Highlights
-
•
The radiomics model showed a good diagnostic effect in HOS patients.
-
•
The clinical model performed well in predicting the Huvos grade in HOS patients.
-
•
The combined model is apt for Huvos grading after preoperative HOS chemotherapy.
Keywords: Logistic models, CT, Osteosarcoma, Neoadjuvant therapy, Machine learning
Abstract
Objective
To develop a model combining clinical and radiomics features from CT scans for a preoperative noninvasive evaluation of Huvos grading of neoadjuvant chemotherapy in patients with HOS.
Methods
183 patients from center A and 42 from center B were categorized into training and validation sets. Features derived from radiomics were obtained from unenhanced CT scans.Following dimensionality reduction, the most optimal features were selected and utilized in creating a radiomics model through logistic regression analysis. Integrating clinical features, a composite clinical radiomics model was developed, and a nomogram was constructed. Predictive performance of the model was evaluated using ROC curves and calibration curves. Additionally, decision curve analysis was conducted to assess practical utility of nomogram in clinical settings.
Results
LASSO LR analysis was performed, and finally, three selected image omics features were obtained.Radiomics model yielded AUC values with a good diagnostic effect for both patient sets (AUCs: 0.69 and 0.68, respectively). Clinical models (including sex, age, pre-chemotherapy ALP and LDH levels, new lung metastases within 1 year after surgery, and incidence) performed well in terms of Huvos grade prediction, with an AUC of 0.74 for training set. The AUC for independent validation set stood at 0.70. Notably, the amalgamation of radiomics and clinical features exhibited commendable predictive prowess in training set, registering an AUC of 0.78. This robust performance was subsequently validated in the independent validation set, where the AUC remained high at 0.75. Calibration curves of nomogram showed that the predictions were in good agreement with actual observations.
Conclusion
Combined model can be used for Huvos grading in patients with HOS after preoperative chemotherapy, which is helpful for adjuvant treatment decisions.
1. Introduction
OS stands as predominant malignant bone tumor prevalent. Its hallmark lies in presence of mesenchymal cells or osteogenic progenitors, which generate osteoid and immature bone [1], [2]. Around 90 % of OS cases are classified as high-grade osteosarcomas (HOSs) [3], [4]. Standard treatment protocol for osteosarcoma typically involves preoperative neoadjuvant chemotherapy followed by definitive surgical resection.achieving a 5-year survival rate ranging between 60 % and 70 %. However, a subset of patients displaying poor tissue response to chemotherapy fail to derive benefit from this preoperative intervention [5]. The treatment of HOS depends on the integration of multiple layers of data and delicate decision-making by the oncologist. Endogenous or acquired drug resistance is a major reason for treatment failure and a poor prognosis. Currently, The postoperative pathological sampling to ascertain the tumor necrosis rate following chemotherapy stands out as the most dependable approach to evaluate the response to neoadjuvant chemotherapy.However, this method is invasive lag and involves complex surgery [6]. Therefore, using noninvasive methods is particularly important to track the response before surgery and obtain key information about the tumor itself.
Artificial intelligence (AI) can facilitate clinical decision-making by quantifying imaging information that cannot be detected by humans. Radiomics emerges as an innovative method harnessing sophisticated imaging characteristics for diverse applications, including lymph node metastasis prediction [7], malignant tumor staging [8], [9], [10], prognosis assessment [11], treatment response prediction [12], and even the identification of tumor biological properties [13], [14]. In contrast to conventional radiological diagnostic methods, radiomics offers the potential to furnish supplementary information and enhance diagnostic reproducibility. So, this study was aimed at developing a radiomics model and combining it with preoperative clinical features for the preoperative noninvasive and personalized determination of the Huvos grade in patients with HOS. We used the model to predict efficacy of chemotherapy in patients with HOS according to radiomics and clinical characteristics extracted from plain computed tomography (CT) scans at the initial diagnosis. We believe that the application of this model may improve treatment efficacy by facilitating the adjustment of the chemotherapy regimen in patients who may not respond to treatment.
2. Materials and methods
2.1. Patients and dataset
Data of patients with HOS treated at Beijing Jishuitan Hospital (center A) and Peking University Shougang Hospital (center B) from June 2018 to June 2022 were retrospectively collected. Incorporated into the study were individuals who fulfilled the specified inclusion criteria:(1) Primary osteosarcoma was confirmed by histopathological examination after surgery; (2) Neoadjuvant chemotherapy (NCT) was received before surgical treatment; (3) Routine CT scan was performed within 10 to 17 days before chemotherapy, and no needle biopsy was performed; (4) The tumor necrosis rate after NCT can be calculated; (5) At least 1 year postoperative follow-up.Exclusion criteria: (1) CT plain scan image quality does not meet the requirements of diagnosis and post-processing; (2) Receiving other anti-tumor related treatments; (3) Incomplete clinical data; (4) The patient has not undergone surgical treatment in our hospital, or the tumor necrosis rate cannot be calculated; (5) No follow-up within 1 year after surgery.Finally, 225 patients from two independent institutions were included. The training and independent validation sets consisted of 183 patients with HOS from center A and 42 patients with HOS from center B, respectively. Table 1 presents the specifics regarding two cohorts.
Table 1.
