Abstract
The E1 helicase of papillomavirus is required, in addition to host cell DNA replication factors, during the initiation and elongation phases of viral episome replication. During initiation, the viral E2 protein promotes the assembly of enzymatically active multimeric E1 complexes at the viral origin of DNA replication. In this study we used the two-hybrid system and chemical cross-linking to demonstrate that human papillomavirus type 11 (HPV11) E1 can self-associate in yeast and form hexamers in vitro in a reaction stimulated by single-stranded DNA. Self-association in yeast was most readily detected using constructs spanning the E1 C-terminal domain (amino acids 353 to 649) and was dependent on a minimal E1-E1 interaction region located between amino acids 353 and 431. The E1 C-terminal domain was also able to oligomerize in vitro but, in contrast to wild-type E1, did so efficiently in the absence of single-stranded DNA. Sequences located between amino acids 191 and 353 were necessary for single-stranded DNA to modulate oligomerization of E1 and were also required, together with the rest of the C terminus, for binding of E1 to the origin. Two regions within the C-terminal domain were identified as important for oligomerization: the ATP-binding domain and region A, which is located within the minimal E1-E1 interaction domain and is one of four regions of E1 that is highly conserved with the large T antigens of simian virus 40 and polyomavirus. Amino acid substitutions of highly conserved residues within the ATP-binding domain and region A were identified that reduced the ability of E1 to oligomerize and bind to the origin in vitro and to support transient DNA replication in vivo. These results support the notion that oligomerization of E1 occurs primarily through the C-terminal domain of the protein and is allosterically regulated by DNA and ATP. The bipartite organization of the E1 C-terminal domain is reminiscent of that found in other hexameric proteins and suggests that these proteins may oligomerize by a similar mechanism.
Papillomaviruses are small DNA viruses that are the etiological agents of benign and malignant lesions of the differentiating mucosal and cutaneous epithelium (reviewed in references 6, 20, 45, and 61). The life cycle of these viruses is closely associated with the differentiation program that occurs in the epithelium. Soon after infection, the viral genome is established as a low-copy-number extrachromosomal episome in the nuclei of infected basal cells. As these infected cells start to differentiate from the basal layer toward the upper portion of the epithelium, the viral episome is replicated and maintained at approximately 50 copies per cell. When the infected cells reach the suprabasal layers, amplification of the viral episome occurs to a high copy number, probably as a result of a change in the mode of DNA replication from a bidirectional theta mode to a rolling-circle mode (11). In these upper layers, capsid proteins are synthesized and viral particles are assembled, and they are eventually shed together with the upper layer of the epithelium (20).
In addition to host cell DNA replication factors, two virally encoded proteins, E1 and E2, are directly involved in replication of the viral genome (5, 21, 53, 57). E1 is a DNA helicase (reviewed in reference 50) that is required during the initiation phase of DNA replication as well as during elongation to unwind the DNA ahead of the replication fork (25). E2 is a sequence-specific DNA-binding protein that functions as both a transcription factor and a replication factor (reviewed in reference 32). E2 binds with high affinity to specific sites located in the viral origin of DNA replication and, by forming a complex with E1, enhances the binding of E1 specifically at the origin (2, 13, 14, 28–30, 33, 39, 42, 57). For bovine papillomavirus (BPV) E1 and E2, it has been shown that this highly sequence-specific E1-E2-ori ternary complex then serves as a precursor for the recruitment of additional E1 molecules and assembly of larger multimeric E1-ori complexes that are able to distort the origin (14, 30, 37). In vitro, multimeric E1-ori complexes can be formed either in the presence of E2, via an E1-E2-ori complex intermediate, or in the absence of E2 if high concentrations of E1 are used (30, 37). Multimeric E1-ori complexes formed with or without E2 appear to be identical, since they make similar contacts with the ori DNA and are equally able to distort the origin (14, 37). In vitro and probably in vivo, formation of a multimeric E1-ori complex is accompanied by displacement of E2 from the origin (30, 37). Hence, E2 catalyzes the formation of the multimeric E1-ori complex and is not retained within it.
Different studies using BPV proteins have investigated the effect of ATP-Mg on the formation of the multimeric E1-ori complex. In the absence of E2, ATP and, to a lesser extent, nonhydrolyzable ATP analogues were found to stimulate the formation of the E1-ori complex by increasing its stability but without affecting how E1 contacts the DNA (29, 37, 43, 44). ATP can also stimulate formation of the E1-ori complex in the presence of E2 (29, 37). One study indicated that ATP hydrolysis is required for E2 to stimulate the formation of multimeric E1-ori complexes and for the concomitant displacement of E2 from the origin (37). ATP hydrolysis was also found to be essential for E1 to distort the origin (14). For human papillomavirus type 11 (HPV11) E1, ATP, but not its hydrolysis, stimulates the formation of the E1-ori complex in the absence of E2 (reference 25 and see below). Previously, we showed that ATP and ADP stimulate the E2-dependent binding of E1 to ori, not by affecting the E1-E2 protein-protein interaction but most probably by enhancing the interaction of E1 with DNA (52). Collectively, these results support a model in which ATP-Mg allosterically promotes the assembly of multimeric E1-ori complexes by facilitating conformational changes in E1 that favor oligomerization of the protein, enhance its interaction with the origin, and facilitate displacement of E2.
Several studies attempted to determine the stoichiometry of E1 within the multimeric E1-ori complex. Earlier reports indicated that in the presence of ATP-Mg, BPV E1 binds the origin as a large complex, presumably a double hexamer (30). A different study suggested that in the absence of ATP, E1 forms a trimeric ring-like structure around the origin (40). Recently, three reports confirmed, by biochemical means and electron microscopy, that BPV E1 (12, 41) and HPV11 E1 (24) can assemble into hexameric structures in a reaction that is stimulated by duplex or single-stranded DNA. Oligomerization of E1 was associated with increased ATPase activity (41) and correlated with unwinding activity (12, 41). Thus, the enzymatically active form of E1 is a hexamer. Electron microscopy revealed that E1 assembles on the origin as bilobed structures, most probably double hexamers needed for bidirectional unwinding (12, 24). The human heat shock proteins Hsp70 and Hsp40 are each able to stimulate the binding of E1 to DNA, the latter by promoting the assembly of double hexamers (24). Together, these findings suggest that oligomerization of E1 on DNA may be accompanied by a reorganization of the enzyme catalytic center to favor helicase activity in a process that can be chaperoned by heat shock proteins.
E1 and E2 also interact with essential host replication factors required for viral genome replication. E2 binds to the human single-stranded DNA-binding protein (HSSB/RPA) (23), while E1 can interact with both HSSB/RPA (19) and the polymerase α primase complex (1, 3, 8, 31, 35). Mechanistic studies have identified a domain of HPV E1, spanning approximately the C-terminal 300 amino acids, that is sufficient for binding to both the p70 subunit of the polymerase α primase complex (1, 31) and the transactivation domain of E2 (19, 31, 34, 48, 52, 59, 60) in a mutually exclusive manner (1, 8, 31). This C-terminal domain of E1 has extensive similarity over four highly conserved regions, termed A, B, C, and D, to the corresponding regions of the large T antigens of simian virus 40 (SV40) and other polyomaviruses (Fig. 1) (7). Conserved regions B and C form part of the ATPase domain and encompass three short sequence motifs, named a, b, and c, that are shared among members of superfamily 3 of nucleoside triphosphate (NTP)-binding proteins (17). Motifs a and b correspond to the Walker A and B boxes, which are involved, respectively, in binding the triphosphate moiety of the substrate nucleotide and in coordinating the Mg2+ ion associated with the nucleotide (54). The exact functions of conserved regions A and D of E1 have not yet been described. Despite the fact that it contains the ATPase catalytic site as well as the E2 and polymerase α primase interaction domains, the C-terminal domain of HPV11 E1 (amino acids 353 to 649) is unable to support viral DNA replication in vitro in a human cell-free system (1). This is most probably because this domain lacks sequences located between amino acids 200 and 300 that are required for binding to the origin (reference 48 and see below) and that may also be required for helicase activity.
FIG. 1.
Domain structure of the HPV11 E1 helicase. The 649-amino-acid HPV11 E1 helicase is diagrammed as a box at the center of the figure. Black boxes labeled A, B, C, and D indicate regions of E1 that are conserved in the large T antigens of SV40 and polyomaviruses. The region of HPV11 E1 that corresponds to the minimal DNA-binding domain mapped in BPV E1 (see the text for references) is also indicated as a black box. Hatched boxes indicate two hydrophilic amino acid sequences, termed HR1 and HR3, within this minimal DNA-binding domain. The locations and consensus amino acid sequence of conserved motifs a, b, and c that are common to members of superfamily 3 of NTP-binding proteins are indicated above E1. A sequence alignment of conserved region A from SV40 T antigen, BPV1, and HPV11 is shown below E1. Highly conserved residues in region A are shaded in black. The two cysteine and two histidine residues in SV40 T antigen that are involved in binding zinc are underlined. Residues that were mutated in this study are indicated by asterisks.
Here we provide evidence that the C-terminal domain of HPV11 E1 (amino acids 353 to 649) is sufficient for oligomerization of the protein in the yeast two-hybrid system and for formation of hexamers in vitro. Two regions within this C-terminal domain have been identified as important for oligomerization, the ATP-binding domain and conserved region A, which lies in a minimal E1-E1 interaction domain (amino acids 353 to 431) identified in yeast. Mutational analysis of these two subdomains indicated that they are required for E1 to oligomerize and to bind to the origin in vitro as well as to support transient HPV DNA replication in vivo. Oligomerization of the C-terminal domain in vitro, unlike that of wild-type E1, was found to occur readily in absence of single-stranded DNA. Collectively, these results suggest that DNA and ATP allosterically regulate the oligomerization of the E1 C-terminal domain.
