Severe, no expressive language | Severe | Severe, no expressive language |
| Delay of motor development | Moderate | Severe | Severe | Moderate | Profound | Moderate | Severe |
| Behavioural abnormalities | Autistic features, hyperphagia | Aggression | − | Autistic features, aggression | − | Stereotypic movements | − |
| Vision impairment | + | n.d. | n.d. | − | − | + | + |
| Hearing impairment | + | n.d. | n.d. | Unilateral deafness | + | − | Profound bilateral SNHL |
| Other organ systems | Micropenis, NASH | Cleft plate | Cardiac malformation | Micropenis, maldescended testis | − | Mild left ventricular enlargement | Thalassaemia minor |
| Dysmorphic facial features | Asymmetry | Asymmetry, reduced left facial movement, broad nasal root | Asymmetry, reduced left facial movement, broad nasal root | Asymmetry, reduced left facial movement, down slanting palpebral fissures, prominent epicanthic folds | Large epicanthal folds, high arched palate | − | Hypotonic facies |
| Outer ear | Unilateral malformation with angulated antihelix, overfolded helix, absent superior crus of antihelix | Bilateral malformation with low set, posteriorly rotated, angulated antihelix, overfolded helix, small ear lobes, absent superior crus of antihelix | Bilateral malformation: posteriorly rotated, left: conchal shelf, right: overfolded helix, small superior crus of antihelix | Unilateral severe dysplasia | Unilateral malformation: microtia, low-set, posteriorly rotated, absent superior crus of antihelix | Bilateral malformation: microtia, low-set, posteriorly rotated, absent superior crus of antihelix | Unilateral malformation: left microtia, dysplasia, low-set, crumpled ear |
| Inner ear | Bilateral SCCs dysplasia | n.d. | n.d. | Bilateral SCC dysplasia | n.d. | n.d. | n.d. |
Compd. compound; m = months; n.d. = not determined; NASH = non-alcoholic steatohepatosis; SCC = semicircular canal; SNHL = sensineural hearing loss; y = years; + = present; − = absent.
aGenBank: NM_032805.3.
bGenBank: NP_116194.
From the four unique ZSCAN10 variants identified, only the stop variant c.1456C>T, p.(Gln486*) was listed in gnomAD (v2.1.1; 17 times in a heterozygous state in the South Asian cohort corresponding to a minor allele frequency of 7 × 10−4 in this subpopulation.). This change was observed in a homozygous state in five affected individuals from Families F2, F4 and F5, originating from neighbouring geographic regions in Iran and Pakistan. Concordant with the reported consanguinity in Families F2 and F5, we detected extended runs of homozygosity (∼2.5 Mb to ∼8.1 Mb) encompassing ZSCAN10 and harbouring the same homozygous common variants over a 1.9 Mb region. These data are in line with a distant shared ancestry among these families.
In silico prediction of loss of ZSCAN10 function
Based on in silico predictions, loss of ZSCAN10 function is the likely pathomechanism for the identified disease alleles (Fig. 1 and Supplementary Fig. 1). The ZSCAN10 transcription unit is composed of six exons including a first non-coding exon. The last exon, exon 6, is very large and contains 66% (518 out of 780 amino acids) of the protein, including all 14 C2H2-type zinc finger motifs. Notably, all identified variants were PTVs located within exon 6, with the most 5′ variant, the 1 bp deletion c.1112del, being located 160 bp downstream of the last exon-exon junction and the most 3′ variant, the 1 bp deletion c.2050del, being located 293 bp upstream of the termination codon. According to the rules governing nonsense-mediated mRNA decay (NMD), the localization of a PTV downstream of the last exon-junction complex (more specifically, no further upstream than 50–55 nucleotides before the last exon-exon junction) and the presence of very long exons lead to reduced NMD efficiency.34,35 However, NMD predictions differ between e.g. the most upstream and downstream PTVs identified (NMDEscPredictor; Supplementary Fig. 1), and the question of whether aberrant transcripts escape NMD or not at endogenous expression levels could not be investigated for lack of a suitable model system. In case aberrant ZSCAN10 mRNAs escape NMD, translation might result in prematurely truncated polypeptides missing 13% to 56% of the C-terminal wild-type protein sequence. This region contains the 14 highly conserved C2H2-type zinc finger motifs (encoded by amino acids p.347–p.779), and even the most downstream PTV c.2050del, p.(His684Thrfs*153) alters nearly a third of them (motifs 11–14). These considerations together with the autosomal recessive interheritance patterns are in line with the hypothesis that loss of ZSCAN10 function is the likely consequence and pathomechanism of the identified disease alleles (Fig. 1).
Altered expression of stem cell marker genes in mouse embryonic stem cells due to loss of ZSCAN10
To further model the loss-of-function consequences in embryonic development, we generated Zscan10−/− mESCs lines using the CRISPR/Cas9 technique with two gRNAs targeting upstream and downstream sequences of exon 2 in which the translational start site (ATG) is located (Fig. 2A). RNA-seq confirmed loss of exon 2 expression in Zscan10−/− mESCs cell lines (Fig. 2A). The expression of other exons was also decreased (Fig. 2A), which might be due to NMD. RNA-seq analysis detected 1310 differentially expressed genes, of which 710 genes were downregulated and 600 genes were upregulated (FDR < 0.05) (Fig. 2B). KEGG pathway analysis revealed that the significantly affected pathway of the downregulated genes is ‘ATP-dependent chromatin remodeling’, while the significantly affected pathway of the upregulated genes is ‘oxidative phosphorylation’. When comparing the differentially expressed genes with ZSCAN10 target genes in mESCs,36 we observed that some of these genes related to stem cell pluripotency and differentiation were dysregulated, including Pou5f1, Sall4, Mtf2, Hoxb13 and Meis2 (Fig. 2B).36 This observation supports the function of ZSCAN10 in maintaining ESC pluripotency and regulating their differentiation.
