Abstract
Herpesvirus saimiri (HVS) is the prototype gamma-2 herpesvirus; it has significant homology to the human gammaherpesviruses Kaposi's sarcoma-associated virus and Epstein-Barr virus and the murine gammaherpesvirus murine herpesvirus 68. HVS causes a persistent asymptomatic infection in its natural host, the squirrel monkey. Both subgroups A and C possess the ability to immortalize common marmoset T lymphocytes to interleukin-2-independent proliferation. However, only subgroup C is capable of transforming human, rabbit, and rhesus monkey lymphocytes in vitro. In addition, HVS can stably transduce a variety of human cell lines where the virus persists as a nonintegrating circular episome. In this study, we have developed a system in which the HVS DNA is stably maintained as a nonintegrated circular episome in the human lung carcinoma cell line A549. Virus production can be reactivated using chemical inducing agents, including tetradecanoyl phorbol acetate and n-butyrate, suggesting that the infection in human A549 cells is latent. To analyze virus gene expression in these stably transduced cells, Northern blot analysis was performed using a series of probes produced from restriction fragments spanning the entire coding region of the HVS genome. This demonstrated that an adjacent set of genes containing open reading frames (ORFs) 71 to 73 are expressed in this stably transduced cell line. Moreover, these genes are transcribed as a polycistronic mRNA species produced from a common promoter upstream of ORF 73. This model may serve as a useful tool in the further analysis of the role of ORFs 71 to 73 in gamma-2 herpesvirus latency.
Herpesvirus saimiri (HVS) is the prototype gamma-2 herpesvirus, or rhadinovirus; it has significant homology to the human gammaherpesviruses Kaposi's sarcoma-associated virus (KSHV) and Epstein-Barr virus (EBV) and the murine gammaherpesvirus murine herpesvirus 68 (MHV-68) (1, 39, 43, 53). HVS was originally isolated from its natural host, the squirrel monkey (Saimiri sciureus), in which it causes a persistent asymptomatic infection. However, HVS infection of other New World primates results in acute T-cell lymphoma within a few weeks (16). Both subgroups A and C possess the ability to immortalize common marmoset T lymphocytes to interleukin-2-independent proliferation (10, 49). However, only subgroup C is capable of transforming human, rabbit, and rhesus monkey lymphocytes in vitro (3, 5–7).
Analysis of the left end of the unique L-DNA of the viral genome of subgroups A and C has revealed variable open reading frames (ORFs) (6, 21, 24), which are essential for T-cell transformation (12). HVS subgroup A contains a single ORF, STP-A (saimiri transforming protein A), whereas subgroup C contains two genes at this position, STP-C, a divergent form of the STP oncogene, and Tip (which encodes tyrosine kinase-interacting protein), which interacts with the tyrosine kinase Lck (4, 25, 37, 57). STP-A interacts with a Src kinase, whereas STP-C associates with cellular Ras (23). Immortalized T cells harbor the viral genome as a persisting high-copy-number nonintegrating episome without production of virus particles (5, 26, 55). Initial analysis of viral gene expression in these cells using Northern blotting suggested that viral gene expression was confined to the transforming genes (14, 15). However, the use of either STP-C or Tip deletion mutants demonstrated that these genes were essential for transformation in cell culture but dispensable for replication or long-term episomal persistence (12).
The HVS genome encodes a number of cellular homologues whose products may play a role in transformation, immune evasion and long-term persistence of the viral episome. These include U RNA homologues, antiapoptotic proteins (ORF16/vBcl2 and ORF71/vFLIP), a cyclin D homologue (ORF72/vCyclin), complement control inhibitory proteins (ORF4/CCPH and ORF15/vCD59), nucleotide metabolism enzymes (ORF2/DHFR and ORF70/TS), a viral superantigen (ORF14/vSag), and cytokine homologues (ORF13/vIL-17 and ORF74/vGCR) (1). Recent analysis of immortalized T cells by subtractive hybridization demonstrated that immediate-early IE14/vSag transcripts were found in abundance (31). IE14/vSag encodes a 50-kDa secreted glycoprotein which binds to major histocompatibility complex class II molecules and stimulates T-cell proliferation (58). Further analysis of the role of ORF14/vSag by using deletion mutants demonstrated that ORF14/vSag is dispensable for virus replication but that its role in transformation, persistence, and pathogenicity is variable depending on the cell type (13, 32).
Interestingly, HVS also has the ability to infect a variety of human cell lines, including hematopoietic lineage, myeloid, fibroblast, and carcinoma-derived cells (19, 46, 47). Although different levels of virus production have been observed in these cell lines, the virus generally persists as a nonintegrating circular episome. Analysis of the HVS genome has identified a cis-acting element which is required for viral episomal maintenance (34). However, no protein homologous to the EBV EBNA 1, which enables the EBV genome to be maintained as a stable episome in latently infected cells (29, 38, 42, 48), has been identified. Recent analysis of primary effusion lymphoma (PEL) cells harboring latent KSHV episomes demonstrated that the latency-associated nuclear antigen (LANA) encoded by ORF 73 colocalized with KSHV DNA. In addition, LANA was necessary and sufficient for the persistence of episomes containing KSHV DNA, suggesting that it tethers KSHV DNA to chromosomes during mitosis, enabling the efficient segregation of KSHV episomes to progeny cells (2). Moreover, LANA associates with histone HI in KSHV-infected BCBL cells, suggesting that the tether mechanism by which episomes are linked to host chromatin occurs via host chromosomal proteins (2, 8).
These results prompted us to investigate which genes are transcribed in a nontransformed, stably transduced human cell line where the virus persists as a nonintegrated episome. In this report, we describe a system in which HVS DNA is stably maintained as a nonintegrated circular episome in the human lung carcinoma cell line A549. We show that virus production can be reactivated using chemical inducing agents, suggesting that the infection in human A549 cells is truly latent. Northern blot analysis demonstrated that an adjacent set of genes containing ORFs 71 to 73 are expressed in this stably transduced cell line. Moreover, these genes are transcribed from a polycistronic mRNA species produced from a common promoter upstream of ORF 73.