MATERIALS AND METHODS
Yeast two-hybrid analysis.
Saccharomyces cerevisiae strain Y153 (MATa leu2-3,112 ura3-52 trp1-901 his3-Δ200 ade2-101 gal4Δ gal80Δ URA3::GAL-lacZ LYS::GAL-HIS3) was used for two-hybrid analysis (10). Transformation of yeast strain Y153 and β-galactosidase assays were performed as described previously (52). β-Galactosidase activity was measured spectrophotometrically (using the optical density at 578 nm [OD578]) with the substrate chlorophenyl-red-β-d-galactopyranoside (Boehringer Mannheim) and calculated using the equation activity (Miller units) = (1,000 × OD578)/(elapsed time [minutes] × 1.5 ml of culture × OD600).
Plasmid constructions. (i) In vitro transcription-translation.
Plasmids used for synthesis of wild-type and truncated HPV-11 E1 in vitro were derived from pTM1 (obtained from Bernard Moss, National Institutes of Health), which carries a T7 promoter and the encephalomyocarditis virus internal ribosome entry site. To construct plasmid pTM1-E1, which expresses wild-type E1, the complete open reading frame (ORF) was amplified by PCR using oligonucleotides 5′-GTACGATCCCATGGCGGACGATTCAGGTACAGAAAAT-3′ and 5′-GTACGATGGGATCCTTATTATAAAGTTCTAACAACTGATCCTGGCAC-3′. The resulting PCR product was digested with NcoI and BamHI (encoded by the two oligonucleotides) and inserted between the NcoI and BamHI sites of plasmid pTM1. Plasmids to express N-terminally truncated E1 proteins in vitro were constructed by amplification of the desired portion of the E1 ORF with specific primers bearing an NcoI site and a BamHI site. PCR products were digested with NcoI and BamHI and inserted between the NcoI and BamHI sites of plasmid pTM1. The sequences of the different primers that were used for PCR will be provided upon request. Plasmid pTM1-FLAG-E1(72–649), which encodes a truncated HPV-11 E1 protein that lacks the N-terminal 71 amino acids but which is tagged at its N-terminus with the FLAG epitope (Met Asp Tyr Lys Asp Asp Asp Asp Lys [Kodak]), was constructed by PCR amplification using an oligonucleotide which encodes the FLAG epitope, 5′-GGGGGCCATGGACTACAAGGACGACGACGACAAGGCGGATGCTCATTATGACTG-3′ (the sequence encoding the FLAG epitope is underlined), and the reverse primer 5′-CCCGGATCCTCATAAAGTTCTAACAACT-3′. Plasmids similar to pTM1-FLAG-E1(72–649) but which encode E1 proteins with a truncated C terminus were constructed by PCR amplification of the desired portion of the E1 ORF with the FLAG epitope primer and the appropriate reverse primer bearing a BamHI restriction site. PCR products were digested with NcoI and BamHI and inserted between the NcoI and BamHI sites of plasmid pTM1. The sequence of the various reverse primers will be provided upon request.
(ii) Yeast two-hybrid system.
Unless described otherwise, HPV-11 E1 DNA fragments were amplified by PCR with specific primers bearing an NcoI site (forward primer) or a BamHI site (reverse primer). PCR products were digested with NcoI and BamHI and inserted, in frame, between the NcoI and BamHI sites of the yeast two-hybrid vectors pAS1 (GAL4 DNA-binding domain) and pACT2 (GAL4 activation domain) (10). Two hybrid plasmids encoding the complete E1 protein (amino acids 1 to 649) were constructed in a similar way, except that the forward primer contained a BamHI site instead of an NcoI site. In this case the PCR product was cut with BamHI and inserted, in frame, into the BamHI sites of pAS1 and pACT2. The sequences of the various forward and reverse primers will be provided upon request. pAS1- and pACT2-derived plasmids encoding E1 residues 353 to 572, 353 to 536, and 353 to 458 were constructed in two steps. In the first step, E1 sequences were amplified by PCR using primers 5′-GGCTGGATCCATGGACAGTCAATTTAAATTAACT-3′ and 5′-CCCGGATCCAGTGTGATGGATATCTGCAG-3′ (pCR3). The templates for PCR were pCR3-derived plasmids expressing a truncated E1 ORF: either E1 amino acids 1 to 572, 1 to 536, or 1 to 458. One of the two oligonucleotides used for PCR amplification hybridizes over codon 353 of E1. The other oligonucleotide (pCR3) hybridizes in the polylinker region of pCR3, downstream of the truncated E1 ORF. The PCR products were digested with NcoI and BamHI and cloned between the NcoI and BamHI sites of pAS1 and pACT2.
(iii) Transient HPV DNA replication.
Plasmids that were used in transient HPV DNA replication assays to express E1 and E2 in transfected cells were all derived from pCR3 (Invitrogen, Carlsbad, Calif.) and have been described previously (52). Plasmid pN9 (27) was obtained from D. McCance (University of Rochester) and contains the complete origin of replication of HPV-11 (nucleotides 7884 to 61) cloned into pBluescriptII SK(+) (Stratagene).
Site-directed mutagenesis.
Site-directed mutagenesis of E1 was performed with the QuickChange site-directed mutagenesis kit (Stratagene) as specified by the manufacturer. For each mutagenesis, a pair of complementary oligonucleotides was used. The sequences of the various oligonucleotide pairs used for mutagenesis will be provided upon request.
E1 origin-binding assays.
The TNT coupled reticulocyte lysate system (Promega) was used to produce the E1 protein by coupled transcription-translation in vitro. The lysate was programmed with 2 μg of the appropriate plasmid per 50 μl of TNT reticulocyte lysate as specified by the manufacturer. When required, the E1 protein was radiolabeled by incorporation of [35S]methionine. We estimated previously (52) by Western blotting of different amounts of E1-containing lysate and of purified E1 as standards that approximately 5 to 20 ng of E1 was synthesized per μl of in vitro transcription-translation reaction mixture. Binding reactions were performed by mixing 30 μl of lysate containing E1, 200 to 400 ng of a 33P-radiolabeled DNA probe, and 7.5 μl of 10× DNA binding buffer (200 mM Tris-HCl [pH 7.6], 1 M NaCl, 10 mM EDTA, 10 mM dithiothreitol [DTT]) in a final volume of 75 μl. Binding reactions were allowed to proceed at the indicated temperature for 90 min. When indicated, ATP (or a related nucleotide) and MgCl2 were added to the binding-reaction mixtures at final concentrations of 5 and 3 mM, respectively. DNA-protein complexes were immunoprecipitated either with the anti-FLAG M2 monoclonal antibody (Eastman Kodak), when using FLAG-tagged E1, or with the K72 polyclonal antibody which was raised in rabbits against a peptide derived from the C-terminal 15 amino acids of HPV11 E1. Before use in immunoprecipitation, the antibodies were prebound to either protein G-Sepharose beads (when using anti-FLAG) or protein A-Sepharose beads (when using K72). Immunoprecipitation of protein-DNA complexes was carried out for 1 h at the binding-reaction temperature. Complexes were washed three times with 200 μl of wash buffer (50 mM Tris [pH 7.6], 100 mM NaCl, 0.1% Triton X-100). DNA present in these complexes was extracted with phenol-chloroform and precipitated with ethanol in the presence of carrier yeast tRNA. The precipitated radiolabeled DNA fragments were resolved on a 5% polyacrylamide Tris-borate-EDTA (TBE) gel and visualized by autoradiography. The radiolabeled probe that was used in these experiments consists of two DNA fragments and was prepared in two steps. In the first step, plasmid pN9 was linearized by digestion with XmaI and the ends were labeled with the Klenow fragment of DNA polymerase I in the presence of 5 μCi of [α-32P]dCTP and 0.1 mM each dTTP, dATP, and dGTP. Labeled DNA was purified on QIAquick PCR purification columns (Qiagen). In the second step, linear radiolabeled pN9 was digested with PvuII to generate two labeled fragments: a 370 bp fragment which contains the HPV-11 origin of replication and a 186-bp control fragment which lacks the origin.
The E2-dependent E1 origin-binding assay was performed as described previously (52). This assay is similar to the E1 origin-binding assay described above, except that 15 μl of in vitro-synthesized HPV11 E2 is added to each binding-reaction mixture.
Cross-linking of in vitro-translated E1.
35S-labeled E1 protein was made by in-vitro transcription-translation (TNT coupled reticulocyte lysate system) as specified by the manufacturer and cross-linked using the sulfhydryl-reacting cross-linker bismaleimidohexane (BMH; Pierce). For each reaction, 12.5 μl of translated E1 was incubated for 1 h at 37°C or at the indicated temperature in the presence or absence of 50 ng of single-stranded DNA (60-mer, corresponding to nucleotides 7902 to 34 of the HPV11 origin) per μl in a final volume of 37.5 μl containing 20 mM Tris (pH 7.6), 100 mM NaCl, 1 mM DTT, 5 mM ATP, and 3 mM MgCl2. Cross-linking was performed by diluting the binding-reaction mixtures 13-fold with 0.1 M phosphate buffer (pH 7.0) containing 100 μM BMH. The cross-linking reactions were stopped after 1 min by addition of DTT to a final concentration of 2.5 mM. E1 proteins were then immunoprecipitated with a polyclonal antibody directed against the C-terminal 14 amino acids of HPV11 and analyzed by gel electrophoresis on a 3% Weber-Osborn polyacrylamide gel (55) and autoradiography.
GST pull-down assay.