Demographic and clinical characteristics of patients with osteosarcoma in the training and validation cohorts.
| Characteristics |
Training cohort |
Validation cohort |
||||
|---|---|---|---|---|---|---|
| pGR group (n = 91) | non-pGR group (n = 92) | P-value | pGR group (n = 22) | non-pGR group (n = 20) | P-value | |
| Sex | 0.834 | 0.569 | ||||
| Male | 62 | 64 | 14 | 11 | ||
| Female | 29 | 28 | 8 | 9 | ||
| Age (mean ± SD, years) | 18.18 ± 11.050 | 19.03 ± 11.152 | 0.522, F = 0.279 | 22.09 ± 14.858 | 16.90 ± 7.813 | 0.170, F = 2.769 |
| ALP level (IU/L) | 0.069 | 0.122 | ||||
| Normal | 73 | 63 | 21 | 16 | ||
| Elevated | 18 | 29 | 1 | 4 | ||
| LDH level (U/L) | 0.258 | 0.58 | ||||
| Normal | 69 | 76 | 19 | 16 | ||
| Elevated | 22 | 16 | 3 | 4 | ||
| Pulmonary metastases | 0.152 | 0.372 | ||||
| Yes | 34 | 44 | 14 | 10 | ||
| No | 57 | 48 | 8 | 10 | ||
| Location of the primary tumor | 0.122 | 0.778 | ||||
| Femur | 47 | 49 | 12 | 9 | ||
| Tibia | 28 | 17 | 4 | 7 | ||
| Fibula | 4 | 5 | 2 | 1 | ||
| Foot | 0 | 1 | 0 | 0 | ||
| Shoulder | 1 | 2 | 0 | 0 | ||
| Humerus | 9 | 8 | 2 | 2 | ||
| Ulna | 1 | 0 | 1 | 0 | ||
| Radius | 0 | 3 | 0 | 0 | ||
| Pelvis | 1 | 7 | 1 | 1 | ||
2.2. Clinical and pathological data
As per the treatment recommendations outlined by the National Comprehensive Cancer Network and consensus of experts from CSCO regarding diagnosis and management of osteosarcoma,multi-drug combination chemotherapy was used with methotrexate, cisplatin, and doxorubicin. Neoadjuvant chemotherapy was performed for 2–3 months before surgery and continued for 6–8 months after surgery.
Following NCT, all patients underwent surgical intervention, with tumor necrosis rate assessed using Huvos grading criteria [15]. In this investigation, a tumor necrosis rate exceeding 90 % served as benchmark for evaluating response to neoadjuvant chemotherapy. A tumor necrosis rate of at least 90 % was deemed indicative of a favorable response to chemotherapy (pGR). Poor chemotherapy response (non-pGR) occurs in tumor necrosis rates of less than 90 % [15], [16].
2.3. CT examination
The imaging data were obtained using 5 different CT scanners: clinical spectral detector CT, 64-multislice CT (MSCT; Philips Healthcare, Netherlands), 256-MSCT (Philips Healthcare, Netherlands), 320-MSCT (Toshiba Medical Systems, Japan), and 64-MSCT (Toshiba Medical Systems, Japan). Scanning parameters were set as follows: tube voltage ranged from 80 to 120 kVp, automatic milliampere-second, pitch of 1.0, and slice thickness of 0.5 mm. All medical images utilized in this investigation adhered to standard DICOM format.
All lesions, defined as VOIs, underwent manual annotation utilizing ITK-SNAP (version 4.0.0; https://www.itksnap.org). The delineated VOIs encompassed the entirety of the lesion, including both osseous lesion and its adjacent soft-tissue mass, along with any cystic necrotic regions within lesion. However, areas of edema and adjacent blood vessels were deliberately excluded from annotations. Subsequent to image preprocessing, radiomics features were extracted from delineated tumor regions on CT images.
Subsequently, pGR, non-pGR groups were classified per the pathological response classifier.
Thirty patients were selected from among the 239 patients by a random sampling method. One resident physician who had worked for 2 years and one chief physician who had worked for 22 years were recruited for VOI determination, and consistency between groups was analyzed. In order to prevent the non-uniformity caused by different centers, different scanning instruments and scanning parameters, all images are normalized.For each VOI, the image intensity range was normalized to 0–255 and resampled to a 1 × 1 × 1 mm3 voxel size to reduce variability in image acquisition from different scanners [17], [18].
2.4. Select features and develop models
Radiomics feature selection process involved utilizing the LASSO algorithm, which identified features appearing in over 75 % of selections (a total of 20 times). Subsequently, a multivariate LR model was employed to establish quantitative radiomics labels for the selected characeristics exhibiting nonzero coefficients. The radiomics score for each patient was derived through a weighted linear combination of coefficients associated with selected features in radiomics label. To address missing data in training set, zero-value method was employed for data preprocessing [19]. Following feature selection, LR algorithm served as foundational component of models. LR, being a widely used algorithm in machine learning, is frequently employed as a benchmark for binary classification tasks. LR model was trained on complete dataset from source A and subsequently evaluated on an independent test set from source B.
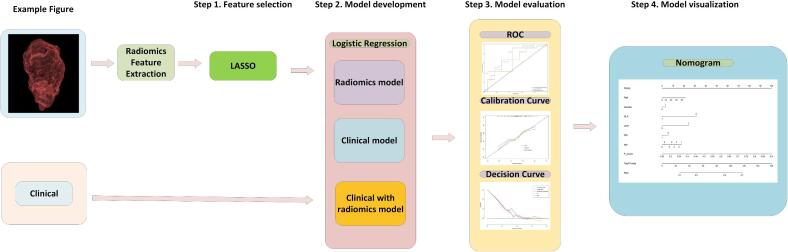
Three distinct models: 1) a clinical model utilizing solely clinical features, 2) a radiomics model derived from conventional plain CT scans, and 3) a composite model integrating independent clinical factors with delta-radiomics signature. Each model was constructed utilizing selected features to establish relationship and weights between features and the likelihood of progressive disease (pGR). Performance evaluation of models encompassed metrics such as accuracy, recall, sensitivity, and specificity. Furthermore, ROC curves, calibration plots, and decision curves were generated to depict model's performance. The final nomogram model was created by amalgamating independent clinical factors with the delta-radiomics signature. Model development, nomogram construction, and generation of calibration and decision curves were executed using R language. The schematic representation of research workflow is depicted in Fig. 1.
