Abstract
Heterozygous RTN2 variants have been previously identified in a limited cohort of families affected by autosomal dominant spastic paraplegia (SPG12-OMIM:604805) with a variable age of onset. Nevertheless, the definitive validity of SPG12 remains to be confidently confirmed due to the scarcity of supporting evidence.
In this study, we identified and validated seven novel or ultra-rare homozygous loss-of-function RTN2 variants in 14 individuals from seven consanguineous families with distal hereditary motor neuropathy (dHMN) using exome, genome and Sanger sequencing coupled with deep-phenotyping.
All affected individuals (seven males and seven females, aged 9–50 years) exhibited weakness in the distal upper and lower limbs, lower limb spasticity and hyperreflexia, with onset in the first decade of life. Nerve conduction studies revealed axonal motor neuropathy with neurogenic changes in the electromyography. Despite a slowly progressive disease course, all patients remained ambulatory over a mean disease duration of 19.71 ± 13.70 years. Characterization of Caenorhabditis elegans RTN2 homologous loss-of-function variants demonstrated morphological and behavioural differences compared with the parental strain. Treatment of the mutant with an endoplasmic/sarcoplasmic reticulum Ca2+ reuptake inhibitor (2,5-di-tert-butylhydroquinone) rescued key phenotypic differences, suggesting a potential therapeutic benefit for RTN2-disorder. Despite RTN2 being an endoplasmic reticulum (ER)-resident membrane shaping protein, our analysis of patient fibroblast cells did not find significant alterations in ER structure or the response to ER stress.
Our findings delineate a distinct form of autosomal recessive dHMN with pyramidal features associated with RTN2 deficiency. This phenotype shares similarities with SIGMAR1-related dHMN and Silver-like syndromes, providing valuable insights into the clinical spectrum and potential therapeutic strategies for RTN2-related dHMN.
Keywords: polyneuropathy, hereditary spastic paraplegia, dHMN, neurodegeneration
Maroofian et al. delineate a new form of autosomal recessive distal hereditary motor neuropathy (dHMN) with pyramidal features associated with RTN2 deficiency, and conclude that the disorder shares similarities with SIGMAR1-related dHMN and Silver-like syndromes.
Introduction
RTN2 (HGNC:10468) encodes RTN2, an endoplasmic reticulum (ER)-shaping protein localized to the ER. In maize, it binds the autophagy protein ATG8a upon ER stress.1 However, its role in ER structure is still unclear. Heterozygous RTN2 variants have previously been associated with progressive hereditary spastic paraplegia (HSP) (SPG12-MIM#604805).2,3 However, this association is yet to be confidently established.
To date, the empirical evidence includes information concerning two extended European families, encompassing a minimum of 23 individuals harbouring a segregating ultra-rare founder frameshift variant. Furthermore, there are reports of seven individuals from seven small families with four missense variants, one nonsense variant, one complete gene deletion and two frameshift variants. The reported phenotype is predominantly pure HSP with a varying age of onset ranging from 5 to 50 years old.2,3
While the co-segregation of the founder RTN2 frameshift variant in two independent extended families strongly supports its association with the HSP phenotype on chromosome 19q13, it is important to clarify that this evidence supports the linkage rather than directly proving the pathogenicity of the variant itself. Furthermore, all the reported variants are found in very low frequency in large human genetic variant databases such as gnomAD, UK-Biobank and the UK's 100 000 Genomes Project. This may be attributed to incomplete or low penetrance of the variants or their lack of pathogenicity.
Here, we identified 14 individuals from seven independent consanguineous families with homozygous loss-of-function (LoF) RTN2 variants presenting with slowly progressive distal motor neuropathy with spasticity and hyperreflexia affecting the upper and lower limbs. Analyses of fibroblasts from a patient homozygous for the RTN2 p.Gly27Ter did not reveal obvious changes in ER structure or the response to ER stress. By generating a Caenorhabditis elegans disease model containing LoF of the worm RTN2 orthologue, ret-1, we identified key behavioural and phenotypic differences in the mutant, which are amenable to drug screening efforts, and confirmed that the inhibition of sarcoplasmic reticulum (SR)/ER Ca2+ reuptake may have some therapeutic benefit in the treatment of diseases involving RTN2 variants.
Materials and methods
Patient ascertainment, clinical assessments and genetic investigation
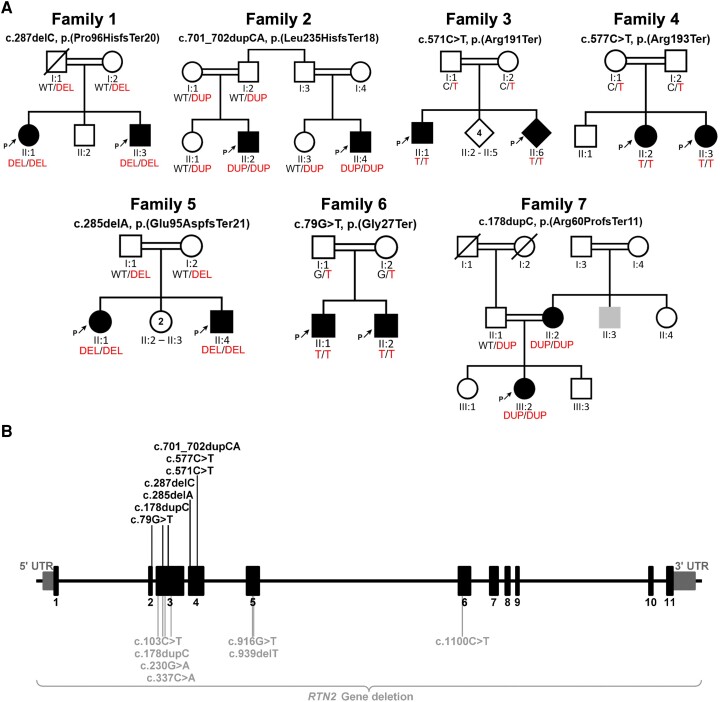
Using exome, genome, and Sanger sequencing, along with extensive data sharing with global clinical and research genetics laboratories, we identified 14 individuals (seven males and seven females) from seven consanguineous families harbouring homozygous LoF RTN2 variants (Fig. 1A). The age range of the affected individuals was 9–35 years for males and 11–50 years for females. Comprehensive data, including detailed clinical descriptions, electrodiagnostic results, brain and muscle imaging, as well as photographic and video documentation of the affected individuals, were collected. All participating families provided written informed consent.