MATERIALS AND METHODS
Viruses, cell culture, and transfections.
HVS (strain A11) and HVS-GFP (56) were propagated in owl monkey kidney (OMK) cells maintained in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% fetal calf serum (FCS). HVS-transformed T cells from cottontop tamarin monkeys, B133 cells (20, 30), were propagated in RPMI 1640 medium (Life Technologies) supplemented with 10% FCS and interleukin-2 (20 U/ml). To generate an A549 cell line stably transduced with HVS, 106 A549 cells were infected with HVS-GFP at a multiplicity of infection (MOI) of 1 and cultured in Dulbecco's modified Eagle's medium–10% FCS in the presence of 600 μg of Geneticin (Life Technologies) per ml. Plasmids used in the transfections were prepared using Qiagen plasmid kits as specified by the manufacturer. Cos-7 cells were seeded at 5 × 105 cells per 35-mm-diameter petri dish 24 h prior to transfection. Transfections were performed using Lipofectamine (Life Technologies) as described by the manufacturer, with 2-μg portions of the appropriate DNAs.
Gardella gel electrophoresis and Southern blot analysis.
Episomal DNA molecules were detected using the Gardella technique as previously described (9, 17). Horizontal gels were prepared in two steps. Initially a 0.75% agarose gel in Tris-borate-EDTA (TBE) buffer was poured. Once it had solidified, 5 cm of the gel was removed and replaced with 0.8% agarose containing 2% sodium dodecyl sulfate and 1 mg of self-digested pronase (Sigma) per ml. Control and HVS stably transduced A549 cells (106) were resuspended in sample buffer (15% Ficoll, 0.01% bromophenol blue) and electrophoresed at 4°C for 2 h at 40 V and then for 18 h at 160 V. DNA was detected by Southern blot analysis as previously described (44). DNA was hybridized with a 32P-radiolabelled random-primed probe specific for the KpnE fragment of the HVS genome, using the Megaprime kit (Amersham) as described by the manufacturer.
Chemical induction and virus recovery assays.
To induce a lytic replication cycle in the stably HVS-transduced A549 cell line, cells were incubated in the presence of either 20 ng of tetradecanoyl phorbol acetate (TPA) (Sigma, Poole, United Kingdom) per ml or 3 mM n-butyrate (Sigma) for 48 h. After chemical induction, virus recovery assays were performed. Serial dilutions of the harvested supernatants were used to infect 106 OMK cells. After 1 h at 37°C, the supernatants were removed and replaced with medium supplemented with 2% FCS and 0.45% (wt/vol) high-viscosity carboxymethyl cellulose (Sigma). The mixtures were transferred to dishes and incubated at 37°C for 5 to 7 days. The infected cells were subsequently fixed in Formol saline solution (0.85% [wt/vol] NaCl, 10% [vol/vol] formaldehyde) and stained with 0.1% (wt/vol) gentian violet, and plaques were counted.
Total-RNA extraction.
Total RNA was extracted from HVS stably transduced A549 cells and OMK cells infected with HVS (strain A11) at a MOI of 1, at various times postinfection. The cells were lysed using Trizol reagent (Life Technologies). Chloroform (0.2 ml) was then added, and the solution was vortex mixed for 20 s and stored at 20°C for 15 min. Samples were centrifuged for 15 min at 4°C, and the aqueous phase containing nucleic acids was precipitated using 0.5 ml of isopropanol. The pellet was then washed with 70% ethanol, resuspended in 50 μl of water, and stored at −70°C.
Northern blot analysis.
Northern blot analysis was performed essentially as described by Sambrook et al. (44). Total RNA was isolated from HVS stably transduced A549 cells or HVS-infected OMK cells and separated by electrophoresis on a 1% denaturing formaldehyde agarose gel. The RNA was transferred to Hybond-N membranes and hybridized with 32P-labeled random primed probes. Probes specific for ORFs 71 to 73 were amplified by PCR using the following primer pairs: ORF71F (5′-CGC GGA TCC GGC AAG GTC ACT TCG CCC TAT CTG) plus ORF71R (5′-CCG GAA TTC CTG TGT TAC ACA TAA CAG ACT), ORF72F (5′-CGC GGA TCC GCT GCA ATG GCA GAT TCA CC) plus ORF72R (5′-CCG GAA TTC GGT CTG CAG TTA GTG TTG TCA G-3), ORF73F (5′-ACG CGT CGA CCC ATC TAT AAT TGC AAC AAA CAC C) plus ORF73R (5′-CCC AAG CTT CAC ATA TAT GAA TGC TAG TGC AC), ORF74F (5′-GCT GGG TAT CTG CTA) plus ORF74R (5′-CCA ATA CAC TAT AGC), and ORF75F (5′-GAA CAT ATG CCA GCT ACA) plus ORF75R (5′-GTC TCT GGA TCT TCA AGC). The PCR (1 cycle of 5 min at 95°C; 30 cycles of 1 min at 95°C, 1 min at 55°C, and 2 min at 72°C; 1 cycle of 10 min at 72°C) was performed using 2 U of Klentaq (Clontech).
Rapid amplification of cDNA ends (RACE).
First-strand cDNA was reverse transcribed using Superscript II reverse transcriptase (Life Technologies) and either an oligo(dT) primer or gene-specific antisense primers: for ORF 71, 5′-GTG TTT CTA ATT GTG GCT; for ORF 72, 5′-CAC AGA TAT CTG TCT AAG; and for ORF 73, 5′-GTC ATC GTC GCC TTG AGG. The 5′ ends of the ORF 71 to 73 genes were located using the Clontech Marathon cDNA amplification kit as specified by the manufacturer. 5′-cDNA was amplified using the nested primers ORF 71 (5′-AAG AGG TTC AGT AAT AGT), ORF 72 (5′-AGC AGA TGC ATC CAG GTA), and ORF 73 (5′-GCA CAT GAC ACT TAC ATT). The amplified cDNAs were cycle sequenced using the fmol DNA-sequencing system (Promega).