Fig. 1.
Schematic representation of the study flow.
3. Statistical analyses
In training set, analysis was conducted by SPSS 20.0, Medcalc 15.0 software, R studio. Normally distributed measurement data were presented as mean ± standard deviation (), skewed data were expressed as median (range). Use Mann-Whitney U to compare group for skewed data and χ2 test for enumeration data. Logistic regression (LR) analysis, initiated with all candidate variables exhibiting a P-value of < 0.05 in the univariate logistic model, was performed to discern clinical and imaging features correlated with progressive disease (pGR). The model was trained using the complete dataset and evaluated on an independent test set. Sensitivity, specificity, and accuracy of various diagnostic approaches were calculated using pathological results as the reference standard. Threshold for statistical significance is P < 0.05.
4. Results
4.1. Festures of patient
Without significant differences observed in response to chemotherapy (pGR and non-pGR), age, sex, tumor diameter, preoperative ALP and LDH levels, presence of new lung metastases, or LPT between two groups. The rates of pGR in training and independent validation cohorts were 49.7 % and 52.4 %, respectively. the inter-cohort difference was not significant (p = 0.756, χ2 = 0.096). ICC analysis showed that the VOI sketched by the two doctors had good consistency (Table 2). In this study, VOI sketched by senior physicians was selected for data analysis.
Table 2.
Consistency analysis of radiomics features.
| Characteristics of radiomics | ICC |
|---|---|
| exponential_ngtdm_Complexity | 0.931(0.779~0.977) |
| wavelet_HH_firstorder_Mean | 0.975(0.914~0.991) |
| wavelet_HH_glcm_ClusterShade | 0.885(0.752~0.933) |
4.2. Outcomes achieved using different models for the prediction and classification of pGR and non-pGR
Overall, 1743 image omics features were extracted based on plain CT scans, and 1233 features with an ICC of > 0.8 between the two physicians were selected for further analysis. LASSO LR analysis was performed, and finally, three selected image omics features were obtained. These characteristics are first-order statistics (wavelet_HH_firstorder_Mean), GLCM feature, specifically wavelet_HH_glcm_ClusterShade, and NGTDM feature, exponential_ngtdm_Complexity, exhibited notable distinctions between pGR and non-pGR groups (necrosis scores of ≥ 90 % and < 90 %, respectively) in training and independent validation sets (P < 0.01). Functionality of P score is depicted below:
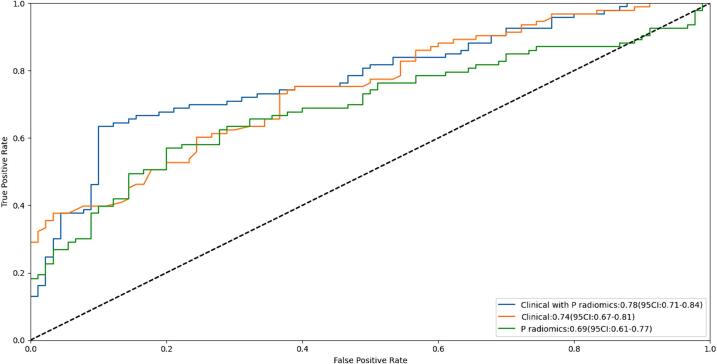
Comparison of AI models constructed using selected characteristics to predict pGR and non-pGR was conducted. ROC curves for training and independent test sets are depicted in Fig. 2, respectively. In training and independent validation cohorts, radiomics model based on plain CT scans demonstrated reasonable AUCs (0.69 [95 % CI: 0.61–0.77] and 0.68 [95 % CI: 0.51–0.85]). Clinical models (including sex, age, pre-chemotherapy ALP and LDH levels, new lung metastases within 1 year after surgery, and incidence) performed well in terms of Huvos grade prediction, with an AUC of 0.74 (95 % CI: 0.67–0.81) for training set. The AUC for independent validation set was 0.70 (95 % CI: 0.51–0.85).The model integrating both radiomics and clinical features exhibited promising predictive performance in training set (AUC: 0.78 [95 % CI: 0.71–0.84]), a finding further validated in independent validation set (AUC: 0.75 [95 % CI: 0.57–0.9]).
Fig. 2.
The receiver operating characteristic (ROC) curves of the training set (A). The ROC curves (B) of the models based on the selected features.
4.3. Visualization and clinical application of artificial intelligence model
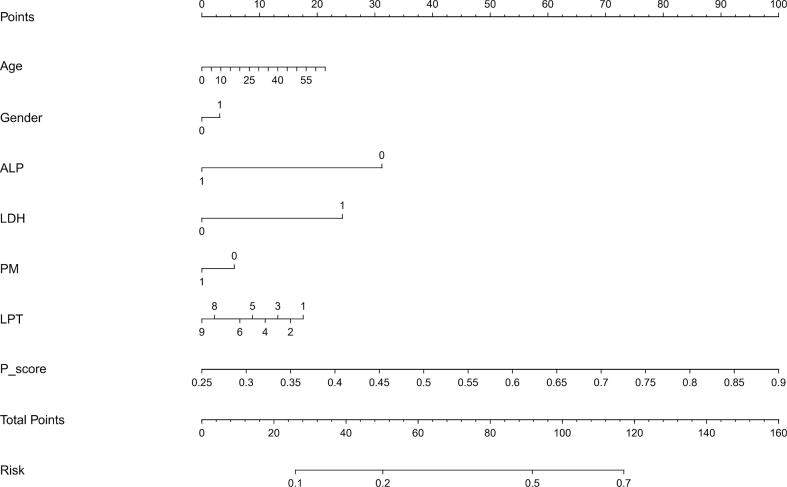
A nomogram was utilized to visually represent AI model for clinical application.Variables including age, sex, ALP and LDH levels, presence of pulmonary metastasis,LPT, and final P_score were utilized in prediction model. Each variable is depicted by a line segment marked with a scale, representing the range of possible values for the feature. The length of line segment expresses the contribution of feature to outcome event. Different values of each feature correspond to specific points. By summing points from all features, the total points for patient are determined.A vertical line is drawn downwards to determine risk for pGR.