Figure 1.
The illustration depicts family pedigrees and the RTN2 gene, highlighting the biallelic and monoallelic variants identified in this study and previous studies respectively. (A) Pedigrees of 14 individuals from seven consanguineous families with segregating homozygous RTN2 variants are shown. (B) The localization of the presented variants across the gene is displayed. With homozygous loss-of-function variant (black) identified in this study shown above the gene schematic, and previously reported heterozygous variants (grey) shown below the gene.
Cell culture and protein analysis including immunocytochemistry and immunoblotting
Human fibroblasts were cultured in Dulbecco’s Modified Eagle medium (DMEM, Gibco, 31966–021) supplemented with 10% fetal bovine serum (Biowest, S1810-500) and 1% penicillin streptomycin (Gibco, 15070–063) and maintained in a humidified atmosphere under 5% CO2 at 37°C.
For immunostaining, cells were seeded on 14 mm coverslips and fixed in 4% paraformaldehyde (PFA) for 15 min. Cells were washed three times, permeabilized with 0.25% Triton X-100 for 10 min and blocked using 5% normal goat serum (BIOZOL, S-1000) for 1 h at room temperature. Coverslips were incubated overnight with primary antibodies: sheep anti-CLIMP63 (1:1000, R&D Systems, AF-7355) and rabbit anti-NOGO (1:1000, Abcam, ab74085) diluted in blocking solution at 4°C. The next day the coverslips were washed three times and incubated with secondary antibodies conjugated with a fluorophore for 1 h at room temperature. After washing three times with 1× phosphate buffered saline (PBS), cells were stained with Hoechst (1:10,000, Invitrogen, H3569) and mounted with fluoromount mounting medium (BIOZOL, SBA-0100-01). Images were acquired with a confocal scanning fluorescence microscope (Zeiss LSM 880) with Airyscan using the Plan-Apochromat 63×/1.4 oil differential interference contrast (DIC) M27 objective and further analysed in ImageJ.
For ER stress induction and the cell viability count, cells were seeded in six-well plates and cultured to 70–80% confluency. Cells were washed with PBS and incubated with new medium supplemented with 5 µg/ml tunicamycin (Santa Cruz) without or in combination with 1 µM MG132 (Calbiochem, 474787-10MG) to inhibit the proteasome. After 48 h, the culture medium was removed, and cells were washed with PBS and trypsinized. All cells were pooled, centrifuged at 800 rpm for 5 min (Heraeus Sepatech Megafuge 2.0R) and resuspended in fresh medium. Cell viability was measured by trypan blue exclusion with an automatic counting device (Bio-Rad TC20 Automatic Cell Counter). For western blot analysis, cells from 10 cm plates were washed twice on ice in ice-cold PBS and lysed directly by adding radioimmunoprecipitation assay (RIPA) lysis buffer. Samples were prepared and subjected to sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting as described previously.4
C. elegans mutant generation and drug response analysis
A homozygous C. elegans mutant strain, ret- (syb4955), containing LoF deletion (17.8 kbp) of the entire coding region of the worm RTN2 orthologue, ret-1, was generated by SunyBiotech (Fuzhou, Fujian, China) using clustered regularly interspaced short palindromic repeats (CRISPR) in an N2 background. Worms were cultured on nematode growth medium at 20°C with Escherichia coli strain OP50 following standard methods.5 Phenotypic differences in baseline morphology, posture and locomotion of ret-1(syb4955) and differences in behaviour as the result of treatment with a panel of bioactive molecules were identified using high-resolution worm tracking coupled with automated behavioural phenotyping. Age synchronized populations of young adult worms were reared and imaged using methods previously described in detail.6 For drug experiments, worms were exposed to compounds for 4 h prior to imaging (detailed protocol: dx.doi.org/10.17504/protocols.io.dm6gpj19dgzp/v1). Videos were acquired and behavioural features extracted using a high-resolution multi-camera array tracker and Tierspy, following methods previously described in detail.7,8
Ethics
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board at University College London (#310045/1571740/37/598). All participating families provided written informed consent.
Results
Clinical phenotyping and characterization
Affected individuals were from consanguineous Iranian (two families), Indian (two families), Egyptian, Tunisian and Pakistani families. One affected sibling of the affected individual from Family 3 did not give consent and was therefore excluded from the detailed analysis (Table 1 and Fig. 2).
Table 1.
