Abstract
Background
Uterine leiomyosarcoma (uLMS) represents one of the most common sarcoma histotypes, demonstrating an overall dismal prognosis. Previous studies reported uLMS to carry recurrent somatic BRCA2 homozygous deletions, related to significant clinical benefits from the use of PARP inhibitors.
Methods
To investigate the prevalence in uLMS of genomic alterations (alt) in BRCA2 and other homologous recombination (HR) and DNA damage response (DDR) genes, cBioPortal was accessed and data were retrieved from studies including pan-sarcoma histologies. HR-/DDR-genes included BRCA1, BRCA2, ATM, BARD1, BRIP1, CHEK1, CHEK2, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM, NBN, PALB2, RAD51C, RAD51D, RAD50, and ATR. Only oncogenic/likely oncogenic alterations were included according to OncoKB.
Clinical Report and Results
We reported a clinical case of a patient affected by a highly pretreated uLMS discussed at the European Institute of Oncology Molecular Tumor Board. A targeted next-generation sequencing panel demonstrated a somatic BRCA2 homozygous deletion (homDel). Upon access to Niraparib, a remarkable response of 15 months was observed before experiencing disease progression. In the genomic query, among 2393 cases, uLMS (n = 193) displayed 9 of all 31 BRCA2alt observed, representing the only sarcoma histotype showing an enrichment in BRCA2alt (4.66%; q < 0.001). All of 9 BRCA2alt were represented by homDel, which related to a high fraction of genome altered.
Conclusion
uLMS displays a significant frequency of somatic BRCA2alt homDel. Considering their dismal prognosis, further investigation is warranted to test the use of PARPi in uLMS, and particularly in the setting of BRCA1/2 alterations.
Keywords: uterine leiomyosarcoma, niraparib, next-generation sequencing, BRCA2 homozygous deletion
This Molecular Tumor Board report describes a remarkably response achieved with the use of Niraparib in a patients affected by uterine leiomyosarcoma carrying a somatic biallelic BRCA2 alteration. In the accompanying genomic analysis, an enrichment of BRCA2 somatic alterations were observed among uterine leiomyosarcoma, confirming their prevalence and potential clinical actionability.
Key points.
Prolonged response to a poly (ADP-ribose) polymerase inhibitor was observed in a patient affected by highly pretreated uterine leiomyosarcoma (uLMS) carrying a somatic, homozygous BRCA2 deletion.
BRCA2 alterations are enriched in uLMS compared to other sarcoma histotypes.
Homozygous deletions account for most BRCA2 alterations in uLMS.
BRCA1/2 homozygous deletions yield high genomic instability.
Further investigation is mandatory for the use of PARPi in uLMS carrying BRCA2 alterations.
Case presentation
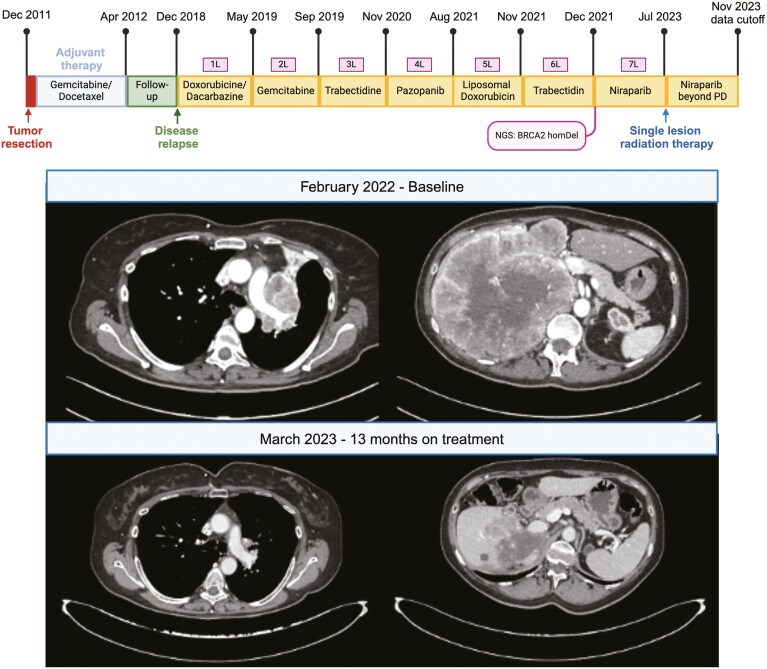
On December 2011, a 60-year-old female was diagnosed with a uterine leiomyosarcoma (uLMS) after undergoing a bilateral hysteroannessiectomy. Following surgery, 4 cycles of adjuvant therapy of gemcitabine with docetaxel were administered until May 2012.