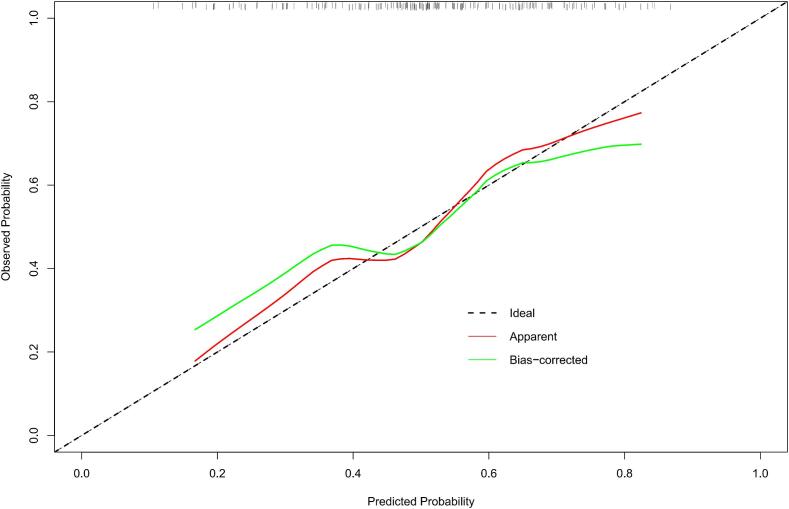
In calibration curve (Fig. 3B) for nomogram, x-axis represents nomogram-predicted probability, while y-axis denotes actual probability of pGR. Black dotted line represents an ideal prediction scenario. Solid red line indicates the performance across the entire cohort. The solid green line, adjusted for bias via bootstrapping (1000 repetitions). Fig. 3B demonstrates that nomogram performance closely approximates that of an ideal prediction scenario.
Fig. 3.
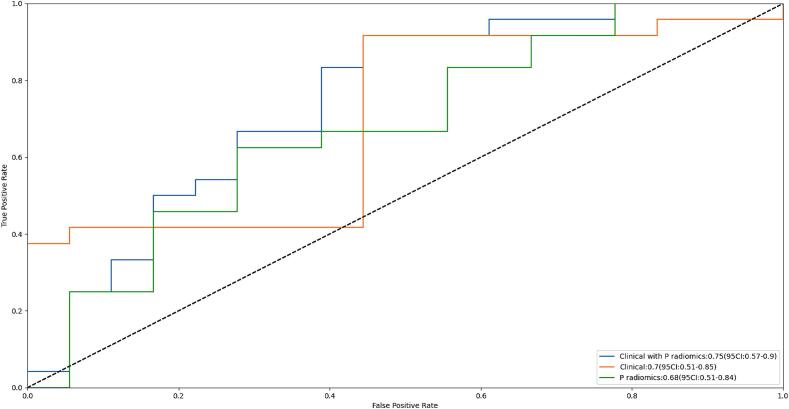
The nomogram for predicting the Huvos grade in patients with high-grade osteosarcoma (HOS) (A), the calibration curve of the nomogram (B) and the decision curve of different artificial intelligence (AI) models for predicting and classifying the Huvos grade (C).
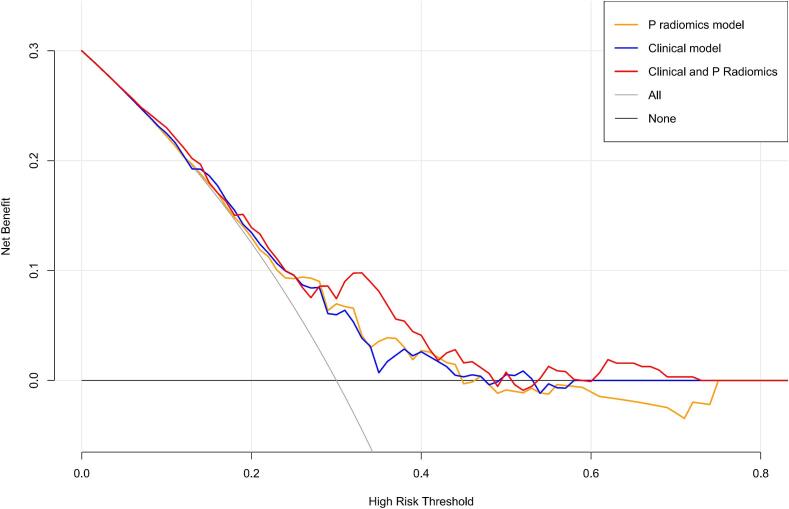
In Fig. 3C, decision curves of radiomics, clinical, and combined models are illustrated. X-axis represents the threshold probability, where Pi denotes the recorded risk probability when various methods reach a certain value, and Pt signifies the threshold at which Pi is stated as positive, prompting specific actions. The y-axis denotes the net benefit (NB) rate, calculated by subtracting pros and cons. Additionally, two gray lines representing extreme cases are included. The horizontal line shows that all samples are negative (Pi < Pt), necessitating no intervention and resulting in an NB rate of 0. Sloping line shows that all samples are positive, requiring intervention in every cases, with NB rate indicated by a backslash.