Clinical features of affected individuals with RTN2-related distal hereditary motor neuropathy
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case 1 | Case 2 | Case 3 | Case 4 | Case 5 | Case 6a | Case 7 | Case 8 | Case 9 | Case 10 | Case 11 | Case 12 | Case 13 | Case 14 | |
| Ethnic origin | Iranian | Iranian | Indian | Tunisian | Indian | Egyptian | Pakistani | |||||||
| Age of onset | 2 | 2 | 1 | 2 | 5 | – | 2 | 6 | 1–2 | 2 | 1–2 | 1–2 | 5 | ∼5 |
| Current age/sex/consanguinity | 27/F/Y | 17/M/Y | 23/M/Y | 18/M/Y | 28/M/Y | – | 11/F/Y | 13/F/Y | 35/M/Y | 45/F/Y | 11/M/Y | 9/M/Y | 16/F/Y | 50/F/Y |
| Presentation | Tiptoe walking | Tiptoe walking | Foot drop, falls, muscle wasting LL | – | Falls, gait disturbance | Gait disturbance, tiptoe walking | Tiptoe walking | Gait disturbance | Falls, foot drop | Tiptoe walking | ||||
| Ambulatory status | Walks independently | Walks independently | Walks independently | – | Walks independently | Walks independently | Walks independently | Walks independently | ||||||
| Muscle weakness | Distal LL > UL, foot drop, weak finger extensors | Distal LL > UL, foot drop, weak finger extensors | Distal LL = UL, foot drop | – | Distal LL > UL, foot drop | Distal LL = UL, foot, drop, weak finger extensors | Distal LL = UL Foot drop |
Distal LL > distal.UL > Proximal LL Weak finger extensors |
Distal LL > distal.UL > Proximal LL Weak finger extensors |
Distal LL > distal.UL > Proximal LL Foot drop Weak finger extensors |
Distal LL > distal.UL > Proximal LL Foot drop Weak finger extensors |
|||
| Muscle atrophy | Distal UL and LL | Distal UL and LL | Distal UL and LL | – | Distal UL and LL | Distal UL and LL | Distal UL and LL | Distal LL | ||||||
| Hyperreflexia | Y | Y | Y | – | Y | Y | Y | Y | ||||||
| Ankle DTRs | Absent | Absent | Absent | – | Present | Absent | Absent | Absent | ||||||
| LL spasticity | Y | Y | Y | – | Y | Y | Y | Y | ||||||
| Babinski sign | Y | Y | Y | – | Y | Y | Y | Y | ||||||
| Movement abnormalities | Action tremor UL | Action tremor UL | N | – | N | N | N | N | ||||||
| Gait | Spastic | Spastic | Spastic | – | Spastic | Spastic | Spastic | Spastic | ||||||
| Sensory deficit | N | N | N | – | N | N | N | N | ||||||
| Bladder dysfunction | N | N | N | – | N | N | N | Urgency | Urgency | |||||
| Motor delay | N | N | N | – | Delayed walking (20 months) | N | N | Delayed walking (2 years) | N | Motor delay | ||||
| Cognitive impairment | N | N | N | – | N | N | N | N | ||||||
| Skeletal system | Pes planus | Pes cavus, hammer toes | Pes planus | – | Pes planus, SBD | Pes planus, hyperlordosis, SBD | Pes planus, hammer toes | Pes planus, hammer toes | Pes cavus | |||||
| Respiratory system | N | N | N | – | N | Bronchial asthma | N | N | ||||||
| Bulbar involvement | N | N | N | – | N | N | N | N | N | |||||
| Visual system | N | N | Congenital cataract | – | N | N | N | N | ||||||
| Electrodiagnostic | Axonal motor neuropathy with acute denervation and chronic neurogenic changes | Axonal motor neuropathy with acute denervation and chronic neurogenic changes | Axonal motor-predominant neuropathy with chronic neurogenic changes | – | Normal at 8 years old | Normal at 7 years old | Axonal motor neuropathy | Axonal motor and sensory neuropathy with chronic neurogenic changes | Chronic axonal motor neuropathy | Axonal motor neuropathy with chronic neurogenic changes | Axonal motor neuropathy with chronic neurogenic changes | |||
| Muscle MRI | Fatty infiltration, muscle atrophy and oedema in calf muscle compartments | NA | NA | NA (Elevated CK) | NA | Normal | NA | |||||||
| Brain MRI | Normal | Normal | NA | Normal | NA | Normal | Mildly reduced volume of posterior periventricular white matter and CC splenium | NA | ||||||
| RTN2 variant | Homozygous c.287del, p.(Pro96HisfsTer20) | Homozygous c.701_702dup, p.(Leu235HisfsTer18) | Homozygous c.571C>T, p.(Arg191Ter) | Homozygous c.577C>T, p.(Arg193Ter) | Homozygous c.285del, p.(Glu95AspfsTer21) | Homozygous c.79G > T, p.(Gly27Ter) | Homozygous c.178dupC, p.(Arg60ProfsTer11) | |||||||
CC = corpus collosum; CK = creatine kinase; ENMG = electroneuromyography; F = female; LD = learning difficulties; LL = lower limb; M = male; N = no; NA = not available; SBD = shoulder blades detachment; UL = upper limb; Y = yes.
aNot reported details due to refused patient consent.
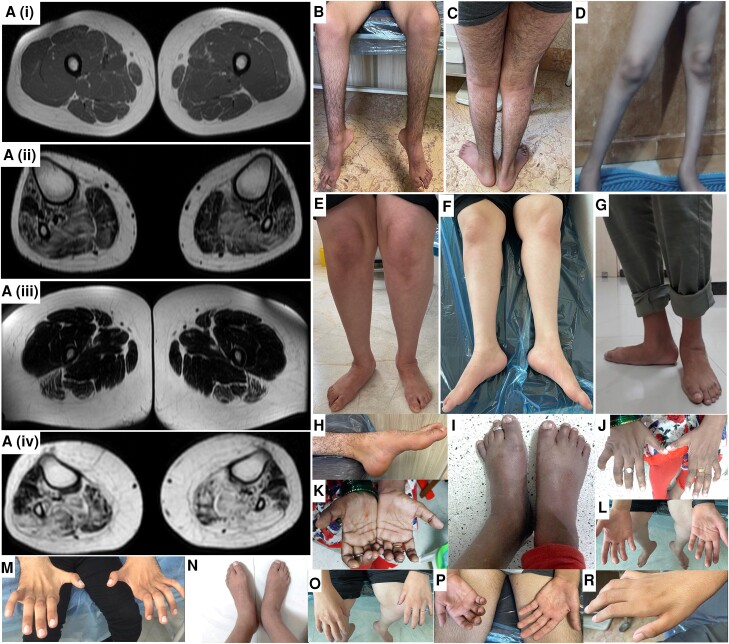
Figure 2.
Images displaying distal limbs and muscle MRI from families with RTN2-related distal hereditary motor neuropathy. Muscle weakness and wasting of the distal lower > upper limbs in affected individuals from Families 1 (A–F and M–R), 2 (G) and 3 (H–L). Axial fat T1-weighted images of the lower limbs in a 17-year-old Iranian boy [A(i and ii)] and his 27-year-old sister from Family 1 [A(iii and iv)]. The thigh muscles of the boy are normal [A(i)]. There is fatty infiltration of his leg muscles with relative sparing of the medial head of gastrocnemius and extensor digitorum longus [A(ii)]. Changes are more severe in his older sister with generalized loss of muscle bulk and fatty infiltration of the hamstrings [A(iii)]. There is more extensive change in her legs, with relative sparing of the medial head of the gastrocnemius, tibialis anterior, extensor digitorum longus and peroneus longus [A(iv)].