Purified glutathione S-transferase (GST) and GST-E2 transactivation domain (TAD) were purified as described previously (52) and immobilized on gluthathione beads (Amersham Pharmacia Biotech) at approximately 1 mg/ml. GST pull-down assays were performed essentially as described previously (52). Briefly, the beads were washed with 20 volumes of buffer A (20 mM Tris [pH 7.6], 1 M NaCl, 4 mM MgCl2, 2 mM DTT, 0.5% NP-40) and then equilibrated with 20 volumes of binding buffer (20 mM Tris [pH 7.6], 50 mM NaCl, 4 mM MgCl2, 2 mM DTT, 0.5% NP-40) containing 5 mg of bovine serum albumin (BSA) per ml followed by 20 volumes of the same buffer containing 1 mg of BSA per ml. For each binding reaction, 20 μl of 35S-labeled in vitro-translated E1 protein was diluted with 180 μl of binding buffer containing 1 mg of BSA per ml and incubated with 40 μl of GST or GST-E2 beads, at room temperature for 1 h. The beads were then washed three times with 20 volumes of binding buffer. Bound proteins were eluted first with 120 μl of high-salt buffer (20 mM Tris [pH 7.6], 500 mM NaCl, 1 mM DTT) and then with 120 μl of high-salt buffer containing 1% sodium dodecyl sulfate (SDS). Eluted proteins were then visualized by SDS-polyacrylamide gel electrophoresis and autoradiography.
Transient HPV DNA replication assay.
The transient HPV DNA replication assay has been described previously (52). Briefly, CHO-K1 cells were transfected with 250 ng of pCR3-E1, 25 ng of pCR3-E2, and 250 ng of pN9 plasmids by using Lipofectamine (Gibco BRL). Cells were harvested 72 h posttransfection, and total DNA was isolated using the QIAmp blood kit (Qiagen). Replicated pN9 plasmid DNA was detected by PCR amplification of a pN9 fragment bearing multiple DpnI restriction sites with DpnI-digested total DNA as a template. As a control, a fragment of the pCR3-E1 plasmid devoid of DpnI restriction sites was amplified in the same PCR. The primers and conditions for PCR have been described previously (52). PCR products were made radioactive by the addition of [α-33P]dCTP to the PCR mixtures and were visualized by agarose gel electrophoresis and autoradiography.
RESULTS
E1-E1 interaction in yeast.
The two-hybrid system was used to test whether HPV11 E1 can self-associate in yeast and to map a domain involved in E1-E1 interaction (Fig. 2). As we noticed previously (52), a fusion protein consisting of the entire E1 molecule (amino acids 1 to 649) fused to the DNA-binding domain of GAL4 was able to activate transcription of the UASGal-LacZ reporter gene in yeast strain Y153 (Fig. 2A). Shorter fusion proteins lacking the N-terminal 71 amino acids of E1 did not activate transcription and therefore could be used to test for interaction with the entire E1 protein fused to the GAL4 activation domain (Fig. 2A). Interaction of these shorter fusion proteins with the entire E1 molecule gave rise to only low, but reproducibly higher than background, levels of β-galactosidase (Fig. 2A and data not shown). These results indicated that HPV11 E1 can self-associate in yeast similarly to what was observed for HPV16 E1 (58). A series of deletion mutants was used to map the interaction domain to the C-terminal region of E1 (amino acids 353 to 649) (Fig. 1A). Self-association of E1 was more readily detected using fusion proteins containing only the C-terminal portion of E1 (amino acids 353 to 649 or 330 to 649) (Fig. 2). A minimal E1-E1 interaction domain capable of interaction with E1(330–649) was mapped between amino acids 353 and 431 (Fig. 2B). A shorter fragment spanning amino acids 353 to 416 of E1 was also active, albeit with reduced efficiency. A C-terminal E1 fragment (amino acids 435 to 649) lacking the minimal E1-E1 interaction domain was unable to associate with E1(330–649) (Fig. 2A and B), although it retained the ability to interact with E2 (52). The minimal E1-E1 interaction domain was able to interact not only with the complete E1 C-terminal domain but also with itself (Fig. 2C). These results indicated that residues 353 to 431 of E1 encode a homotypic E1-E1 interaction domain.
FIG. 2.
E1-E1 interaction in the yeast two-hybrid system. A diagram of the HPV11 E1 protein is shown at the top of each panel. Black boxes labeled A, B, C, and D indicate regions of E1 that are conserved in the large T antigens of SV40 and polyomaviruses. The region of HPV11 E1 that corresponds to the minimal DNA-binding domain mapped in BPV E1 (see the text for references) is also indicated as a black box (panel A). Numbers on the right of the figure indicate the levels of β-galactosidase activity measured in yeast cells cotransformed with two different plasmids expressing a GAL4–DNA-binding domain (BD) fusion protein and a GAL4–activation domain (AD) fusion protein. The amino acids at the N and C terminus of each truncated E1 proteins are indicated. (A) The indicated truncated GAL4-BD-E1 fusion proteins were tested for interaction with a GAL4-AD-E1 fusion protein composed of the entire E1 protein (amino acids 1 to 649) or with the GAL4-AD alone. (B) The indicated truncated GAL4-AD-E1 fusion proteins were tested for interaction with a GAL4-BD-E1 fusion protein composed of amino acids 330-649 of E1 or with the GAL4-AD alone. (C) Self-association of E1 fragments in yeast. Three different E1 fragments were tested for self-association by being expressed concurrently in the same yeast strain as a fusion to the GAL4-AD and as a fusion to the GAL4-BD. For comparison, each GAL4-E1-BD fusion was also tested for interaction with GAL4-AD-E1(330–649) or with the GAL4-AD alone. n.d., not done.
Domains of E1 required for binding to the viral origin in vitro.
To substantiate the results obtained in yeast, we set out to identify regions of E1 involved in oligomerization in vitro, using an assay that detects the binding of E1 to the HPV origin. By analogy to BPV-1 E1, we anticipated that oligomerization of E1 would occur upon binding to the origin (see the introduction for references). In this assay, HPV-11 E1 protein was synthesized by coupled transcription-translation in a rabbit reticulocyte lysate and incubated with a mixture of two radioactive DNA fragments, one of which contained the HPV-11 origin. E1 protein-DNA complexes were allowed to form and were then immunoprecipitated with an antibody against E1. The coprecipitated DNA was visualized by gel electrophoresis and autoradiography. In these experiments, a series of truncated E1 proteins (Fig. 3D) were used in addition to the wild-type protein, in order to define the minimal domain capable of binding to the origin. All E1 proteins were expressed at similar levels (data not shown). Three observations were made. First, using wild-type E1, only small amounts of E1-ori complexes were formed under the conditions of the assay (Fig. 3A). This in vitro-synthesized E1 protein is functional however, since we previously demonstrated that its binding to the origin can be stimulated by E2 (52) and that it is active in supporting HPV DNA replication in vitro in a cell-free system (1). The second observation we made was that a mutant E1 protein lacking the N-terminal 71 residues had increased affinity (approximately fivefold) for the origin compared to the affinity of the wild-type protein (Fig. 3A). As a control for specificity, we showed that binding of the truncated E1 protein to the origin was affected by the two double-amino-acid substitutions K286A R288A and A292L R293E (Fig. 3B). Similar amino acid substitutions were shown to abolish the binding of BPV-E1 to its cognate origin (51). Specificity was also demonstrated by showing that binding of E1(72–649) to the origin was affected by a triple mutation in the E1-binding site (Fig. 3C). This triple point mutation was shown previously, by DNase I footprinting analysis, to affect the binding of wild-type HPV11 E1 to the origin (49). The last observation that we made was that the smallest E1 protein that could bind to the origin was composed of amino acids 191 to 649 (Fig. 3D). A requirement for amino acids 191 to 353 was anticipated, given that this region is analogous to the BPV E1 minimal DNA-binding domain (4, 22, 38, 51) and that amino acid substitutions within this region of HPV11 E1 abolish binding to the origin (Fig. 3B). However, this region is clearly not sufficient, as demonstrated by the fact that deletions at the C terminus of E1 abolished binding to the origin (Fig. 3D). The simplest interpretation of these results, which is supported by results presented below, is that stable interaction of E1 with the origin requires a DNA-binding surface (located approximately within residues 200 to 300) in addition to the C-terminal domain (amino acids 353 to 649). Since the C-terminal domain contains both the ATPase catalytic site and the minimal E1-E1 interaction domain, we set out to investigate the role of these two regions in binding of E1 to the origin.
FIG. 3.
Binding of in vitro-synthesized E1 to the viral origin of DNA replication in vitro. (A) Protein-DNA complexes were formed with wild-type E1 (amino acids 1 to 649) or a truncated E1 (amino acids 72 to 649) or in the absence of E1 (−E1). E1-DNA complexes were immunoprecipitated with an antibody directed against E1, and the coprecipitated DNA, along with 0.5% of the amount of probe used in each binding reaction, was visualized by electrophoresis and autoradiography. An arrow indicates the probe fragment that contains the origin. (B) Effect of amino acid substitutions in the DNA-binding domain on interaction of E1(72–649) with the origin. Binding reactions were carried out as described for panel A. (C) Effect of a triple-nucleotide mutation in the E1-binding site of the HPV11 origin on interaction of E1(72–649) with the origin. Binding reactions were carried out as described for panel A. (D) Mapping of the domain of E1 required for binding to the viral origin in vitro. Truncated E1 proteins were synthesized in vitro and assayed for binding to the viral origin as described in panel A. N-terminally truncated proteins were immunoprecipitated using a polyclonal antibody directed against the C-terminal 14 amino acids of E1. C-terminally truncated proteins were tagged at their N terminus with the FLAG epitope and immunoprecipitated using an anti-FLAG monoclonal antibody. WT, wild type.
Role of ATP, and of the ATP-binding domain, in binding of E1 to the origin in vitro.