Benefits of three models were higher than that indicated by the extreme curve, and the combined model had the largest and safest Pt value range.
5. Discussion
We developed a radiomics model combining clinical risk factors and radiomic features from CT scans for a noninvasive, personalized assessment of the Huvos grade before surgery in patients with HOS.
Although CT examination has been used as one of the important examination methods for early diagnosis and evaluation of HOS due to its advantages of rapid, economical and convenient, clinical application value of conventional imaging features in evaluation of HOS Huvos classification is limited. Literature shows that Huvos grading has no correlation with the number and distribution of tumor blood vessels under microscope. In addition, the relationship between texture analysis of CT enhanced images and Huvos grading lacks scientific medical explanation. Therefore, in theory, it is more medical logical to use CT plain scan images to evaluate Huvos grading. CT plain scan does not need contrast agent injection, and is easier to acquire than CT enhanced scan.
Radiomics features offer insights into three-dimensional volume, capturing its heterogeneity and providing a view of tumor to guide treatment decisions. Moreover, this noninvasive approach facilitates continuous monitoring of tumor dynamics and the ongoing response to treatment [20], [21]. Previous studies of radiomics have mainly focused on establishing malignant tumor staging [9], [10], predicting lymph node metastasis [22], [23], obtaining a differential diagnosis [24], [25], and predicting the tumor phenotype [26], [27]. The pathological response of tumor cells to neoadjuvant chemotherapy is mainly reflected in tumor cell necrosis, apoptosis, differentiation and fibrosis, and this pathological change often affects the gray value of CT image pixels. Therefore, it is feasible to track the response of HOS neoadjuvant chemotherapy non-invasculatively based on texture analysis of CT plain scan images before surgery.
In this study, the developed predictive features provided additional information on pathological response after neoadjuvant chemotherapy. However, The primary challenge for integration of radiomics into clinical practice lies in ensuring reliability and reproducibility of developed models. This reliability is primarily influenced by two factors: segmentation and variability in image acquisition [28], [29]. In this study, The sketch of VOI was independently completed by two physicians in a double-blind manner. Although the two physicians' image knowledge and diagnostic experience were inconsistent, ICC analysis showed that the VOI sketched by the two physicians was in good agreement (ICC > 0.8), which indicated that different physicians could reach a relatively unanimous opinion on VOI segmentation when interpreting and analyzing the same image data. The segmentation differences caused by human subjective factors are reduced. Uniformity is a first-order characteristic parameter, which is mainly used to describe local and global characteristics of image data. ICC analysis results show that uniformity has the highest reliability (ICC = 0.975), indicating that uniformity has a high degree of consistency when measured at different times or by different observers. It also indicates that although the data in this study come from different centers, different sampling devices and different scanning parameters, the uniformity is highly repeatable [30]. The reliability of texture features is relatively low, because different centers, different sampling devices and different scanning parameters may lead to changes in the relative position and gray level relationship between pixels, so the performance of texture features may also be affected, resulting in poor repeatability. It is a complex and important task to establish standard CT scanning techniques and parameters and to improve the stability and repeatability of image omics feature extraction results. In the future, the author will further study the relevant technology and optimize the process to provide support for improving the value and reliability of imaging omics in medical diagnosis and treatment.
Previous radiomics studies mostly used single-center imaging data [6], [31]. However, radiomics features are easily affected by different CT acquisition parameters, and these single-center studies did not consider whether the CT acquisition equipment would hinder the transmission of radiomics features between different imaging centers. In this study, plain CT data were obtained from two centers and different hospitals were considered for the collection of CT parameters. Furthermore, the omics characteristics of images in this study were captured before the parameters in the standardized treatment were combined with clinical indicators, and independent validation was simultaneously adopted to evaluate the clinical and omics label value in clinical application. Therefore, we believe that results of study are generalizable and important for clinical application.
In this study, 3D VOI was applied to target lesion segmentation of CT images. Compared with 2D single or multiple sections, 3D segmentation can retain more spatial information, better capture complex structures and relationships, reduce distortion and distortion, increase the degree of differentiation and recognition of tumor heterogeneity, and truly reflect the biological characteristics of tumors. It is important to note that how to design efficient algorithms and achieve automated 3D segmentation is very challenging.
In this study, a total of 1743 imaging omics features of 7 categories based on 10 kinds of image preprocessing were included. 1233 features with ICC > 0.8 between 2 physicians were selected for further analysis, and 3 features corresponding to the smallest diagnostic error were selected through 20 LASSO regression operations to form imaging omics labels. The logistic regression prediction model was established. The AUC values for training set and independent verification set are 0.69 (95 % CI: 0.61–0.77) and 0.68 (95 % CI: 0.51–0.85). These findings suggest that model exhibits high stability and low redundancy.Radiomic features can reveal the internal heterogeneity of the tumor and reflect a series of pathological features of the tumor after treatment, such as changes in cell functional status, intra-tumor necrosis, tumor microenvironment, vascularization and cell density [32], [33]. The feature pool is divided into intensity statistical feature, geometric feature, texture feature and Baud sign.Intensity statistical features quantify statistical distribution of voxel intensities within tumor region. Geometric features characterize three-dimensional shape attributes of tumor region. Texture features are computed using various matrices such as GLCM, GLSZM, GLRLM, and NGTDM, elucidating relation and distribution of voxel spatial intensities within tumor region. Additionally, wavelet transform is employed to diminish image noise and enhance image sharpness,and the transformed high-order features can be used to describe the internal structure and texture of the tumor, as well as the differences between normal and diseased tissues [34]. Uniformity can reflect the heterogeneity within the tumor and can be used to monitor the therapeutic effect of the tumor. High uniformity suggests that the more complex the biological behavior of the tumor, the worse the prognosis and the lower the necrosis rate of neoadjuvant chemotherapy. GLCM texture features are used to describe the spatial relationship between pixels in an image, and provide information about the variation, distribution and local texture structure of pixels [35]. NGTDM texture features can capture structure, edge and texture information in images, better reveal local texture structure in images, and be more sensitive to subtle changes in tumors [36]. In conclusion, the non-invasive first-order features and texture features selected in this study can provide important disease information for patients with HOS, real-time monitoring of the disease, and individualized prognosis assessment. This kind of assessment is also more convenient and effective.
