Abstract
Despite being an innocuous commensal of human skin and mucous membranes, Staphylococcus epidermidis, infects surgical wounds and causes infections through biofilm formation. This study evaluates, in a time-dependent experiment, the self-dispersion of S. epidermidis CIP 444 biofilm when formed on borosilicate glass (hydrophilic) and polystyrene (hydrophobic) surfaces, using physical and molecular approaches. During a seven-day period of incubation, absorbance measurement revealed a drop in biofilm optical density on both studied surfaces on day 4 (0.043–0.035 nm/cm2, polystyrene), (0.06–0.053 nm/cm2, borosilicate glass). Absorbance results were correlated with crystal violet staining that showed a clear detachment from day 4. The blue color increases again on day 7, with an increase in biofilm optical density indicating the regeneration of the biofilm. Changes in gene expression in the S. epidermidis biofilm were assessed using a real-time reverse transcription-polymerase chain reaction. High expression of agr genes was detected on days 4 and 5, confirming our supposition of dispersion in this period, autolysin genes like atlE1 and aae were upregulated from day 3 until day 6 and the genes responsible for slime production and biofilm accumulation, were upregulated on days 4, 5, and 6 (ica ADBC) and on days 5, 6 and 7 (aap), indicating a dual process taking place. These findings suggest that S. epidermidis CIP 444 biofilms disperse at day 4 and reform at day 7. Over the course of the seven-day investigation, 2-ΔΔCt results showed that some genes in the biofilm were dramatically enhanced while others were significantly decreased as compared to planktonic ones.
Keywords: S. epidermidis, Biofilm, Self-dispersion, agr, icaADBC
1. Introduction
Staphylococci are gram-positive bacteria that were first identified in 1878. They have been linked to infections at various body sites, more recently to infecting implanted medical devices [1]. Staphylococcus epidermidis, a coagulase-negative staphylococcus, has been considered a major nosocomial pathogen despite being a normal component of the human flora, primarily of the skin and mucous membranes. It is capable of forming biofilm, infecting surgical wounds, and causing infections associated with indwelling medical devices [2]. Globally, it causes 50–70 % of catheter-related infections and 30–43 % of joint prosthetic infections [3,4]. The infection rate, however, eventually rises to 100 % for transient implants like venous catheters [5].
Biofilms are the typical way that bacteria inhabiting medical equipment gather and spread. Biofilm matrix polymers are chemically diverse in nature and include polysaccharides, proteins, lipids, and teichoic acids. Other substances released from dead cells are also believed to contribute to the biofilm matrix, including DNA, which is known as extracellular DNA (eDNA). The stages of biofilm establishment consist of formation, development, and dispersal. Each stage incorporates a wide set of genes that control the whole process. Staphylococci are threatening pathogens in both acute and chronic human infections because of their capacity to appropriately regulate a wide arsenal of virulence factors, such as toxins, degradative enzymes, antimicrobial resistance genes, and immune evasion mechanisms, and to successfully secure infectious niches in a wide range of organ systems [1,6].
The adhesion between the bacterial cells and the surface is mediated by polysaccharide adhesin (PS/A), the bacterial surface-associated proteins SSP-1 and SSP-2, autolysin E, and probably bhp, a protein similar to Bap in Staphylococcus aureus. It has been proposed that autolysin genes atlE1 and aae promote S. epidermidis adherence to immobilized plasma proteins vitronectin, fibronectin, and fibrinogen and that they may be responsible for triggering the development of biofilm on conditioned biomaterial surfaces [7,8]. To form the biofilm matrix, the second step involves the production of polysaccharide intercellular adhesin (PIA) or, according to its chemical composition, poly-N-acetylglucosamine (PNAG) [9], assuring the accumulation of multilayered bacterial cell clusters. The N-acetylglucosaminyl moieties in PIA are 1–6 linked [10]. PIA biosynthesis is encoded by the ica locus in S. epidermidis, which comprises the icaA, icaD, icaB, and icaC genes [11]. Upstream of the ica operon is the negative regulator (repressor) icaR gene, which is transcribed in the opposite direction of the icaADBC operon [12]. Additional molecule which contribute to biofilm formation especially in the case of PIA-independent biofilms, is the accumulation-associated