After a negative follow-up, a relapse was observed with disease recurrence in the lungs and adrenal glands on December 2018. Subsequent treatments included doxorubicin plus dacarbazine; gemcitabine; trabectedin; pazopanib; liposomal doxorubicin; and trabectedin rechallenge, ultimately showing progressive disease on December 2021 (Figure 1).
Figure 1.
Patient oncological history and target lesions assessment.
On January 25, 2022, with no additional standard treatments available, patients received a comprehensive genomic profiling with next-generation sequencing (NGS; see Methods), whose report was referred to the European Institute of Oncology (IEO) Molecular Tumor Board (MTB). NGS was performed on the primary tumor tissue dated to the time of surgery, with genomic signatures and alterations (alt) reported in Table 1. Of note, a BRCA2 deletion was found, whose somatic origin was confirmed by a negative germline test. Considering the rationale for BRCA2 actionability and the absence of molecular alterations suggestive of primary resistance to poly (ADP-ribose) polymerase inhibitors (PARPi), an indication to the off-label use of PARPi was recommended by the MTB, which patient received from February 2022. Targeted treatment with Niraparib resulted tolerable and showed a durable radiological partial response which lasted until June 2023 (Figure 1), when disease progression occurred in a single liver lesion. Subsequent radiation therapy was administered to the progressing lesion on July 2023, with patient still receiving Niraparib 4 months after local radiation therapy.
Table 1.
Alterations found in the targeted NGS panel. VUS, variants of uncertain significance.
| Genes | Alteration | Annotation | ||
|---|---|---|---|---|
| Protein | Coding | TRANSCRIPT ID | ||
| BRCA2 | \ | Loss | \ | Pathogenic |
| ATRX | \ | c.5273-1G>A | NM_000489 | Pathogenic |
| C17orf39 | \ | Amplification | \ | Pathogenic |
| NCOR2 | \ | 6980-100_7023del144 | NM_006312 | Pathogenic |
| RB1 | \ | Loss exons 18-27 | Pathogenic | |
| TP53 | p.P278S | c.832C>T | NM_000546 | Pathogenic |
| EPHA7 | \ | Loss | \ | VUS |
| ERBB4 | p.N465K | c.1395C>A | NM_005235.3 | VUS |
| FLCN | \ | Amplification | \ | VUS |
| FLT4 | p.R1070H | \ | \ | VUS |
| MAP2K4 | Amplification | VUS | ||
| MLL2 | p.L2973P and p.P692T | \ | \ | VUS |
| SPEN | p.S2841G | \ | \ | VUS |
Methods
Next-generation sequencing platform and MTB at the European Institute of Oncology
In the presented case, blood-based FoundationOneHEME1 was used for genomic analysis. Multiplex Ligation-dependent Probe Amplification (MLPA)2 was performed on peripheral blood for germline BRCA2 testing.
IEOs MTB includes oncologists, molecular pathologists, molecular biologists, geneticists, radiotherapists, and pharmacologists, as previously reported.3
Patient discussed at the MTB whose case is reported provided informed consent. The present work was approved by the IEO internal review board and was conducted in accordance with the principles of the Declaration of Helsinki and with the principles of good clinical practice.
cBioportal genomic analysis to investigate the prevalence of alterations affecting homologous recombination/DNA damage response genes among sarcoma histologies
In our genomic analysis, our primary aim was to investigate the prevalence of BRCA2 and other homologous recombination (HR)/DNA damage response (DDR) alterations in uLMS as compared to other sarcoma histotypes. cBioPortal4,5 was queried for publicly available genomic and clinical data using the cBioPortalR package.6 Data were extracted from studies including uLMS, filtered for patients duplicated across selected repositories. HR-/DDR-genes selected in the genomic query and subsequent analysis included BRCA1, BRCA2, ATM, BARD1, BRIP1, CHEK1, CHEK2, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM, PALB2, RAD51C, RAD51D, RAD50, NBN, and ATR. Oncogenic and likely oncogenic alterations were included in the analysis according to OncoKB.7.