Among various diagnostic models examined, the integrated diagnostic model demonstrated the highest diagnostic efficacy and was comparatively superior to other models. This combined model effectively amalgamates both patient's clinical information and radiomics characteristics extracted from CT plain scans, resulting in a comprehensive diagnostic approach. ROC curve analysis revealed that combined model exhibited superior performance with higher AUC values, sensitivity, specificity, and accuracy in training set and independent validation set. Notably, in training set, the AUC reached as high as 0.78, surpassing predictive performance of Huvos classification, clinical model, and imaging omics model. These findings suggest that combined diagnosis serves as a crucial tool for evaluating efficacy of chemotherapy for osteosarcoma. By enhancing accuracy and providing a more comprehensive and precise diagnostic foundation for clinical imaging, combined diagnosis has potential to improve assessment outcomes significantly.
At present, the status of ALP and LDH in pre-treatment blood tests can be easily obtained at hospitalization, and these clinical indicators may have a certain suggestive role in the Huvos grade of osteosarcoma, so we included them in our study. ALP is one of the signature products of bone metabolism and can reflect the overall activity of bone. LDH exists in cytoplasm of cells and is responsible for generating a large amount of ATP in process of glycolysis to provide energy for tissue cells. The nomogram shows that ALP contributes more to the total number of points, indicating that ALP can be used as a common clinical indicator to evaluate Huvos grade of osteosarcoma. Of course, this prediction needs to be verified by large sample and multi-center data.
Although the application methods and emphasis of clinical model and radiomic feature model are different, they can complement and combine with each other in practical applications. For example, the introduction of radiomic features into clinical models can improve prediction accuracy of the models; At the same time, radiomic signature models can also be combined with patient clinical data to more fully assess a patient's disease status. In conclusion, both clinical model and radiomic feature model are important medical prediction models, and their relationship is complementary to each other, which can provide strong support for medical research and clinical practice.
In this study, clinical radiomics nomogram, constructed utilizing CT radiomics features and clinical characteristics, exhibited superior predictive capabilities for HOS patients. ROC curve analysis was used to assess performance of combined according to AUC value, sensitivity, specificity, and accuracy, both in training set and the independent validation set. Remarkably, in training set, the AUC reached 0.78, surpassing performance of clinical model and a simplistic imaging omics label forecast.Further evaluation based on calibration curve with high consistency analysis showed that the predicted and measured values indicated that the nomogram achieved the goal of predicting the Huvo’s grade of HOS patients well, and the results were further confirmed in the independent validation set (AUC: 0.75). To address clinical utility concern and illustrate relation between benefits and risks associated with different cut-off points across various models, decision curve analysis was implemented [37], [38]. In our study, all three models demonstrated benefits exceeding those of extreme curve. Furthermore, compared to both clinical model and the radiomics model, combined model exhibited the widest and safest range of Pt values. Decision curve generated from nomogram showcased favorable clinical utility. Noninvasive tool visually represents diagnostic model and can serve as a valuable reference for physicians.
5.1. Deficiency
Firstly, retrospective nature of our analysis solely included patients meeting predefined inclusion criteria, potentially introducing selection bias. Future investigations should encompass better prospective studies conducted in multicentric settings with more patient cohort to verify reliability and reproducibility. Secondly, it's worth noting that standard for pretreatment imaging in osteosarcoma patients in some hospitals is magnetic resonance imaging MRI, not CT, thereby potentially limiting generalizability of our findings to clinical practice. Thirdly, while our study compared models with different features, we did not explore different algorithms utilizing the same features. As such, it remains uncertain whether alternative algorithms may better suit this task, a question that warrants exploration in future research endeavors.
6. Conclusion
The amalgamation of radiomics features extracted from plain CT scans and clinical features holds significant potential for individualized pathological response assessment (Huvos grading) in patients with HOS following preoperative chemotherapy. This combined approach can offer valuable insights for guiding adjuvant treatment decisions tailored to the specific needs of each patient.
Funding
1. Beijing Hospitals Authority Clinical Medicine Development of Special Funding (NO.ZYLX202107); 2. Beijing Jishuitan Hospital Research Funding (NO.ZR202315).
CRediT authorship contribution statement
Fan Yang: Writing – original draft, Methodology, Data curation, Conceptualization. Ying Feng: Formal analysis, Data curation. Pengfei Sun: Formal analysis, Conceptualization. Alberto Traverso: Writing – review & editing, Validation, Supervision. Andre Dekker: Validation, Supervision. Bin Zhang: Validation, Resources. Zhen Huang: Methodology, Data curation. Zhixiang Wang: Writing – review & editing, Writing – original draft, Supervision, Software, Project administration. Dong Yan: Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Zhixiang Wang, Email: zhixiang.wang@maastro.nl, zhwang93@163.com.
Dong Yan, Email: bmuyandong@126.com.