Primer extension analysis.
Primer extension was performed essentially as described by Sambrook et al. (44). First, a 21-bp oligonucleotide primer (5′-CGT CTT CTC TTG CGA GGA CTT ACA) homologous to the ORF 73 coding region was radiolabeled using [γ-32P]ATP. Then 3 μg of RNA was mixed with 300 ng of the radiolabeled primer. The samples were boiled for 5 min and snap-chilled on ice. To the reaction mixtures were added 1 mM (each) dATP, dTTP, dCTP, and dGTP; buffer (final concentrations, 50 mM KCl, 10 mM Tris-HCl [pH 8.3], and 1.5 mM MgCl2); and 1 μl of Superscript II reverse transcriptase (Life Technologies) in a final volume of 20 μl. The reaction was performed at 42°C for 60 min, and primer extension products were resolved on a 6% acrylamide–7 M urea gel. After electrophoresis, the gel was dried and bands were visualized by exposure to X-ray film.
Plasmid constructs.
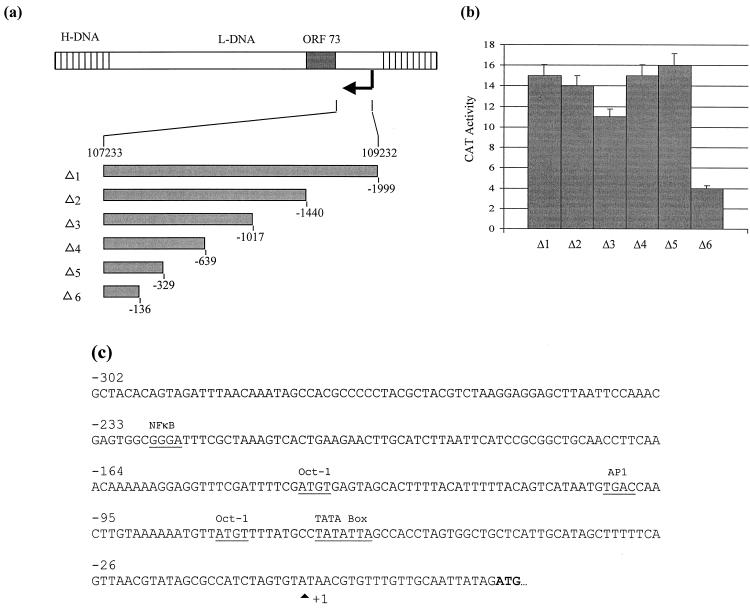
The 5′ ORF 73 promoter deletion series was produced by a PCR-based method using a series of forward primers, Δ1 (5′-AAA CTG CAG CCC AGA GAG CTG GAC ACT), Δ2 (5′-AAA CTG CAG CCA TGC AGC CAT GCG CTG), Δ3 (5′-AAA CTG CAG CAC CAT CAC ATG AGG AAG), Δ4 (5′-AAA CTG CAG CAC ATA TAT GAA TGC TAG), Δ5 (5′-AAA CTG CAG GTG GCT ACA CAG TA), and Δ6 (5′-AAA CTG CAG CAG TCA TAA TGT GAC C), and a reverse primer (5′-CGC GGA TCC CCA TCT ATA ATT GCA ACA AAC). These oligonucleotides incorporated PstI or SalI restriction sites for convenient cloning of the PCR products. Each fragment was inserted into the eukaryotic chloramphenicol acetyltransferase (CAT) reporter vector, pCATBasic (Promega), to derive p73Δ1-5.
CAT assay.
Cell extracts were prepared 48 h after transfection and incubated with [14C]chloramphenicol in the presence of acetyl coenzyme A as described previously (18). The percent acetylation of chloramphenicol was quantified by scintillation counting (Packard) of appropriate regions of the thin-layer chromatography plate.
RESULTS
Production of the HVS stably transduced A549 cell line.
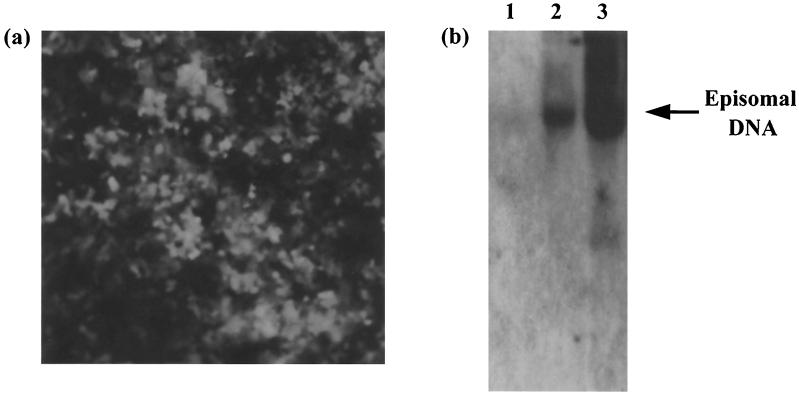
We have previously constructed a recombinant HVS, HVS-GFP, expressing the green fluorescent protein (GFP) and the neomycin resistance genes under the control of the immediate-early cytomegalovirus and simian virus 40 promoters, respectively (56). Analysis of this virus has demonstrated infection of a wide variety of human cell lines (47, 56). In addition, these cell lines are able to support episomal persistence of viral genomes, in agreement with previous observations in human hematopoietic cells and epithelial cells (19, 46). To generate a lung carcinoma A549 cell line which contained HVS as a nonintegrated circular episome, 106 A549 cells were infected with HVS-GFP and cultured in the presence of Geneticin. After 2 weeks, only cells which had been successfully transduced remained viable, with 100% exhibiting the GFP phenotype when analyzed by fluorescence microscopy (Fig. 1a). The cells continued to grow and express the transgene in the presence of selection for over 12 months, suggesting that the infection in this cell type is highly stable. To verify the existence of virus genomes in a circular episomal form, untransduced and HVS stably transduced cells were analyzed by Gardella gel electrophoresis and Southern blot analysis as previously described (9, 17). An HVS-transformed T-cell line from cottontop tamarin monkeys, which contains HVS as a circular nonintegrated episome, was also analyzed as a suitable control. Results show an episomal band clearly visible in the stably transduced A549 and transformed cells (Fig. 1b), demonstrating that HVS is present in a nonintegrated circular form in these A549 cells.
