Abstract
Herpes simplex virus (HSV) ICP27 is an essential and multifunctional regulator of viral gene expression that modulates RNA splicing, polyadenylation, and nuclear export. We have previously reported that ICP27 causes the cytoplasmic accumulation of unspliced α-globin pre-mRNA. Here we examined the effects of a series of ICP27 mutations that alter important functional regions of the protein on the processing and nuclear transport of α-globin and HSV ICP0 RNA. The results demonstrate that ICP27 mutants that are impaired for growth in noncomplementing cells, including mutants in the N- and C-terminal regions, are defective in the accumulation of α-globin pre-mRNA. Unexpectedly, several mutants that are competent to repress the expression of reporter genes in transient transfection assays failed to accumulate unspliced RNA, implying that different mechanisms are responsible for transrepression and pre-mRNA accumulation. Several mutants caused a marked increase in the length and heterogeneity of the α-globin mRNA poly(A) tail, suggesting that ICP27 may directly or indirectly affect the regulation of poly(A) polymerase. ICP27 was also required for the accumulation of multiple ICP0 intron-bearing transcripts, but this effect displayed a mutational sensitivity profile different from that of accumulation of unspliced α-globin RNA. Moreover, unlike spliced and unspliced α-globin RNAs, which were efficiently exported to the cytoplasm, spliced and intron-containing ICP0 transcripts were predominantly nuclear in localization, and ICP27 was not required for nuclear retention of the spliced message. We propose that these transcript- and ICP27 allele-specific differences may be explained by the presence of a strong cis-acting ICP27 response element in the α-globin transcript.
Herpes simplex virus (HSV) is the prototypical member of the Herpesviridae, a large group of enveloped nuclear DNA viruses that infect a wide range of metazoan organisms. Like all herpesviruses, HSV displays both lytic and latent modes of interaction with its natural human host (reviewed in reference 46). The HSV lytic cycle involves a complex genetic program encompassing a variety of transcriptional and posttranscriptional controls (reviewed in references 13, 46, and 63): expression of most cellular genes is strongly suppressed, and three temporal classes of viral genes are sequentially activated in a regulatory cascade. Five immediate-early (IE) genes are expressed first, through the transactivation function of the virion protein VP16 in combination with cellular factors. Four of the IE gene products (ICP0, ICP4, ICP22, and ICP27) are nuclear regulatory proteins that orchestrate the timely expression of the early (E) and late (L) genes.
The IE protein ICP27 is essential for the viability of the virus in cultured cells, and ICP27 homologs are present in all of the mammalian and avian herpesvirus genomes that have been characterized to date. ICP27 plays a fundamental and multifunctional role in the viral life cycle that has yet to be completely defined. HSV type 1 (HSV-1) ICP27 null mutants display reduced levels of some E and most L mRNAs and are defective in viral DNA replication (12, 26, 30, 41, 42, 48, 53, 59). In addition, they fail to efficiently suppress cellular gene expression (16, 18, 48). The modes of action of ICP27 in these various processes are not completely clear; however, it is becoming increasingly apparent that ICP27 functions primarily to modulate the posttranscriptional processing and transport events that are required to convert nuclear primary transcripts into functional mRNA molecules in the cytoplasm. It has been recognized for some time that ICP27 can activate or repress the expression of reporter genes driven by HSV promoters in transient cotransfection assays (17, 30, 41, 42, 45, 53, 58). Although it was initially thought that the activation and repression was at the transcriptional level, it is now clear that ICP27 exerts these effects at least in part by modulating the processes of polyadenylation and splicing (18, 27, 28, 37, 39, 51, 52). Rather than being promoter dependent, activation of gene expression results from the enhancement of the selection and cleavage of weak poly(A) sites (27–29, 52). The repression function correlates with the presence of introns in the reporter genes (52), a finding that led to the view that ICP27 represses expression of intron-bearing genes by inhibiting RNA splicing. A significant amount of data has supported the notion that ICP27 impairs or modulates splicing: (i) ICP27 colocalizes with and redistributes snRNPs in HSV-infected cell nuclei (37); (ii) ICP27 coimmunoprecipitates with splicing factors that react with anti-Sm antisera and appears to alter the phosphorylation status of some of these proteins (50); and (iii) nuclear extracts prepared from cells infected with wild-type HSV carry out in vitro splicing reactions less efficiently than those prepared from uninfected cells, and this reduction requires ICP27 (18). It has been suggested that inhibition of splicing by ICP27 is responsible for the delayed shutoff of cellular gene expression that occurs during HSV infection (52). This is an appealing idea because unlike cellular genes, the majority of HSV genes do not contain introns, and thus HSV gene expression would be relatively resistant to inhibition. Consistent with this hypothesis, ICP27 mutants fail to induce the decline in the levels of cellular mRNAs characteristic of infection with wild-type HSV (16, 18).
ICP27 is a nuclear/cytoplasmic shuttling protein (32, 38, 49, 55) and has been shown to bind RNA through a region rich in arginine and glycine residues, the RGG box (33). A model has emerged recently suggesting that these activities mediate the cytoplasmic accumulation of intronless viral L RNAs in a fashion similar to that for the human immunodeficiency virus (HIV) Rev protein (49, 55, 57). ICP27 may also play a role in increasing the stability of certain RNAs (1).
In keeping with its multifunctional role in viral gene expression, the 512-residue ICP27 protein is composed of multiple functional regions that confer several possibly independent properties on the protein. For example, ICP27's nuclear/cytoplasmic shuttling ability is conferred by a leucine-rich nuclear export signal (NES) at the N terminus similar to that of the HIV Rev protein (49), in combination with multiple nuclear localization signals (NLS), including a strong NLS localized to amino acids 110 to 137 (31). The RGG box corresponds to residues 138 to 152 (33). The C-terminal half of the molecule has been implicated in the above-mentioned effects on polyadenylation (the activation function) and splicing (the repression function) (17, 30, 42, 45), although residues in the N terminus also appear to contribute to activation (45) and repression (44). Not surprisingly in view of the complex functions it fulfills, ICP27 has been shown to interact with numerous other viral and cellular proteins. Physical interactions with the main viral transactivator, ICP4, have been documented (36). Furthermore, it has recently been shown that ICP27 can self-associate (65). Interactions with proteins of the splicing apparatus (50), hnRNP K and casein kinase 2 (61), have also been demonstrated, all of which may contribute in various ways to the functions of ICP27.
We have been investigating how ICP27 affects mRNA processing and nuclear export of the transcript encoded by the cellular α-globin gene. Normally silent in cells of nonerythroid lineage, the α-globin gene is induced during HSV infection by the actions of the IE proteins ICP0, ICP4, and ICP22, leading to accumulation of correctly initiated RNAs (6). We have recently reported that ICP27, while having little effect on the levels of spliced α-globin RNA, causes cytoplasmic accumulation of unspliced α-globin pre-mRNA (5). This observation was surprising, for two reasons. First, a large body of evidence demonstrates that transcripts of most intron-bearing cellular genes must be processed by the splicing apparatus in order to access the nuclear export machinery (2, 7–9, 14, 19, 20, 22, 25, 35, 47). The process of splicing itself appears to be required, as evidenced by the finding that cDNA copies of many intron-bearing genes fail to direct the accumulation of stable cytoplasmic RNA, and this defect can be rescued by placing a heterologous intron in the transcription unit (14). Thus, our finding that ICP27 promotes cytoplasmic accumulation of unspliced α-globin RNA suggested that ICP27 provides a novel splicing-independent pathway for RNA export. Second, previous studies by other investigators have been interpreted to indicate that ICP27 induces nuclear retention of intron-bearing transcripts of the HSV genes encoding ICP0 and UL15 (18, 39, 49). This effect, ascribed to a global inhibition of splicing mediated by ICP27, was taken to suggest that the RNA transport function of ICP27 distinguishes between transcripts arising from intron-bearing and intronless genes and is capable of transporting only the latter. In contrast, our data were more compatible with the hypothesis that ICP27 stimulates splicing-independent transport of a specific subset of RNAs, irrespective of the presence or absence of introns.
Our hypothesis that ICP27 causes the accumulation of α-globin pre-mRNA by inducing a splicing-independent RNA transport system (rather than by inhibiting the process of splicing per se) raised the possibility that accumulation of unspliced α-globin RNA could be uncoupled by mutation from the previously described transrepression function of ICP27, which has been attributed to direct inhibition of splicing. In addition, the contrast between our results and those previously reported for intron-bearing transcripts of the HSV ICP0 and UL15 genes suggested that ICP27 can discriminate between different intron-bearing RNAs. Here we present the results of experiments that test these predictions, by examining the effects of a large panel of ICP27 mutations on the processing and transport of α-globin and ICP0 RNAs.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Vero cells were propagated in DMEM containing 5% FBS. The HSV-1 wild-type strain used in this study was KOS1.1 (24). The HSV strains bearing mutated versions of the ICP27 gene are shown in Fig. 1. Mutants d27-1, n59R, n263R, n406R, and n504R (42, 45), d1-2 (44), d3-4 and d4-5 (31), and M11, M15, and M16 (43) have been described elsewhere. Construction of the d1-5, d2-3, d5-6, and d6-7 viruses (S. A. Rice, V. Leong, and C. Guy, unpublished data) was analogous to that of the d3-4 and d4-5 viruses (31). The M1X and M2X viruses were constructed as follows. Plasmid pM1 (43) was modified by inserting an NheI linker having stop codons in all three reading frames into the engineered XhoI site in the ICP27 gene at codons 11 and 12. A recombinant HSV bearing this version of ICP27 was generated and designated M1X. This virus produces a truncated ICP27 protein that reacts with monoclonal antibody H1113 (which recognizes residues 109 to 137) but not H1119 (which recognizes residues 1 to 11) (S. A. Rice, unpublished data; 31). Thus, we assume that translation begins at the downstream AUG at codon 50 and the truncated ICP27 protein lacks its N-terminal 49 amino acids. Virus M2X was constructed in a similar fashion except that the NheI linker was inserted at the XhoI site of pM2 (43), which is at codons 63 and 64. This virus does not make any detectable ICP27 protein (Rice, unpublished). All ICP27 mutant strains were propagated in V27 cells, which are derivatives of Vero cells engineered to express ICP27 upon infection with HSV (42). V27 cells were maintained in DMEM containing 5% FBS and 100 μg of Geneticin (G418; Gibco-BRL) per ml. Infections of HeLa cells (∼80% confluent) were carried out at a multiplicity of 10 PFU/cell, and in some experiments phosphonoacetic acid (PAA; 300 μg/ml) was included in the medium to inhibit viral DNA replication.