Figure 2.
Characterization of Zscan10 in mouse endothelial stem cells and its functional impact. (A) Top: Schematic diagram showing the strategy to knock out the Zscan10 gene in mouse endothelial stem cells (mESCs). Two gRNAs targeting upstream (gRNA1) and downstream (gRNA2) of Zscan10 exon 2 were used to knock out exon 2 of the Zscan10 gene in mESCs. Bottom: RNA-sequencing (RNA-seq) data coverage plot showing the expression of the Zscan10 gene in wild-type and Zscan10 knockout mESCs (Zscan10−/−). Exon 2 of Zscan10 is lost in Zscan10−/− cell lines (grey shaded area). (B) Volcano plot showing the differentially expressed genes (DEGs) in Zscan10−/− mESCs compared with control mESCs, including 710 downregulated genes and 600 upregulated genes. The genes regulated by Zscan10 and related to pluripotency and differentiation of ESCs are labelled. FC = fold-change; FDR = false discovery rate. (C) Top: Schematic diagrams showing the structures of wild-type ZSCAN10 (ZSCAN10 full) and ZSCAN10 c.1456C>T variant (expressing only the first 485 amino acids of the ZSCAN10 protein) expression vectors. Bottom: Representative western blot showing expression of ZSCAN10 full and ZSCAN10 485 protein. (D) Chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR) shows the binding affinity of ZSCAN10 full, ZSCAN10 485 mutated protein on the Pou5f1 promoter in different vector transfected mESCs. Empty vector transfected mESCs were used as a negative control. ****P < 0.0001. (E) RNA-seq data coverage plot showing the expression of the Pou5f1 gene in wild-type and Zscan10−/− mESCs. Chr17 = chromosome 17. (F) Representative images of immunofluorescence staining show the subcellular distribution of ZSCAN10 full and ZSCAN10 485 proteins. The wild-type ZSCAN10 protein (ZSCAN10 full) is located mainly in the nucleus, whereas the mutant ZSCAN10 protein (ZSCAN10 485) is located mainly in the cytoplasm.
Impact of human ZSCAN10 protein truncating variant on subcellular distribution and DNA binding
Next, we sought to investigate the mechanism by which the identified variants could affect the biological function of ZSCAN10. We first generated a wild-type human ZSCAN10 expression vector and a mutant ZSCAN10 expression (c.1456C>T) vector (Fig. 2C). The ZSCAN10 c.1456C>T variant was found in many families in our cohort (Fig. 1) and was therefore considered representative of the pathomechanism. Western blot analysis confirmed the expression of both the full-length ZSCAN10 vector (ZSCAN10 full) and the c.1456C>T variant ZSCAN10 (ZSCAN10 485) vector (Fig. 2C). As POU5F1 is a critical target gene of ZSCAN10 (also known as ZFP206),36 we next performed ChIP-qPCR to show ZSCAN10 binding on mouse Pou5f1 gene promoter using an anti-flag antibody and mESCs transfected with different ZSCAN10 vectors. We observed that wild-type ZSCAN10 (ZSCAN10 full) strongly bound to Pou5f1 promoter but the mutant ZSCAN10 (ZSCAN10 485) lost its binding activity on the Pou5f1 promoter (Fig. 2D). RNA-seq analysis also confirmed decreased expression of Pou5f1 in Zscan10−/− mESCs (Fig. 2E and Supplementary material). In addition, we performed immunofluorescence staining to determine the subcellular distribution of the wild-type and mutant ZSCAN10. This analysis revealed that the wild-type ZSCAN10 (ZSCAN10 full) was mainly localized in nuclei, whereas the mutant ZSCAN10 (ZSCAN10 485) protein was mainly distributed in the cytoplasm. This observation supports the notion that the ZSCAN10 c.1456C>T variant disrupts the nuclear transportation of the encoded truncated protein, which could also explain why the mutant ZSCAN10 protein has lost DNA binding to the Pou5f1 promoter. These experimental data further substantiate the predicted detrimental impact of the identified variants and that loss-of-function is the likely pathomechanism underlying ZSCAN10-associated disease.