protein (aap) which is a cell wall-associated protein of approximately 220 kDa, and its S. aureus orthologue, sasG [13]. Unknown regulatory mechanisms govern the transcription of aap. A transcriptomics investigation of S. epidermidis biofilms at various developmental stages indicated that aap was down-regulated at the transcriptional level in late-stage biofilms, indicating that its function may be limited to the early phases of biofilm creation and may not be involved in the maintenance of S. epidermidis biofilms [14]. Quorum sensing stands out as one of the most thoroughly investigated and, likely, most significant regulatory mechanisms for the control of pathogenesis. Through cell-cell communication, quorum sensing is a form of environment- and population density-dependent gene regulation [15]. The accessory gene regulator (agr) system and the luxS system are two regulatory systems found in staphylococci. In Gram-positive bacteria, agr is regarded as the prototype quorum-sensing regulator system and luxS's function as a quorum-sensing system in staphylococci is less clear [16]. In several animal models of acute infection caused by S. aureus, the up-regulation of virulence factors by agr is required for disease progression [17,18]. The agr locus contains four genes: agrB, agrD, agrC, and agrA [19]. Autoinducing peptide (AIP), a peptide precursor of agr's extracellular quorum signal, is encoded by the agrD transcript [20]. The thiolactone modification, C-terminal cleavage, and export of the AIP are all carried out by the transmembrane endopeptidase encoded by the agrB gene [21,22]. agrC is a transmembrane protein that is phosphorylated upon the binding of AIP. agrA, the related response regulator, forms a two-part signal transduction system encoded by the agrC and agrA genes [[23], [24], [25]]. After maturation to the three-dimensional structure [26], disruptive forces take place through enzymes (e.g. proteases and nucleases) that degrade biofilm polymers, and surfactant-like molecules, like the staphylococcal phenol-soluble modulins (PSMs), which disrupt non-covalent interactions [27,28]. This disruption causes biofilm detachment. During an infection, bacterial cells will detach from the biofilm and colonize other sites. In these cases, auto-dispersed bacterial cells will form a fresh biofilm, thus spreading and worsening the infection [29]. PSMs and proteases, two main biofilm structuring and dispersal factors, are controlled by the accessory gene regulator system (agr). Enhanced agr activity causes upregulation of many exoproteins, such as the delta toxin which belongs to (PSMs) family, promoting biofilm detachment and downregulation of surface-associated proteins [30].
Biofilm dispersion is a complicated stage incorporating a network of signals that are not fully understood. Two main strategies are followed by biofilms during detachment: active dispersal, in which the biofilm cells respond to an anti-biofilm stimulus (nutrient starvation or sequestration, dispersal signal release, quorum sensing inhibition, or stringent response interference) by actively degrading the matrix, thereby releasing planktonic cells and passive dispersal, in which an external force (sharp debridement) causes the complete or partial destruction of the biofilm [30]. We focus our research in this study on the auto-dispersion stage of S. epidermidis biofilm in a time-dependent manner on different surfaces used in the medical field, detecting the period of time during which biofilm disintegrates with the assessment of gene expression.
2. Materials and methods
2.1. Bacterial strain
In this research, the gram-positive bacterium S. epidermidis CIP 444 (109563) was investigated. It is a clinical strain isolated from a patient with an infected implanted device who was being treated at the Mignot Hospital in Versailles, France. This strain is a reference strain, extensively described in earlier investigations, and incorporated into the Institute Pasteur's collection of microorganisms in 2007.
Biofilm formation on different surfaces S. epidermidis CIP 444 was cultured in tryptic soy broth (TSB) from Himedia (Mumbai, India) for 18 h at 37 °C. For biofilm formation, two different surfaces, slightly polar hydrophilic glass (Borosilicate, 50 mL, Pyrex, Germany) and hydrophobic plastic (Polystyrene, 60 mm, Plastilab, Lebanon) were used. Both variants were supplemented with 7 mL of TSB supplemented with 0.25 % (wt/vol) glucose, inoculated with 1 × 106 CFU/mL from the preculture and incubated at 37 °C for different periods of time, from 1 to 7 days. The experiment was carried out in triplicate and with three independent repetitions.