Consistent features included lower limb spasticity, hyperreflexia, spastic gait and weakness of both the distal upper and lower limbs (lower > upper) with an early age of onset of between 1 and 6 years. Typically, affected individuals presented with early gait disturbances (tiptoe walking, frequent falls). Foot deformities were common and included pes cavus, hammertoes or pes planus. Atrophy of distal upper and lower limb muscles, along with weakness, prominent in finger extensors, was seen in nearly all individuals (Fig. 2B–P). Sensory deficits and cranial nerve involvement were not reported in any cases. Mild motor delay was reported in three individuals and urinary urgency in one.
Nerve conduction studies confirmed axonal motor neuropathy with neurogenic changes on electromyography. In two individuals, sensory nerve conduction in the lower limbs was also reported to be abnormal. Muscle MRI in two affected siblings of Family 1 performed at the ages of 17 and 27 years showed muscle atrophy, inflammation and fatty infiltration, particularly of the calf muscles (Fig. 2A). However, hand and thigh muscle MRI images were normal in two affected siblings from Family 6 at the ages of 9 and 11 years. Brain MRI images from Families 1 and 2 were reported to be normal, while one individual (Individual 13) from Family 7 showed some non-specific neuroimaging findings.
All participants had a slowly progressive disease course and remained ambulatory at the time of the investigation; the oldest participant is currently 50 years of age. None of the affected individuals died prematurely or had any vision, hearing, swallowing or respiratory problems.
While a comprehensive neurological examination was not conducted, it is noteworthy that all parents and carrier siblings exhibited no observable neurological symptoms.
Genetic findings
Exome and genome sequencing revealed the following homozygous LoF variants in RTN2 (NM_005619.5): frameshift variants c.287del p.(Pro96HisfsTer20) and c.701_702dup p.(Leu235HisfsTer18) in Families 1 and 2, respectively; a stop-gain variant, c.571C>T p.(Arg191Ter), in Family 3; a stop-gain variant, c.577C>T p.(Arg193Ter), in Family 4; a frameshift variant, c.285del p.(Glu95AspfsTer21), in Family 5; a stop-gain variant, c.79G>T p.(Gly27Ter), in Family 6; and a frameshift variant, c.178dupC p.(Arg60ProfsTer11), in Family 7. Sanger sequencing segregation analysis demonstrated the complete co-segregation of all variants with the affected status within their respective families (Fig. 1A and B). Notably, these variants were located within sizable regions of homozygosity. Furthermore, their presence was notably absent or occurred at extremely low allele frequencies across diverse genetic variant frequency databases, including but not limited to gnomAD (versions V2, V3 and V4), UK Biobank, Queen Square Genomics, Centogene and the 100 000 Genome Project. The aggregate number of alleles assessed across these databases exceeded 2 000 000 (Supplementary Table 1).
Functional characterization
Because of the prominent role of ER shaping proteins in ER structure, we stained fibroblasts from Patient XYZ, who was homozygous for the RTN2 p.G27Ter, and control fibroblasts for the ER sheet marker CLIMP63 and the ER tubule marker RTN4 (NOGO)9 (Supplementary Fig. 2A–D). Surprisingly, both the CLIMP63- and the NOGO-labelled relative cell areas did not differ between the patient and control cells. In agreement, the abundance of CLIMP63 and NOGO did not differ in the protein lysates of patient and control fibroblasts (Supplementary Fig. 2E and F). Moreover, cell viability did not differ in steady state or upon induction of ER stress with tunicamycin (Supplementary Fig. 2G–I), suggesting that major ER functions remained intact without RTN2.
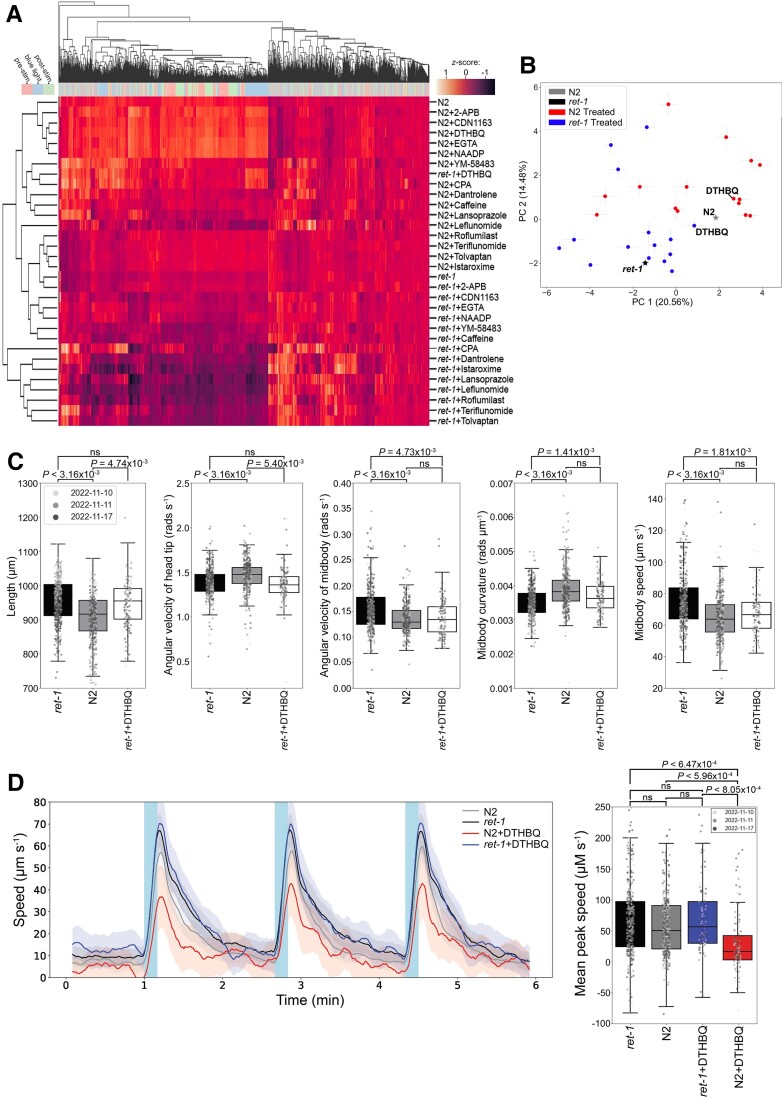
Deletion of the C. elegans RTN2 orthologue (ret-1) caused no developmental delay in worms (data not shown). Comparison of the ret-1 LoF mutant, ret-1(syb4955), and the parental strain (N2) revealed significant differences in 3345 morphological, postural and locomotive behavioural features (of 8829 in total) extracted from videos of worms crawling across an agar surface (Fig. 3). Key phenotypic behavioural differences between the strains were as follows: ret-1(syb4955) worms were longer, had decreased angular velocity of the head, increased angular velocity of the midbody and increased curvature across the body compared with N2. Although there was no significant difference in the overall baseline speed of ret-1(syb4955) and N2 (data not shown), individual body parts, primarily the head and midbody, exhibited increased speed in ret-1 LoF worms compared with N2.