The results presented above indicated that sequences spanning the ATPase domain of E1 are required for E1 to form a stable complex with the origin. These results, together with our previous observation that the E2-dependent binding of HPV11 E1 to the origin is stimulated by ATP-Mg (52), prompted us to investigate more directly the role of ATP in binding of HPV11 E1 to the origin. Using the origin-binding assay described above, we tested the effect of supplementing the binding-reaction mixtures with ATP and Mg at 5 and 3 mM, respectively. We also investigated the effect of temperature by performing the binding reactions at three different temperatures (4, 23, and 37°C). In the absence of supplemented ATP-Mg, the binding of E1(72–649) to the origin was reduced dramatically at 37°C (Fig. 4A). This inhibition by high temperature could be relieved by the addition of ATP-Mg (Fig. 4A). At lower temperatures, (23 and 4°C), ATP-Mg had only a modest effect. Different types of nucleotides, in combination with magnesium, were tested for their ability to stimulate the binding of E1 to the origin at 37°C. ADP, but not AMP or adenosine, could substitute for ATP (Fig. 4B). Similarly, the three other ribonucleotides (CTP, GTP, and UTP) and all four deoxyribonucleotides (dATP, dCTP, dGTP, and TTP) could also stimulate binding to the origin (Fig. 4C). Two nonhydrolyzable ATP analogues, ATP-γ-S and GTP-γ-S, were also stimulatory, albeit to a lesser extent than ATP, indicating that binding of the substrate, but not its hydrolysis, is necessary for E1 to bind to the origin (Fig. 4C).
FIG. 4.
Effect of temperature and nucleotides on the binding of E1 to the viral origin. (A) Binding of E1(72–649) to the viral origin was performed at three different temperatures (4, 23, and 37°C) and in the presence (+ATP) or absence (−ATP) of 5 mM ATP supplement. (B) Binding of E1 to the origin was performed in the absence of nucleotide (−nuc.) or in the presence of either adenosine (adeno.), AMP, ADP, ATP or the indicated NTP at a concentration of 5 mM (C).
To further characterize the effect of ATP-Mg, amino acid substitutions in the ATP-binding domain of E1 were tested for their effect on the binding of E1 to the origin in vitro (Fig. 5). These amino acid substitutions lie in one of three motifs, termed a, b, and c, that characterize the ATP-binding domains of E1 and other members of superfamily 3 of NTP-binding proteins (Fig. 1). As mentioned in the introduction, motif a (phosphate-binding loop [P-loop]), together with motif b, is involved in binding ATP as a magnesium chelate. The exact function of conserved motif c is unknown, but it has been suggested that it may also participate in binding ATP. Another mutant E1 protein was also tested in which a highly conserved residue, F509, which lies between motifs b and c (17) and whose function is unknown, was mutated. We reported previously that all of these substitutions, with the exception of K484E and K484Q, have little effect on the ability of E1 to form a complex with E2 at the origin (52), indicating that they do not alter dramatically the conformation of E1. Binding of these mutant proteins to the origin was assayed at 23 and 37°C. Reaction mixtures for reactions performed at 37°C were supplemented with ATP-Mg. Most substitutions in motif a (P-loop) reduced or abolished the binding of E1 to the origin. Several of these substitutions, such as P479S, K484A, K484I, and K484R, had a more drastic effect at 37°C. This is consistent with our observation presented above that the stimulatory effect of ATP-Mg is maximal at high temperature (Fig. 4A). Replacement of conserved residues in motifs b and c had a less profound effect on E1 binding to the origin, whereas replacement of phenylalanine at position 509 had no effect (Fig. 5A). Collectively, the results presented above support the notion that binding of ATP-Mg, but not its hydrolysis, is essential for optimal binding of E1 to the origin.
FIG. 5.
Effect of substitutions in the ATP-binding domain and in conserved region A on the binding of E1 to the viral origin. In vitro-synthesized E1 (72–649), or mutant derivatives bearing a substitution in the ATP-binding domain (A) or in conserved region A (B) were tested for binding to the viral origin at the indicated temperature and as described in the legend to Fig. 3.
Role of conserved region A in binding of E1 to the origin.
The minimal E1-E1 interaction domain (amino acids 353 to 416) that we identified in the yeast two-hybrid system encompasses conserved region A, one of four regions of high sequence similarity between the E1 proteins of various papillomaviruses and the large T antigens of SV40 and other polyomaviruses (7) (Fig. 1). To determine if this conserved region is essential for E1 to form a stable complex with the origin, six independent amino acid substitutions were created in this domain (Fig. 1). Four of the six substitutions affect residues that are invariant between papillomaviruses and polyomaviruses (N389A, A390G, F393A, and Q399A) (Fig. 1). The other two substitutions (F378A and Y380A) affect conserved hydrophobic residues, which in large T antigen, but not in E1, form part of a zinc-binding finger (Fig. 1) that is required for T-antigen oligomerization (26, 36). Although this zinc finger motif is not conserved in papillomaviruses, F378 and Y380 lie in a region of E1 which, like the analogous region in large T, is predicted to fold into an alpha helix (31). Binding of these mutant E1(72–649) proteins to the origin was assayed both at 23°C in the absence of ATP-Mg and at 37°C in the presence of ATP-Mg (5 and 3 mM). Under both sets of conditions, the results were very similar. Three of the substitutions, Y380A, N389A, and F393A, drastically reduced the binding of E1 to the origin (Fig. 5B). Two other substitutions, A390G and Q399A, were also deleterious and allowed only a modest amount of binding to the origin. Only one substitution, F378A, had little effect on the binding of E1 to the origin. The deleterious effect of most of the substitutions in conserved region A supports the notion that this region of E1 is required for stable interaction with the origin.
Detection of E1 oligomers by cross-linking in vitro.
To determine more directly the oligomerization status of E1 in vitro, we used a cross-linking assay with the sulfhydryl-reacting cross-linker BMH. Single-stranded DNA was used in this assay since it has been shown recently to promote the oligomerization of BPV-1 E1 (12, 41). In this cross-linking assay (see Materials and Methods), in vitro-synthesized and 35S-labeled E1(72–649) protein was incubated in the presence or absence of a single-stranded DNA oligonucleotide for 1 h at 37°C and then subjected to cross-linking with BMH. After stopping the cross-linking reaction, the E1 protein was immunoprecipitated and analyzed by gel electrophoresis and autoradiography. Under these conditions, single-stranded DNA greatly stimulated the oligomerization of E1 (Fig. 6A). Five different protein bands corresponding to oligomers of E1 were observed in addition to monomeric E1. These oligomeric E1 species, when compared with cross-linked phosphorylase b as molecular weight markers (Sigma), migrated at the expected positions for dimers, trimers, tetramers, pentamers, and hexamers (data not shown). This was also observed when cross-linking experiments were performed with full-length E1 or with truncated E1 proteins [E1(191–649) and E1(353–649) (see below)], thus ruling out the notion that proteins from the reticulocyte lysate are part of these complexes. Similar results were also obtained with single-stranded DNA oligonucleotides that are not derived from the HPV origin, indicating that stimulation of ori binding by single-stranded DNA is sequence independent (data not shown).
FIG. 6.
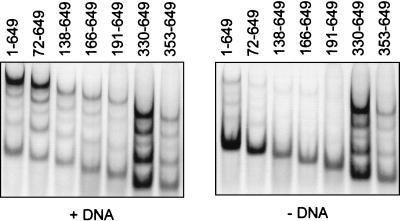
Cross-linking of radiolabeled E1(72–649). (A) In vitro-synthesized 35S-labeled E1(72–649) was cross-linked with BMH in the presence (+) or absence (−) of single-stranded DNA. Cross-linking reactions were terminated by the addition of excess DTT. E1 proteins were then immunoprecipitated and analyzed on a Weber-Osborn 3% polyacrylamide gel followed by autoradiography (see Materials and Methods). The positions of E1 oligomers corresponding in size to monomers, dimers, trimers, tetramers, pentamers, and hexamers (1 to 6, respectively) of E1 are indicated by arrows. Formation of E1 oligomers was stimulated by single-stranded DNA and was detected only in the presence of the cross-linking agent BMH. (B) Time course analysis of the formation of E1 oligomers. Radiolabeled wild-type E1(1–649) was cross-linked with BMH at 37°C at the indicated times after addition of single-stranded DNA (+DNA) or at similar times in the absence of single-stranded DNA (−DNA). The reaction products were analyzed as described for panel A. A reaction performed without the cross-linking agent BMH (No X-L) was included as a control. (C) Similar time course analysis as in panel B but performed at 23 and 4°C in the presence of single stranded DNA.
Next we investigated the oligomerization status of wild-type E1 (amino acids 1 to 649) by cross-linking (Fig. 6B). In this case, we also studied the kinetics of oligomer formation by performing the cross-linking at different time points following the addition of DNA to the reaction mixture. At very early time points, some E1 oligomers could be detected even in the absence of single-stranded DNA (Fig. 6B). These oligomers probably formed during synthesis of E1 in the reticulocyte lysate. Addition of single-stranded DNA greatly stimulated the formation of higher-order oligomers, in a time-dependent fashion. At 37°C, the formation of hexamers was rapid and was almost complete 15 to 30 min after the addition of DNA (Fig. 6B). After 30 min, monomers and hexamers were the two predominant species in the reaction. At intermediate time points, formation of smaller oligomeric species corresponding in size to trimers, tetramers, and pentamers was observed. The abundance of these smaller oligomers later decreased concomitantly with the appearance of hexamers, suggesting that they may represent intermediates in the formation of complete hexamers. We also studied the oligomerization of E1 at the lower temperatures of 23 and 4°C. At 23°C, oligomerization appeared to proceed more slowly than at 37°C (Fig. 6C). Oligomerization was even slower at 4°C, where trimers of E1 were the predominant species at most time points (Fig. 6C). At this temperature, higher-order oligomers, primarily tetramers, were formed only at later time points. Together, these results indicated that HPV11 E1, like BPV1 E1 (12, 41), has the capacity to form hexamers on binding to single-stranded DNA and that this reaction is optimal at the physiological temperature. Finally, in similar cross-linking experiments, we found that the E1 proteins of HPV6, HPV16, HPV18, and cottontail rabbit papillomavirus are also capable of forming oligomers (data not shown).