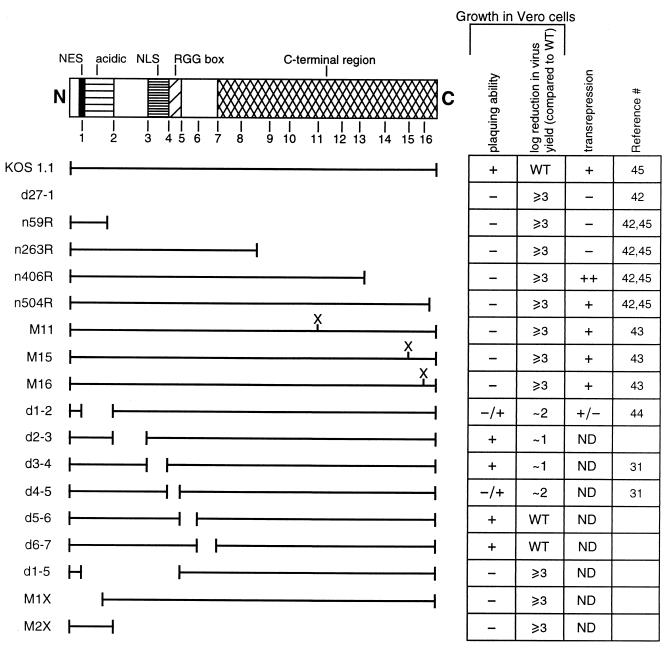
FIG. 1.
HSV-1 recombinants encoding altered forms of ICP27. Shown is a schematic diagram of the ICP27 polypeptide and the approximate positions of the NES, N-terminal acidic region, NLS, RGG box and C-terminal region. The positions of 16 XhoI sites engineered in a family of mutant ICP27 plasmids (43) are indicated. Mutants M11, M15, and M16 contain one or two altered amino acids as a consequence of the engineered XhoI sites at positions 11, 15, and 16, respectively. The sites were also used to construct the in-frame deletion mutants shown. The portion of the ICP27 polypeptide that is encoded in each mutant virus is shown by the horizontal lines. The growth characteristics of each mutant virus in Vero cells are indicated by their ability to form plaques and their virus yield relative to the yield of the wild-type (WT) virus. The ability of the ICP27 mutants to carry out the transrepression function of ICP27, as defined by transient transfection assays using reporter genes, is indicated. ND, not determined.
Isolation of RNA and Northern blot analysis.
Total RNA was harvested from infected HeLa cells in 100-mm-diameter dishes using the Trizol reagent (Gibco-BRL). Poly(A)+ RNA was selected from total RNA using an Oligotex mRNA isolation kit (Qiagen). Nuclear and cytoplasmic RNA fractions were isolated from infected HeLa cells in 60-mm-diameter dishes using an RNeasy purification kit (Qiagen). Briefly, infected cells were trypsinized, pelleted, and lysed in a buffer containing 0.5% NP-40, followed by centrifugation to pellet the nuclei. Cytoplasmic RNA was prepared from the supernatant according to the protocol given in the RNeasy handbook. The pelleted nuclei from the cell fractionation protocol were processed as described in the RNeasy handbook for whole cells, to give the nuclear RNA fraction. In some experiments, RNA was treated with RNase H in the presence of oligo(dT) as previously described (5) to remove the poly(A) tail prior to Northern blot analysis. RNA samples (10 μg of total RNA or the corresponding cell equivalent of nuclear or cytoplasmic RNA) were electrophoresed on a 1 or 1.5% agarose-formaldehyde gel followed by blotting to a Genescreen membrane (NEN). The blot was hybridized to radiolabeled probes specific for various RNA sequences. The probe used to detect α-globin transcripts was a 1.5-kb PstI fragment bearing the entire human α2-globin gene purified from pUCα2 (6). All double-stranded ICP0 probes were fragments purified from plasmid pSHZ, which bears the ICP0 open reading frame and surrounding sequences (34). The ICP0 exon/intron probe was a 1,078-bp NcoI-XhoI fragment; the ICP0 intron probe was a 165-bp PshAI fragment; the ICP0 upstream probe was a 160-bp SphI-DrdI fragment (see Fig. 4A). To prepare the ICP0 intron-specific riboprobe, plasmid pBS-9 was constructed by cloning a 165-bp PshAI fragment from pSHZ (blunt ended with the large fragment of Escherichia coli DNA polymerase I) into the SmaI site of the vector pBluescript II SK+ (Stratagene). A strand-specific riboprobe was obtained by transcribing EcoRI-linearized pBS-9 with T3 RNA polymerase in the presence of [32P]CTP, followed by purification on a NucTrap column (Stratagene). To detect thymidine kinase (TK) transcripts, a 662-bp SstI/SmaI fragment from plasmid pTK173 (60) was used. All hybridizations with double-stranded DNA probes or the riboprobe were carried out in Church buffer (11) at 65°C. To probe for the U3 small nucleolar RNA (snoRNA), a radiolabeled oligonucleotide specific for U3 was used as previously described (5); this hybridization was done in ExpressHyb (Clontech) according to the user manual.
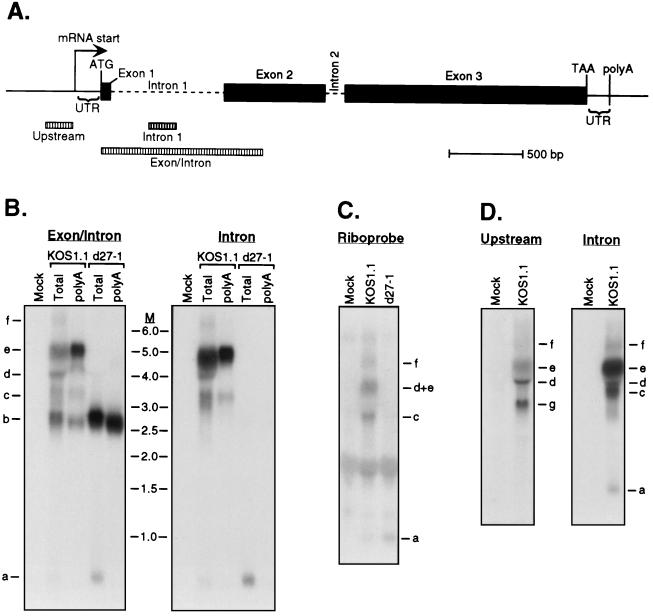
FIG. 4.
Northern blot analysis detects more than one ICP0 intron-bearing transcript in KOS1.1-infected HeLa cells, and their accumulation requires ICP27. (A) Schematic diagram of the HSV-1 ICP0 gene. Exon sequences are indicated by filled boxes, and introns are marked by dashed lines. Positions of the mRNA start site, ATG start codon, TAA stop codon, 5′ and 3′ untranslated regions, and the poly(A) cleavage site are shown. The fragments specific for upstream sequences, intron 1 sequences, and exon plus intron sequences that were used to prepare radiolabeled probes for Northern blot analysis are indicated by hatched bars. (B) Total RNA or poly(A)+ RNA was prepared from mock-infected, KOS1.1-infected, or d27-1-infected HeLa cells at 6 h postinfection. Northern blot analysis was conducted using the intron-specific probe (right), and then the membrane was reprobed with the exon/intron probe (left). Transcripts were given the designations a to f for ease of description in the text. M, RNA size markers in kilobases. (C) Northern blot of total RNA from mock-infected, KOS1.1-infected, or d27-1-infected HeLa cells using the ICP0 intron 1 strand-specific riboprobe. (D) Northern blot of total RNA from mock-infected or KOS1.1-infected HeLa cells using the upstream probe (left) and the intron 1 probe (right).
RESULTS
ICP27 mutations that impair virus replication prevent accumulation of unspliced α-globin RNA.