Figure 3.
Loss-of-function ret-1 (RTN2 homologue) Caenorhabditis elegans phenotyping and drug screen data. (A) Hierarchical clustering of behavioural fingerprints of untreated wild-type (N2) and C. elegans RTN2 orthologue, ret-1, loss-of-function (LoF) strains, alongside treatment with the panel of 15 bioactive compounds. The heatmap shows the entire set of 8289 behavioural features extracted by Tierpsy for untreated worms and worms exposed to 100 μM of each compound for 4 h. The top dendrogram shows the relationship of the individual extracted features within the entire feature set, with the tracking period bar denoting when during image acquisition the feature was extracted: pre-stimulation (pink), during stimulation with blue light (blue) and post-stimulation (green). The left dendrogram shows the phenotypic similarity of the worms and the colour map (top left) represents the normalized z-score of the features. Despite the strains generally clustering separately from each other, treatment of ret-1 LoF mutant with 2,5-di-tert-butylhydroquinone (DTHBQ) caused this strain to cluster alongside N2, suggesting phenotypic rescue of the mutant. (B) Position of N2 and ret-1 LoF worms in phenospace with respect to the top principal components in the behavioural feature set upon drug treatment. Untreated worms are denoted by stars and treated N2 or treated ret-1(syb4955). Error bars represent the standard error of the mean. Treatment of the ret-1 LoF strain with DTHBQ moved the mutant towards the untreated wild-type in phenomic space. (C) Key morphological, postural and locomotive features were significantly different between the untreated ret-1 LoF mutant (black boxes) and untreated N2 (grey boxes). From left to right: ret-1 mutants are longer; have decreased angular velocity of the head; increased angular velocity of the midbody; decreased midbody curvature; and increased midbody speed. White boxes show ret-1(syb4955) exposed to 100 μM DTHBQ for 4 h, rescuing of 3/5 of the key behavioural features. (D) Overall speed of ret-1 LoF mutant and N2 worms after 4 h exposure to DTHBQ. Treatment of N2 with DTHBQ resulted in an attenuation of the photophobic escape response (increased speed) following stimulation of the worms with three 10 s bursts of high-intensity blue light 100 s apart (denoted by blue shaded regions on the line plot), whereas the ret-1 LoF mutant showed resistance to the effects of DTHBQ. The mean peak speed (box plot, right) was calculated as an average of the maximum detected speed across the three independent pulses of blue light. Individual points marked on all box plots are average values from multiple worms (n = 3) in a single well. The different point colours indicate data from independent experimental days. P-vales were calculated using block permutation t-tests (n = 10 000 permutations). Permutations were shuffled randomly within, but not between, the independent days of image acquisition in order to control for day-to-day variation in the experiments and corrected for multiple comparisons using the Benjamini-Hochberg procedure to control the false discovery rate at 5%. P > 0.05 was considered not statistically significant (ns).
We performed a phenotypic comparison of worms treated with a panel of 15 bioactive molecules known to be involved in intracellular calcium signalling pathways (Fig. 3). Hierarchical clustering the behavioural fingerprints of ret-1(syb4955) and N2 treated with the various compounds (all at a concentration of 100 µM) across the entire feature set revealed five distinct phenotypic clusters (Fig. 3A). There was a defined separation of the parental/mutant strains in phenomic space, although N2 treated with leflunomide or ret-1(syb4955) treated with cyclopiazonic acid (CPA) clustered as outliers. Treatment with 2-aminoethyl diphenylborinate (2-APB), CDN1163, egtazic acid (EGTA) or nicotinic acid adenine dinucleotide phosphate (NAADP) caused little change in the overall behaviour of wild-type/mutant worms. However, treatment with lansoprazole, leflunomide, roflumilast, teriflunomide, tolvaptan or istaroxime led to large behavioural differences, pushing N2 towards the mutant and ret-1(syb4955) further away from the parental strain in phenospace.
Importantly, we found that treating the ret-1 LoF mutant with 2,5-di-tert-butylhydroquinone (DTBHQ), an ER/SR Ca2+ reuptake inhibitor, caused the strain to cluster closer to N2 (Fig. 3A), resulting in a reduced number of significant behavioural differences between the mutant and N2 (1072 out of 8289). This finding was further supported by the top principal components extracted from the behavioural feature set (Fig. 3B), where there was clear movement of the ret-1 LoF mutant towards untreated N2 with respect to both principal components. Furthermore, we note that treatment with DTHBQ rescued 3/5 of the key phenotypic differences observed between N2 and ret-1(syb4955) (Fig. 3C). We noted that treatment of N2 with DTHBQ caused significant attenuation of the photophobic escape response (defined as an increase in speed following exposure of worms to bursts of high intensity blue light), whereas there was no significant difference in the photophobic escape response of untreated N2/ret-1(syb4955) or the mutant strain treated with DTHBQ (Fig. 3D). We also noted that the ret-1 LoF mutant demonstrated resistance to severe attenuation of the photophobic escape response upon treatment with 2-APB [an inositol trisphosphate (IP3) receptor and transient receptor potential (TRP) channel inhibitor; Supplementary Fig. 1]. Overall, these findings demonstrated clear movement of the disease model strain towards the healthy parental strain in overall phenospace upon treatment with DTHBQ.