References
- 1.Isakoff M.S., Bielack S.S., Meltzer P., Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eaton B.R., Schwarz R., Vatner R., Yeh B., Claude L., Indelicato D.J., Laack N. Osteosarcoma. Pediatr. Blood Cancer. 2021;68(Suppl 2) doi: 10.1002/pbc.28352. [DOI] [PubMed] [Google Scholar]
- 3.Letaief F., Khrouf S., Yahiaoui Y., Hamdi A., Gabsi A., Ayadi M., Mezlini A. Prognostic factors in high-grade localized osteosarcoma of the extremities: the tunisian experience. J. Orthop. Surg. (Hong Kong) 2020;28 doi: 10.1177/2309499020974501. [DOI] [PubMed] [Google Scholar]
- 4.Kim W., Han I., Lee J.S., Cho H.S., Park J.W., Kim H.S. Postmetastasis survival in high-grade extremity osteosarcoma: a retrospective analysis of prognostic factors in 126 patients. J. Surg. Oncol. 2018;117:1223–1231. doi: 10.1002/jso.24963. [DOI] [PubMed] [Google Scholar]
- 5.Yao Z., Tan Z., Yang J., Yang Y., Wang C., Chen J., Zhu Y., Wang T., Han L., Zhu L., Yang Z. Prognostic nomogram for predicting 5-year overall survival in Chinese patients with high-grade osteosarcoma. Sci. Rep. 2021;11:17728. doi: 10.1038/s41598-021-97090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Ge Y., Gao Q., Zhao F., Cheng T., Li H., Xia Y. Machine learning-based radiomics nomogram with dynamic contrast-enhanced MRI of the osteosarcoma for evaluation of efficacy of neoadjuvant chemotherapy. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.758921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang C., Huang M., Li S., Chen J., Yang Y., Qin N., Huang D., Shu J. Radiomics model of magnetic resonance imaging for predicting pathological grading and lymph node metastases of extrahepatic cholangiocarcinoma. Cancer Lett. 2020;470:1–7. doi: 10.1016/j.canlet.2019.11.036. [DOI] [PubMed] [Google Scholar]
- 8.Peeken J.C., Spraker M.B., Knebel C., Dapper H., Pfeiffer D., Devecka M., Thamer A., Shouman M.A., Ott A., von Eisenhart-Rothe R., Nusslin F., Mayr N.A., Nyflot M.J., Combs S.E. Tumor grading of soft tissue sarcomas using MRI-based radiomics. EBioMedicine. 2019;48:332–340. doi: 10.1016/j.ebiom.2019.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin X., Zhao S., Jiang H., Jia F., Wang G., He B., Jiang H., Ma X., Li J., Shi Z. A radiomics-based nomogram for preoperative T staging prediction of rectal cancer. Abdom Radiol (NY) 2021;46:4525–4535. doi: 10.1007/s00261-021-03137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao G., Rong W.C., Hu Y.C., Shi Z.Q., Yang Y., Ren J.L., Cui G.B. MRI radiomics analysis for predicting the pathologic classification and TNM staging of thymic epithelial tumors: a pilot study. AJR Am. J. Roentgenol. 2020;214:328–340. doi: 10.2214/AJR.19.21696. [DOI] [PubMed] [Google Scholar]
- 11.Wang T., She Y., Yang Y., Liu X., Chen S., Zhong Y., Deng J., Zhao M., Sun X., Xie D., Chen C. Radiomics for survival risk stratification of clinical and pathologic stage IA pure-solid non-small cell lung cancer. Radiology. 2022;302:425–434. doi: 10.1148/radiol.2021210109. [DOI] [PubMed] [Google Scholar]
- 12.Shin J., Seo N., Baek S.E., Son N.H., Lim J.S., Kim N.K., Koom W.S., Kim S. MRI radiomics model predicts pathologic complete response of rectal cancer following chemoradiotherapy. Radiology. 2022;303:351–358. doi: 10.1148/radiol.211986. [DOI] [PubMed] [Google Scholar]
- 13.Tagliafico A.S., Piana M., Schenone D., Lai R., Massone A.M., Houssami N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast. 2020;49:74–80. doi: 10.1016/j.breast.2019.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bodalal Z., Trebeschi S., Nguyen-Kim T.D.L., Schats W., Beets-Tan R. Radiogenomics: bridging imaging and genomics. Abdom Radiol (NY) 2019;44:1960–1984. doi: 10.1007/s00261-019-02028-w. [DOI] [PubMed] [Google Scholar]
- 15.Bacci G., Bertoni F., Longhi A., Ferrari S., Forni C., Biagini R., Bacchini P., Donati D., Manfrini M., Bernini G., Lari S. Neoadjuvant chemotherapy for high-grade central osteosarcoma of the extremity histologic response to preoperative chemotherapy correlates with histologic subtype of the tumor. Cancer. 2003;97:3068–3075. doi: 10.1002/cncr.11456. [DOI] [PubMed] [Google Scholar]
- 16.Coffin C.M., Lowichik A., Zhou H. Treatment effects in pediatric soft tissue and bone tumors: practical considerations for the pathologist. Am. J. Clin. Pathol. 2005;123:75–90. doi: 10.1309/h0d4vd760nh6n1r6. [DOI] [PubMed] [Google Scholar]
- 17.U.J. Nyúl L.G., Zhang, New variants of a method of MRI scale standardization, IEEE Trans Med Imaging 19(2000)143-150, 10.1109/42.836373. [DOI] [PubMed]
- 18.Shafiq-Ul-Hassan M., Zhang G.G., Latifi K., Ullah G., Hunt D.C., Balagurunathan Y., Abdalah M.A., Schabath M.B., Goldgof D.G., Mackin D., Court L.E., Gillies R.J., Moros E.G. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017;44:1050–1062. doi: 10.1002/mp.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCartney K., Bub K.L., Burchinal M. Best practices in quantitative methods for developmentalists. Monogr. Soc. Res. Child Dev. 2006;71:1–145. doi: 10.1111/j.1540-5834.2006.07103001.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen H., Zhang X., Wang X., Quan X., Deng Y., Lu M., Wei Q., Ye Q., Zhou Q., Xiang Z., Liang C., Yang W., Zhao Y. MRI-based radiomics signature for pretreatment prediction of pathological response to neoadjuvant chemotherapy in osteosarcoma: a multicenter study. Eur. Radiol. 2021;31:7913–7924. doi: 10.1007/s00330-021-07748-6. [DOI] [PubMed] [Google Scholar]