Statistical analysis
In the genomic analysis, categorical variables were reported as absolute number and proportion, and continuous variables as median and interquartile range. Categorical variables were compared using the Fisher’s exact test or chi-squared test, as appropriate. Bartlett test and Shapiro-Wilk test were used to assess variances and normal distributions, respectively. Non parametrical test for continuous variable included the Wilcox test and Kruskal-Wallis test. Dunn’s test was used for multiple pairwise comparisons after a significant Kruskal-Wallis test. False discovery rate was used for multiple comparisons. All tests were performed using a 2-sided significance level of <.05. Statistical analysis was performed using R Software version 4.3.2.8
Genomic analysis
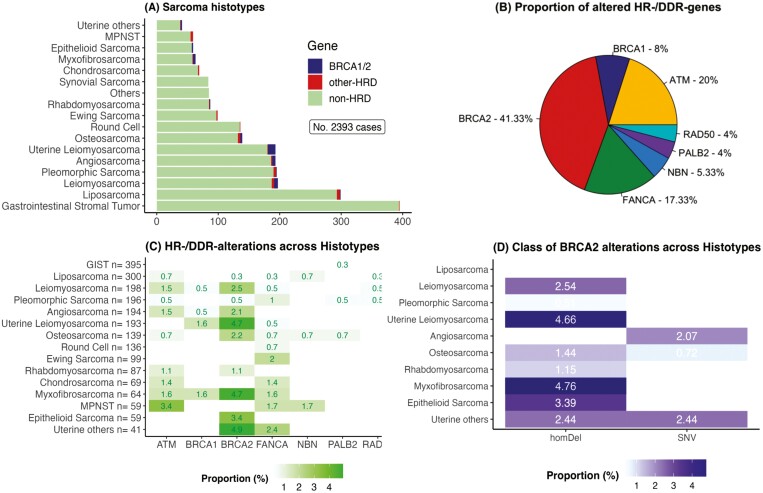
A total of 2393 patients affected by sarcoma were retrieved, among which uLMS represented the sixth most common histotype (n = 193, 8.07%; Figure 2A). Across histotypes, 75 HR-/DDR-genealt in 70 cases were observed, with BRCA2 showing the highest frequency (31/75, 41.33%; Figure 2B). LMS accounted for 48.65% (18 of 37) of all BRCA1/2alt, with 14 of 18 (77.78%) represented by BRCA2alt (Figures 2C and 3). uLMS showed a higher proportion of BRCA1/2alt compared to nonuterine Leiomyosarcoma (non-uLMS; 6.21% vs. 3.04%, P = .21). Across histotypes, both uLMS (6.21%; q < 0.01) and myxofibrosarcoma (6.35%; q = 0.01) showed an enrichment of BRCA1/2alt, while only uLMS showed enrichment in BRCA2alt when excluding BRCA1alt (uLMS 4.66%, q < 0.001; Figure 2D).
Figure 2.
Distribution of genes and classes of HR-/DDR-alterations across sarcoma histotypes. homDel, homozygous deletion; MPNST, malignant peripheral nerve sheath tumor; NO, number; SNV, single-nucleotide variant.