2.2. Physical examination
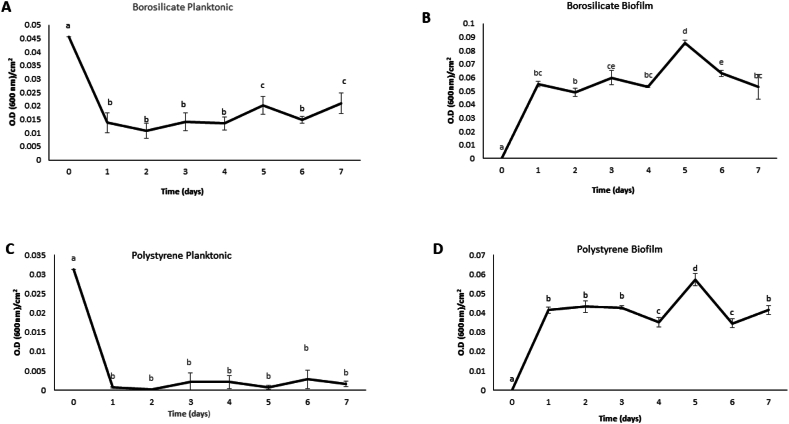
The supernatant (containing planktonic bacteria) was separated daily from the biofilm layer grown on the surfaces. The latter were divided into two sets. In the first set, the biofilm was fixed by heating at 80 °C and stained for 10 min with 2 mL crystal violet (0.1 % wt/vol), then the stained was discarded and the sample was washed three times with distilled water to remove the excess of the dye. However, the second set of biofilm and planktonic bacteria were examined discretely for absorbance at 600 nm using a UV–Vis spectrophotometer (GENESYS 10 UV, Thermo Electron Corporation, United States). The area of both studied surfaces was determined, and the obtained optical density was calculated as OD per cm2 as shown in Fig. 1(A-D) to compare the biofilm formation and dispersion on the two surfaces.
Fig. 1.
Optical density measurement at 600 nm per cm2 for both samples planktonic and biofilm of S. epidermidis CIP 444 growing on borosilicate (A, B) and polystyrene (C, D) surfaces. The area of the studied surfaces was calculated for comparison. The biofilm and planktonic were separated on a daily basis and optical density was measured for each, for a period of seven days using a UV–Vis spectrophotometer then converted as OD per cm2. The letters (a, b, c) represent significant differences in the biofilm density during the seven days on the two surfaces after applying ANOVA followed by Duncan's multiple range test (at p level <0.05). Error bars indicate standard deviations of the means.
Optimization for viable cell count. Sonication was an essential step to disintegrate the biofilm sample in order to determine the viable count. In order, to exclude the destructive effect of sonication on bacterial viability, pre-cultured planktonic suspensions of S. epidermidis CIP 444 were subjected to sonication for periods of increasing duration (1 min, 5 min, 10 min, 15 min) and at an increased sonication frequency (20, 25, 30, 35, and 40 kHz). Sonication was performed on an ultrasonic cleaner (Wisd, Daihan, Korea) at 25 °C, and ice was added to minimize the effect of temperature on the cell viability. The bacteria were then stained with methylene blue for Neubauer chamber visualization under a microscope to check cell aggregates before performing a viable count. The results of viable counting determined the optimum duration and frequency that are suitable for biofilm disintegration without causing cell death.
Based on our trials concerning the minimum frequency, it was confirmed that there is no significant effect of using ultrasound at a frequency of 20 kHz on the number of viable bacteria for 5 min. This frequency was fixed, and the number of bacteria was evaluated three times, repeatedly every 5 min, over a total period of 15 min. A Duncan multiple range test was done to determine, more precisely, the perfect time of sonication. A significant difference in bacterial count only after 15 min of sonication. Therefore, a period of 10 min was used in the study. After fixing the sonication time, trials were made to choose the best frequency to separate cells while avoiding the loss of viable ones. The proposed frequencies were 20, 25, 30, 35, and 40 kHz. The number of bacteria was evaluated with a repetition of three times at the above-mentioned frequencies. This work scenario was repeated two times, respectively. The highest common frequency between both trials, which did not reflect a significant difference in the bacterial count, was 30 kHz. Henceforth, this value was chosen for the continuation of the experiment.
Colony Forming Units (CFU). Following optimization, biofilm and planktonic cultures were sonicated, and viable counts were examined with the pour plate method by adding a volume of 100 μL from the sonicated culture after a series of dilutions on a polystyrene plate containing tryptic soy agar (TSA) (Himedia, India). Three plates per dilution were inoculated then incubated at 37 °C for 24 h, and the concentration of alive bacteria in CFU/mL was determined according to the formula: number of colonies x dilution factor/volume of the culture plate.