Clinical phenotype
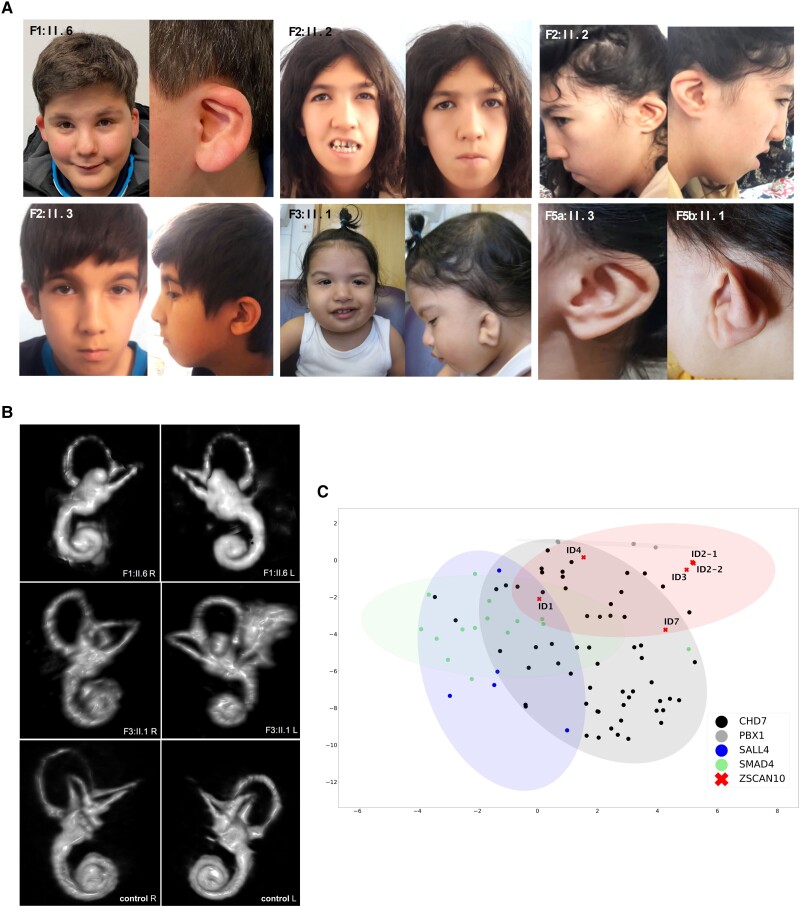
Pregnancy and birth measurements were reportedly normal in all individuals. However, due to variable congenital abnormalities including outer ear (7/7) and inner ear malformations (2/2 MRI datasets available for reanalysis) with variable hearing impairment (4/5), microgenitalia (2/7), heart defects (1/7) and cleft palate (1/7), all individuals underwent further medical examination after birth. Common dysmorphic features noted in most of the affected individuals (5/7) were facial asymmetry with unilaterally reduced facial movements or hypotonic facies, down slanting palpebral fissures and prominent epicanthic folds (Fig. 3). The dysmorphic facial features were highly consistent among the affected individuals, although of variable degrees. In Individuals F1:II.6 and F3:II.1 bilateral semicircular canal (SCC) dysplasia was diagnosed by cranial MRI (Fig. 3 and Table 1), but no other cerebral anomalies were identified. Evaluation of MRI data for soft tissues and bones revealed a subtle osseous asymmetry of the skull and midface. Uni- or bilateral hearing impairment or complete sensorineural hearing loss (SNHL) was confirmed in four individuals (4/5), however, vertigo or imbalance were not reported. Motor development of the children was markedly delayed. Individual F4:II.2 did not achieve unsupported walking. All affected individuals had a mild to severe cognitive impairment. Five of them showed delayed or minimal speech development and two individuals did not develop expressive language. Individuals F1:II.6 and F5A:II.3 exhibited behavioural abnormalities with autistic features, and Individual F1:II.6 developed excessive hyperphagia. Laboratory findings were unremarkable besides transiently elevated transaminases in Individual F1:II.6.
Figure 3.
Facial dysmorphisms and neuroimaging findings in subjects with ZSCAN10 deficiency. (A) Dysmorphic features in ZSCAN10 deficiency include facial asymmetry (Individual F1:II.6 and F3:II.1) with reduced facial movements, outer ear malformations, down slanting palpebral fissures and prominent epicanthic folds. (B) MRI-based 3D reconstruction of the inner ear. Right (R) and left (L) inner ear reconstructions showing bilateral aplasia of the horizontal semicircular canals (SCC) and dysplasia of the vestibule in Individual F1:II.6 and SCC dysplasia in Individual F3:II.1 as well as a healthy individual as the control. (C) 2D visualization of 90 images from individuals with pathogenic variants in ZSCAN10, CHD7, PBX1, SALL4 and SMAD4 by t-distributed stochastic neighbour embedding.
GestaltMatcher analysis
To validate the clinical finding of facial asymmetry in ZSCAN10 deficiency, we used the GestaltMatcher approach, an encoder for portraits.25 This analysis recapitulated the facial asymmetry as a clinical feature in all individuals. We observed that the subjects with ZSCAN10 deficiency were not highly similar to each other, except Individuals F2:II.2 and F2:II.3, who are siblings (Fig. 3). Comparing subjects with ZSCAN10 deficiency with other genetic disorders associated with string interactors of ZSCAN10 (PBX1: OMIM #617641; SALL4: OMIM #607323; and SMAD4: OMIM #139210) or suggested interacting factors (CHD7: OMIM #214800)37 (Supplementary Table 2), the clusters of individuals harbouring pathogenic variants in ZSCAN10 and CHD7 partially overlapped (Fig. 3). Individuals F1:II.6 and F3:II.1 showed a high degree of similarity and Individual F5b:II.1 also showed a certain degree of similarity to individuals with CHD7-associated CHARGE syndrome. This finding is consistent with the initial assessment of clinicians who considered CHARGE syndrome as a differential diagnosis. We then hypothesized that the similarity between ZSCAN10 and CHD7 might result from the phenotypic feature of facial asymmetry. To investigate this aspect further, we designed an experiment for this particular phenotypic feature (Supplementary material). After training the model, we tested it on the validation set of 50 images and six images of subjects with ZSCAN10 deficiency. The classification accuracy of the validation set was 82% (41/50 correct), indicating that the network learned to recognize the phenotypic feature of facial asymmetry. Moreover, all six ZSCAN10 frontal images were correctly classified.