We previously reported that expression of functional ICP27 is necessary and sufficient for cytoplasmic accumulation of unspliced polyadenylated α-globin mRNA in HSV-1-infected HeLa cells (5). To investigate which regions of the ICP27 protein mediate pre-mRNA accumulation, we examined the α-globin transcripts in cells infected with a panel of HSV-1 recombinants in which each encodes a different mutated version of ICP27 (Fig. 1). The ICP27 protein can be divided into several functional regions. At the amino terminus is the leucine-rich NES (amino acids 5 to 17) (49), which partially overlaps an acidic region extending from amino acids 12 to 63 (44). The strong NLS at amino acids 110 to 137 (31) is immediately adjacent to the RGG box (residues 138 to 152) (33), which mediates RNA binding. The C-terminal region (from amino acids 262 to 512), conserved in all mammalian and avian herpesvirus ICP27 homologs (1), is particularly sensitive to mutations, and this half of the protein is required for the transactivation and transrepression functions of ICP27 (17). However, sequences located in the N-terminal portion of the polypeptide also play a role in these functions (44, 45). The mutations analyzed include a null mutant having a deletion of most of the ICP27 open reading frame (d27-1), N-terminal (M1X) and C-terminal (n59R, n263R, n406R, n504R, and M2X) deletions, clustered point mutations that alter one or two amino acids in the C-terminal region (M11, M15, and M16), and in-frame deletions within the coding sequence (d1-2, d2-3, d3-4, d4-5, d5-6, d6-7, and d1-5) (Fig. 1). The latter were engineered to precisely remove certain segments of the ICP27 protein; for example, d1-2 deletes the acidic region, and d3-4 and d4-5 remove the NLS and the RGG box, respectively. Detailed phenotypic analyses of many of these mutant viruses have been previously reported (see reference list in Fig. 1). The mutants can be grouped into three categories, based on their ability to replicate in Vero cells: (i) mutants d27-1, n59R, n263R, n406R, n504R, M11, M15, M16, d1-2, d4-5, d1-5, M1X, and M2X are growth defective, being unable to form plaques or forming plaques at greatly reduced efficiency; (ii) mutants d2-3 and d3-4 are growth deficient, being able to form plaques but having reduced yields in single-cycle growth assays; and (iii) mutants d5-6 and d6-7 are replication competent, being indistinguishable from the wild-type virus.
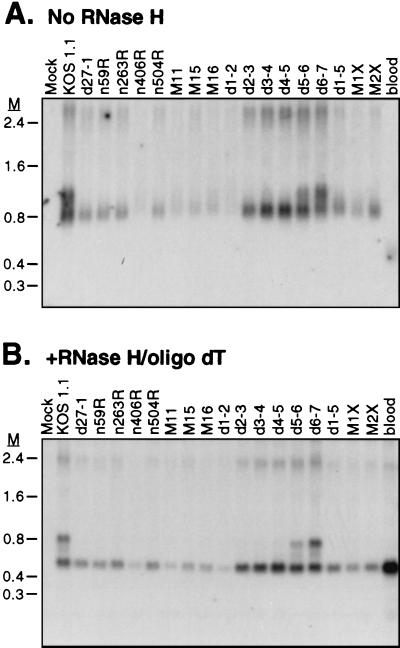
HeLa cells were infected with each mutant virus in the presence of PAA, and total RNA was harvested at 6 h postinfection. The RNA samples were either left untreated (Fig. 2A) or treated with RNase H in the presence of oligo(dT) to remove the poly(A) tail (Fig. 2B) and then analyzed for the presence of α-globin transcripts by Northern blot hybridization. As we have observed previously (5), removal of the poly(A) tail resulted in a considerable sharpening of the α-globin bands (discussed further below). Deadenylated RNA extracted from cells infected with the wild-type virus (KOS1.1) displayed three species of α-globin RNA (ca. 580, 700, and 840 nucleotides [nt]), with the 700-nt RNA being considerably less abundant than the other two (Fig. 2B). We have previously shown that these RNAs correspond to fully spliced (576 nt), partially spliced (688 and 718 nt), and unspliced (835 nt) α-globin transcripts that start at the α-globin promoter and are processed at the α-globin poly(A) site (5). Cells infected with the ICP27 null virus, d27-1, accumulated only the fully spliced species, confirming our previous finding that ICP27 is required for the accumulation of unspliced RNA (5). All of the ICP27 mutant viruses induced accumulation of the fully spliced mRNA, although we note that the signal intensity was somewhat reduced in infections with several of the mutant viruses (particularly n406R, M11, and d1-2). We have not yet determined the basis for this reduction, but it was seen in multiple experiments. In contrast, only two of the mutants, d5-6 and d6-7, showed accumulation of the unspliced α-globin transcript (Fig. 2B); all of the other mutants were as defective as d27-1 in this respect. These data demonstrate first that the ICP27 mutants that are defective for virus replication in cultured cells fail to accumulate unspliced α-globin RNA, while those that are replication competent display wild-type levels of unspliced RNA. Second, multiple nonoverlapping segments of ICP27, including the N- and C-terminal regions, are required for accumulation of α-globin pre-mRNA. Third, and somewhat surprisingly, several of the mutant forms of ICP27 that have been previously shown to retain the ability to repress reporter gene expression in transient transfection experiments (n406R, n504R, M11, M15, M16, and d1-2 [Fig. 1]) nevertheless fail to accumulate unspliced α-globin RNA. It has been suggested that the transrepression function of ICP27 is a consequence of impaired splicing by ICP27, because repression correlates with the presence of introns in the reporter gene (16, 18, 52). Our findings argue that the accumulation of unspliced transcripts by ICP27 can be uncoupled by mutation from the repression function.
FIG. 2.
Effect of ICP27 mutants on accumulation of α-globin pre-mRNA. HeLa cells were infected with the indicated ICP27 mutant viruses at a multiplicity of 10 in the presence of PAA. Total RNA was harvested 6 h postinfection, and 10 μg was either left untreated (A) or treated with RNase H in the presence of oligo(dT) to remove the poly(A) tails (B). α-Globin transcripts were detected by Northern blot analysis. M, RNA markers in kilobases. The α-globin transcript in RNA isolated from blood is shown as a control.
Several ICP27 mutations markedly increase the length and heterogeneity of the α-globin mRNA poly(A) tail.
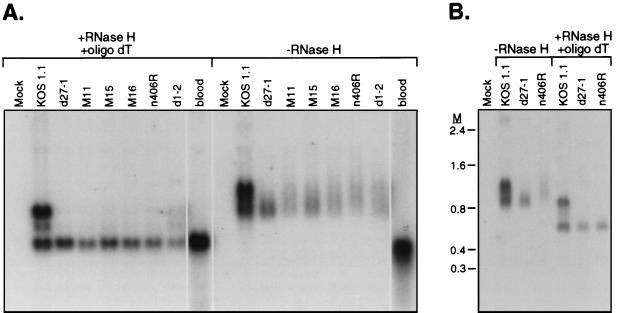
The poly(A) tails of most mRNAs vary in length from ca. 200 to 250 nt. Spliced α-globin mRNA is only ca. 580 nt long [excluding the poly(A) tail], and the heterogeneity in the poly(A) tail therefore significantly influences its electrophoretic mobility, leading to diffuse bands on a Northern blot. This can be observed quite clearly with the RNA from KOS1.1-infected cells (Fig. 2A): without prior deadenylation, the unspliced, partially spliced, and spliced bands were broad and poorly resolved. Likewise, the spliced mRNA signal from the d27-1 infection was quite diffuse. In both cases, these relatively diffuse bands collapsed into sharper bands of the predicted sizes after the poly(A) tails were removed by treatment with RNase H in the presence of oligo(dT) (compare Fig. 2A and B). Remarkably, several of the ICP27 mutants gave rise to exceptionally diffuse (and more slowly migrating) signals when RNA retaining the poly(A) tail was examined (Fig. 2A, mutants n406R, M11, M16, d1-2, and to a lesser extent M15). In fact, without removal of the poly(A) tail, we were unable to definitively determine whether the signals observed corresponded to spliced or unspliced RNA or to a mixture of the two as in the KOS1.1 sample. However, in the n406R, M11, M15, and M16 samples, the heterogeneity was eliminated by treatment with RNase H-oligo(dT), giving rise to a single sharp band corresponding to spliced RNA (Fig. 2B). This effect is shown more clearly in the experiment depicted in Fig. 3A (-RNase H), where RNA from cells infected with M11, M15, M16, n406R, and d1-2 were directly compared to the d27-1 sample. The M11, M15, M16, n406R, and d1-2 RNAs all migrated more slowly than the d27-1 sample and gave rise to a broad indistinct signal. However, with the exception of RNA from cells infected with mutant d1-2, removing the poly(A) tail resolved the majority of the signal into a discrete 580-nt band that comigrated with the spliced mRNA band in KOS1.1-infected cells and in blood RNA (Fig. 3A, +RNase H +oligo dT). By far the most likely explanation of these data is that the mutations in the n406R, M11, M15, and M16 viruses greatly increase the length and heterogeneity of the poly(A) tail of spliced α-globin mRNA. In the case of mutant d1-2, deadenylation of the RNA revealed detectable quantities of unspliced and partially spliced transcripts, although in lower amounts than in KOS1.1 RNA, which at least partly accounts for the broad α-globin signal in the untreated sample. The most striking example of the poly(A) extension effect is evident with the n406R mutant (Fig. 3B). In cells infected with n406R, the α-globin RNA is significantly higher in molecular weight than the d27-1 counterpart, and the signal extends almost to the position of the unspliced transcript present in KOS1.1-infected cells, which is 260 nt longer than the spliced mRNA (Fig. 3B, -RNase H). We estimate from these data that the poly(A) tail of α-globin mRNA in cells infected with this mutant ranges in length from ∼275 to 520 nt, markedly longer and more heterogeneous than the relatively discrete 200- to 250-nt tail observed in d27-1 infections. This extension phenomenon is clearly a consequence of expressing the altered ICP27 protein, as the effect was not seen with either wild-type or ICP27-null virus. Moreover, a marker-rescued virus in which the n406R ICP27 allele was replaced with the wild-type allele did not display the extension phenotype (data not shown). Although we do not know the mechanism by which some mutant forms of ICP27 cause extension of the poly(A) tail, these data suggest that ICP27 may be intimately involved with the polymerization of the poly(A) tail in addition to its previously documented effects on poly(A) site selection and usage (27–29).