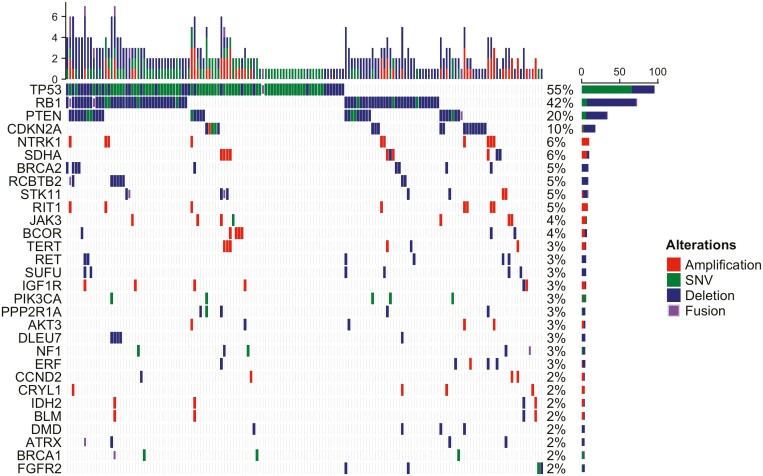
Figure 3.
Oncoprint of genomic alterations in uterine leiomyosarcoma. SNV, single-nucleotide variant.
BRCA1/2 alt classes were unevenly distributed, with homDel representing most of BRCA1/2alt (70.27%, 26 of 37, P < .001) and BRCA2alt (80.65%, 25 of 31, P < .001). Of note, all 9 BRCA2alt in uLMS consisted in homDel.
Tumors carrying BRCA1/2alt showed higher fraction genome altered (FGA) compared to HR-/DDR-wild-type tumors (0.32 [interquartile range, IQR, 0.21-0.52] vs. 0.16 [IQR 0.04-0.34]; P < .01) but not compared to non-BRCA1/2 HR-/DDR-alterations (vs. 0.29 [IQR 0.09-0.46]; P = .33). HomDel in BRCA2 yielded higher FGA compared to BRCA2 single-nucleotide variants (0.409 [IQR 0.29-0.56] vs. 0.014 [IQR0.005-0.128], P = .003).
Discussion
In the presented case, we reported a long-lasting response to PARPi in a patient affected by a highly pretreated uLMS carrying a somatic BRCA2 homozygous deletion.
Our observation is consistent with previous findings. In a case series of Seligson and colleagues,9 prolonged responses to olaparib were observed among 4 highly pretreated uLMS, with 3 of them carrying somatic BRCA2 deletions and 1 showing a truncating BRCA2 alteration. Similarly, prolonged responses to PARPi were observed among 4 uLMS demonstrating BRCA2 homDel in the study of Hensley et al,10 with other similar studies corroborating the remarkable efficacy of PARPi in this setting of disease.11,12
Several trials testing the use of PARPi in sarcomas are currently ongoing. Preliminary results of the phase II TOMAS2 trial did not demonstrate benefits from the addition of olaparib to trabectedin among 130 patients affected by sarcomas, with a 6-month PFS rate of 32% (95% CI 22%-46%) as compared to 28% (95% CI 19%-42%) in the control group (P = 0.122).13 Of note, despite preclinical evidence of trabectedin to enhance the activity of olaparib irrespective of HR-/DDR-alterations,14 no biomarker was considered for patients inclusion in the study, which could have led to a low number of cases ultimately showcasing predictive biomarkers of PARPi efficacy. Indeed, as we observed in our analysis, alterations in HR-/DDR-genes occur infrequently among sarcomas, not suggesting the indiscriminate use of PARPi in sarcomas, either alone or with chemotherapy, might yield clinical benefits and cost-effective treatment strategies.
In our analysis, across sarcoma histotypes, BRCA2 was found to be the most commonly altered HR-/DDR-gene. Noteworthy, of all BRCA2alt observed among histotypes, 29.03% (9 of 31) occurred in uLMS, found in 4.66% of cases, in line with previous reports.10,15 Specifically, in uLMS BRCA2alt represented 60% (9 of 15) of all HR-/DDR-genes defects. Therefore, HR-pathway alterations in uLMS are predominantly driven by BRCA2alt, which occur with a relevant frequency.