FIG. 1.
(a) Production of HVS stably transduced A549 cell line. A549 cells were infected with HVS-GFP at a MOI of 1 and cultured in the presence of Geneticin. After 2 weeks, only cells which had been successfully transduced remained viable, with 100% exhibiting the GFP phenotype when analyzed by fluorescence microscopy. (b) Gardella gel and Southern blot analysis of A549 cells (lane 1), HVS stably transduced A549 cells (lane 2), and HVS-transformed B133 T cells (lane 3). Episomal and linear DNAs were separated by electrophoresis, transferred to nitrocellulose, and hybridized with a radiolabeled 32P-labeled random-primed probe specific for the KpnE fragment of the HVS genome.
Virus production can be reactivated in HVS stably transduced A549 cells.
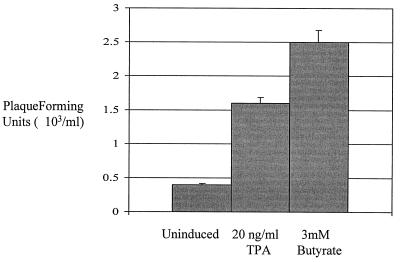
To determine if HVS episomes were maintained in a latent state in the stably transduced A549 cells, reactivation of virus production was attempted. To induce a lytic replication cycle, stably transduced A549 cells were incubated in the presence of either 20 ng of TPA per ml or 3 mM n-butyrate for 48 h. After chemical induction, virus recovery assays were performed. Serial dilutions of the control uninduced and induced harvested supernatants were used to infect 106 OMK cells, and plaque assays were performed. A significant increase in PFU was observed from supernatants taken from the chemically induced cell lines (Fig. 2). A very low level of virus production was observed in the uninduced cell line, suggesting that a small amount of spontaneous replication does occur in the stably transduced A549 cell line (Fig. 2). However, the induction experiment does suggest that in the stably transduced A549 cells, HVS is maintained as a latent nonintegrated circular episome.
FIG. 2.
A lytic replication cycle can be induced from the HVS stably transduced A549 cell line. HVS-infected A549 cells were incubated in the presence of either no addition (lane 1), 20 ng of TPA per ml (lane 2), or 3 mM n-butyrate (lane 3) for 48 h. After chemical induction, virus recovery assays were performed. The numbers of plaques formed are shown in graphical format: the variations between three replicate assays are indicated.
Identification of gene expression in HVS stably transduced A549 cells.
To identify which genes were expressed when the virus is maintained as a nonintegrated circular episome in stably transduced A549 cells, Northern blot analysis was performed. Total RNA was extracted from the HVS-GFP stably transduced A549 cell line. As controls, total RNA was extracted from lytic infections of OMK cells at 16 and 24 h postinfection and from uninfected A549 cells. Total RNA was separated by gel electrophoresis on a 1% denaturing formaldehyde-agarose gel, transferred to a nylon membrane, and hybridized with radiolabeled probes specific for a series of EcoRI or KpnI fragments spanning the entire L-DNA region of HVS (strain A11) (33).
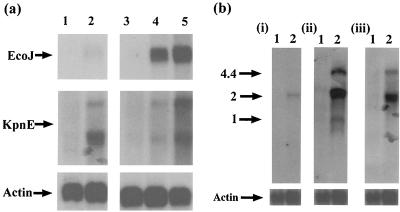
Northern blot analysis using RNA harvested from a lytic infection showed strong hybridization with all the HVS genomic restriction fragments, indicating a high level of global gene expression in the lytic replication cycle (data not shown). However, transcription during the latent infection of A549 cells was limited to genes within the KpnE restriction fragment. Two transcripts approximately 2 and 4 kb in length were identified in the stably transduced A549 cells. Although very weak, transcripts were faintly observed using probes specific for the highly expressed lytic gene ORF 57 in the latently infected samples (Fig. 3a). We believe that this may be due to a very low level of spontaneous lytic replication in a subpopulation of the A549 cells. These results suggest that the HVS genes expressed when the virus is maintained as a nonintegrated circular episome in the stably transduced A549 cell line are confined to the KpnE fragment, which contains ORFs 71 to 75.
FIG. 3.
(a) Transcription mapping of the HVS genome in latently infected A549 cells. Total RNA was extracted from A549 cells (lane 1), HVS-stably transduced A549 cells (lane 2), OMK cells (lane 3), 16-h-infected OMK cells (lane 4), and 24-h-infected OMK cells (lane 5) and then separated by gel electrophoresis on a 1% denaturing formaldehyde-agarose gel. The RNA was transferred to Hybond-N membranes and hybridized with radiolabeled probes specific for EcoJ (i) and KpnE (ii). (b) Total RNA was extracted from A549 cells (lane 1) and HVS-stably transduced A549 cells (lane 2) and separated by gel electrophoresis on a 1% denaturing formaldehyde-agarose gel. The RNA was transferred to Hybond-N membranes and hybridized with radiolabeled probes specific for ORF 71 (i), ORF 72 (ii), and ORF 73 (iii).
ORFs 71 to 73 are expressed in A549 cells stably transduced with recombinant HVS.