FIG. 3.
Several ICP27 mutants cause an increase in the length and heterogeneity of the α-globin 3′ poly(A) tail. (A) Total RNA was harvested from HeLa cells infected with the indicated ICP27 mutant viruses in the presence of PAA at 6 h postinfection. The RNA was analyzed for the presence of α-globin transcripts before or after removal of the poly(A) tail by treatment with RNase H and oligo(dT). The α-globin transcript in RNA isolated from blood is shown as a control. (B) Total RNA was isolated from HeLa cells that were either mock infected or infected with HSV-1 strain KOS1.1, d27-1, or n406R in the presence of PAA. α-Globin transcripts were detected by Northern blot hybridization before or after removal of the poly(A) tail by treatment with RNase H and oligo(dT). M, RNA markers in kilobases.
Multiple intron-bearing ICP0 transcripts are present in HSV-infected HeLa cells.
It has been suggested in the literature that ICP27 induces the delayed shutoff of host protein synthesis by virtue of its ability to inhibit RNA splicing (16, 18, 52). However, we have previously shown that although ICP27 induces accumulation of unspliced α-globin RNA, it does not detectably inhibit accumulation of the fully spliced globin transcript (5). Moreover, the data presented in Fig. 2 demonstrate that accumulation of unspliced α-globin RNA can be uncoupled from ICP27's repression function (which is thought to reflect inhibition of splicing). The ICP0 gene is one of the few HSV-1 genes that contain introns (Fig. 4A), and it has been reported that ICP27 causes the accumulation of ICP0 pre-mRNA in infected cells (18, 39, 49). These data have been interpreted to indicate that ICP27 inhibits the splicing of HSV transcripts as well as cellular RNAs. Given our results, we wished to confirm that ICP27 induces accumulation of unspliced ICP0 RNA in our experimental system and examine the effects of our panel of ICP27 mutations on this process. In particular, we wished to determine if accumulation of unspliced RNA could be uncoupled from the repression function of ICP27 with this transcript as well.
We examined the ICP0 transcripts present in HSV-infected HeLa cells by Northern blot hybridization (Fig. 4B). It should be noted that previous studies used in situ hybridization (39) or RNase protection (16, 18) to detect unspliced ICP0 RNA; although these methods are useful, they do not give an overview of the number and structures of the transcripts detected. Total and poly(A)+ RNA was isolated from cells infected in the presence of PAA with wild-type KOS1.1 or the ICP27 null d27-1 at 6 h postinfection. We used two probes to analyze the ICP0 transcripts (Fig. 4A). The exon/intron probe extends from the beginning of exon 1 through the first intron into the 5′ portion of exon 2 and should detect all ICP0 transcripts. In contrast, the intron probe detects only unspliced RNAs retaining intron 1. Total RNA extracted from cells infected with KOS1.1 contained at least six transcripts that hybridized to the exon/intron probe (labeled a to f in Fig. 4B). These ranged in size from ca. 800 nt (transcript a) to ca. 7 kb (transcript f). Transcript a was also detected with the intron-specific probe, and this small RNA was not recovered in the poly(A)+ RNA fraction. We therefore conclude that it corresponds to the excised intron 1 RNA previously observed by other investigators (4). In contrast, transcripts b, c, and e (and possibly d) were all recovered in the poly(A)+ fraction. Transcript b (∼2,700 to 2,800 nt) is the size predicted for ICP0 mRNA and did not hybridize to the intron probe. Taken together, these data indicate that transcript b corresponds to spliced, polyadenylated ICP0 mRNA. The larger transcripts (c to f) all hybridized to the intron probe and thus contain at least a portion of intron 1. RNA from d27-1-infected cells displayed only transcripts a and b (free intron 1 and fully spliced mRNA, respectively). Thus, ICP27 was required for the accumulation of all of the larger intron-bearing transcripts, in accord with the observations of others.
We examined the nature of these intron-bearing RNAs in more detail. Transcript c (∼3,500 nt) is the size predicted for RNA that initiates at the ICP0 gene promoter, bears both introns, and terminates at the designated ICP0 poly(A) site (Fig. 4A). Additional evidence supporting that assignment is presented below. The remaining RNA species (d to f) are all significantly larger, and notably, one of these (transcript e) is considerably more abundant than transcript c. It is unclear if RNAs d and e are separate species, since there is some distortion at this position on the gel due to the presence of the 28S rRNA. All of these transcripts (with the possible exception of d) hybridized to a strand-specific riboprobe derived from intron 1 (Fig. 4C), demonstrating that they are transcribed from the same DNA strand as ICP0 mRNA. In principle, these larger intron-bearing transcripts might initiate upstream of the ICP0 mRNA start site and/or terminate downstream of the poly(A) site. To investigate these possibilities, duplicate samples of total RNA from KOS1.1-infected cells were run on the same gel and blotted to a Genescreen membrane. One half of the membrane was probed with a labeled DNA fragment derived from sequences located immediately upstream of the ICP0 mRNA start site (Fig. 4A), and the other half was probed with the intron 1 probe (Fig. 4D). The upstream probe did not detect band a, b, or c, consistent with our designation of these RNAs as the excised intron 1, spliced mRNA, and ICP0 pre-mRNA initiating at the ICP0 promoter, respectively. The upstream probe did, however, detect bands d, e, and f, and also illuminated an additional transcript labeled g. These data demonstrate that the larger intron-containing transcripts (d to f) initiate upstream of the ICP0 promoter and read through at least a portion of the ICP0 intron 1. The transcriptional polarity of transcript g has not yet been determined. We have not determined exactly where transcripts d to f initiate or terminate.
Taken together, these data show that several transcripts bearing at least part of ICP0 intron 1 accumulate in HeLa cells infected with wild-type HSV-1. The origin of some of these RNAs is not clear, but several initiate upstream of the ICP0 promoter. Importantly, accumulation of all of these intron-containing transcripts requires the ICP27 protein, under these conditions of infection. Transcript c is the most likely candidate for bona fide ICP0 pre-mRNA (i.e., the pre-mRNA that is spliced to produce translatable ICP0 mRNA). It is worth pointing out that ICP0 transcript c is much less abundant relative to spliced ICP0 mRNA than the α-globin pre-mRNA is relative to spliced α-globin mRNA: the relative band intensity of RNA c is considerably lower than its α-globin counterpart; in addition the ICP0 exon/intron probe used is composed mostly of intron sequences (only 29% of the length of the probe is homologous to the exon sequences), and thus the band intensity of the mRNA species is underrepresented three- to fourfold relative to the intron-bearing transcripts. In contrast, the α-globin probe used is 70% exonic, and the unspliced α-globin signal is often as intense as that of the fully spliced RNA (Fig. 2, 3, and 6). Thus, compared to α-globin pre-mRNA, ICP0 transcript c is a very minor species. These considerations suggest that ICP27 causes the accumulation of α-globin pre-mRNA much more efficiently than ICP0 pre-mRNA.
FIG. 6.
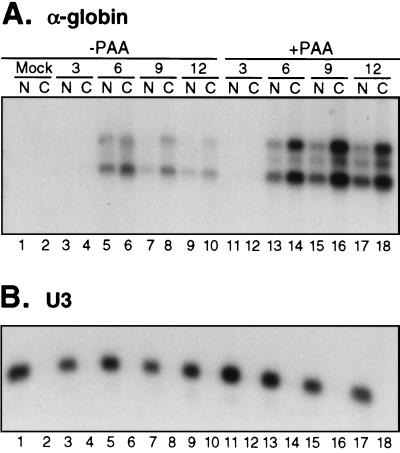
Cytoplasmic accumulation of unspliced and spliced α-globin RNA is independent of viral DNA replication. HeLa cells were mock infected or infected with HSV-1 strain KOS1.1 in the absence or presence of PAA. At 3, 6, 9, or 12 h postinfection, nuclear (N) or cytoplasmic (C) RNA fractions were prepared. RNA from an equal number of cells was analyzed by Northern blotting for the presence of α-globin transcripts (A) or the U3 snoRNA (B).
Effect of ICP27 mutations on accumulation of ICP0 intron-bearing transcripts.