Zscan10 knockout mouse model
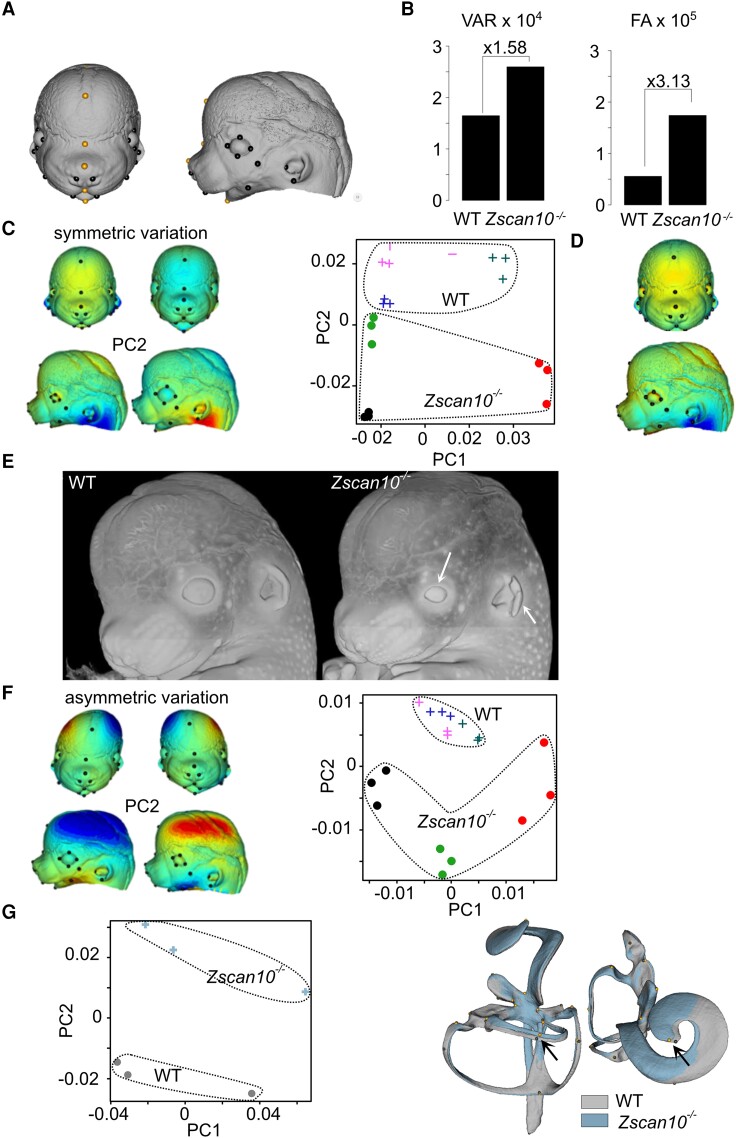
Guided by clinical findings in human, we performed expanded imaging analyses to study the involvement of Zscan10 in murine embryonic development. We used mice homozygous for a gene targeting the Zscan10 tm2a allele (Zscan10tm2a(EUCOMM)Wtsi) (Supplementary material).28 We acquired 3D facial datasets from three homozygous Zscan10-deficient mice at embryonic Day 14.5 and three matched wild-type controls.29 Symmetric and asymmetric shape variations were obtained and observed differences (both within FA or between individuals) were large and significant using Fisher’s F-test (Fig. 4B). Principal component analysis (PCA) of symmetric shape variation showed that PC2, which describes skull shape, relative eye size and ear opening, consistently opposed Zscan10-deficient to wild-type mice (Fig. 4C–F). We next assessed the ear malformation phenotype from the generalized Procrustes analysis of 22 landmarks on the 3D inner-ear model. We detected a mild difference between Zscan10-deficient and wild-type mice, with PC2 accounting for 20% of the variance corresponding to a misalignment of one of the semicircular canals and a shortening of the cochlea (Fig. 4G).
Figure 4.
Facial dysmorphism in Zscan10-deficient mouse embryos. (A) Twenty-six surface landmarks (10 paired in black plus six unpaired in orange) were affixed in triplicates on three independent wild-type (WT) and three Zscan10−/− embryonic Day (E)14.5 embryos. (B) Among-individual Procrustes variance32 and fluctuating asymmetry (FA). Variance (VAR) estimates were multiplied by 104 and FA by 105. (C) Principal components of symmetric variation. Left: Coloured meshes correspond to variation (negative–positive) along PC1 (left) or PC2 (right). Blue corresponds to contraction and red to expansion compared with the opposite side, i.e. the shape on left corner corresponds to the shape inferred for the negative PC2 and indicates the deviation from the shape on its right; for instance, in the knockout (KO) there is a relative opening of the ear compared with the WT. Right: Colours correspond to individuals (three replicates per individual): filled circles represent KO individuals and crosses represent WT individuals. (D) Differences between symmetric averages. Shape corresponds to the least-squared (LS) means of the KO and the colours show the expansion38 or contraction (blue) from the WT. Shape changes are very similar to PC2 (α = 18°) from C. (E) Representative head volumes for WT and Zscan10−/− E14.5 embryos showing smaller eye size and ear opening (white arrows). (F) Principal components of asymmetric variation indicate that Zscan10-deficient and WT mice appear to have very different asymmetry patterns, with Zscan10-deficient mice displaying larger and more fluctuating deviations. (G) Principal components of inner ear reconstitution. Average reconstitutions were computed and overlayed (grey for WT and blue for Zscan10−/−) showing a mild difference between Zscan10-deficient and WT mice with PC2 accounting for 20% of the variance corresponding to a misalignment of one of the semicircular tubes and a shortening of the cochlea (black arrow).
Discussion
The identification of four different bi-allelic PTVs in ZSCAN10 in seven affected members of five families establishes ZSCAN10 as a gene confidently implicated in this syndromic neurodevelopmental disorder. Highly consistent phenotypic features include global developmental delay, behavioural abnormalities and variable facial asymmetry with outer and inner ear malformations leading to profound SNHL. Although the expression of ZSCAN10 decreases in the later stages of human embryonic development,13-16 residual expression in the brain and testis may be associated with phenotypic features that include cognitive impairment and, in some cases, microgenitalia and maldescended testis (Table 1).