The C terminus of E1 is sufficient for oligomerization in vitro.
To map a domain of E1 that is sufficient for oligomerization, a series of N-terminally truncated E1 proteins were tested by cross-linking for their ability to oligomerize in the presence or absence of single-stranded DNA. The results presented in Fig. 7 indicated that a C-terminal fragment of E1 encompassing residues 353 to 649 was sufficient to form oligomers in vitro. This result was entirely consistent with our previous observation that this fragment of E1 is also able to self-associate in the yeast two-hybrid system (Fig. 2). Interestingly, the levels of oligomerization of E1(330–649) and E1(353–649) in vitro were substantial even in the absence of single-stranded DNA and were not further increased by the addition of single-stranded DNA. The fact that the C terminus of E1 oligomerizes substantially even in the absence of single-stranded DNA in vitro provides a plausible explanation of why this domain could readily self-associate in the yeast two-hybrid system (Fig. 2), in contrast to the complete protein. Finally, the fact that oligomerization of E1(191–649) was stimulated by single-stranded DNA, in contrast to oligomerization of E1(330–649), suggests that residues between amino acids 191 and 330 may play a role in conferring stimulation by single-stranded DNA (see below).
FIG. 7.
Cross-linking of truncated E1 proteins. The indicated truncated E1 proteins were synthesized and 35S labeled in vitro and then cross-linked, as described in the legend to Fig. 6, in the presence (+) or absence (−) of single-stranded DNA. Cross-linked oligomers were detected as described in the legend to Fig. 6.
Effect of amino acid substitutions in the ATP-binding domain on oligomerization of E1.
To investigate the role of the ATP-binding domain in oligomerization of E1, the cross-linking assay was used to test the effect of amino acid substitutions that change highly conserved residues implicated in ATP binding. These substitutions were the same as those used in the experiment in Fig. 5A. With the exception of the F509A substitution, all the substitutions reduced E1(72–649) oligomerization to various degrees (Fig. 8A). Substitutions in motif a had the greatest effect. Substitutions in motifs b or c reduced oligomer formation but did not completely abolish it. Together, the deleterious effects of these substitutions support the notion that the ATP-binding domain of E1 is essential for oligomerization and that ATP is an allosteric effector of this process. Alternatively, these deleterious substitutions could affect the ability of E1 to bind to the origin indirectly by affecting the proper folding or stability of the entire C-terminal domain. However, the fact that all of the mutant E1 proteins that are defective in oligomerization, except K484E and K484Q, retain the ability to bind cooperatively to E2 at the origin (52), suggest that these substitutions do not alter dramatically the overall structure of the C-terminal domain.
FIG. 8.
Effect of amino acid substitutions in the ATP-binding domain and in conserved region A on E1 oligomerization. (A and B) 35S-labeled E1(72–649) proteins carrying a substitution in one of three motifs of the ATP-binding domain (A) or in conserved region A (B) were cross-linked in the presence of single-stranded DNA and analyzed as described in the legend to Fig. 6. (C) Effect of the F393A substitution in region A on the oligomerization of E1(353–649). 35S-labeled E1(353–649) and E1(353–649)F393A, as well as E1(72–649) as a control, were cross-linked in the presence or absence of single-stranded DNA and analyzed as described in the legend to Fig. 6. WT, wild type.
Effect of amino acid substitutions in conserved region A on oligomerization of E1.
We next tested by cross-linking the effect of amino acids in conserved region A of E1 on oligomerization of the protein. The six mutant E1(72–649) proteins that had been tested previously for their ability to bind to the HPV origin (Fig. 5B) were used in these experiments. These mutant proteins were synthesized by in vitro translation and cross-linked in the presence of single-stranded DNA as described above. Three of the six mutant proteins tested, Y380A, N389A, and F393A, were severely defective in this assay (Fig. 8A). As expected, these were the same three mutant proteins that were severely defective in binding to the HPV origin (Fig. 5B). These results reinforce the notion that conserved region A of E1 is required for oligomerization. In principle, mutations in conserved region A could either prevent the association between E1 monomers or affect oligomerization indirectly, by affecting the binding of E1 to single-stranded DNA, or both. We investigated these possibilities by determining the effect of one of the deleterious substitutions, F393A, on the oligomerization of E1(353–649), since this fragment of E1 oligomerizes independently of single-stranded DNA. As can be seen in Fig. 8C, we found that the F393A substitution also prevented oligomerization of the E1 C-terminal domain, suggesting that this amino acid change affects the interaction between E1 monomers.
Effect of substitutions in conserved region A of E1 on the interaction with E2.
We had previously found that a region spanning amino acids 435 to 649 of E1 was sufficient for interaction with E2 in the yeast two-hybrid system but that a longer domain composed of residues 353 to 649 and encompassing conserved region A was required for binding to E2 in vitro (52). One interpretation of these results is that amino acids 435 to 649 of E1 contain most or all of the E2-binding surface but that amino acids 353 to 434 can influence the conformation of this E2-binding domain. To determine if conserved region A of E1 is required for interaction with E2 in vitro, we investigated the effect of amino acid substitutions in this region on the binding of 35S-labeled in vitro-translated E1(72–649) to the TAD of E2 fused to GST. We showed previously that under these assay conditions, the interaction of E1 with E2 is resistant to high salt concentrations (500 mM NaCl) and ethidium bromide, suggesting that their interaction is specific and not artifactually mediated by nucleic acids present in the E1-containing reticulocyte lysate (52). The results presented in Fig. 9 indicate that all five region A E1(72–649) mutant proteins were able to bind to the E2 TAD, albeit in some cases with slightly reduced efficiency. The greatest effect was seen for E1 F393A, whose binding to the E2 TAD was reduced approximately twofold. Collectively, these results indicate that the highly conserved residues in region A may contribute to but are not essential for the binding of E1 to E2.
FIG. 9.
Effect of substitutions in conserved region A on interaction of E1 with the transactivation domain of E2. GST pull-down assays were used to test the interaction of in vitro-synthesized E1(72–649), either wild type (WT) or carrying the indicated substitution in conserved region A, with the transactivation domain of E2 fused to GST (GST-E2) or with GST alone. In these experiments, luciferase (Luc.) was used as a control. E1 protein bound to GST-E2 or GST was eluted sequentially with 0.5 M NaCl and 1% SDS. For each binding reaction, 1/40 of the input protein and 1/10 of the NaCl and SDS eluates were analyzed on a gel and the amount of E1 protein in each lane was quantified with a PhosphorImager. The total amount of E1 protein eluted by each condition is given as a percentage of the total input protein below each lane.
Effect of substitutions in conserved region A of E1 on E2-dependent binding to the origin.
The results presented above indicated that most amino acid substitutions in conserved region A affect oligomerization and binding of E1 to the origin without having a dramatic effect on interaction with E2. It has been shown previously that certain E1 mutant proteins defective in DNA binding could be stimulated to bind to the origin by E2 (51). We therefore tested the effect of E2 on the binding of the various region A mutant E1 proteins to the HPV ori. For these experiments, full-length (amino acids 1 to 649) E1 mutant proteins as well as E2 were synthesized by in vitro translation and tested for binding to the origin as described previously (52). The results shown in Fig. 10A indicate that the three region A substitutions Y380A, N389A, and F393A, which affected the binding of E1 to the origin in the absence of E2 (Fig. 6), also have a deleterious effect in the presence of E2. In contrast, the binding to the origin of the two mutant proteins E1 A390G and E1 Q399A, which was weak but detectable in the absence of E2 (Fig. 5B), could be stimulated by E2. Not surprisingly, E1 F378A, which could bind to the origin like wild-type E1 (Fig. 5B), was also able to bind to the origin with E2.
FIG. 10.
Effect of substitutions in conserved region A on E2-dependent binding of E1 to the origin in vitro and on transient DNA replication in vivo. (A) E2-dependent binding of E1 to the origin. DNA-protein complexes were assembled without E1 (−E1), or with either wild-type (WT) E1 or the indicated mutant E1 proteins (amino acids 1 to 649) carrying a substitution in conserved region A. Complexes were immunoprecipitated with an antibody directed against E1, and the coprecipitated DNA was visualized by electrophoresis and autoradiography. (B) Transient HPV DNA replication. The amount of replicated origin-containing plasmid (ori) or of an internal control plasmid (E1-expressing plasmid [E1]) was detected by quantitative PCR from genomic DNA isolated from transfected CHO cells. These cells were transfected with the origin-containing plasmid pN9 and with either a plasmid expressing E1 (−E2) or a combination of plasmids expressing E2 and E1 (either wild type [1 to 649] or the indicated mutant E1 proteins). Shown in the figure is an autoradiogram of a 1% agarose gel used to analyzed the ori and E1 PCR products.
Effect of substitutions in conserved region A of E1 on transient HPV DNA replication.