Of note, we observed all BRCA2alt in uLMS being represented by homDel, involving the structural deletion of both alleles. Biallelic alterations in HR-/DDR-genes are increasingly recognized as a genomic biomarker of HRD and PARPi sensitivity, and particularly for homozygous deletions preventing the occurrence of BRCA1/2 reversal alterations.16–20 Albeit germline HR-/DDR-alterations generally relate to a higher proportion of biallelic compared to monoallelic alterations, in a pancancer analysis uLMS exhibited the highest frequency of BRCA2 somatic biallelic alterations.21 In the same study, all BRCA2 alterations consisted of homDel of somatic origin,21 as we observed in our case report and genomic analysis. Accordingly, in uLMS, biallelic BRCA2alt, mainly consisting in structural variants, occurs at relevant biallelic rates despite of their somatic origin.
Besides alterations in HR-/DDR-genes, previous studies reported 25%-30% of uLMS to carry a COSMIC mutational signature 3, which acts as a genomic surrogate of HRD and PARPi responsiveness.12 Regardless, few data are available to relate signatures of HRD with PARPi sensibility in uLMS. In the study of Dall and colleagues, all 13 of 58 uLMS subjected to whole-genomic sequencing displayed a COSMIC mutational signature 3, with one patient receiving PARPi demonstrating a minor response at 4 months before interrupting the treatment due to toxicity.12 In a single-arm, phase II trial evaluating the combination of olaparib plus temozolomide in 22 uLMS, despite no BRCA1/2alt were observed, 50% of cases demonstrated HRD by RAD51 assay,22 which correlated with prolonged PFS from olaparib plus temozolomide (PFS 11.2 vs. 5.4 months; P = .05).23 Additionally, in the TOMAS2 trial, while no benefits were observed from the addition of olaparib to trabectedin in the prespecified subgroup analysis of LMS (n = 130), in an exploratory analysis a higher benefit was observed from olaparib among LMS showing HRD, defined as a level of Genomic Instability Score above the median (6-month PFS rate of 46 [95% CI 26%-83%] vs. 20% [95% CI 6%-69%], P = .053).13 Altogether, these data suggest a larger cohort of patients affected by uLMS, and possibly non-uLMS, could potentially benefit from the use of PARPi. Accordingly, substantial rationale exists for the design of clinical trials leveraging on HRD-related biomarkers for testing PARPi in uLMS in a biomarker-driven strategy.
It must be noted that our work presents some limitations. Our retrospective, exploratory analysis included a limited number of patients showing HR-/DDR-genes alterations, and thus our results should be interpreted with caution. In addition, in the genomic analysis, we could not discriminate between somatic and germline genomic alterations. Moreover, we could not distinguish allele-specific status of HR-/DDR-genes alterations, as no access to raw sequencing data was available. Lastly, our retrieved data lacked information about anti-neoplastic treatments and clinical follow-up for patients included in the genomic query.
Conclusion
Our presented case corroborates similar findings reporting the remarkable efficacy of PARP inhibitors in the context of somatic BRCA2 homozygous deletions in uLMS. In addition, our genomic analysis underscores the prevalence of BRCA2 alterations in uLMS, emphasizing the importance of genomic profiling to detect the subgroup of patients affected by uLMS which might potentially benefit from the use of PARPi. Accordingly, further research to test the use of PARPi in uLMS is demanded. Furthermore, our findings highlight the infrequent occurrence of HR-/DDR-gene alterations in sarcomas, advocating for a refined patient selection strategy in clinical trials testing the use of PARPi across sarcoma histotypes.
Acknowledgments
L.B.B. contributed with conception, data retrieval, statistical analysis, and literature research, and drafted the first version of the manuscript. G.C. contributed with conception and supervision, and drafted the first version of the manuscript. All authors provided critical revisions to the manuscript and accepted the final version. Figures and tables were created with R software version 4.3.2.
Contributor Information
Luca Boscolo Bielo, Division of New Drugs and Early Drug Development for Innovative Therapies, European Institute of Oncology, IRCCS, Milan, Italy; Department of Oncology and Hemato-Oncology, University of Milan, Milan, Italy.
Matteo Repetto, Early Drug Development Service, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Edoardo Crimini, Division of New Drugs and Early Drug Development for Innovative Therapies, European Institute of Oncology, IRCCS, Milan, Italy; Department of Oncology and Hemato-Oncology, University of Milan, Milan, Italy.