To further ascertain the pattern of gene expression observed in the stably transduced A549 cells, Northern blot analysis was repeated, with the same membrane, using probes specific for ORFs 71 to 75. The results are shown in Fig. 3b. Hybridization with a probe specific for ORF 71 detected only a single transcript of approximately 2 kb in the stably transduced A549 cells. The probe specific for ORF 73 detected the 2- and 4.4-kb transcripts, and a probe specific for ORF 72 detected three transcripts of 2, 4.4, and 1 kb. No signals were observed with probes specific for ORF 74 or 75 (data not shown). Therefore, gene expression which could be identified using Northern blot analysis was limited to ORFs 71 to 73 when HVS was maintained as a nonintegrated circular episome in the stably transduced A549 cell line.
Mapping the transcripts of ORFs 71 to 73.
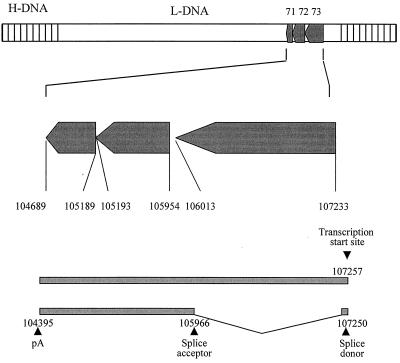
To characterize the multiple transcripts identified by Northern blot analysis, 5′ and 3′ RACE was performed using an oligo(dT) and gene-specific primers for ORFs 71 to 73. Analysis of the resulting PCR products revealed the transcription initiation start site was the same for ORFs 71, 72, and 73, situated 24 bp upstream of the ORF 73 initiation codon, at bp 107257 of the published sequence (1) (Fig. 4). Sequencing of the PCR products demonstrated that these transcripts were produced from a polycistronic mRNA species comprising the 4.4-kb transcript identified by the Northern blot analysis. In addition, the polycistronc mRNA species can be spliced, removing a single intron of 1,290 bp with a splice donor site at bp 107250 and an acceptor site at bp 105966 of the published sequence (1), to yield the 2-kb bicistronic mRNA containing ORFs 71 and 72 (Fig. 4). In addition, analysis demonstrated both transcripts terminated 30 bp downstream of a consensus AAUAAA polyadenylation signal at bp 104395 of the published sequence, 294 bp downstream of the ORF 71 termination codon. This suggests that both transcripts are processed at this consensus polyadenylation signal.
FIG. 4.
Schematic representation of the genomic organization of the ORF 71 to 73 region of HVS based on 5′ and 3′ RACE and primer extension analysis. The numbers indicate nucleotide positions based on the published sequence (1).
To confirm the ORF 73 transcription initiation site identified by 5′ RACE, primer extension analysis was also performed. Total RNA was isolated from untransduced A549 cells and HVS stably transduced cells and hybridized to a 32P-labeled primer homologous to the ORF 73 coding region. Primer extension was then performed, and the results are shown in Fig. 5. A primer extension product of 133 bp was observed using RNA harvested from the HVS stably transduced cell line but not from the control A549 cells, mapping the transcription start site of ORF 73 to bp 107257 of the published sequence (1), confirming the data obtained in the above 5′-RACE experiment.
FIG. 5.
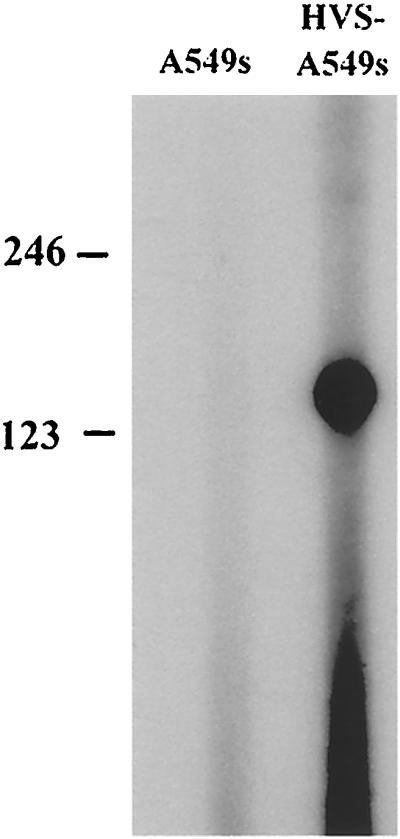
Primer extension analysis of the ORF 73 transcription initiation start site. Total RNA was isolated from A549 cells (lane 1) or HVS-infected A549 cells (lane 2) and hybridized to a 32P-labeled oligomer homologous to the ORF 73 coding region. Primer extension was performed, and the primer extension products were run on a 6% acrylamide–7 M urea gel and visualized by exposure to X-ray film.
Analysis of the ORF 71 to 73 regulatory region.
To determine which region upstream of the transcription initiation site contains a functional promoter from which the polycistronic mRNA transcript is produced, a series of deletion constructs from the putative promoter region were cloned upstream of a CAT reporter gene in pCAT-Basic (Fig. 6a). Each promoter construct was assayed for activity in transiently transfected Cos-7 cells. These were used due to the low transfection efficiency of the stably transduced A549 cells. Transfection efficiency was normalized using a control plasmid, pCMVβ (Clontech), expressing β-galactosidase. The results of three independent experiments are shown in Fig. 6b. A 1,999-bp fragment containing the sequence from bp 107233 to 109232 of the published sequence (1) resulted in 15% CAT activity. Similar results were observed using truncated promoter constructs, up to bp 107562. This suggests that the minimal functional promoter was contained within the sequence 329 bp upstream of the putative initiation site, i.e., bp 107233 to 107562 of the published sequence (1). To identify potential transcription factor binding sites within this putative ORF 73 promoter, MatInspector v2.2 was utilized (40). The putative promoter sequence reveals the presence of a TATA box and two potential Oct-1 binding sites (Fig. 6c).
FIG. 6.