The mutant E1 proteins carrying substitutions in conserved region A were tested for their ability to support the replication of an origin-containing plasmid, together with E2, in transiently transfected CHO cells. In this assay, the amount of newly replicated DNA was measured by quantitative PCR amplification of a fragment of the origin-containing plasmid, from DpnI-digested total cellular DNA (see Materials and Methods). To control for transfection efficiency and variations in the recovery of genomic DNA, the amount of E1 expression vector contained in total DNA was also determined. This was accomplished in the same PCR but with a different pair of primers, which amplify a fragment devoid of DpnI sites. As can be seen in Fig. 10B, three of the E1(1–649) mutant proteins, F378A, A390G, and Q399A, were capable of supporting HPV DNA replication, albeit at reduced levels compared to wild-type E1 in the case of A390G and Q399A. Three of the E1 mutants, Y380A, N389A, and F393A, were unable to support replication. These results indicated that conserved region A of E1 is required for transient HPV DNA replication. In general, the ability of the E1 mutants to support transient HPV DNA replication correlated well with their ability to bind to the origin with E2 (Fig. 10A) and to oligomerize in presence of single-stranded DNA (Fig. 8B). A potential caveat in these experiments is that the stability of the various E1 mutant proteins, compared to that of wild-type E1, could not be assessed due to the low levels of expression of E1 (data not shown). It is therefore possible that the low level of replication observed with some mutant proteins is partially due to protein instability.
DISCUSSION
In this study, we investigated the ability of the HPV11 E1 helicase to oligomerize in the yeast two-hybrid system and in vitro by chemical cross-linking. A C-terminal domain of E1 (amino acids 353 to 649) was identified that is sufficient for self-interaction in yeast and for formation of hexamers in vitro. Two regions within this C-terminal domain have been identified as important for oligomerization: conserved region A, which is part of the minimal E1-E1 interaction domain identified in the yeast two-hybrid system, and the ATP-binding domain. Amino acid substitutions of conserved residues within those two subdomains were identified that reduce the oligomerization of E1. These deleterious substitutions also affect the ability of E1 to bind to the HPV origin in vitro and to support transient HPV DNA replication in transfected cells. These results are discussed below.
Domain of HPV11 E1 required for stable binding to the origin.
On the basis of previous studies using purified E1 (see the introduction), we anticipated that oligomerization of E1 would be required for stable binding to the origin. By deletion analysis, we identified a truncated E1 protein encompassing residues 191 to 649 that retains the ability to bind to the origin and hence is likely to contain the E1 oligomerization domain in addition to sequences required for contacting DNA. These mapping results are consistent with our previous observation that this truncated E1 (191 to 649) is able to support HPV DNA replication in a human cell-free system, albeit with reduced efficiency compared to wild-type E1 (1). Our mapping results are also in agreement with those of a previous study in which GST-E1 fusion proteins were used to locate the DNA-binding domain of HPV11 E1 to residues 186 to 649 (48). The origin-binding domain of BPV E1, in contrast to that of HPV E1, was mapped to a much smaller region of the protein, located approximately between amino acids 150 and 300 (4, 22, 38, 51). Although we cannot rule out that the difference in mapping data between BPV E1 and HPV E1 is due to differences in assay conditions, we think this possibility is unlikely, given that one of these studies (38) used a DNA coimmunoprecipitation assay very similar to ours to measure the binding of in vitro-synthesized BPV E1 to its cognate origin. Two conserved short hydrophilic regions within this domain, termed HR1 and HR3 (Fig. 1) (15), have been identified as essential for DNA-binding (15, 51). The region in HPV11 E1 that corresponds to the minimal DNA-binding domain of BPV1 E1 is contained within the larger E1 DNA-binding domain identified in this study and that of Sun et al. (48). Furthermore, we have shown that two double-amino-acid substitutions in HR3 abolish the binding of HPV11 E1 to the origin (Fig. 3B), indicating that HPV11 E1 and BPV E1 bind to the origin in an analogous manner. The finding that the C-terminal oligomerization domain of HPV11 E1 is required for origin- binding suggests that this protein may bind as an oligomer, in contrast to the BPV1 E1 minimal DNA-binding domain, which binds stably to the BPV origin as two monomers (4).
It was suggested previously that in contrast to BPV E1, HPV11 E1 may bind to the viral origin only weakly (9, 24, 49). This may explain why we detected only low binding of in vitro-synthesized HPV11 E1 to the origin (Fig. 2A). During the course of this work, we found unexpectedly that a truncated E1 protein lacking the N-terminal 71 amino acids has increased affinity for the origin. In other studies, we showed that this in vitro-synthesized truncated protein is as active as wild-type E1 in interacting with E2 and the host polymerase α primase, as well as in supporting cell-free HPV DNA replication (1, 52). Although the identification of this truncated protein has made it possible to develop an E1-origin-binding assay using in vitro-synthesized E1, we do not know whether this increased binding to the origin is due to a genuine increased affinity for the origin, an increased stability of the truncated E1-DNA complex, or other reasons.
Domain of HPV11 E1 required for oligomerization.
Using the yeast two-hybrid system as well as chemical cross-linking of in vitro-synthesized E1 protein, we determined that oligomerization occurs primarily via the C-terminal domain of the protein (amino acids 353 to 649). Our results do not rule out any involvement of the N terminus of E1 in oligomerization. Rather, we found that N-terminally truncated E1 proteins are generally less efficient than the wild-type protein at forming complete hexamers (Fig. 7). We found that oligomerization of the C-terminal domain, unlike that of wild-type E1, was substantial even in the absence of single-stranded DNA. This may explain why oligomerization in the yeast two-hybrid system was easier to detect with the E1 C-terminal domain than with larger E1 constructs (Fig. 2). In addition, this result suggests that in the context of the complete protein, sequences within the N-terminal half of E1 may affect oligomerization of the C-terminal domain. By deletion analysis, we found that amino acids 191 to 330 of E1 are necessary for oligomerization to be stimulated by single-stranded DNA in vitro. Interestingly, this region of E1 corresponds to the minimal ori-binding domain identified in BPV E1 (4, 22, 38, 51). This raises the possibility that in vivo, residues 191 to 330 of E1 may also modulate the oligomerization of E1 upon binding to DNA, either to double-stranded ori DNA or to a region of single-stranded DNA created in a structurally distorted origin (41). Regardless of the precise mechanism, allosteric regulation of oligomerization by DNA binding may serve as a mechanism to prevent the assembly of E1 monomers into enzymatically active hexamers prior to their recruitment to the HPV origin.
The pathway by which E1 hexamers assemble onto single-stranded DNA in vitro may bear some resemblance to the one taking place in vivo in a region of single-stranded DNA within a distorted origin. Kinetic analysis suggests that E1 hexamers are assembled in a stepwise manner in vitro. At various time points during the assembly of E1 hexamers, we observed the accumulation of trimers, tetramers, and, to a lesser extent, pentamers. With time, all of these smaller species disappeared concomitantly with the accumulation of hexamers. These observations suggest that these smaller species are not “dead-end” complexes but true intermediates in the assembly of E1 hexamers. These results are consistent with a model in which hexamers are formed by the sequential addition of E1 monomers, at least under our experimental conditions.
Role of the ATP-binding domain in origin binding and oligomerization.
Because the C-terminal oligomerization domain of E1 is needed for ori binding and because this portion of E1 encompasses the ATP-binding domain, we investigated the effect of ATP on the interaction of E1 with the origin. We found that supplementing the binding-reaction mixture with ATP-Mg (at 5 and 3mM) stimulated the binding of E1 to ori primarily at 37°C. At this temperature, in absence of supplemented ATP-Mg, binding to ori was very weak. Interestingly, we previously observed similar effects of ATP-Mg and temperature on the E2-dependent binding of E1 to ori (52). The results presented in this study confirmed our earlier suspicion (52) that ATP-Mg enhances the binding of E1 to ori at 37°C rather than affecting the E1-E2 protein-protein interaction per se. This stimulatory effect of ATP-Mg on origin binding was also observed with purified recombinant E1 protein (reference 25 and data not shown). In our assay, ADP or nonhydrolyzable ATP analogues could substitute for ATP, indicating that ATP binding rather than its hydrolysis is required to stimulate the binding of E1 to ori. Other nucleotides or deoxynucleotides were also stimulatory, indicating that the ATP-binding pocket in E1 can accommodate a wide variety of nucleotides. The observation that binding of E1 to ori is temperature sensitive and that this temperature sensitivity can be relieved by ATP-Mg is consistent with the notion that ATP-Mg may be facilitating a conformational change in E1.
The role of the ATP-binding domain was confirmed by mutational analysis of highly conserved residues implicated in ATP binding. Of the three ATP-binding motifs in E1, the P-loop was the most strongly affected by amino acid substitutions. Two classes of P-loop substitutions were identified. One class includes substitutions K484E and K484Q, which abolish the binding of E1 to ori at both 23 and 37°C. We previously reported that these two substitutions affect the binding to E2 and suggested that these proteins may be misfolded (52). Substitutions in the second class were more informative. Substitutions such as K484A, K484R, or P479S reduced the binding of E1 to ori at 23°C and nearly completely abolished it at 37°C, despite the presence of high concentrations of ATP-Mg. In a previous study we showed that these substitutions have little effect on the interaction of E1 with E2 (52), indicating that they do not dramatically alter the conformation of E1. The simplest interpretation of these results is that these substitutions affect the ability of E1 to bind ATP, which may be required allosterically to enhance binding to the origin, particularly at 37°C. In principle, ATP-Mg may increase formation of the E1-origin complex or increase its stability once formed, or both. Further experiments are needed to address these different possibilities. However, we note that previous studies with BPV E1 indicated that ATP-Mg substantially increases the half-life of the E1-ori complex (29, 37, 44). The origin-binding assay that we used in this study requires that the E1-ori complexes formed in the binding reaction be stable enough to withstand the immunoprecipitation step and the subsequent washes. For this reason, this type of assay might be more susceptible to the stabilizing effect of ATP-Mg than would an assay in which a cross-linking agent is used to stabilize the E1-DNA complexes.