Carmen Belli, Division of New Drugs and Early Drug Development for Innovative Therapies, European Institute of Oncology, IRCCS, Milan, Italy.
Elisabetta Setola, Melanoma, Sarcoma and Rare Tumors Oncology Department, European Institute of Oncology (IEO) IRCCS, Milan, Italy.
Gabriella Parma, Department of Gynecology, European Institute of Oncology (IEO) IRCCS, Milan, Italy.
Nicola Fusco, Department of Oncology and Hemato-Oncology, University of Milan, Milan, Italy; Division of Pathology, IEO, European Institute of Oncology IRCCS, Milan, Italy.
Massimo Barberis, Division of Pathology, IEO, European Institute of Oncology IRCCS, Milan, Italy.
Elena Guerini Rocco, Department of Oncology and Hemato-Oncology, University of Milan, Milan, Italy; Division of Pathology, IEO, European Institute of Oncology IRCCS, Milan, Italy.
Antonio Marra, Division of New Drugs and Early Drug Development for Innovative Therapies, European Institute of Oncology, IRCCS, Milan, Italy.
Nicoletta Colombo, Department of Gynecology, European Institute of Oncology (IEO) IRCCS, Milan, Italy; Department of Medicine and Surgery, University of Milan-Bicocca, Milan, Italy.
Giuseppe Curigliano, Division of New Drugs and Early Drug Development for Innovative Therapies, European Institute of Oncology, IRCCS, Milan, Italy; Department of Oncology and Hemato-Oncology, University of Milan, Milan, Italy.
Funding
None declared.
Conflicts of interest
M.R. received travel expenses reimbursement from Sanofi. E.G.-R. has received honoraria and/or advisory fees and/or research funding from AstraZeneca, Exact Sciences, Novartis, Roche, and ThermoFisher. N.C. reports personal fees from AstraZeneca, MSD, Roche, Tesaro, GSK, Clovis Oncology, PharmaMar, Pfizer, Amgen, Novartis, Biocad, and Immunogen. G.C. received honoraria for speaker’s engagement: Roche, Seattle Genetics, Novartis, Lilly, Pfizer, Foundation Medicine, NanoString, Samsung, Celltrion, BMS, MSD; Honoraria for providing consultancy: Roche, Seattle Genetics, NanoString; Honoraria for participating in Advisory Board: Roche, Lilly, Pfizer, Foundation Medicine, Samsung, Celltrion, Mylan; Honoraria for writing engagement: Novartis, BMS; Honoraria for participation in Ellipsis Scientific Affairs Group; Institutional research funding for conducting phase I and II clinical trials: Pfizer, Roche, Novartis, Sanofi, Celgene, Servier, Orion, AstraZeneca, Seattle Genetics, AbbVie, Tesaro, BMS, Merck Serono, Merck Sharp Dome, Janssen-Cilag, Philogen, Bayer, Medivation, and Medimmune. The remaining authors have no conflict of interest to declare.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. He J, Abdel-Wahab O, Nahas MK, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood. 2016;127(24):3004-3014. 10.1182/blood-2015-08-664649` [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stuppia L, Antonucci I, Palka G, Gatta V.. Use of the MLPA assay in the molecular diagnosis of gene copy number alterations in human genetic diseases. Int J Mol Sci . 2012;13(3):3245-3276. 10.3390/ijms13033245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Repetto M, Crimini E, Boscolo Bielo L, et al. Molecular tumour board at European Institute of Oncology: report of the first three year activity of an Italian precision oncology experience. Eur J Cancer. 2023;183:79-89. 10.1016/j.ejca.2023.01.019 [DOI] [PubMed] [Google Scholar]
- 4. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1-pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cerami E, Gao J, Dogrusoz U, et al. The cBio Cancer Genomics Portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401-404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Karissa Whiting. cbioportalR: Browse and Query Clinical and Genomic Data from cBioPortal. R package version 110. https://www.karissawhiting.com/cbioportalR/,https://github.com/karissawhiting/cbioportalR. Accessed November 3,2023. [Google Scholar]