(a) Diagrammatic representation of the deletion series of the ORF 73 promoter. A series of 5′ mutants were constructed by PCR amplification and ligated into pCAT-Basic. (b) Cos-7 cells were cotransfected with 2 μg of p73Δ1–6 in the presence of a transfection control plasmid, pCMVβ. Cells were harvested at 48 h posttransfection, and the cell extracts were assayed for CAT activity. Percentages of acetylation were calculated by scintillation counting of the appropriate regions of the chromatography plate and are shown in graphical format: the variations among three replicate assays are indicated. (c) Potential transcription factor binding sites within the ORF 73 promoter. The putative transcription start site and transcription factor binding sites are highlighted.
DISCUSSION
In this study, we have investigated viral gene expression in a stably transduced A549 cell line, where HVS persists as a nonintegrated episome, albeit under selection. We have demonstrated that virus production can be reactivated using both TPA and butyrate, suggesting that the virus is maintained in a latent state. Analysis of gene expression by Northern blot analysis, using RNA extracted from these stably transduced cells, has demonstrated that three genes are expressed at high levels, ORFs 71 to 73. Moreover, these genes are transcribed from a polycistronic mRNA species produced from a common promoter upstream of ORF 73.
These results are different from the gene expression observed in HVS-immortalized T cells. Northern blotting and subtractive hybridization techniques have demonstrated that the transforming STP and Tip genes and the immediate-early IE14/vSag genes are expressed (14, 15, 31). Although we have not detected the expression of other virus genes using Northern blot analysis, it cannot be excluded that a more sensitive method may identify additional virus gene expression. However, it is interesting that viral gene expression observed in the HVS stably transduced A549 cell line has similarities to gene expression during latent infections with other gammaherpesviruses, particularly KSHV (11, 29, 45, 50, 54). A recent survey of KSHV gene transcription in the PEL cell line showed that a region spanning ORFs 71 to 73 is expressed and that separate mRNAs for ORF 73 and ORF 72 are generated from a common latency-specific promoter (11). This result is further supported by the work of Sarid et al. (45) and Talbot et al. (50), who detected two transcripts of approximately 6.0 and 2.0 kb in BC-1 cells and PEL cell lines, respectively. The larger transcript encodes the ORF 73, ORF 72, and ORF K13 products, while the smaller transcript encodes only the ORF 72 and ORF K13 products but not the ORF 73 product. Although similar expression is observed in the HVS stably transduced A549 cells, it must be noted that the HVS transcripts are produced from a promoter that does not correspond precisely to the promoters mapped expressing the latent KSHV genes (11, 45). At present we are determining if a functional promoter is present in the corresponding position in the HVS genome.
Previous analysis of the HVS ORF 71 and ORF 72 genes has shown that they encode a FLICE antiapoptosis inhibitory protein (vFLIP) and a homologue of the cellular type D cyclin, respectively (22, 52). V-cyclin associates with cdk6, and this complex is capable of directing phosphorylation of the retinoblastoma protein (22). Expression of these two gene products in the latent stably transduced A549 cells may serve to protect virus-infected cells from death receptor-induced apoptosis and drive host cell transit through the pRB-controlled G1 checkpoint, enabling viral DNA synthesis to occur. At present, the role of the ORF 73 gene product has yet to be conclusively determined.
ORF 73 is also transcribed in KSHV and MHV-68 latent infections (11, 45, 50, 54). Sera from KSHV-infected individuals react in immunofluorescence studies with LANA in latently infected BCBL cell lines, giving a characteristic speckled immunofluorescence pattern. In Western blot analysis of nuclear extracts from the BCBL cell line BC-1, patient sera have been shown to react with a 222- and 234-kDa doublet band, termed the latent nuclear antigen (LNA). It has been shown that LNA is encoded by ORF 73 of KSHV and that antibodies to this protein produce the speckled nuclear immunofluorescence pattern characteristic of LANA. This suggests that the ORF 73 LNA product is at least a component of LANA (27, 28, 41).
It is interesting that ORF 73 is contained in a region of the HVS genome which is poorly conserved among gammaherpesviruses. Equine herpesvirus 2, for example, lacks an ORF 73 homologue (51), and the ORF 73 homologues of bovine herpesvirus 4 and MHV-68 do not contain the internal acidic repeat region found in the ORF 73 of KSHV and HVS (36, 53). A similar acidic domain is found in the latent EBV EBNA-1 protein, which inhibits recognition of this antigen by cytotoxic T lymphocytes (35). In addition, the speckled nuclear staining pattern of ORF 73-encoded LANA is similar to that observed with EBNA-2 and EBNA-LP, which are the earliest proteins expressed in B lymphocytes newly infected with EBV and are essential for B-lymphocyte transformation (reviewed in reference 29). Furthermore, LANA colocalizes with KSHV DNA in dots in interphase nuclei and along mitotic chromosomes, and LANA is required for the persistence of episomes containing a specific KSHV cis-acting region. Moreover, LANA associates with histone HI in KSHV-infected BCBL cells, suggesting that LANA tethers KSHV DNA to chromosomes via host chromosomal proteins during mitosis, allowing the segregation of the KSHV episomes to progeny cells (2, 8). Whether HVS ORF 73 plays a role in host cell transformation, episomal persistence or cell cycle regulation is unknown and further investigation of the role of the ORF 73 gene product in HVS episomal maintenance is required.
In conclusion, we have developed an in vitro model which may be valuable in the study of genes involved in episomal maintenance of gamma-2 herpesviruses, based on the nontransformed lung carcinoma cell line A549. The stably transduced cell line contains HVS as a nonintegrated episome which is efficiently segregated to progeny cells. Analysis of viral gene expression in this model has shown that three genes, ORFs 71 to 73, are expressed as a polycistronic mRNA from a common promoter upstream of ORF 73. This model may serve as a useful tool in the further analysis of the role of ORFs 71 to 73 in gamma-2 herpesvirus latency. In particular, due to the lack of a permissive cell culture system for KSHV and the fact that it is possible to produce recombinant HVS mutants, we believe that this HVS model may serve as a useful tool in the further analysis of the role of ORFs 71 to 73 in KSHV latency.