Discussion
The distal hereditary motor neuropathies represent a clinically and genetically heterogeneous spectrum of rare neuromuscular disorders characterized by length-dependent predominantly motor neuropathy, manifesting as distal muscle weakness and atrophy.10-13 This spectrum is proposed to exhibit a continuum rather than discrete entities, demonstrating both clinical and genetic overlap with axonal Charcot-Marie-Tooth disease (CMT2), juvenile amyotrophic lateral sclerosis, HSP, spinocerebellar ataxia and, in certain instances, distal myopathy. These observations suggested a potential shared molecular pathogenesis among these hereditary neurological disorders.10-13
Variable pyramidal tract signs of hyperreflexia and spasticity with distal motor neuropathy can be found in association with pathogenic variants in several genes.10-12 Monoallelic pathogenic variants in BSCL2 (HGNC:15832) encoding an ER membrane protein lead to a wide spectrum of highly variable neurological disorders ranging from pure dHMN type V without spasticity and CMT2 to Silver syndrome (SPG17-MIM#270685) and ‘juvenile amyotrophic lateral sclerosis-mimics’.14 Monoallelic pathogenic SETX (HGNC:445; MIM#608465) variants can also manifest as dHMN accompanied with pyramidal signs.15
Additionally, heterozygous pathogenic variants of KIF5A (HGNC:6323), SPAST (HGNC:11233) and ATL1 (HGNC:11231) can lead to Silver-like syndrome presentations in SPG10, SPG4 and SPG3A, respectively.16-18REEP1 (HGNC:25786) variants are found in patients who occasionally present with features overlapping with dHMN-V. In these cases, early and selective involvement of the thenar and dorsalis interosseus I muscle can be seen.19 Spasticity can also rarely be observed in dominant dHMN caused by pathogenic variants of HSPB1 (HGNC:5246).20
Biallelic pathogenic variants of SIGMAR1 (HGNC:8157), encoding a transmembrane ER protein involved in ER-mitochondrial signalling, have recently been associated with autosomal recessive dHMN and Silver-like syndrome in several families.21-23 The phenotype associated with SIGMAR1, and one of the most frequent HSP subtypes, SPG11 (HGNC:11226), can also mimic juvenile amyotrophic lateral sclerosis and might be misdiagnosed as such.21,24 Furthermore, biallelic COQ7 (HGNC:2244) variants were also recently described to cause dHMN with and without pyramidal signs.25
In this report, we have delineated a new subtype of recessive dHMN with pyramidal features associated with RTN2 deficiency. The distinct and unusual clinical feature of the novel RTN2-disorder is finger extensor weakness, mimicking the phenotype of SIGMAR1-disorder. Furthermore, the phenotypic presentation mimics Silver-like diseases. However, in contrast to patients with biallelic variants in SIGMAR1, those with biallelic RTN2 pathogenic variants have a consistent spastic involvement of the legs in early childhood. Furthermore, unlike Silver-like syndromes, all affected individuals (apart from one) showed a predominantly distal lower limb weakness with early foot deformities and gait abnormalities at disease onset instead of early upper limb involvement. Interestingly, none of the heterozygous family members had any symptoms related to spasticity or neuropathy, which might be explained by incomplete or very low penetrance or a controversial role of the variants in the monoallelic state.
The muscle MRI findings in four individuals reported here, suggested an imaging phenotype consistent with length-dependent polyneuropathies in older individuals, specifically greater lower than upper limb involvement and more severe involvement distally. In the thighs, the hamstrings appeared to be involved first, while in the lower legs, the medial head of gastrocnemius and extensor digitorum longus were relatively spared. The imaging changes progressed with age and disease stage, with normal findings in the affected individuals aged 9 and 11 years from Family 6 and relatively mild changes and more severe changes in the individuals aged 17 years and 27 years from Family 1, respectively. However, confirmation of any phenotype/genotype correlation will require the analysis of MRI images from more patients.
The exact pathogenicity of RTN2-related disorder is poorly understood. RTN2 is known to be localized to the ER and involved in the dynamic remodelling of the ER.1,4 In addition, RTN2 interacts with FAM134B, a protein mediating ER-phagy. FAM134B LoF is associated ER expansion and the accumulation of misfolded proteins, and it sensitizes cells to undergo apoptosis and causes recessive hereditary sensory and autonomic neuropathy type IIB, which can present with motor symptoms such as spasticity and distal weakness.4,26-28 Our preliminary analysis, however, did not unveil major alterations to the ER structure or the response to ER stress as previously observed with FAM134B LoF.4,27
The ER plays critical roles in several cellular processes, including calcium homeostasis, and several monogenic neurodegenerative disorders, particularly those affecting upper or lower motor neurons with a variable degree of sensory involvement, have been associated with pathogenic variants in genes including ATL1, ATL3 (HGNC:24526), REEP1, REEP2 (HGNC:17975), SPAST, RTN2, ARL6IP1 (HGNC:697) and LNPK (HGNC:21610), which encode ER-resident membrane-shaping proteins characterized by reticulon homology domains.4,29 More recently, deletion of the Drosophila RTN2 orthologue was shown to alter ER organization and the function of presynaptic terminals significantly.30 Despite little change in resting Ca2+ storage capacity, this variant led to major reductions in activity-evoked Ca2+ fluxes in the cytosol, ER lumen and mitochondria. Interestingly, cytosolic and mitochondrial Ca2+ responses in transheterozygous larvae were similar to those in larvae harbouring the wild-type RTN2 orthologue, further suggesting a pathogenic role for total loss of the protein.
Here, we generated a C. elegans RTN2 orthologous (ret-1) LoF mutant and characterized morphological and behavioural differences, which could be rescued via inhibition of ER/SR Ca2+ reuptake. These data support findings from Drosophila functional studies that suggest changes in Ca2+ signalling pathways contribute towards HSP pathology,30 and as such we identified a potential benefit of inhibiting Ca2+ reuptake in the ER/SR for the treatment of RTN2-related disorders. Moreover, characterization of the behavioural differences of C. elegans ret-1 LoF mutants means they are well suited for future high-throughput compound screens to further elucidate the molecular underpinnings of this rare neurological disorder and identify additional candidate targets for treatment. Our findings delineate a new form of autosomal recessive dHMN with pyramidal features associated with RTN2 deficiency.
Supplementary Material
Acknowledgement
We are grateful for the essential support from affected individuals and families.
Contributor Information
Reza Maroofian, Centre for Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Payam Sarraf, Department of Neuromuscular Diseases, Iranian Centre of Neurological Research, Neuroscience Institute, Tehran University of Medical Sciences, Tehran 1416753955, Iran; Department of Neurology, Imam Khomeini Hospital Complex, Tehran University of Medical Sciences, Tehran 1416753955, Iran.