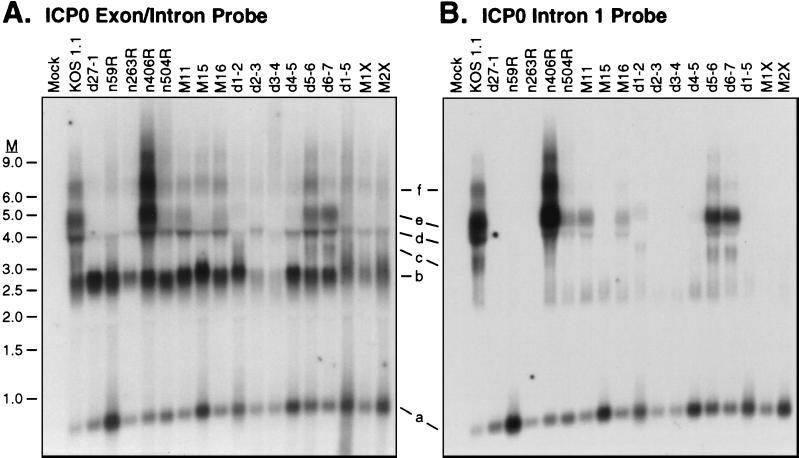
The foregoing data confirmed that ICP27 is required for accumulation of intron-bearing transcripts of both the α-globin and ICP0 genes. We next wished to determine if accumulation of the intron-containing ICP0 transcripts exhibits the same mutational sensitivity spectrum as accumulation of unspliced α-globin pre-mRNA. Total RNA harvested from cells infected with the panel of ICP27 mutants in the presence of PAA was analyzed by Northern blot hybridization using the ICP0 exon/intron (Fig. 5A) and intron 1 (Fig. 5B) probes. Transcripts were given the same designations (a to f) as in Fig. 4. Fully spliced ICP0 mRNA was present in all of the samples, as was the excised intron 1 (Fig. 5A, bands b and a, respectively). Several of the mutants (d2-3 and d3-4 in particular) displayed reduced levels of these RNAs; we have not determined the basis for this reduction. Unlike the results obtained with α-globin RNA, we noted several different phenotypes with respect to the transcripts containing intron 1 sequences (Fig. 5B). First, the majority of the mutant viruses (n59R, n263R, M15, d2-3, d3-4, d4-5, d1-5, M1X, and M2X) behaved like the ICP27-null mutant, d27-1: in these infections, no intron-containing transcripts were evident. Second, d5-6 and d6-7 displayed the same spectrum of intron-bearing RNAs as the wild-type virus. d5-6 and d6-7 are the only mutants analyzed that are not growth impaired in Vero cells, and they also efficiently accumulate α-globin pre-mRNA. Third, four mutants (n406R, n504R, M11, and M16) accumulated the intron-containing RNAs that initiate upstream of the ICP0 promoter (RNAs d to f) but failed to display the intron-containing transcript that likely initiates at the ICP0 promoter (RNA c). Mutant n406R displayed the most striking example of this phenotype, in that transcripts d to f were overproduced relative to wild-type KOS1.1. Finally, in the d1-2 infection, all of the intron-bearing RNAs were lacking, but a faint new band that migrates slightly higher than transcript c appeared. We have not investigated the nature of this transcript further. We conclude from these data that the effects of ICP27 on accumulation of intron-bearing RNAs are both transcript and ICP27 allele specific. While d5-6 and d6-7 display the wild-type phenotype, several other ICP27 mutants distinguish between ICP0 transcript c and α-globin pre-mRNA on one hand (no accumulation) and ICP0 transcripts d to f on the other (accumulation). Possible explanations for these findings are presented below (see Discussion).
FIG. 5.
Effect of ICP27 mutants on accumulation of ICP0 intron-bearing transcripts. HeLa cells were infected with the indicated viruses at a multiplicity of 10 in the presence of PAA, and total RNA was harvested 6 h postinfection. Ten micrograms of each RNA was subjected to Northern blot analysis using the ICP0 intron 1 probe (B). The membrane was subsequently reprobed with the ICP0 exon/intron probe (A). Positions of transcripts a to f are indicated. M, RNA size markers in kilobases.
Subcellular localization of α-globin and ICP0 transcripts.
We previously reported that the unspliced α-globin pre-mRNA that accumulates as a consequence of ICP27 expression is efficiently exported into the cytoplasm (5). This finding was surprising because, as reviewed in the introduction, intron-bearing transcripts are normally confined to the nucleus, and substantial evidence indicates that such pre-mRNAs must be processed by the splicing apparatus in order to access the nuclear export machinery. Based on our data, we proposed that ICP27 induces a splicing-independent pathway for nuclear export, one that transports specific transcripts irrespective of the presence or absence of introns. However, it has been previously observed that ICP27 causes the accumulation, preferentially in the nucleus, of intron-bearing transcripts of the HSV ICP0 and UL15 genes, an effect that was ascribed to inhibition of splicing (18, 39, 49). This has led to the suggestion that the ICP27-induced RNA transport system globally distinguishes between transcripts of intronless and intron-bearing genes, transporting the former and blocking splicing and export of the latter (49). We therefore compared the effects of ICP27 on the subcellular localization of α-globin and ICP0 pre-mRNA in more detail, in an attempt to resolve the apparent discrepancies between our data and the interpretations that currently prevail in the literature.
A major factor that could influence the nuclear/cytoplasmic distribution of mRNA and pre-mRNA is the onset of viral DNA replication. We have routinely blocked viral DNA replication with PAA in all of our previous studies of ICP27's effects on α-globin pre-mRNA metabolism, for two reasons. First, expression of the chromosomal α-globin gene declines after 6 h postinfection, like that of viral E genes, and this decline is prevented by blocking viral DNA replication (6). Second, and more important, ICP27-null mutants display a marked defect in viral DNA replication, and the various ICP27 mutants that we have analyzed differ considerably in the ability to support DNA replication. The onset of DNA replication profoundly affects viral gene expression and leads to pronounced changes in nuclear organization. We included PAA to eliminate these potentially confounding secondary effects, which are predicted to vary substantially between the ICP27 mutants. In contrast, the previous study indicating that ICP27 causes nuclear retention of ICP0 pre-mRNA did not control for the effects of viral DNA replication (39). We therefore examined the subcellular localization of α-globin and ICP0 RNAs, in the presence or absence of a blockade of viral DNA replication imposed by PAA (Fig. 6 and 7).
FIG. 7.
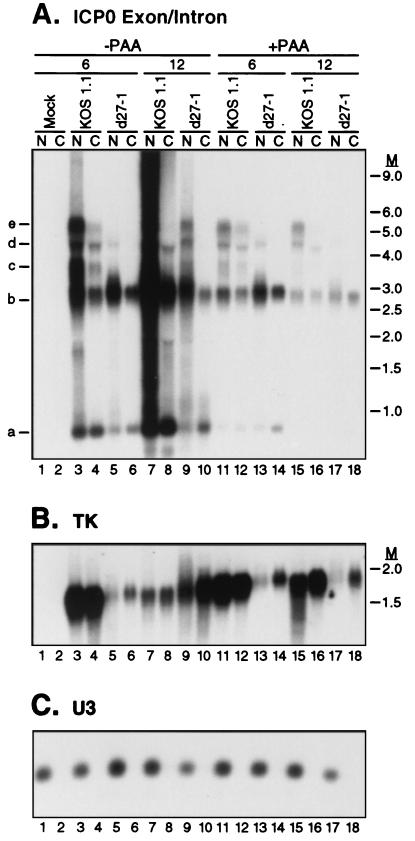
Nuclear retention of all ICP0 transcripts is dramatically enhanced by viral DNA replication but is independent of ICP27. HeLa cells were mock infected or infected with HSV-1 strain KOS1.1 or d27-1 in the absence or presence of PAA. At 6 or 12 h postinfection, nuclear (N) or cytoplasmic (C) RNA fractions were prepared. RNA from an equal number of cells was analyzed by Northern blotting for the presence of ICP0 transcripts using the ICP0 exon/intron probe (A), TK transcripts (B), or the U3 snoRNA (C). M, RNA size markers in kilobases.
In the first such experiment, nuclear and cytoplasmic RNA was harvested at various times after infection with wild-type KOS1.1, and deadenylated samples were scored for the presence of spliced and unspliced α-globin RNA by Northern blot hybridization (Fig. 6A). As a control, the same membrane was probed with an oligonucleotide specific for the U3 snoRNA, an exclusively nuclear RNA (Fig. 6B). The U3 snoRNA was detected only in the nuclear RNA samples, indicating that the cytoplasmic RNA was free of nuclear RNA contamination. As reported previously (6), α-globin transcript levels peaked at 6 h postinfection in the absence of PAA, and at this time point a significant fraction of the pre-mRNA and mRNA was cytoplasmic (lanes 5 and 6). As the infection progressed, the combined pre-mRNA and mRNA signal declined, and both species became increasingly cytoplasmic (lanes 7 to 10). This progressive shift is consistent with the hypothesis that α-globin RNA synthesis declines at later times, while nuclear export of the preexisting transcripts continues. The overall decline in the α-globin signal likely stems from turnover in the cytoplasm. Consistent with this interpretation, the decline is largely eliminated by inactivating the virion host shutoff protein vhs (P. Cheung and J. R. Smiley, unpublished data). In the presence of PAA, the levels of α-globin RNA increased over time, and the unspliced and spliced transcripts efficiently gained access to the cytoplasm at all time points (lanes 11 to 18). These data demonstrate that PAA does not greatly influence the nuclear export of α-globin mRNA or pre-mRNA. In particular, the unspliced RNA is largely cytoplasmic in both the presence and absence of the drug.