ACKNOWLEDGMENTS
We thank Helmut Fickenscher for providing the library of HVS-11 genomic clones and the transformed B133 cell line and for helpful advice.
This work was supported in parts by grants from Medical Research Council, Yorkshire Cancer Research, Candlelighter's Trust, and the Wellcome Trust. A.W. and D.J.G. are the recipients of an MRC fellowship and Ph.D. studentship, respectively. M.S.G. is a Wellcome Trust Entry Level Clinical Research Fellow.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas M E, Chatis P A, Kaye K M. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]
- 3.Behrend K, Jung J, Boyle T, DiMaio M, Mungal S, Desrosiers R, Lyerly K. Phenotypic and functional consequences of herpesvirus saimiri infection of human CD8+ cytotoxic T lymphocytes. J Virol. 1993;67:6317–6321. doi: 10.1128/jvi.67.10.6317-6321.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biesinger B, Tsygankov A, Fickenscher H, Emmrich F, Fleckenstein B, Bolen J, Broker B. The product of the herpesvirus saimiri open reading frame 1 (tip) interacts with T cell-specific kinase p56lck in transformed cells. J Biol Chem. 1995;270:4729–4734. doi: 10.1074/jbc.270.9.4729. [DOI] [PubMed] [Google Scholar]
- 5.Biesinger B, Muller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biesinger B, Trimble J, Desrosiers R C, Fleckenstein B. The divergence between two oncogenic herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology. 1990;176:505–514. doi: 10.1016/0042-6822(90)90020-r. [DOI] [PubMed] [Google Scholar]
- 7.Broker B, Tsygankov A, Muller-Fleckenstein I, Guse A, Chitaev N, Biesinger B, Fleckenstein B, Emmrich F. Immortalisation of human T cell clones by herpesvirus saimiri. Signal transduction analysis reveals functional CD3, CD4 and IL-2 receptors. J Immunol. 1993;51:1184–1192. [PubMed] [Google Scholar]
- 8.Cotter M A, Robertson E S. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma. Virology. 1999;264:254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- 9.Decker L L, Klaman L D, Thorley-Lawson D A. Detection of the latent form of Epstein-Barr virus DNA in the peripheral blood of healthy individuals. J Virol. 1996;70:3286–3289. doi: 10.1128/jvi.70.5.3286-3289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desrosiers R C, Silva D, Waldron L, Letvin N. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J Virol. 1986;57:701–705. doi: 10.1128/jvi.57.2.701-705.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duboise S M, Guo J, Czajak S, Desrosiers R C, Jung J U. STP and Tip are essential for herpesvirus saimiri oncogenicity. J Virol. 1998;72:1308–1313. doi: 10.1128/jvi.72.2.1308-1313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duboise S M, Guo J, Czajak S, Lee H, Veazey R, Desrosiers R C, Jung J U. A role for herpesvirus saimiri orf14 in transformation and persistent infection. J Virol. 1998;72:6770–6776. doi: 10.1128/jvi.72.8.6770-6776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fickenscher H, Biesinger B, Knappe A, Wittman S, Fleckenstein B. Regulation of the herpesvirus saimiri oncogene stpC, similar to that of T-cell activation genes, in growth-transformed human T lymphocytes. J Virol. 1996;70:6012–6019. doi: 10.1128/jvi.70.9.6012-6019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fickenscher H, Bokel C, Knappe A, Biesinger B, Meinl E, Fleischer B, Fleckenstein B, Broker B M. Functional phenotype of transformed human alpha/beta and gamma/delta T cells determined by different subgroup C strains of herpesvirus saimiri. J Virol. 1997;71:2552–2262. doi: 10.1128/jvi.71.3.2252-2263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleckenstein, B., and R. C. Desrosiers. Herpesvirus saimiri and herpesvirus ateles, p. 253–332. In B. Roizman (ed.), The herpesviruses, vol. 1. Plenum Press, New York, N.Y.
- 17.Gardella T, Medveczky P, Sairenji T, Mulder C. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyl-transferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grassmann R, Fleckenstein B. Selectable recombinant herpesvirus saimiri is capable of persisting in a human cell line. J Virol. 1989;63:1818–1821. doi: 10.1128/jvi.63.4.1818-1821.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiller C, Wittmann S, Slavin S, Fickenscher H. Functional long-term thymidine kinase suicide gene expression in human T cells using a herpesvirus saimiri vector. Gene Ther. 2000;7:664–674. doi: 10.1038/sj.gt.3301158. [DOI] [PubMed] [Google Scholar]
- 21.Jung J U, Trimble J J, King N W, Biesinger B, Fleckenstein B W, Desrosiers R C. Identification of transforming genes of subgroup A and C strains of herpesvirus saimiri. Proc Natl Acad Sci USA. 1991;88:7051–7055. doi: 10.1073/pnas.88.16.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung J U, Stager M, Desrosiers R C. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung J U, Desrosiers R C. Association of the viral oncoprotein STP-C488 with cellular Ras. Mol Cell Biol. 1995;15:6506–6512. doi: 10.1128/mcb.15.12.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung J U, Desrosiers R C. Identification and characterization of the herpesvirus saimiri oncoprotein, STP-C488. J Virol. 1991;65:6953–6960. doi: 10.1128/jvi.65.12.6953-6960.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung J U, Lang S, Jun T, Roberts T M, Veillette A, Desrosiers R C. Downregulation of Lck-mediated signal transduction by tip of herpesvirus saimiri. J Virol. 1995;69:7814–7822. doi: 10.1128/jvi.69.12.7814-7822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaschaka-Dierich C, Werner F J, Bauer I, Fleckenstein B. Structure of nonintegrated circular herpesvirus saimiri and herpesvirus ateles genomes in tumor cell lines and in vitro-immortalized cells. J Virol. 1982;44:295–310. doi: 10.1128/jvi.44.1.295-310.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kedes D H, Lagunoff M, Renne R, Ganem D. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J Clin Investig. 1997;100:2606–2610. doi: 10.1172/JCI119804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kellam P, Boshoff C, Whitby D, Matthews S, Weiss R A, Talbot S J. Identification of a major latent nuclear antigen LNA-1, in the human herpesvirus 8 genome. J Hum Virol. 1997;1:19–29. [PubMed] [Google Scholar]