- 21.Huang Q., Chen C., Lou J., Huang Y., Ren T., Guo W. Development of a nomogram for predicting the efficacy of preoperative chemotherapy in osteosarcoma. Int. J. Gen. Med. 2021;14:4819–4827. doi: 10.2147/IJGM.S328991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu J., Deng Y., Liu T., Zhou J., Jia X., Xiao T., Zhou S., Li J., Guo Y., Wang Y., Zhou J., Chang C. Lymph node metastasis prediction of papillary thyroid carcinoma based on transfer learning radiomics. Nat. Commun. 2020;11:4807. doi: 10.1038/s41467-020-18497-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J., Li L., Zhe X., Tang M., Zhang X., Lei X., Zhang L. The diagnostic performance of machine learning-based radiomics of DCE-MRI in predicting axillary lymph node metastasis in breast cancer: a meta-analysis. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.799209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai Y., Yin P., Mao N., Sun C., Wu J., Cheng G., Hong N. Differentiation of pelvic osteosarcoma and ewing sarcoma using radiomic analysis based on T2-Weighted images and contrast-enhanced T1-weighted images. Biomed. Res. Int. 2020;2020:9078603. doi: 10.1155/2020/9078603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yin P., Zhi X., Sun C., Wang S., Liu X., Chen L., Hong N. Radiomics models for the preoperative prediction of pelvic and sacral tumor types: a single-center retrospective study of 795 cases. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.709659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan D.C., Obuchowski N.A., Kessler L.G., Raunig D.L., Gatsonis C., Huang E.P., Kondratovich M., McShane L.M., Reeves A.P., Barboriak D.P. Metrology standards for quantitative imaging biomarkers. Radiology. 2015;277:813–825. doi: 10.1148/radiol.2015142202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polan D.F., Brady S.L., Kaufman R.A. Tissue segmentation of computed tomography images using a random forest algorithm: a feasibility study. Phys. Med. Biol. 2016;61:6553–6569. doi: 10.1088/0031-9155/61/17/6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B., Tian J., Dong D., Gu D., Dong Y., Zhang L., Lian Z., Liu J., Luo X., Pei S., Mo X., Huang W., Ouyang F., Guo B., Liang L., Chen W., Liang C., Zhang S. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin. Cancer Res. 2017;23:4259–4269. doi: 10.1158/1078-0432.CCR-16-2910. [DOI] [PubMed] [Google Scholar]
- 30.He L., Huang Y., Ma Z., Liang C., Liang C., Liu Z. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci. Rep. 2016;6:34921. doi: 10.1038/srep34921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao S., Su Y., Duan J., Qiu Q., Ge X., Wang A., Yin Y. Radiomics signature extracted from diffusion-weighted magnetic resonance imaging predicts outcomes in osteosarcoma. J Bone Oncol. 2019;19 doi: 10.1016/j.jbo.2019.100263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlo C.A., Di Paolo P.L., Chaim J., Hakimi A.A., Ostrovnaya I., Russo P., Hricak H., Motzer R., Hsieh J.J., Akin O. Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology. 2014;270:464–471. doi: 10.1148/radiol.13130663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khaleel S., Katims A., Cumarasamy S., Rosenzweig S., Attalla K., Hakimi A.A., Mehrazin R. Radiogenomics in clear cell renal cell carcinoma: a review of the current status and future directions. Cancers (Basel) 2022;14:2085. doi: 10.3390/cancers14092085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huynh E., Coroller T.P., Narayan V., Agrawal V., Hou Y., Romano J., Franco I., Mak R.H., Aerts H.J. CT-based radiomic analysis of stereotactic body radiation therapy patients with lung cancer. Radiother. Oncol. 2016;120:258–266. doi: 10.1016/j.radonc.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 35.Fujima N., Homma A., Harada T., Shimizu Y., Tha K.K., Kano S., Mizumachi T., Li R., Kudo K., Shirato H. The utility of MRI histogram and texture analysis for the prediction of histological diagnosis in head and neck malignancies. Cancer Imaging. 2019;19:5. doi: 10.1186/s40644-019-0193-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J., Lian T., Chen H., Wang X., Quan X., Deng Y., Yao J., Lu M., Ye Q., Feng Q., Zhao Y. Pretreatment prediction of relapse risk in patients with osteosarcoma using radiomics nomogram based on CT: a retrospective multicenter study. Biomed Res. Int. 2021;2021 doi: 10.1155/2021/6674471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang L., Wang Q., Cui T., Huang J., Shi N., Jin H. Reporting of coronavirus disease 2019 prognostic models: the transparent reporting of a multivariable prediction model for individual prognosis or diagnosis statement. Ann. Transl. Med. 2021;9:421. doi: 10.21037/atm-20-6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong J., Hu Y., Zhang G., Xing Y., Ding D., Ge X., Pan Z., Yang Q., Yin Q., Zhang H., Zhang H., Yao W. An updated systematic review of radiomics in osteosarcoma: utilizing CLAIM to adapt the increasing trend of deep learning application in radiomics. Insights Imaging. 2022;13:138. doi: 10.1186/s13244-022-01277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]