- 7. Chakravarty D, Gao J, Phillips S, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017;1(1):1-16. 10.1200/po.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2022. https://www.R-project.org. Accessed November 3, 2023. [Google Scholar]
- 9. Seligson ND, Tang J, Jin DX, et al. Drivers of genomic loss of heterozygosity in leiomyosarcoma are distinct from carcinomas. NPJ Precis Oncol. 2022;6(1):29. 10.1038/s41698-022-00271-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hensley ML, Chavan SS, Solit DB, et al. Genomic landscape of uterine sarcomas defined through prospective clinical sequencing. Clin Cancer Res. 2020;26(14):3881-3888. 10.1158/1078-0432.CCR-19-3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Seligson ND, Kautto EA, Passen EN, et al. BRCA1/2 functional loss defines a targetable subset in leiomyosarcoma. Oncologist. 2019;24(7):973-979. 10.1634/theoncologist.2018-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dall G, Vandenberg CJ, Nesic K, et al. Targeting homologous recombination deficiency in uterine leiomyosarcoma. J Exp Clin Cancer Res. 2023;42(1):112. 10.1186/s13046-023-02687-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. D’Ambrosio L, Merlini A, Brunello A, et al. LBA91 TOMAS2: a randomized phase II study from the Italian Sarcoma Group (ISG) of trabectedin plus olaparib (T+O) or trabectedin (T) in advanced, metastatic, or unresectable soft tissue sarcomas (STS) after failure of standard treatments. Ann Oncol. 2023;34:S1332. [Google Scholar]
- 14. Pignochino Y, Capozzi F, D’Ambrosio L, et al. PARP1 expression drives the synergistic antitumor activity of trabectedin and PARP1 inhibitors in sarcoma preclinical models. Mol Cancer. 2017;16(1):86. 10.1186/s12943-017-0652-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nacev BA, Sanchez-Vega F, Smith SA, et al. Clinical sequencing of soft tissue and bone sarcomas delineates diverse genomic landscapes and potential therapeutic targets. Nat Commun. 2022;13(1):3405. 10.1038/s41467-022-30453-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van der Wijngaart H, Hoes LR, van Berge Henegouwen JM, et al. Patients with biallelic BRCA1/2 inactivation respond to olaparib treatment across histologic tumor types. Clin Cancer Res. 2021;27(22):6106-6114. 10.1158/1078-0432.CCR-21-1104 [DOI] [PubMed] [Google Scholar]
- 17. Sokol ES, Pavlick D, Khiabanian H, et al. Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol. 2020;442(4):442-465. 10.1200/po.19.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stover EH, Konstantinopoulos PA, Matulonis UA, Swisher EM.. Biomarkers of response and resistance to DNA repair targeted therapies. Clin Cancer Res. 2016;22(23):5651-5660. 10.1158/1078-0432.CCR-16-0247 [DOI] [PubMed] [Google Scholar]
- 19. Boscolo Bielo L, Trapani D, Repetto M, et al. Variant allele frequency: a decision-making tool in precision oncology? Trends Cancer. 2023;9(12):1058-1068. 10.1016/j.trecan.2023.08.011 [DOI] [PubMed] [Google Scholar]
- 20. Harvey-Jones E, Raghunandan M, Robbez-Masson L, et al. Longitudinal profiling identifies co-occurring BRCA1/2 reversions, TP53BP1, RIF1 and PAXIP1 mutations in PARP inhibitor-resistant advanced breast cancer. Ann Oncol. 2024;35(4):364-380. 10.1016/j.annonc.2024.01.003 [DOI] [PubMed] [Google Scholar]
- 21. Jonsson P, Bandlamudi C, Cheng ML, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571(7766):576-579. 10.1038/s41586-019-1382-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Castroviejo‐Bermejo M, Cruz C, Llop‐Guevara A, et al. A RAD 51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med. 2018;10(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ingham M, Allred JB, Chen L, et al. Phase II study of olaparib and temozolomide for advanced uterine leiomyosarcoma (NCI Protocol 10250). J Clin Oncol. 2023;41(25):4154-4163. 10.1200/JCO.23.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.