- 29.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2396. [Google Scholar]
- 30.Knappe A, Hiller C, Niphuis H, Fossiez F, Thurau M, Wittmann S, Kuhn E M, Lebecque S, Banchereau J, Rosenwirth B, Fleckenstein B, Heeney J, Fickenscher H. The interleukin-17 gene of herpesvirus saimiri. J Virol. 1998;72:5797–5801. doi: 10.1128/jvi.72.7.5797-5801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knappe A, Hiller C, Thurau M, Wittman S, Hofmann H, Fleckenstein B, Fickenscher H. The superantigen-homologous viral immediate-early gene ie14/vsag in herpesvirus saimiri-transformed human T cells. J Virol. 1997;71:9124–9133. doi: 10.1128/jvi.71.12.9124-9133.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knappe A, Thurau M, Niphuis H, Hiller C, Wittman S, Kuhn E M, Rosenwirth B, Fleckenstein B, Heeney J, Fickenscher H. T-cell lymphoma caused by herpesvirus saimiri C488 independently of ie14/vsag, a viral gene with superantigen homology. J Virol. 1998;72:3469–3471. doi: 10.1128/jvi.72.4.3469-3471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knust E, Schirm S, Dietrich W, Bodemer W, Kolb E, Fleckenstein B. Cloning of herpesvirus saimiri DNA fragments representing the entire L-region of the genome. Gene. 1983;25:281–289. doi: 10.1016/0378-1119(83)90232-9. [DOI] [PubMed] [Google Scholar]
- 34.Kung S H, Medveczky P G. Identification of a herpesvirus saimiri cis-acting DNA fragment that permits stable replication of episomes in transformed T cells. J Virol. 1996;70:1738–1744. doi: 10.1128/jvi.70.3.1738-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levitskaya J, Coram M, Levitsky V, Imreh S, Stigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 36.Lomonte P, Bublot M, van Santen V, Keil G M, Pastoret P-P, Thiry E. Analysis of bovine herpesvirus 4 genomic regions located outside the conserved gammaherpesvirus gene blocks. J Gen Virol. 1995;76:1835–1841. doi: 10.1099/0022-1317-76-7-1835. [DOI] [PubMed] [Google Scholar]
- 37.Lund T, Medveczky M, Neame P, Medveczky P. A herpesvirus saimiri membrane protein required for interleukin-2 independence forms a stable complex with p56lck. J Virol. 1996;70:600–606. doi: 10.1128/jvi.70.1.600-606.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein-Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niepel F, Albrecht J C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector—new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rainbow L, Platt G M, Simpson G R, Sarid R, Gao S-J, Stoiber H, Herrington C S, Moore P S, Schulz T F. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J Virol. 1997;71:5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo J J, Bohenzhy R A, Chein M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequences of the Kaposi's sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Sarid R, Wiezorek J S, Moore P S, Chang Y. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J Virol. 1999;73:1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simmer B, Alt M, Buckreus I, Berthold S, Fleckenstein B, Platzer E, Grassman R. Persistence of selectable herpesvirus saimiri in various human haematopoietic and epithelial cell lines. J Gen Virol. 1991;72:1953–1958. doi: 10.1099/0022-1317-72-8-1953. [DOI] [PubMed] [Google Scholar]
- 47.Stevenson A J, Cooper M, Griffiths J C, Gibson P G, Whitehouse A, Jones E F, Kinsey S E, Markham A F, Meredith D M. Assessment of herpesvirus saimiri as a potential human gene therapy vector. J Med Virol. 1999;57:269–277. [PubMed] [Google Scholar]
- 48.Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphocytes transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szomolanyi E, Medvecsky P, Mulder C. In vitro immortalization of marmoset cells with three subgroups of herpesvirus saimiri. J Virol. 1987;61:3485–3490. doi: 10.1128/jvi.61.11.3485-3490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 51.Telford E A R, Watson M S, Aird H C, Perry J, Davison A J. The DNA sequence of equine herpesvirus 2. J Mol Biol. 1995;249:520–528. doi: 10.1006/jmbi.1995.0314. [DOI] [PubMed] [Google Scholar]
- 52.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory protein (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 53.Virgin H W, IV, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Virgin H W, IV, Presti R M, Li X-Y, Liu C, Speck S H. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J Virol. 1999;73:2321–2332. doi: 10.1128/jvi.73.3.2321-2332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Werner F, Bornkamm G W, Fleckenstein B. Episomal viral DNA in a herpesvirus saimiri-transformed lymphoid cell line. J Virol. 1977;22:794–803. doi: 10.1128/jvi.22.3.794-803.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitehouse A, Stevenson A J. Gene regulation in herpesvirus saimiri and its implication towards the development of a novel gene therapy vector. Gene Ther Mol Biol. 1999;3:35–44. [Google Scholar]
- 57.Wiese N, Tsyganov A, Klauenberg U, Bolen J, Fleischer B, Broker B. Selective activation of T cell kinase p56lck by herpesvirus saimiri protein Tip. J Biol Chem. 1996;271:847–852. doi: 10.1074/jbc.271.2.847. [DOI] [PubMed] [Google Scholar]
- 58.Yao Z, Maraskovsky E, Spriggs M K, Cohen J I, Armitage R J, Alderson M R. Herpesvirus saimiri open reading frame 14, a protein encoded by a T lymphotrophic herpesvirus, binds to MHC class II molecules and stimulates T cell proliferation. J Immunol. 1996;56:3260–3266. [PubMed] [Google Scholar]