It was reported previously that the human heat shock proteins Hsp70 and Hsp40 can independently and additively stimulate the binding of purified HPV11 E1 to the origin (24). The stimulatory effect of Hsp70, unlike that of Hsp40, was shown to require ATP hydrolysis. Although we cannot rule out the possibility that heat shock proteins present in the reticulocyte lysate enhance the binding of E1 to ori, our result that nonhydrolyzable ATP analogues can stimulate ori binding suggest that this stimulation does not involve Hsp70. Moreover, the negative effect of substitutions in the ATP-binding domain of E1 suggests that ATP-Mg exerts its effect by binding directly to E1. We also determined more directly by chemical cross-linking that the integrity of the ATP-binding domain of E1 is required for oligomerization. All of the E1 mutant proteins that were defective in binding to ori at 37°C could not be cross-linked into oligomers in the presence of single-stranded DNA. These mutant proteins all bear substitutions in the P-loop. Amino acid substitutions in the other two motifs of the ATPase domain, which reduced but did not completely abolish the binding to ori at high temperatures, also reduced the oligomerization of E1 in the cross-linking assay. The good correlation between the effect of amino acid substitutions in the DNA-binding assay and in the cross-linking assay is consistent with the notion that oligomerization of E1 may be required for stable binding to the origin and underscores the role of the ATP-binding domain in this process.
Role of conserved region A in origin binding and oligomerization.
In this study, we provided evidence that conserved region A of E1 is required, in addition to the ATP-binding domain, for oligomerization of the protein. This region of E1, spanning approximately amino acids 353 to 431, was the smallest domain that could self-associate in the yeast two-hybrid system, suggesting that it contains an E1-E1 interaction interface. We also found that this small domain, as a purified fusion protein with thioredoxin, can inhibit in trans the binding of in vitro-synthesized E1 to ori (data not shown), thereby providing additional evidence that this domain is able to interact with E1.
To assess further the importance of conserved region A in the oligomerization of E1, we substituted some highly conserved residues of this domain and tested the ability of the resulting mutant E1(72–649) proteins to bind to the origin and to oligomerize as measured by cross-linking. With the exception of F378A, all substitutions were deleterious to various extents. Three substitutions, Y380A, N389A, and F393A, greatly reduced the ability of E1(72–649) to oligomerize on single-stranded DNA, to bind to the origin either alone or in combination with E2, and to support transient HPV DNA replication in cells. The remaining two substitutions, A390G and Q399A, had an intermediate effect in all assays. Thus, the ability of these mutant E1 proteins to oligomerize on single-stranded DNA correlated well with their ability to bind to ori in vitro and to support transient HPV DNA replication in cells. As controls, we showed that the mutant E1 proteins bearing substitutions in region A appear to be folded correctly, since their affinity for E2 was not reduced significantly (less than twofold [Fig. 9]). These mutant E1 proteins were also able to bind to the p70 subunit of polymerase α primase (data not shown) in agreement with the fact that the region A substitutions, with the exception of Q399A, lie outside of the minimal domain needed for interaction with p70 (amino acids 397 to 583 in HPV16 E1 [31]). Taken together, our results suggest that the region A substitutions affect primarily the oligomerization of E1 and support the argument that conserved region A is part of an E1-E1 interaction domain. Finally, as an additional control, we showed by cross-linking that the F393A substitution also affects the oligomerization of the E1 C-terminal domain (amino acids 353 to 649), which oligomerizes independently of single-stranded DNA. This result rules out the possibility that this substitution affects oligomerization indirectly by affecting the binding to single-stranded DNA. Collectively, our results reinforce the notion that region A plays a role in oligomerization and in binding of E1 to the origin. In this context, the inability of region A mutant proteins to bind to the origin may be due to a defect in oligomerization, which may be a prerequisite for formation of a stable complex with the origin. Alternatively, or in addition, it is formally possible that region A is required for stable binding of E1 to the origin, because this region is directly involved in contacting DNA.
Similarities in the organization of the oligomerization domain of E1 and related proteins.
It was shown recently that the C-terminal domain of SV40 T antigen (amino acids 259 to 708), which is homologous to the C-terminal domain of E1, is sufficient for binding to single-stranded DNA, probably as an hexamer (56). The high degree of sequence and functional similarity between T antigen and E1, in particular the fact that both proteins assemble at the origin as double-hexamer structures in a reaction that is stimulated by ATP, suggests that these proteins may oligomerize by a similar mechanism. It is intriguing that in the context of T antigen, conserved region A is located immediately C-terminal to a zinc-binding motif that is essential for oligomerization of the protein (Fig. 6A) (26, 36). This zinc finger motif is conserved in the T antigens of SV40 and polyomavirus but not in any papillomavirus E1 proteins. By analogy to papillomavirus E1, this zinc finger domain might be part of a larger oligomerization domain in T antigen that would include conserved region A and the ATPase domain. The proximity of this zinc-binding domain to conserved region A raises the possibility that the two regions may be conformationally coupled and suggests a mechanism by which the zinc finger region may affect the oligomerization of T antigen. More generally, the fact that sequences needed for oligomerization are located immediately N-terminal to the ATPase domain in both E1 and T antigen provides further evidence that the functional domains of these proteins are organized in a similar manner. This bipartite organization also closely resembles that of the Rep78 hexameric helicase of adeno-associated virus (46), which also belongs to superfamily 3 of NTPases. Indeed, recent studies indicated that the Rep78 oligomerization domain involves both the ATPase domain and a putative helical region located just upstream (46). The regions of E1 and T antigen encompassing conserved region A, despite not being obviously similar in sequence to Rep78, are also predicted to be α-helical (31), suggesting that these proteins may oligomerize in a similar manner. As pointed out previously (18), oligomerization domains have also been localized N-terminal to the ATPase domain of other structurally related hexameric proteins including the phage T7 gene 4 helicase (18) and RecA (47). This conserved arrangement of functional domains suggests that E1 and these related proteins may all oligomerize by a similar mechanism, which may generally apply to other hexameric NTPases.
The identification of two distinct functional domains of E1 involved in oligomerization, namely, conserved region A and the ATP-binding domain, agrees well with a structural model of the E1 C-terminal domain proposed by Masterson et al. (31). These authors suggested that the helical region contained within amino acids 353 to 435 and encompassing conserved region A may form a separate domain of the protein, which in different helicases is separated from the ATPase domain by an unstructured linker region of variable size and sequence (31). This suggestion was based in part on the fact that region A is conserved only among T antigens and papillomavirus E1, in contrast to the ATP-binding domain, which is shared among all members of superfamily 3 of NTPases. Our observation that amino acids 353 to 431 of E1 are able to self-associate in the yeast two-hybrid system supports the prediction that this region of E1 encodes a separate functional domain.
ACKNOWLEDGMENTS
Steve Titolo and Alex Pelletier contributed equally to this work.
We thank Steve Mason for critical reading of the manuscript.
REFERENCES
- 1.Amin, A. A., S. Titolo, A. Pelletier, D. Fink, M. G. Cordingley, and J. Archambault. Identification of domains of the HPV11 protein required for DNA replication in vitro. Virology, in press. [DOI] [PubMed]
- 2.Blitz I L, Laimins L A. The 68-kilodalton E1 protein of bovine papillomavirus is a DNA binding phosphoprotein which associates with the E2 transcriptional activator in vitro. J Virol. 1991;65:649–656. doi: 10.1128/jvi.65.2.649-656.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonne-Andrea C, Santucci S, Clertant P, Tillier F. Bovine papillomavirus E1 protein binds specifically DNA polymerase alpha but not replication protein A. J Virol. 1995;69:2341–2350. doi: 10.1128/jvi.69.4.2341-2350.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen G, Stenlund A. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J Virol. 1998;72:2567–2576. doi: 10.1128/jvi.72.4.2567-2576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiang C-M, Ustav M, Stenlund A, Ho T F, Broker T R, Chow L T. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc Natl Acad Sci USA. 1992;89:5799–5818. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow L T, Broker T R. Small DNA tumor viruses. In: Nathanson N, editor. Viral pathogenesis. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 267–301. [Google Scholar]
- 7.Clertant P, Seif I. A common function for polyoma virus large-T and papillomavirus E1 proteins? Nature. 1984;311:276–279. doi: 10.1038/311276a0. [DOI] [PubMed] [Google Scholar]
- 8.Conger K L, Liu J-S, Kuo S-R, Chow L T, Wang T S-F. Human papillomavirus DNA replication: interactions between the viral E1 protein and two subunits of human polymerase α primase. J Biol Chem. 1999;274:2696–2705. doi: 10.1074/jbc.274.5.2696. [DOI] [PubMed] [Google Scholar]
- 9.Cooper C S, Upmeyer S N, Winokur P L. Identification of single amino acids in the human papillomavirus 11 E2 protein critical for the transactivation and replication functions. Virology. 1998;241:312–322. doi: 10.1006/viro.1997.8941. [DOI] [PubMed] [Google Scholar]
- 10.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 11.Flores E R, Lambert P F. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J Virol. 1997;71:7167–7179. doi: 10.1128/jvi.71.10.7167-7179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fouts E T, Egelman E H, Botchan M R. Biochemical and electron microscopic image analysis of the hexameric E1 helicase. J Biol Chem. 1999;274:4447–4458. doi: 10.1074/jbc.274.7.4447. [DOI] [PubMed] [Google Scholar]