Thomas J O’Brien, Institute of Clinical Sciences, Imperial College London, London SW7 2AZ, UK; MRC Laboratory of Medical Sciences, London W12 0HS, UK.
Mona Kamel, Department of Pediatrics, Neurology and Metabolic Division, Kasr Alainy Faculty of Medicine, Cairo University, Cairo 4240310, Egypt.
Arman Cakar, Centre for Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Neuromuscular Unit, Istanbul Faculty of Medicine, Istanbul University, Istanbul 34093, Turkey.
Nour Elkhateeb, Department of Clinical Genetics, Cambridge University Hospitals NHS Foundation Trust, Cambridge CB2 0QQ, UK.
Tracy Lau, Centre for Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Siddaramappa Jagdish Patil, Division of Medical Genetics, Mazumdar Shaw Medical Center, Narayana Hrudayalaya Hospital, Bangalore 560099, India.
Christopher J Record, Centre for Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Alejandro Horga, Centre for Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Miriam Essid, LR18SP04, Department of Child and Adolescent Neurology, National Institute Mongi Ben Hmida of Neurology, University of Tunis El Manar, Tunis, 1007, Tunisia.
Laila Selim, Department of Pediatrics, Neurology and Metabolic Division, Kasr Alainy Faculty of Medicine, Cairo University, Cairo 4240310, Egypt.
Hanene Benrhouma, LR18SP04, Department of Child and Adolescent Neurology, National Institute Mongi Ben Hmida of Neurology, University of Tunis El Manar, Tunis, 1007, Tunisia.
Thouraya Ben Younes, LR18SP04, Department of Child and Adolescent Neurology, National Institute Mongi Ben Hmida of Neurology, University of Tunis El Manar, Tunis, 1007, Tunisia.
Giovanni Zifarelli, CENTOGENE GmbH, Rostock 18055, Germany.
Alistair T Pagnamenta, NIHR Oxford Biomedical Research Centre, Centre for Human Genetics, University of Oxford, Oxford OX3 9DU, UK.
Peter Bauer, CENTOGENE GmbH, Rostock 18055, Germany.
Mukhran Khundadze, Institute of Human Genetics, Jena University Hospital, Friedrich Schiller University, Jena 07747, Germany.
Andrea Mirecki, Institute of Human Genetics, Jena University Hospital, Friedrich Schiller University, Jena 07747, Germany.
Sara Mahmoud Kamel, Department of Radiology, Cairo University, Cairo 12613, Egypt.
Mohamed A Elmonem, Department of Clinical and Chemical Pathology, Kasr Alainy Faculty of Medicine, Cairo University, Cairo 12613, Egypt.
Ehsan Ghayoor Karimiani, Molecular and Clinical Sciences Institute, St. George’s, University of London, London SW17 0RE, UK; Innovative Medical Research Center, Mashhad Branch, Islamic Azad University, Mashhad 9187147578, Iran.
Yalda Jamshidi, Molecular and Clinical Sciences Institute, St. George’s, University of London, London SW17 0RE, UK.
Amaka C Offiah, Division of Clinical Medicine, School of Medicine & Population Health, University of Sheffield, Sheffield S10 2RX, UK.
Alexander M Rossor, Centre for Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Ilhem Ben Youssef-Turki, LR18SP04, Department of Child and Adolescent Neurology, National Institute Mongi Ben Hmida of Neurology, University of Tunis El Manar, Tunis, 1007, Tunisia.
Christian A Hübner, Institute of Human Genetics, Jena University Hospital, Friedrich Schiller University, Jena 07747, Germany; Center for Rare Diseases, Jena University Hospital, Friedrich Schiller Universität, Jena 07747, Germany.
Pinki Munot, Dubowitz Neuromuscular Centre, Great Ormond Street Hospital NHS Trust, London WC1N 3JH, UK.
Mary M Reilly, Centre for Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
André E X Brown, Institute of Clinical Sciences, Imperial College London, London SW7 2AZ, UK; MRC Laboratory of Medical Sciences, London W12 0HS, UK.
Sara Nagy, Centre for Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK; Department of Neurology, University Hospital Basel, University of Basel, Basel 4031, Switzerland.
Henry Houlden, Centre for Neuromuscular Diseases, UCL Queen Square Institute of Neurology, London WC1N 3BG, UK.
Data availability
Clinical or functional data may be shared with any qualified investigator upon request. Genome sequencing data for Family 5 is available in the National Genomic Research Library (https://doi.org/10.6084/m9.figshare.4530893.v7).
Funding
Our study is funded by the Wellcome Trust, the Medical Research Council (grant MC-A658-5TY30 to A.E.X.B.), the Multiple System Atrophy Trust, the National Institute for Health Research, UCLH Biomedical Research Centre, the Michael J Fox Foundation for Parkinson’s Research (MJFF), the Biotechnology and Biological Sciences Research Council (BBSRC), the Fidelity Trust, Rosetrees Trust, Ataxia UK, Brain Research UK, Sparks Great Ormond Street Hospital Charity, Alzheimer’s Research UK (ARUK) and CureDRPLA. Furthermore, our study received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (Grant agreement No. 714853 to A.E.X.B.). M.M.R. is grateful to the Medical Research Council (MRC MR/S005021/1), the National Institute of Neurological Disorders and Stroke and Office of Rare Disease Research (U54NS065712 and 1UOINS109403-01 and R21TROO3034), Muscular Dystrophy Association (MDA510281) and the Charcot-Marie-Tooth Association (CMTA) for their support. C.A.H. was funded by Deutsche Forschungsgemeinschaft (HU 800/15-1) and A.M. by the DAAD.