2.3. Real-time PCR for genes related to biofilm formation and dispersion of S. epidermidis CIP 444 (109563)
q-PCR primer design and validation. Based on each gene's function in S. epidermidis biofilm formation and dispersion, distinct primer sets have been developed for each gene. Based only on the coding sequence of the respective genes, primers were designed to amplify a 50–150 bp segment and were synthesized by Macrogen (Korea). Primer-dimer or self-complementarities were evaluated using the Primer 3 software. Before starting the experiment, primers were optimized using gradient PCR to determine the ideal annealing temperature. Table 1 lists the primer pairings for the chosen genes.
Table 1.
List of reverse and forward primers for the genes used in the study.
| Gene | Primers 5’ → 3′ | Function |
|---|---|---|
|
aae-F aae-R |
5′-GAGGAGGATTTTAAAGTGC-3′ 5′-AACATGACCATAGTAACC-3′ |
Surface-associated protein with bacteriolytic and adhesive properties autolysin/adhesins potentially involved in colonization. |
|
aap-F aap-R |
5′-GCACCAGCTGTTGTTGTACC-3′ 5′-GCATGCCTGCTGATAGTTCA-3′ |
Accumulation-associated protein (AAP) is involved in protein-associated biofilm formation and accumulation |
|
atlE-F atlE-R |
5′- TGT CCT GCT TTC ACG TAT GA -3′ 5′- TTTACGTGGTTAAGGTTTCT-3′ |
Autolysin/adhesins involved in primary adherence and attachment |
|
ica ADBC-F icaADBC-R |
5′-TTATCAATG CCGCAGTTGTC-3′ 5′- GTTTAACGCGAGTGCGCTAT-3′ |
Gene cluster encoding the production of polysaccharide intercellular adhesin (PIA), which mediates the intercellular adherence of bacteria and the accumulation of multilayer biofilm |
|
agrA-F agrA-R |
5′- AAGCGGGGAAGTAATTCAGT-5′-ATCGTGTCGAACTACTTGCA-3' |
S. epidermidis accessory gene regulator system. Enhanced agr activity causes upregulation of many exoproteins, such as the d-toxin, promoting biofilm detachment and downregulation of surface-associated proteins. |
|
agrB-F agrB-R |
5′-TTCGTTTAGGGATGCAGGTA-3′ 5′-ATCCTCTGTACGTGTGCCAT-3′ |
|
|
agrC-F agrC-R |
5′-CATCAATATCGCATTCATCG-3′ 5′-ATTTCCACTATTAGCGCCAA-3′ |
|
|
agrD-F agrD-R |
5′CACTACAATCTTGGAATTTATTGGT-3′ 5′-AACTGCTTGGTCTTCATGGTC-3′ |
|
|
luxS-F luxS-R |
5′-ACACCGCTGCGATACTGCCGCCGGT-3′ 5′-AGCCGGTGCGGCAGCCAAACGGTGA-3′ |
Quorum sensing system |
|
bhp-F bhp-R |
5′-TCCTCAGTAGCAGCAAGCAA-3′ 5′-GAAGTAAACCATTCAGATAGCGACA-3′ |
Bap homologue protein involved in accumulation in a PIA independent biofilm |
|
16S rRNA-F 16S rRNA-R |
5′- GGGCTACACACGTGCTACAA-3′ 5′-GTACAAGACCCGGGAACGTA-3′ |
16 S ribosomal RNA/housekeeping genes |
Total RNA isolation.S. epidermidis CIP 444 biofilm was formed on polystyrene plates over 7 days, as mentioned so far. After being separated from the supernatant, the biofilm was vigorously pipetted each day using RNase-free water to collect it. The cells were centrifuged at 13,000 g for 5 min at 4 °C. The pellet was kept separate from the supernatant and pipetted into a 100 μL lysis solution containing 10 mg of Tris, 1 mg of EDTA, and 1 mg of SDS. Along with 20 μL of lysozyme (1 mg/mL) and 60 μL of lysostaphin (1 mg/mL), 10 μL of proteinase-K (1 mg/mL) was added. Following a 30-min incubation period, 1 mL of QIAzol lysis reagent (Qiagen, Germany) was used to extract RNA from the mixture. 300 μL of chloroform was then added, and the mixture was centrifuged for 15 min at 13000 g. In the last stage, 500 μL of isopropanol was used to precipitate the RNA that had been extracted from the top layer. The Genova Nano Micro-Volume Spectrophotometer (Life Science, Jenway, UK) was used to measure the total RNA yield and purity of the purified RNA after it had been air dried and washed three times with 1 mL of 75 % ethanol. Protein contamination was indicated by the absorbance ratio A260/280, whereas contamination with polysaccharides, phenol, or chaotropic salts was indicated by the absorbance ratio A260/230. Using the previously described approach, RNA was also extracted for planktonic bacteria.