ZSCAN10 is conserved in Boreoeutheria with 71.21% sequence identity shared between Homo sapiens (NP_116194.1) and Mus musculus (NP_001028597.2). The human ZSCAN10 gene has the same number of exons as the mouse orthologue, including five translated exons and one untranslated first exon. In line with the localization of the PTVs identified in affected individuals, exon 6 and thus the zinc finger C2H2 domains were targeted in the mouse model investigated in this study.28 In a previously published mouse model, the Zscan10 knockout was induced by inserting a vector into intron 1 upstream of the ATG start codon.38 Both murine models had a C57BL/6 background and were expected to result in a complete knockout. Although they showed significant phenotypic overlap, the observed embryonic lethality in homozygous knockouts was higher in our study (Supplementary material), possibly due to dietary differences or residual amounts of ZSCAN10 derived from alternative transcripts. Owing to embryonic lethality, we cannot provide a detailed comparison to the adult mouse phenotype that was published previously.38 The behavioural alterations reported by Kraus et al.38 in homozygous mutant mice showed similarities to the human phenotype but comparison remains difficult. Other morphological abnormalities in this mouse model (e.g. organ and eye malformations) were not observed in our clinical cohort. Unfortunately, analysis of facial asymmetry and inner ear were not performed in adult mice.38 However, findings on outer and inner ear malformations as well as facial asymmetry observed in the mouse model used in this study showed striking similarities to the human phenotype of ZSCAN10 deficiency. This suggested a high degree of conservation of involved transcriptional networks between mice and humans.
To date, the exact function of ZSCAN10 is still unclear. Its interaction with SOX2 and OCT4 suggests an involvement in the pluripotency of embryonic stem cells.12,14,39 Therefore quantification of ZSCAN10 transcripts has been proposed as a highly sensitive marker for the detection of undifferentiated human induced pluripotent stem cells (iPSCs), rendering this assay an approach for quality control of iPSC-derived cell therapy products.40 Our observations in mESCs outlined that loss of ZSCAN10 dysregulates several genes associated with pluripotency and differentiation of ESCs. Nevertheless, there is evidence suggesting that ZSCAN10 might be dispensable for pluripotency of mESCs.41
Human ZSCAN10 has been characterized as a regulator of the exosome complex and has a proposed role in regulating glutathione homeostasis in aged iPSCs.17,42 Recently, an upregulation of ZSCAN10 expression in glioma cells was associated with an OCT4-dependent oncogene with excessive cell proliferation, and the authors proposed a role in the Wnt/β-catenin signalling pathway.43 Our functional studies in mESCs carrying a representative human protein truncating variant clearly indicated a critical effect on ZSCAN10 function altering its subcellular localization and impairing DNA binding, in particular to the string interacting genes. These in vitro observations strongly suggested that the identified bi-allelic variants in ZSCAN10 lead to a loss-of-function by activating the NMD pathway and/or altering the subcellular distribution of a potentially resulting truncated ZSCAN10 protein.
Using the GestaltMatcher approach, we confirmed facial asymmetry as a recognizable core feature of ZSCAN10 deficiency. This suggested that clinical diagnosis may be supported by an artificial intelligence approach. In addition, the phenotypic overlap with other molecularly defined syndromes could provide insights into shared pathomechanisms. While OCT4 and NANOG have not been implicated in human disease, heterozygous de novo variants in SOX2 cause syndromic microphthalmia-3 (MCOPS3; OMIM #206900) characterized by eye malformations as well as extraocular manifestations including intellectually disability and hearing loss (Supplementary Table 4).44 Interestingly, while the overlap between ZSCAN10- and SOX2-associated presentations seems to be currently limited to facial asymmetry in humans, mice homozygous for a gene trap Zscan10 allele were shown to phenocopy an eye malformation previously reported for Sox2 hypomorphs.38 In addition to the confirmed ZSCAN10 interaction partners, which are established (SOX2) or bona fide candidate disease genes (OCT4, NANOG), gene–disease associations have also been described for other interactors of ZSCAN10 annotated in STRING-DB. This includes the transcription factors PBX1 (congenital anomalies of kidney and urinary tract syndrome with or without hearing loss, abnormal ears, or developmental delay; OMIM #617641), SMAD4 (Myhre syndrome; OMIM #139210) and SALL4 (Duane-radial ray syndrome; OMIM #607323) (Supplementary Table 4).26 Common features of these disorders comprise developmental delay, asymmetric facial features and organ malformations. For PBX1-associated disease, malformations of the outer ears have been reported, which are reminiscent of the abnormalities observed in ZSCAN10 deficiency.45 Notably, our investigation of Zscan10−/− mESCs showed a dysregulation of Sall4 outlining their string interaction. Furthermore, as also suggested by GestaltMatcher, the affected subjects share a high degree of facial similarity with CHD7-associated CHARGE syndrome (OMIM #214800; Fig. 3), which often comprises dysplasia of the SCCs.46 So far, no direct functional link between ZSCAN10 and CHD7 has been established in humans. However, it has been shown that CHD7 co-localizes with the transcription factors OCT4, SOX2 and NANOG, which are known interaction partners of ZSCAN10.47 In mice, Zscan10 and Chd7 are both reported to maintain pluripotency during early embryonic development as interacting factors within the same transcriptional network.37,48 Comparable to the Zscan10 knockout mouse model, Chd7 mice also show morphological abnormalities as well as embryonic lethality.49,50 Although further investigations using appropriate model systems are needed to define the molecular cascades that cause pathology, our study further supports the importance of ZSCAN10 in embryonic development.