Competing interests
G.Z. and P.B. are employees of CENTOGENE. The other authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Zhang X, Ding X, Marshall RS, et al. Reticulon proteins modulate autophagy of the endoplasmic reticulum in maize endosperm. Elife. 2020;9:e51918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Montenegro G, Rebelo AP, Connell J, et al. Mutations in the ER-shaping protein reticulon 2 cause the axon-degenerative disorder hereditary spastic paraplegia type 12. J Clin Invest. 2012;122:538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tian W, Zheng H, Zhu Z, Zhang C, Luan X, Cao L. New phenotype of RTN2-related spectrum: Complicated form of spastic paraplegia-12. Ann Clin Transl Neurol. 2022;9:1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Foronda H, Fu Y, Covarrubias-Pinto A, et al. Heteromeric clusters of ubiquitinated ER-shaping proteins drive ER-phagy. Nature. 2023;618:402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stiernagle T. Maintenance of C. elegans. WormBook. 2006;11:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rosenhahn E, O’Brien TJ, Zaki MS, et al. Bi-allelic loss-of-function variants in PPFIBP1 cause a neurodevelopmental disorder with microcephaly, epilepsy, and periventricular calcifications. Am J Hum Genet. 2022;109:1421–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barlow IL, Feriani L, Minga E, et al. Megapixel camera arrays enable high-resolution animal tracking in multiwell plates. Commun Biol. 2022;5:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin K, Yao Z, van Velthoven CTJ, et al. Cell-type specific molecular signatures of aging revealed in a brain-wide transcriptomic cell-type atlas. bioRxiv. 2023. doi: 10.1101/2023.07.26.550355. [DOI] [Google Scholar]
- 9. Hubner CA, Kurth I. Membrane-shaping disorders: A common pathway in axon degeneration. Brain. 2014;137(Pt 12):3109–3121. [DOI] [PubMed] [Google Scholar]
- 10. Beijer D, Baets J. The expanding genetic landscape of hereditary motor neuropathies. Brain. 2020;143:3540–3563. [DOI] [PubMed] [Google Scholar]
- 11. Zambon AA, Pini V, Bosco L, et al. Early onset hereditary neuronopathies: An update on non-5q motor neuron diseases. Brain. 2023;146:806–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frasquet M, Rojas-Garcia R, Argente-Escrig H, et al. Distal hereditary motor neuropathies: Mutation spectrum and genotype-phenotype correlation. Eur J Neurol. 2021;28:1334–1343. [DOI] [PubMed] [Google Scholar]
- 13. Cakar A, Maroofian R, Parman Y, Reilly MM, Houlden H. Novel and nano-rare genetic causes of paediatric-onset motor neuronopathies. Brain Commun. 2024;6:fcae003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramos-Lopes J, Ribeiro J, Laco M, Alves C, Matos A, Costa C. A De Novo BSCL2 gene S90L mutation in a progressive tetraparesis with urinary dysfunction and corpus Callosum involvement. J Pediatr Genet. 2021;10:253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hadjinicolaou A, Ngo KJ, Conway DY, et al. De novo pathogenic variant in SETX causes a rapidly progressive neurodegenerative disorder of early childhood-onset with severe axonal polyneuropathy. Acta Neuropathol Commun. 2021;9:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nam DE, Yoo DH, Choi SS, Choi BO, Chung KW. Wide phenotypic spectrum in axonal Charcot-Marie-Tooth neuropathy type 2 patients with KIF5A mutations. Genes Genomics. 2018;40:77–84. [DOI] [PubMed] [Google Scholar]
- 17. Orlacchio A, Patrono C, Gaudiello F, et al. Silver syndrome variant of hereditary spastic paraplegia: A locus to 4p and allelism with SPG4. Neurology. 2008;70:1959–1966. [DOI] [PubMed] [Google Scholar]
- 18. Ivanova N, Claeys KG, Deconinck T, et al. Hereditary spastic paraplegia 3A associated with axonal neuropathy. Arch Neurol. 2007;64:706–713. [DOI] [PubMed] [Google Scholar]
- 19. Maroofian R, Behnam M, Kaiyrzhanov R, Salpietro V, Salehi M, Houlden H. Further supporting evidence for REEP1 phenotypic and allelic heterogeneity. Neurol Genet. 2019;5:e379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katz M, Davis M, Garton FC, et al. Mutations in heat shock protein beta-1 (HSPB1) are associated with a range of clinical phenotypes related to different patterns of motor neuron dysfunction: A case series. J Neurol Sci. 2020;413:116809. [DOI] [PubMed] [Google Scholar]
- 21. Ma MT, Chen DH, Raskind WH, Bird TD. Mutations in the SIGMAR1 gene cause a distal hereditary motor neuropathy phenotype mimicking ALS: Report of two novel variants. Neuromuscul Disord. 2020;30:572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Horga A, Tomaselli PJ, Gonzalez MA, et al. SIGMAR1 mutation associated with autosomal recessive silver-like syndrome. Neurology. 2016;87:1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gregianin E, Pallafacchina G, Zanin S, et al. Loss-of-function mutations in the SIGMAR1 gene cause distal hereditary motor neuropathy by impairing ER-mitochondria tethering and Ca2+ signalling. Hum Mol Genet. 2016;25:3741–3753. [DOI] [PubMed] [Google Scholar]
- 24. Khani M, Shamshiri H, Fatehi F, et al. Description of combined ARHSP/JALS phenotype in some patients with SPG11 mutations. Mol Genet Genomic Med. 2020;8:e1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rebelo AP, Tomaselli PJ, Medina J, et al. Biallelic variants in COQ7 cause distal hereditary motor neuropathy with upper motor neuron signs. Brain. 2023;146:4191–4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cakar A, Bagirova G, Durmus H, Uyguner O, Parman Y. Phenotypic features of RETREG1-related hereditary sensory autonomic neuropathy. J Peripher Nerv Syst. 2023;28:351–358. [DOI] [PubMed] [Google Scholar]
- 27. Khaminets A, Heinrich T, Mari M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. [DOI] [PubMed] [Google Scholar]
- 28. Kurth I, Pamminger T, Hennings JC, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet. 2009;41:1179–1181. [DOI] [PubMed] [Google Scholar]
- 29. Sree S, Parkkinen I, Their A, Airavaara M, Jokitalo E. Morphological heterogeneity of the endoplasmic reticulum within neurons and its implications in neurodegeneration. Cells. 2021;10:970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perez-Moreno JJ, Smith RC, Oliva MK, et al. Drosophila SPG12 ortholog, reticulon-like 1, governs presynaptic ER organization and Ca2+ dynamics. J Cell Biol. 2023;222:e202112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Clinical or functional data may be shared with any qualified investigator upon request. Genome sequencing data for Family 5 is available in the National Genomic Research Library (https://doi.org/10.6084/m9.figshare.4530893.v7).