- 13.Frattini M G, Laimins L A. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc Natl Acad Sci USA. 1994;91:12398–12402. doi: 10.1073/pnas.91.26.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillette T G, Lusky M, Borowiec J A. Induction of structural changes in the bovine papillomavirus type 1 origin of replication by the viral E1 and E2 proteins. Proc Natl Acad Sci USA. 1994;91:8846–8850. doi: 10.1073/pnas.91.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez A, Bazaldua-Hernandez C, West M, Woytek K, Wilson V G. Identification of a short, hydrophilic amino acid sequence critical for origin recognition by the bovine papillomavirus E1 protein. J Virol. 2000;74:245–253. doi: 10.1128/jvi.74.1.245-253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopalakrishnan V, Sheahan L, Khan S A. DNA replication specificity and functional E2 interaction of the E1 proteins of human papillomavirus types 1a and 18 are determined by their carboxyl-terminal halves. Virology. 1999;256:330–339. doi: 10.1006/viro.1999.9665. [DOI] [PubMed] [Google Scholar]
- 17.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 18.Guo S, Tabor S, Richardson C C. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 protein is critical for hexamer formation. J Biol Chem. 1999;275:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- 19.Han Y, Loo Y-M, Militello K T, Melendy T. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus large T antigen, with human replication protein A. J Virol. 1999;73:4899–4907. doi: 10.1128/jvi.73.6.4899-4907.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howley P M. Papillomavirinae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 2045–2076. [Google Scholar]
- 21.Kuo S-R, Liu J-S, Broker T R, Chow L T. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J Biol Chem. 1994;269:24058–24065. [PubMed] [Google Scholar]
- 22.Leng X, Ludes-Meyer J H, Wilson V G. Isolation of an amino-terminal region of bovine papillomavirus type 1 E1 protein that retains origin binding and E2 interaction capacity. J Virol. 1997;71:848–852. doi: 10.1128/jvi.71.1.848-852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R, Botchan M. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 24.Liu J-S, Kuo S-R, Makov A M, Cyr D M, Griffith J D, Broker T R, Chow L T. Human Hsp70 and Hsp40 chaperone proteins facilitate human papillomavirus-11 E1 protein binding to the origin and stimulate cell-free DNA replication. J Biol Chem. 1998;273:30704–30712. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- 25.Liu J-S, Kuo S-R, Broker T R, Chow L T. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J Biol Chem. 1995;270:27283–27291. doi: 10.1074/jbc.270.45.27283. [DOI] [PubMed] [Google Scholar]
- 26.Loeber G, Stenger J E, Ray S, Parsons R E, Anderson M E, Tegtmeyer P. The zinc finger region of simian virus 40 large T antigen is needed for hexamer assembly and origin melting. J Virol. 1991;65:3167–3174. doi: 10.1128/jvi.65.6.3167-3174.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu J Z-J, Sun Y-N, Rose R C, Bonnez W, McCance D J. Two E2 binding sites (E2BS) alone or one E2BS plus an A/T-rich region are minimal requirements for the replication of the human papillomavirus type 11 origin. J Virol. 1993;67:7131–7139. doi: 10.1128/jvi.67.12.7131-7139.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lusky M, Fontane E. Formation of the complex of bovine papillomavirus E1 and E2 proteins is modulated by E2 phosphorylation and depends upon sequence within the carboxy terminus of E1. Proc Natl Acad Sci USA. 1991;88:6363–6367. doi: 10.1073/pnas.88.14.6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lusky M, Hurwitz J, Seo Y-S. Cooperative assembly of the bovine papilloma virus E1 and E2 proteins on the replication origin requires an intact E2 binding site. J Biol Chem. 1993;268:15795–15803. [PubMed] [Google Scholar]
- 30.Lusky M, Hurwitz J, Seo Y-S. The bovine papillomavirus E2 protein modulates the assembly of but is not stably maintained in a replication-competent multimeric E1-replication origin complex. Proc Natl Acad Sci USA. 1994;91:8895–8899. doi: 10.1073/pnas.91.19.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masterson P J, Stanley M A, Lewis A P, Romanos M A. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J Virol. 1998;72:7407–7419. doi: 10.1128/jvi.72.9.7407-7419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride A, Myers G. The E2 proteins. In: Myers G, Baker C, Münger K, Sverdrup F, McBride A, Bernard H-U, editors. Human papillomaviruses 1997. Los Alamos, N.M: Los Alamos National Laboratory; 1997. pp. 54–73. [Google Scholar]
- 33.Mohr I J, Clark R, Sun S, Androphy E J, MacPherson P, Botchan M R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 34.Moscufo N, Sverdrup F, Breiding D E, Androphy E J. Two distincts regions of the BPV1 E1 replication protein interact with the activation domain of E2. Virus Res. 1999;65:141–154. doi: 10.1016/s0168-1702(99)00113-6. [DOI] [PubMed] [Google Scholar]
- 35.Park P, Copeland W, Yang L, Wang T, Botchan M R, Mohr I. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc Natl Acad Sci USA. 1994;91:8700–8704. doi: 10.1073/pnas.91.18.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose P E, Schaffhausen B S. Zinc-binding and protein-protein interactions mediated by the polyomavirus large T antigen zinc finger. J Virol. 1995;69:2842–2849. doi: 10.1128/jvi.69.5.2842-2849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders C M, Stenlund A. Recruitment and loading of the E1 initiator protein: an ATP-dependent process catalysed by a transcription factor. EMBO J. 1998;17:7044–7055. doi: 10.1093/emboj/17.23.7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarafi T R, McBride A A. Domains of the BPV-1 E1 replication protein required for origin-specific DNA binding and interaction with the E2 transactivator. Virology. 1995;211:385–396. doi: 10.1006/viro.1995.1421. [DOI] [PubMed] [Google Scholar]
- 39.Sedman J, Stenlund A. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sedman J, Stenlund A. The initiator protein E1 binds to the bovine papillomavirus origin as a trimeric ring-like structure. EMBO J. 1996;15:5085–5092. [PMC free article] [PubMed] [Google Scholar]
- 41.Sedman J, Stenlund A. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J Virol. 1998;72:6893–6897. doi: 10.1128/jvi.72.8.6893-6897.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedman T, Sedman J, Stenlund A. Binding of the E1 and E2 proteins to the origin of replication of bovine papillomavirus. J Virol. 1997;71:2887–2896. doi: 10.1128/jvi.71.4.2887-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo Y-S, Muller F, Lusky M, Hurwitz J. Bovine papillomavirus (BPV)-encoded E1 protein contains multiple activities required for BPV DNA replication. Proc Natl Acad Sci USA. 1993;90:702–706. doi: 10.1073/pnas.90.2.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seo Y-S, Müller F, Lusky M, Gibbs E, Kim H-Y, Phillips B, Hurwitz J. Bovine papilloma virus (BPV)-encoded E2 protein enhances binding of E1 to the BPV replication origin. Proc Natl Acad Sci USA. 1993;90:2865–2869. doi: 10.1073/pnas.90.7.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah K V, Howley P M. Papillomaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. New York, N.Y: Raven Press; 1996. pp. 2077–2109. [Google Scholar]
- 46.Smith R H, Kotin R M. The Rep78 product of adeno-associated virus (AAV) self associates to form a hexameric complex in the presence of AAV ori sequences. J Virol. 1997;71:4461–4471. doi: 10.1128/jvi.71.6.4461-4471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Storey R M, Weber I T, Steitz T A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 48.Sun Y, Han H, McCance D J. Active domains of human papillomavirus type 11 E1 protein for origin replication. J Gen Virol. 1998;79:1651–1658. doi: 10.1099/0022-1317-79-7-1651. [DOI] [PubMed] [Google Scholar]
- 49.Sun Y-N, Lu J Z-J, McCance D J. Mapping of HPV-11 E1 binding site and determination of other important cis elements for replication of the origin. Virology. 1996;216:219–222. doi: 10.1006/viro.1996.0050. [DOI] [PubMed] [Google Scholar]
- 50.Sverdrup F, Myers G. The E1 proteins. In: Myers G, Baker C, Munger K, Sverdrup F, McBride A, Bernard H-U, editors. Human papillomaviruses 1997. Los Alamos, N.M: Los Alamos National Laboratory; 1997. pp. 37–53. [Google Scholar]
- 51.Thorner L K, Lim D A, Botchan M R. DNA-binding domain of bovine papillomavirus type 1 E1 helicase: structural and functional aspects. J Virol. 1993;67:6000–6014. doi: 10.1128/jvi.67.10.6000-6014.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Titolo S, Pelletier A, Sauve F, Brault K, Wardrop E, White P W, Amin A, Cordingley M G, Archambault J. Role of the ATP-binding domain of the human papillomavirus type 11 helicase in E2-dependent binding to the origin. J Virol. 1999;73:5282–5293. doi: 10.1128/jvi.73.7.5282-5293.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ustav M, Stenlund A. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 1991;10:449–457. doi: 10.1002/j.1460-2075.1991.tb07967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the a- and b-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- 56.Wu C, Edgil D, Simmons D T. The origin DNA-binding and single-stranded DNA-binding domains of simian virus 40 large T antigen are distinct. J Virol. 1998;72:10256–10259. doi: 10.1128/jvi.72.12.10256-10259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang L, Li R, Mohr I, Clark R, Botchan M R. Activation of BPV-1 replication in vitro by the transcription factor E2. Nature. 1991;353:628–633. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 58.Yasugi T, Vidal M, Sakai H, Howley P M, Benson J D. Two classes of human papillomavirus type 16 E1 mutants suggest pleiotropic conformational constraints affecting E1 multimerization, E2 interaction, and interaction with cellular proteins. J Virol. 1997;71:5942–5951. doi: 10.1128/jvi.71.8.5942-5951.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yasugi T, Benson J D, Sakai H, Vidal M, Howley P M. Mapping and characterization of the interaction domains of human papillomavirus type 16 E1 and E2 proteins. J Virol. 1997;71:891–899. doi: 10.1128/jvi.71.2.891-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zou N, Liu J-S, Kuo S-R, Broker T R, Chow L T. The carboxyl-terminal region of the human papillomavirus type 16 E1 protein determines E2 protein specificity during DNA replication. J Virol. 1998;72:3436–3441. doi: 10.1128/jvi.72.4.3436-3441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.zur Hausen H, de Villiers E M. Human papillomaviruses. Annu Rev Microbiol. 1994;48:427–447. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]