Supplementary Material
Acknowledgements
First and foremost, all authors thank the families for participating in the study. Individual F3:II.1 was collected as part of the 100 000 Genomes Project. Testing of this patient was made possible through access to the data and findings generated by the 100 000 Genomes Project. The 100 000 Genomes Project is managed by Genomics England Limited (a wholly owned company of the Department of Health and Social Care). The 100 000 Genomes Project is funded by the National Institute for Health Research and NHS England. The Wellcome Trust, Cancer Research UK and the Medical Research Council have also funded research infrastructure. The 100 000 Genomes Project uses data provided by patients and collected by the National Health Service as part of their care and support. We thank Dr Fabrice Prin for facilitating access to 3D datasets downloaded from the DMDD website.
Contributor Information
Lucia Laugwitz, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany; Department of Neuropediatrics, Developmental Neurology and Social Pediatrics, University of Tübingen, Tübingen 72076, Germany.
Fubo Cheng, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany.
Stephan C Collins, Inserm UMR1231, Université de Bourgogne, Dijon Cedex 21070, France.
Alexander Hustinx, Institute for Genomic Statistics and Bioinformatics, University Hospital Bonn, Rheinische Friedrich-Wilhelms-Universität Bonn, Bonn 53127, Germany.
Nicolas Navarro, Biogeosciences, UMR 6282 CNRS, EPHE, Université de Bourgogne, Dijon 2100, France; EPHE, PSL University, Paris 75014, France.
Simon Welsch, Department of General Pediatrics, Neonatology and Pediatric Cardiology, Medical Faculty, Heinrich-Heine-University, Düsseldorf 40225, Germany.
Helen Cox, West Midlands Regional Clinical Genetics Service and Birmingham Health Partners, Birmingham Women’s and Children’s Hospitals NHS Foundation Trust, Birmingham B15 2TG, UK.
Tzung-Chien Hsieh, Institute for Genomic Statistics and Bioinformatics, University Hospital Bonn, Rheinische Friedrich-Wilhelms-Universität Bonn, Bonn 53127, Germany.
Aswinkumar Vijayananth, Institute for Genomic Statistics and Bioinformatics, University Hospital Bonn, Rheinische Friedrich-Wilhelms-Universität Bonn, Bonn 53127, Germany.
Rebecca Buchert, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany.
Benjamin Bender, Diagnostic and Interventional Neuroradiology, Radiologic Clinics, University of Tübingen, Tübingen 72076, Germany.
Stephanie Efthymiou, Department of Neuromuscular Disorders, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
David Murphy, Department of Clinical and Movement Neurosciences, UCL Queen Square Institute of Neurology, University College London, London WC1N 3BG, UK.
Faisal Zafar, Pediatric Neurology, Children’s Hospital, Multan 60000, Pakistan.
Nuzhat Rana, Pediatric Neurology, Children’s Hospital, Multan 60000, Pakistan.
Ute Grasshoff, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany; Center for Rare Disease, University of Tübingen, Tübingen 72072, Germany.
Ruth J Falb, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany.
Mona Grimmel, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany.
Annette Seibt, Department of General Pediatrics, Neonatology and Pediatric Cardiology, Medical Faculty, Heinrich-Heine-University, Düsseldorf 40225, Germany.
Wenxu Zheng, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany.
Hamid Ghaedi, Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran 1985717443, Iran.
Marie Thirion, Inserm UMR1231, Université de Bourgogne, Dijon Cedex 21070, France.
Sébastien Couette, Biogeosciences, UMR 6282 CNRS, EPHE, Université de Bourgogne, Dijon 2100, France; EPHE, PSL University, Paris 75014, France.
Reza Azizimalamiri, Department of Pediatric Neurology, Golestan Medical, Educational, and Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz 6135715794, Iran.
Saeid Sadeghian, Department of Pediatric Neurology, Golestan Medical, Educational, and Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz 6135715794, Iran.
Hamid Galehdari, Department of Biology, Faculty of Science, Shahid Chamran University of Ahvaz, Ahvaz 6135783151, Iran.
Mina Zamani, Department of Biology, Faculty of Science, Shahid Chamran University of Ahvaz, Ahvaz 6135783151, Iran; Narges Medical Genetics and Prenatal Diagnosis Laboratory, Kianpars, Ahvaz 6155689467, Iran.
Jawaher Zeighami, Narges Medical Genetics and Prenatal Diagnosis Laboratory, Kianpars, Ahvaz 6155689467, Iran.
Alireza Sedaghat, Narges Medical Genetics and Prenatal Diagnosis Laboratory, Kianpars, Ahvaz 6155689467, Iran; Diabetes Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz 6135715794, Iran.
Samira Molaei Ramshe, Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran 1985717443, Iran.
Ali Zare, Department of Medical Genetics, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran 1985717443, Iran.
Behnam Alipoor, Department of Laboratory Sciences, Faculty of Paramedicine, Yasuj University of Medical Sciences, Yasuj 7591741417, Iran.
Dirk Klee, Department of Pediatric Radiology, Medical Faculty, Institute of Radiology, Heinrich-Heine-University, Düsseldorf 40225, Germany.
Marc Sturm, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany; Genomics England, Queen Mary University of London, London EC1M 6BQ, UK.
Stephan Ossowski, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany; NGS Competence Center Tübingen (NCCT), University of Tübingen, Tübingen 72076, Germany.
Henry Houlden, Department of Neuromuscular Disorders, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Olaf Riess, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany; Center for Rare Disease, University of Tübingen, Tübingen 72072, Germany.
Dagmar Wieczorek, Medical Faculty and University Hospital Düsseldorf, Institute of Human Genetics, Heinrich-Heine-University Düsseldorf, Düsseldorf 40225, Germany.