We next examined the effects of ICP27 and PAA on the subcellular distribution of ICP0 transcripts at 6 and 12 h postinfection (Fig. 7A). We also probed the same samples for HSV TK mRNA (Fig. 7B), which is derived from an intronless viral gene. Previous studies have indicated that TK mRNA contains multiple cis-acting elements that mediate its nuclear export, likely via interactions with the cellular protein hnRNP L (25). Controls confirmed that, as expected, U3 snoRNA was recovered exclusively in the nuclear fraction (Fig. 7C). The profiles of ICP0 transcripts at 6 h postinfection were similar in the presence and absence of PAA (Fig. 7A): transcripts a to e, as defined in Fig. 4, were readily discernible in the KOS1.1 samples, and only transcripts a and b were evident in d27-1 RNA (the faint higher-molecular-weight band in the d27-1 lanes 5 and 13 was not detected with the intron 1 probe [data not shown]). It is interesting that transcript c, the presumed bona fide ICP0 pre-mRNA, was not apparent in KOS1.1-infected cells at 12 h postinfection, in either the absence or presence of PAA (lanes 8, 15, and 16). The d27-1 RNA harvested 12 h postinfection from cells infected in the absence of PAA (lane 9) displayed the intron-bearing transcripts d and e, in addition to the intron 1 RNA and the spliced message (RNAs a and b, respectively). Thus, unlike at early times of infection, the accumulation of these intron-bearing transcripts was not strictly dependent on ICP27 expression at late times under replication-competent conditions. The significance of this observation is unknown at present. Significant quantities of each of the ICP0 transcripts could be detected in the cytoplasm in cells infected with KOS1.1 in the absence of PAA, as has been observed previously (18, 49); however, the majority of RNAs b to e was recovered in the nucleus (Fig. 7A, lanes 3, 4, 7, and 8). This nuclear retention was greatly exaggerated at 12 h postinfection. Notably, retention was not confined to the intron-bearing RNAs but was exhibited by the spliced mRNA as well (transcript b). Furthermore, a similar degree of nuclear retention of the spliced mRNA was observed in d27-1 infections (Fig. 7A, lanes 5, 6, 9, and 10). Interestingly, the exception to the nuclear retention phenotype was band a, which corresponds to excised intron 1. A significantly greater proportion of this RNA than of the other transcripts was cytoplasmic, independent of the presence or absence of ICP27. Cytoplasmic accumulation of ICP0 intron 1, by unknown mechanisms, has been described previously (4). ICP0 transcript levels were considerably reduced in the presence of PAA (lanes 11 to 18), especially at 12 h postinfection, and modest levels of nuclear retention were observed. These results indicate that in HeLa cells, ICP0 transcripts are predominantly nuclear regardless of the presence or absence of intron sequences. This retention is not obviously mediated by ICP27, since the spliced mRNA is preferentially located in the nucleus even in the absence of functional ICP27.
The membrane shown in Fig. 7A was then stripped and reprobed for TK sequences (Fig. 7B). In KOS1.1-infected cells, TK RNA was distributed approximately equally between the nucleus and cytoplasm at all time points (lanes 3, 4, 7, 8, 11, 12, 15, and 16). However, in the d27-1 infection, the RNA was preferentially cytoplasmic (lanes 5, 6, 9, 10, 13, 14, 17, and 18), an effect that was especially pronounced when viral DNA replication was disabled by PAA. In addition, the amount of TK RNA was substantially reduced in d27-1 infections. One explanation of these results is that the previously described cellular export pathway for the TK transcript is saturated in cells infected with the wild-type virus. However, these data raise the possibility that ICP27 may participate in regulating the nuclear/cytoplasmic distribution of TK transcripts.
Taken together, the data presented above demonstrate that in HSV-infected HeLa cells, α-globin transcripts are exported to the cytoplasm irrespective of the presence or absence of an intron, while conversely both spliced and unspliced ICP0 RNAs are preferentially recovered in the nuclear fraction. Moreover, retention of spliced ICP0 mRNA does not require ICP27.
DISCUSSION
Effects of ICP27 mutations on accumulation of unspliced α-globin and ICP0 pre-mRNA.
We have used a panel of HSV-1 recombinants bearing altered versions of the ICP27 gene to examine which regions of ICP27 are important for the accumulation of unspliced transcripts of two intron-bearing genes, α-globin and ICP0. Our results revealed a striking correlation between the ability of the various mutant forms of ICP27 to support virus replication on noncomplementing cells and accumulation of unspliced α-globin RNA (compare Fig. 1 and 2). Mutations that inactivate or delete the NES, the N-terminal acidic region, the NLS, the RGG box, and the conserved C-terminal half of the protein all eliminated pre-mRNA accumulation. This finding argues that all of these regions, which are essential for ICP27 function, participate in the accumulation of α-globin pre-mRNA. Surprisingly, accumulation of α-globin pre-mRNA did not correlate with the previously described transrepression function of ICP27, that is, its ability to inhibit expression of intron-bearing reporter genes in cotransfection assays. Several mutants that are fully competent for repressing reporter genes (Fig. 1) nevertheless fail to accumulate detectable levels of unspliced α-globin pre-mRNA. Previous studies utilizing a more limited set of ICP27 mutations suggested that accumulation of unspliced RNA and transrepression are operationally equivalent consequences of a global inhibition of splicing by ICP27 (18, 52). Our data provide new and significant information, by demonstrating that these effects can be separated by mutation. One possible explanation to reconcile these observations is the following. Although the repression function of wild-type ICP27 appears to be intron dependent (52), it has not been determined if this is also the case for the repression-competent ICP27 mutants that have been analyzed in our study. Indeed, Sandri-Goldin and Mendoza have reported intron-independent inhibition of reporter gene expression by an ICP27 mutant, S23, that is defective in gene activation (52). This mutant, as well as several other mutants in the activation domain, is transdominant to the wild-type protein, reducing late gene expression and viral yields during coinfection with wild-type virus (54). This suggests the mutant protein is dysfunctional rather than nonfunctional, interfering with normal RNA processing possibly by the formation of inactive heterodimers with wild-type ICP27 or by competing with the wild-type protein for substrates or interacting proteins. Repression-competent dysfunctional proteins such as S23 may thus cause decreased gene expression by mechanisms that are quite distinct from those used by the native ICP27 protein. In this context, it is interesting that several of the ICP27 mutants that are repression competent (n406R, M11, M15, and M16) display the poly(A) tail extension effect, a phenotype not exhibited by the wild-type virus or by the ICP27-null virus (discussed further below). It is possible that this aberrant polyadenylation could lead to reduced expression of certain reporter genes (i.e., repression). Further studies are required to test this hypothesis.
Does ICP27 induce accumulation of α-globin pre-mRNA simply by interfering with RNA splicing? If the accumulation of unspliced RNA were due solely to a decrease in the efficiency of splicing, then one would expect KOS1.1 to induce lower levels of spliced mRNA than d27-1, accompanied by a concomitant increase in the amounts of unspliced RNA. However, we have consistently observed approximately equal band intensities for the fully spliced mRNA in KOS1.1 and d27-1 infections, and indeed KOS1.1 often shows slightly higher levels of the spliced product than d27-1 (Fig. 2; see also reference 5). Recent studies of the Epstein-Barr virus homolog of ICP27, EB2, have produced similar observations (3): EB2 promotes cytoplasmic accumulation of unspliced transcripts of at least one intron-bearing reporter construct without reducing the levels of fully spliced mRNA. To account for these data, we have suggested that ICP27 acts to rescue a subset of α-globin pre-mRNAs that would normally be degraded, rather than inhibiting splicing per se (5). We speculated that ICP27 accomplishes this by dissociating transcripts from nonproductive interactions with the spliceosome, thereby preventing their degradation and promoting their polyadenylation and nuclear export (5) (see below). We further hypothesized that these activities are key to ICP27's ability to promote splicing-independent export of HSV L mRNAs (5). It is interesting that HIV Rev appears to act in a similar fashion to promote nuclear export of unspliced HIV mRNAs (40). If ICP27 indeed functions in this or a similar manner, then it seems likely that multiple properties of the protein (for example, RNA binding, interactions with the spliceosome, interactions with the polyadenylation machinery, nuclear export of RNA cargo, and reimportation of ICP27 into the nucleus) would be required. Inactivating any one of these functions could prevent accumulation of unspliced α-globin RNA. Considered in this light, it is not surprising that the integrity of most of the protein is required for this effect.
We obtained a substantially more complex picture when we examined the effects of the various mutant forms of ICP27 on accumulation of transcripts that retain ICP0 intron 1. The complexity stems in large part from the rather surprising multiplicity of intron-bearing transcripts that were observed. Multiple ICP0 transcripts bearing the intron 1 sequence have not been reported previously, and although it is possible that they are HeLa cell specific, it is more probable that the Northern blot analysis gives a more precise view of the number and sizes of transcripts than do RNase protection or in situ hybridization experiments. Although ICP27 was required for accumulation of all these RNAs early in infection, only one (transcript c) appears to represent unspliced ICP0 pre-mRNA initiated at the ICP0 promoter and processed at the ICP0 poly(A) site. The remainder of the RNAs (transcripts d to f) initiate somewhere upstream of the ICP0 promoter, and we have yet to map their 3′ ends. Thus, although accumulation of these RNAs requires ICP27, it is not yet clear that this effect is due to inhibition of splicing or rescue of the intron-containing transcript from the spliceosome. It is possible that another function of ICP27 could result in accumulation of these RNAs. For example, some or all of these RNAs might initiate at the promoter of the upstream ICP34.5 gene (or another upstream promoter) and read through upstream poly(A) sites in an ICP27-dependent manner into ICP0 sequences. In keeping with this notion, ICP27 has clearly documented effects on regulating gene expression by modulating the choice between alternative poly(A) sites (15, 27–29). In a particularly relevant example, most transcripts of the UL24 gene ordinarily read through the UL24 poly(A) site and are instead processed at the poly(A) site of the downstream UL26 gene; however, when ICP27 is inactivated, virtually all of the UL24 transcripts utilize the promoter-proximal UL24 poly(A) site (15). An analogous mechanism might explain the ICP27-dependent accumulation of transcripts d to f. Other possibilities are that transcripts d to f arise from an upstream promoter that is activated by ICP27, or that ICP27 mediates some form of alternative splicing. Thus, some or all of ICP0 transcripts d to f may arise as a consequence of functions of ICP27 that are not equivalent to transcript rescue or inhibition of constitutive splicing.