Ryan Gavin, West Midlands Regional Genetics Laboratory, Central and South Genomic Laboratory Hub, Birmingham B15 2TG, UK.
Reza Maroofian, Department of Neuromuscular Disorders, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Peter Krawitz, Institute for Genomic Statistics and Bioinformatics, University Hospital Bonn, Rheinische Friedrich-Wilhelms-Universität Bonn, Bonn 53127, Germany.
Binnaz Yalcin, Inserm UMR1231, Université de Bourgogne, Dijon Cedex 21070, France.
Felix Distelmaier, Department of General Pediatrics, Neonatology and Pediatric Cardiology, Medical Faculty, Heinrich-Heine-University, Düsseldorf 40225, Germany.
Tobias B Haack, Institute of Medical Genetics and Applied Genomics, University of Tuebingen, Tübingen, 72076, Germany; Center for Rare Disease, University of Tübingen, Tübingen 72072, Germany.
Data availability
Sequence datasets have been generated and contributed by different study sites and have not been deposited in a public repository due to varying local consent regulations. Depersonalized data and additional experimental data that this study is based on, can be provided upon request. All variants have been deposited into ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under Institute of Medical Genetics and Applied Genomics, University of Tübingen, including SUB11294153, SUB11294175, SUB11294201 and SUB11294215. RNA-seq data generated for this work have been deposited to GEO under accession number GSE234680 (token: sdghqywezbexpol).
Web resources
The URLs for the data presented herein are as follows:
gnomAD server, https://gnomad.broadinstitute.org
The Human Protein Atlas, https://www.proteinatlas.org
GTEx Portal, https://gtexportal.org
Online Mendelian Inheritance in Man (OMIM), https://www.omim.org
Ensembl Variant Effect Predictor, https://www.ensembl.org/Homo_sapiens/Tools/VEP
Combined Annotation Dependent Depletion, https://cadd.gs.washington.edu
UCSC, https://genome.ucsc.edu (UCSC Genome Browser on Human, GRCh38/hg38 Assembly)
STRING-DB, https://string-db.org (v11.5)
NMDEscPredictor, https://nmdprediction.shinyapps.io/nmdescpredictor/
Funding
T.B.H. was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—418081722, 433158657. The study was supported by a grant of the German Research Foundation/Deutsche Forschungsgemeinschaft (DI 1731/2-3) and by a grant from the ‘Elterninitiative Kinderkrebsklinik e.V.’ (Düsseldorf; #701900167) to F.D. This study was partially supported by a grant of Deutsche Forschungsgemeinschaft (CH2339/5-1, AOBJ690451) to F.C. The mouse studies were supported by grants from the French National Research Agency, the National Institute of Health and Medical Research and the European Union through European Regional Development Fund programs (WDR) to B.Y. D.W. is member of the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability ERN-ITHACA [EU Framework Partnership Agreement ID: 3HP-HP-FPA ERN-01-2016/739516]. F.D. is a member of the European Reference Network for Rare Hereditary Metabolic Disorders MetabERN.
Competing interests
B.B. is Co-Founder and Shareholder of AIRAmed GmbH. The other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Sheridan E, Wright J, Small N, et al. Risk factors for congenital anomaly in a multiethnic birth cohort: An analysis of the Born in Bradford study. Lancet. 2013;382:1350–1359. [DOI] [PubMed] [Google Scholar]
- 2. Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: A meta-analysis of population-based studies. Res Dev Disabil. 2011;32:419–436. [DOI] [PubMed] [Google Scholar]
- 3. Kochinke K, Zweier C, Nijhof B, et al. Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet. 2016;98:149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deciphering Developmental Disorders Study . Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaplanis J, Samocha KE, Wiel L, et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586:757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Izumi K. Disorders of transcriptional regulation: An emerging category of multiple malformation syndromes. Mol Syndromol. 2016;7:262–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nowick K, Gernat T, Almaas E, Stubbs L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci U S A. 2009;106:22358–22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al-Naama N, Mackeh R, Kino T. C2H2-Type zinc finger proteins in brain development, neurodevelopmental, and other neuropsychiatric disorders: Systematic literature-based analysis. Front Neurol. 2020;11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ. Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet. 2004;36:1053–1055. [DOI] [PubMed] [Google Scholar]
- 11. Dias C, Estruch SB, Graham SA, et al. BCL11A haploinsufficiency causes an intellectual disability syndrome and dysregulates transcription. Am J Hum Genet. 2016;99:253–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang W, Walker E, Tamplin OJ, Rossant J, Stanford WL, Hughes TR. Zfp206 regulates ES cell gene expression and differentiation. Nucleic Acids Res. 2006;34:4780–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brandenberger R, Wei H, Zhang S, et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat Biotechnol. 2004;22:707–716. [DOI] [PubMed] [Google Scholar]
- 14. Wang ZX, Kueh JL, Teh CH, et al. Zfp206 is a transcription factor that controls pluripotency of embryonic stem cells. Stem Cells. 2007;25:2173–2182. [DOI] [PubMed] [Google Scholar]
- 15. Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. [DOI] [PubMed] [Google Scholar]
- 16. Yu XF, Kim JH, Jung EJ, Jeon JT, Kong IK. Cloning and characterization of cat POU5F1 and NANOG for identification of embryonic stem-like cells. J Reprod Dev. 2009;55:361–366. [DOI] [PubMed] [Google Scholar]
- 17. Skamagki M, Correia C, Yeung P, et al. ZSCAN10 expression corrects the genomic instability of iPSCs from aged donors. Nat Cell Biol. 2017;19:1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Consortium GT. The genotype-tissue expression (GTEx) project. Nat Genet. 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Froukh T, Nafie O, Al Hait SAS, et al. Genetic basis of neurodevelopmental disorders in 103 Jordanian families. Clin Genet. 2020;97:621–627. [DOI] [PubMed] [Google Scholar]