As was the case for α-globin, the only ICP27 mutants that exhibited a wild-type phenotype with respect to the accumulation of ICP0 intron-bearing RNAs were d5-6 and d6-7, which are not growth impaired in Vero cells. However, in marked contrast to the findings with α-globin, several of the growth-defective mutants (n406R, n504R, M11, and M16) also accumulated ICP0 intron-containing RNAs. It is interesting and probably significant that these mutants accumulated only those intron-bearing transcripts that initiate upstream of the ICP0 promoter and failed to display transcript c, which is likely the precursor of ICP0 mRNA. A possible explanation for this finding is based on the notion, described above, that the intron-bearing transcripts d to f arise through another function of ICP27. Perhaps only transcript c accumulates via the transcript rescue function proposed above for α-globin pre-mRNA. Supporting this hypothesis, only this transcript displayed the same mutational sensitivity spectrum as α-globin pre-mRNA. If this interpretation is correct, then the implication is that the transcript rescue function likely operates to some degree on the ICP0 precursor RNA as well as α-globin. However, as noted in Results, accumulation and transport of transcript c are very inefficient compared to the response of α-globin pre-mRNA. One interpretation is that ICP27 acts through specific cis-acting sequence elements and that those present in ICP0 RNA fail to mediate an efficient response (discussed further below). This is in keeping with the finding that ICP27 did not detectably bind ICP0 spliced or unspliced RNA in the nucleus or cytoplasm as measured by in vivo UV cross-linking of HSV-1-infected cells (49).
Poly(A) tail extension by certain mutant forms of ICP27.
The observation that certain mutant forms of ICP27 cause a significant change in the heterogeneity in length of the α-globin mRNA poly(A) tail was surprising and points to a previously unappreciated attribute of ICP27. It has long been recognized that ICP27 increases the efficiency of poly(A) site selection at weak poly(A) sites (27–29). This enhancement of 3′ processing has been correlated with the transactivation function of ICP27, i.e., its ability to stimulate reporter genes in cotransfection assays, and seems to participate in stimulating late gene expression (52). To our knowledge, however, this is the first report of effects of ICP27 on the length of the poly(A) tail. The fact that these effects are not observed in wild-type or ICP27-null infections suggests that the phenotype is manifest as a consequence of expressing an altered protein that interferes with a normal pathway. Many mutants of this kind are dominant to the function of the wild-type protein; in fact, Smith et al. have identified several activation-defective ICP27 mutants that exhibit transdominance to the wild-type protein (54). Preliminary data indicate that n406R, which displays the most striking poly(A) extension phenotype, may be partially dominant negative (K. S. Ellison, S. A. Rice, and J. R. Smiley, unpublished data). The poly(A) tail length of mammalian mRNAs is normally limited to approximately 250 nt, by a mechanism that involves the switching of poly(A) polymerase (PAP) from a processive mode of polymerization to a distributive mode (62; see reference 64 for a recent review). This switch is regulated by interactions between PAP, the RNA molecule, the cleavage and polyadenylation specificity factor, and poly(A) binding protein II. The poly(A) chain is elongated processively only when PAP is stimulated by both the cleavage and polyadenylation specificity factor and poly(A) binding protein II bound to short poly(A) tails: disruption of these interactions ensues at a chain length of ∼250 nt by a mechanism that is as yet unclear (62). Our findings imply that this length control mechanism is fundamentally subverted by the presence of some mutated versions of ICP27. One possibility is that PAP retains its processivity throughout the course of the reaction, perhaps through direct interactions with the altered form of ICP27.
It is interesting that four of the five mutant forms of ICP27 that display the poly(A) tail extension effect (excluding d1-2 [47]) have been previously shown to be defective in ICP27's activation function, and that the activation function has been correlated with enhanced usage of some, but not all, poly(A) sites. We have so far failed to detect a convincing effect of the n406R mutation on the length of the poly(A) tails of HSV ICP0 and TK mRNAs, although the data are not yet definitive (K. S. Ellison, R. Verity, and J. R. Smiley, unpublished data). It will therefore be interesting to determine which, if any, additional viral and cellular RNAs display this effect and to learn whether all of these are substrates for ICP27-induced nuclear export.
Subcellular localization of α-globin and ICP0 transcripts.
We have previously suggested that there are two modes for nuclear export of α-globin transcripts in HSV-infected cells: a splicing-dependent pathway for the spliced mRNA (presumably the same as that utilized for transport of transcripts derived from most intron-bearing cellular genes), and an alternative, splicing-independent pathway that requires ICP27 for the egress of unspliced pre-mRNA (5). This hypothesis was based on the observations that (i) ICP27 induces the cytoplasmic accumulation of unspliced α-globin RNA, while efficient export of the spliced mRNA is ICP27 independent, and (ii) ICP27 does not detectably alter the levels or rate of production of fully spliced mRNA (5). Here we demonstrate that cytoplasmic accumulation of spliced and unspliced α-globin transcripts occurs throughout the course of infection and in the presence or absence of viral DNA replication.
The ICP27-dependent accumulation of unspliced transcripts of two intron-containing HSV genes, ICP0 and UL15, has been attributed to splicing inhibition by ICP27 (18, 39, 49), and one study has suggested that ICP27 causes the retention of these unspliced transcripts in the nucleus (39). We observed both unspliced (intron-bearing) and spliced ICP0 transcripts in the cytoplasm, as has been previously reported (18, 49), but the bulk of the ICP0 transcripts were retained in the nucleus, an effect that was significantly enhanced when viral DNA replication was allowed to proceed. However, the spliced mRNA was retained as well as the intron-bearing RNAs (Fig. 7A). Furthermore, the nuclear/cytoplasmic distribution of the spliced ICP0 mRNA was not substantially altered in the absence of ICP27 (Fig. 7A), particularly when one takes into account the fact that DNA replication is impaired during infection with d27-1. Thus, although we confirm that ICP0 transcripts display enhanced nuclear retention relative to α-globin, this was not caused by the presence of introns in the retained RNA or by expression of ICP27. In keeping with our results, Clements and colleagues found by in situ hybridization that ICP0 transcripts were predominantly nuclear, using an exon-specific probe that would not distinguish between mRNA and pre-mRNA, and suggested that nuclear retention was conferred by ICP0 exon sequences (39). The indirect negative effects of deleting ICP27 on DNA replication were not taken into account in this and other earlier studies, however, which may account for the discrepancies in our interpretations regarding the requirement for ICP27.
Taken in combination, the data presented in this report suggest that ICP27 has different effects on ICP0 and α-globin transcripts with respect to both nuclear export and pre-mRNA accumulation. Unspliced α-globin RNA accumulates to relatively high levels and is transported very efficiently, while ICP0 transcript c accumulates inefficiently and remains largely nuclear. We have previously noted that the proposed transcript rescue function of ICP27 is akin to the mechanism of action of the HIV Rev protein. Rev enhances the intranuclear stability of unspliced and partially spliced HIV transcripts and promotes their nuclear export by binding to a specific cis-acting sequence, the Rev response element (see recent reviews in references 10, 40, and 57). Several naturally intronless genes require cis-acting sequences for splicing independent nuclear export (21, 23, 25). These considerations lead us to speculate that ICP27-dependent nuclear export of RNAs relies on the presence of specific cis-acting RNA sequences. According to this model, RNA molecules containing one or more optimal ICP27 response elements would be rescued and transported more efficiently (α-globin and some viral L RNAs) than those with weak or nonconsensus elements (ICP0 RNA). If such is the case, we may be able to identify the putative transport element by its ability to confer ICP27 responsiveness on a nonresponsive gene. It is interesting that the TK transcript, which has been shown to possess multiple cis-acting transport elements that bind the cellular hnRNP L protein (25), exhibits some ICP27-dependent nuclear retention (Fig. 7B). This could reflect competition by ICP27 for a factor that is common to both export pathways.
While this paper was under review, Soliman and Silverstein published a study indicating that nucleocytoplasmic shuttling of ICP27 is prevented when viral DNA replication is blocked by PAA or mutations in components of the viral DNA replication machinery (56). At first glance, this observation seems to conflict with our proposal that ICP27 mediates cytoplasmic accumulation of unspliced α-globin RNA through its shuttling activity (because this globin pre-mRNA accumulates in the cytoplasm in the absence of viral DNA replication). However, Soliman et al. previously reported that ICP27 requires an RNA cofactor to shuttle (55) and suggested that ICP27 must bind to HSV L RNAs in order to efficiently move into the cytoplasm (55, 56). According to this view, DNA replication stimulates shuttling indirectly, by allowing accumulation of the requisite L mRNAs. If this model is correct, then it seems likely that ICP27 would be capable of shuttling in the absence of viral DNA replication, provided that a suitable RNA cargo (such as α-globin RNA) was available. In this context, it should be noted that others have observed shuttling of ICP27 in the absence of viral L RNAs (32, 49), and ICP27 shuttles in interspecies heterokaryon assays under conditions that almost certainly preclude viral DNA replication (32).
Interrelationship between splicing, polyadenylation, RNA nuclear export, and ICP27.
As more of the biochemical details of mRNA synthesis, processing, and transport become elucidated, it is becoming increasingly evident that the various steps in these processes (transcription, 5′ capping, splicing, polyadenylation, and nuclear export) are not independent but rather are tightly interwoven and functionally interconnected (see reviews in references 57 and 64). Mounting evidence suggests that splicing greatly stimulates polyadenylation and vice versa, and that polyadenylation in turn is required for nuclear export (64). It seems likely that ICP27 is intimately involved with many of these processes. Our finding that certain mutant forms of ICP27 apparently alter some of these pathways suggests that these mutants may be invaluable tools for probing both the mechanisms of ICP27 action in HSV gene regulation and the basic operation of the cellular RNA processing and transport machinery.
ACKNOWLEDGMENTS
We thank Rob Maranchuk and Holly Saffran for excellent technical assistance.
This research was supported by an Establishment Grant from the Alberta Heritage Foundation for Medical Research (AHFMR) to J.R.S. and a grant from the National Institutes of Health (AI42737) to S.A.R. J.R.S. was a Terry Fox Senior Scientist of the NCI(C). S.A.R. was an AHFMR Senior Scholar.
REFERENCES
- 1.Brown C R, Nakamura M S, Mosca J D, Hayward G S, Straus S E, Perera L P. Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J Virol. 1995;69:7187–7195. doi: 10.1128/jvi.69.11.7187-7195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchman A R, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buisson M, Hans F, Kusters I, Duran N, Sergeant A. The C-terminal region but not the Arg-X-Pro repeat of Epstein-Barr virus protein EB2 is required for its effect on RNA splicing and transport. J Virol. 1999;73:4090–4100. doi: 10.1128/jvi.73.5.4090-4100.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter K L, Roizman B. Alternatively spliced mRNAs predicted to yield frame-shift proteins and stable intron 1 RNAs of the herpes simplex virus 1 regulatory gene alpha 0 accumulate in the cytoplasm of infected cells. Proc Natl Acad Sci USA. 1996;93:12535–12540. doi: 10.1073/pnas.93.22.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung P, Ellison K S, Verity R, Smiley J R. Herpes simplex virus ICP27 induces cytoplasmic accumulation of unspliced polyadenylated alpha-globin pre-mRNA in infected HeLa cells. J Virol. 2000;74:2913–2919. doi: 10.1128/jvi.74.6.2913-2919.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung P, Panning B, Smiley J R. Herpes simplex virus immediate-early proteins ICP0 and ICP4 activate the endogenous human α-globin gene in nonerythroid cells. J Virol. 1997;71:1748–1793. doi: 10.1128/jvi.71.3.1784-1793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiou H C, Dabrowski C, Alwine J C. Simian virus 40 late mRNA leader sequences involved in augmenting mRNA accumulation via multiple mechanisms, including increased polyadenylation efficiency. J Virol. 1991;65:6677–6685. doi: 10.1128/jvi.65.12.6677-6685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooke C, Alwine J C. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooke C, Hans H, Alwine J C. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol Cell Biol. 1999;19:4971–4979. doi: 10.1128/mcb.19.7.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 11.Elgadi M M, Smiley J R. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J Virol. 1999;73:9222–9231. doi: 10.1128/jvi.73.11.9222-9231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett R D. The products of herpes simplex virus type 1 (HSV-1) immediate early genes 1, 2, and 3 can activate HSV-1 gene expression in trans. J Gen Virol. 1986;67:2507–2513. doi: 10.1099/0022-1317-67-11-2507. [DOI] [PubMed] [Google Scholar]
- 13.Everett R D. The regulation of transcription of viral and cellular genes by herpesvirus immediate-early gene products. Anticancer Res. 1987;7:589–604. [PubMed] [Google Scholar]
- 14.Hamer D H, Leder P. Splicing and the formation of stable RNA. Cell. 1979;18:1299–1302. doi: 10.1016/0092-8674(79)90240-x. [DOI] [PubMed] [Google Scholar]
- 15.Hann L E, Cook W J, Uprichard S L, Knipe D M, Coen D M. The role of herpes simplex virus ICP27 in the regulation of UL24 gene expression by differential polyadenylation. J Virol. 1998;72:7709–7714. doi: 10.1128/jvi.72.10.7709-7714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardwicke M A, Vaughan P J, Sekulovich R E, O'Conner R, Sandri-Goldin R M. The regions important for the activator and repressor functions of herpes simplex virus type 1 α protein ICP27 map to the C-terminal half of the molecule. J Virol. 1989;63:4590–4602. doi: 10.1128/jvi.63.11.4590-4602.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hentschel C C, Birnstiel M L. The organization and expression of histone gene families. Cell. 1981;25:301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, Carmichael G C. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Carmichael G G. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc Natl Acad Sci USA. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Wimler K M, Carmichael G G. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Z M, Yen T S. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes R G, Munyon W H. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J Virol. 1975;16:275–283. doi: 10.1128/jvi.16.2.275-283.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Mertz J E. HnRNP L binds a cis-acting RNA sequence element that enables intron-independent gene expression. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 26.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 deletion mutant exhibits altered patterns of transcription and are DNA deficient. J Virol. 1989;63:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLauchlan J, Loney P C, Sandri-Goldin R M, Clements J B. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J Virol. 1992;66:6939–6945. doi: 10.1128/jvi.66.12.6939-6945.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLauchlan J, Simpson S, Clements J B. Herpes simplex virus induces a processing factor that stimulates poly(A) site usage. Cell. 1989;59:1093–1105. doi: 10.1016/0092-8674(89)90765-4. [DOI] [PubMed] [Google Scholar]
- 30.McMahan L, Schaffer P A. The repressing and enhancing functions of the herpes simplex virus regulatory protein ICP27 map to the C-terminal regions and are required to modulate viral gene expression very early in infection. J Virol. 1990;64:3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mears W E, Lam V, Rice S A. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J Virol. 1995;69:935–947. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mears W E, Rice S A. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology. 1998;242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 33.Mears W E, Rice S A. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J Virol. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nabel G J, Rice S A, Knipe D M, Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988;239:1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- 35.Niwa M, Rose S D, Berget S M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 36.Panagiotidis C A, Lium E K, Silverstein S J. Physical and functional interactions between herpes simplex virus immediate-early proteins ICP4 and ICP27. J Virol. 1997;71:1547–1557. doi: 10.1128/jvi.71.2.1547-1557.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phelan A, Carmo-Fonseca M, McLauchlan J, Lamond A I, Clements J B. A herpes simplex virus type-1 immediate early gene product IE63 regulates small ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phelan A, Clements J B. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J Gen Virol. 1997;78:3327–3331. doi: 10.1099/0022-1317-78-12-3327. [DOI] [PubMed] [Google Scholar]
- 39.Phelan A, Dunlop J, Clements J B. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J Virol. 1996;70:5255–5265. doi: 10.1128/jvi.70.8.5255-5265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 41.Rice S A, Knipe D M. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J Virol. 1988;62:3814–3823. doi: 10.1128/jvi.62.10.3814-3823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice S A, Knipe D M. Genetic evidence for two distinct transactivation functions of the herpes simplex virus α protein ICP27. J Virol. 1990;64:1704–1715. doi: 10.1128/jvi.64.4.1704-1715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice S A, Lam V. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J Virol. 1994;68:823–833. doi: 10.1128/jvi.68.2.823-833.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice S A, Lam V, Knipe D M. The acidic amino-terminal region of herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J Virol. 1993;67:1778–1787. doi: 10.1128/jvi.67.4.1778-1787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice S A, Su L, Knipe D M. Herpes simplex virus alpha protein ICP27 possesses separable positive and negative regulatory activities. J Virol. 1989;63:3399–3407. doi: 10.1128/jvi.63.8.3399-3407.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 47.Ryu W S, Mertz J E. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J Virol. 1989;63:4386–4394. doi: 10.1128/jvi.63.10.4386-4394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sacks W R, Greene C C, Ashman D P, Schaffer P A. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sandri-Goldin R M, Hibbard M K. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J Virol. 1996;70:108–118. doi: 10.1128/jvi.70.1.108-118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandri-Goldin R M, Mendoza G E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 53.Sekulovich R E, Leary K, Sandri-Goldin R M. The herpes simplex virus type 1 α protein ICP27 can act as a trans-repressor or a trans-activator in combination with ICP4 and ICP0. J Virol. 1988;62:4510–4522. doi: 10.1128/jvi.62.12.4510-4522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith I, Sekulovich R, Hardwicke M, Sandri-Goldin R. Mutations in the activation region of herpes simplex virus regulatory protein ICP27 can be trans dominant. J Virol. 1991;65:3656–3666. doi: 10.1128/jvi.65.7.3656-3666.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soliman T M, Sandri-Goldin R M, Silverstein S J. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soliman T M, Silverstein S J. Herpesvirus mRNAs are sorted for export via Crm1-dependent and -independent pathways. J Virol. 2000;74:2814–2825. doi: 10.1128/jvi.74.6.2814-2825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 58.Su L, Knipe D M. Herpes simplex virus α protein ICP27 can inhibit or augment viral gene transactivation. Virology. 1989;170:496–504. doi: 10.1016/0042-6822(89)90441-8. [DOI] [PubMed] [Google Scholar]
- 59.Uprichard S L, Knipe D M. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J Virol. 1996;70:1969–1980. doi: 10.1128/jvi.70.3.1969-1980.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Varmuza S L, Smiley J R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985;41:793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- 61.Wadd S, Bryant H, Filhol O, Scott J E, Hsieh T-Y, Everett R D, Clements J B. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J Biol Chem. 1999;274:28991–28998. doi: 10.1074/jbc.274.41.28991. [DOI] [PubMed] [Google Scholar]
- 62.Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J Biol Chem. 1995;270:2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- 63.Ward P L, Roizman B. Herpes simplex genes: the blueprint of a successful human pathogen. Trends Genet. 1994;10:267–274. doi: 10.1016/0168-9525(90)90009-u. [DOI] [PubMed] [Google Scholar]
- 64.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhi Y, Sciabica K S, Sandri-Goldin R M. Self-interaction of the herpes simplex virus type 1 regulatory protein ICP27. Virology. 1999;257:341–351. doi: 10.1006/viro.1999.9698. [DOI] [PubMed] [Google Scholar]