- 20. Mencacci NE, Kamsteeg EJ, Nakashima K, et al. De Novo mutations in PDE10A cause childhood-onset chorea with bilateral striatal lesions. Am J Hum Genet. 2016;98:763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Magin TM, McWhir J, Melton DW. A new mouse embryonic stem cell line with good germ line contribution and gene targeting frequency. Nucleic Acids Res. 1992;20:3795–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng F, Zheng W, Liu C, et al. Intronic enhancers of the human SNCA gene predominantly regulate its expression in brain in vivo. Sci Adv. 2022;8:eabq6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng F, Zheng W, Barbuti PA, et al. DYT6 mutated THAP1 is a cell type dependent regulator of the SP1 family. Brain. 2022;145:3968–3984. [DOI] [PubMed] [Google Scholar]
- 25. Hsieh TC, Bar-Haim A, Moosa S, et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat Genet. 2022;54:349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 28. Skarnes WC, Rosen B, West AP, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dickinson ME, Flenniken AM, Ji X, et al. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weninger WJ, Geyer SH, Martineau A, et al. Phenotyping structural abnormalities in mouse embryos using high-resolution episcopic microscopy. Dis Model Mech. 2014;7:1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schlager S, Rüdell A. Sexual dimorphism and population affinity in the human zygomatic structure-comparing surface to outline data. Anat Rec (Hoboken). 2017;300:226–237. [DOI] [PubMed] [Google Scholar]
- 32. Bonfante B, Faux P, Navarro N, et al. A GWAS in Latin Americans identifies novel face shape loci, implicating VPS13B and a Denisovan introgressed region in facial variation. Sci Adv. 2021;7:eabc6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Klingenberg CP, Barluenga M, Meyer A. Shape analysis of symmetric structures: Quantifying variation among individuals and asymmetry. Evolution. 2002;56:1909–1920. [DOI] [PubMed] [Google Scholar]
- 34. Teran NA, Nachun DC, Eulalio T, et al. Nonsense-mediated decay is highly stable across individuals and tissues. Am J Hum Genet. 2021;108:1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindeboom RG, Supek F, Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat Genet. 2016;48:1112–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu HB, Kunarso G, Hong FH, Stanton LW. Zfp206, Oct4, and Sox2 are integrated components of a transcriptional regulatory network in embryonic stem cells. J Biol Chem. 2009;284:31327–31335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buecker C, Srinivasan R, Wu Z, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kraus P, Yu VS, B H, et al. Pleiotropic functions for transcription factor zscan10. PLoS One. 2014;9:e104568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Papatsenko D, Darr H, Kulakovskiy IV, et al. Single-cell analyses of ESCs reveal alternative pluripotent cell states and molecular mechanisms that control self-renewal. Stem Cell Reports. 2015;5:207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shi H, Feng T, Wang R, Wu L, Gu Y. Universal markers for hiPSCs residue detection. Front Biosci (Landmark Ed). 2022;27:239. [DOI] [PubMed] [Google Scholar]
- 41. Yamane M, Fujii S, Ohtsuka S, Niwa H. Zscan10 is dispensable for maintenance of pluripotency in mouse embryonic stem cells. Biochem Biophys Res Commun. 2015;468:826–831. [DOI] [PubMed] [Google Scholar]
- 42. Barcena C, Lopez-Otin C. A fruitful liaison of ZSCAN10 and ROS on the road to rejuvenation. Nat Cell Biol. 2017;19:1012–1013. [DOI] [PubMed] [Google Scholar]
- 43. Jiang Y, Huang H, Zhu X, et al. ZSCAN10 promotes cell proliferation, upregulates OCT4 expression, and activates Wnt/beta-catenin signaling in glioma. Int J Clin Exp Pathol. 2019;12:700–710. [PMC free article] [PubMed] [Google Scholar]
- 44. Williamson KA, Yates TM, FitzPatrick DR.. SOX2 Disorder. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, eds. Genereviews®. University of Washington; 1993-2024. [PubMed] [Google Scholar]
- 45. Le Tanno P, Breton J, Bidart M, et al. PBX1 haploinsufficiency leads to syndromic congenital anomalies of the kidney and urinary tract (CAKUT) in humans. J Med Genet. 2017;54:502–510. [DOI] [PubMed] [Google Scholar]
- 46. da Costa Monsanto R, Knoll RM, de Oliveira Penido N, et al. Otopathologic abnormalities in CHARGE syndrome. Otolaryngol Head Neck Surg. 2022;166:363–372. [DOI] [PubMed] [Google Scholar]
- 47. Schnetz MP, Handoko L, Akhtar-Zaidi B, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bajpai R, Chen DA, Rada-Iglesias A, et al. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hurd EA, Capers PL, Blauwkamp MN, et al. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome. 2007;18:94–104. [DOI] [PubMed] [Google Scholar]
- 50. Collins SC, Vancollie VE, Mikhaleva A, et al. Characterization of two mouse Chd7 heterozygous loss-of-function models shows dysgenesis of the corpus Callosum and previously unreported features of CHARGE syndrome. Int J Mol Sci. 2022;23:11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence datasets have been generated and contributed by different study sites and have not been deposited in a public repository due to varying local consent regulations. Depersonalized data and additional experimental data that this study is based on, can be provided upon request. All variants have been deposited into ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under Institute of Medical Genetics and Applied Genomics, University of Tübingen, including SUB11294153, SUB11294175, SUB11294201 and SUB11294215. RNA-seq data generated for this work have been deposited to GEO under accession number GSE234680 (token: sdghqywezbexpol).