Real-time PCR. Using the manufacturer's instructions of the iScriptTM cDNA Synthesis Kit (Bio-Rad, USA), the total RNA was reverse transcribed. With a final volume of 10 μL, reverse transcription was performed under the following reaction conditions: enzyme activation (85 °C, 5 min), primer annealing (25 °C, 5 min), and reverse transcription (50 °C, 15 min). In a 96-well plate, real-time PCR experiments were conducted using a BIORAD CFX 96 real-time system (CFX 1000, United States) with iTaq TM Universal SYBR Green Supermix (BIO-RAD, United States), a 10 μL mixture comprising 1.5 μL (x 2) reverse and forward primers, 1 μL cDNA, and 5 μL SYBR Green master mix. The qPCR conditions consist of a 3-min initial denaturation at 95 °C, 40 cycles of denaturation and annealing for 30 s at 95 °C, and 1 min at the optimum annealing temperature, respectively. Then followed by an elongation stage lasting 30 s at 72 °C. After every qPCR study, a melting curve was generated to confirm the amplification of a single product.
The housekeeping gene 16S rRNA of S. epidermidis was used to standardize the expression levels of all examined genes. The findings are shown as threshold cycles (Ct), and the delta (Ct) method was used to compute the transcript levels: ΔCt(gene) - ΔCt (reference). The following fold change was computed for each gene's expression based on the planktonic bacteria (control): 2-(ΔΔCt), where ΔΔCt is equal to ΔCt (control) - ΔCt (sample). At least three separate samples were used for the experiment.
Statistical analysis. Statistical analysis was done using IBM SPSS, version 23. Analysis of variance ANOVA was applied in order to determine the significant changes in biofilm formation for each day. Afterwards, analysis was done for optical density, and then for gene expression with respect to days. Duncan's multiple range test was applied as post hoc test (in the function One Way ANOVA in SPSS software) for pairwise comparison of means. Significant difference is usually shown by the use of letters such as a, b, and c, as well as combinations such as ab, bc, etc in case of similarities between treatments in different groups. These letters are frequently used to denote groups or categories that, according to the statistical analysis of the data, differ considerably from one another (shown as different columns in Duncan test table in SPSS). Therefore, different letters will indicate a significant difference in optical density of the biofilm, viable count or genetic expression.
Using the ‘R’ program, Version 3.4. (R Foundation for Statistical Computing, Vienna, Austria) [31], Pearson's correlation (r) was generated for all studied genes. Genes are said to be positively correlated, when r ≥ 0.5 and negatively correlated when r ≥ -0.5 and heat map was done for gene expression in biofilm cells as folds (log10) compared to planktonic using function heat map 2 in ‘R’. All assays were conducted in triplicates.
3. Results and discussion
Optical density measurement. Planktonic and biofilm cultures were separated as previously described. The absorbance of each part was measured at 600 nm (Fig. 1A-D) according to the area of the used surface. Results of optical density/cm2 revealed that planktonic cells growing on the two surfaces with an initial OD of 0.045 and 0.031 decreased after 24 h of inoculation to 0.014 and 0 for the borosilicate glass and polystyrene surfaces, respectively. For the biofilm, an increase in OD from 0 to 0.041 (polystyrene) and 0.055 (borosilicate) was detected (Fig. 1B and D). OD remained approximately stable until day 4, where it decreased (0.035 polystyrene, 0.053 borosilicate glass), and then rose on day 5 to 0.057 (polystyrene), and 0.086 (borosilicate). This decline is due to dead cells lysing (autolysis), which led to a drop in cellular density (concentration). Hence, it indicates that dispersion is taking place in the biofilm at this period; furthermore, the OD value rose at day 5, which was explained by biofilm disintegration that increases particles in the suspension, and thus more light will be scattered by particles, so diffraction will be intensified, which in turn will be recorded on the detector as a higher “OD”. In a study concerning the response of P. aeruginosa biofilms to Cephalosporin-3′-diazeniumdiolates (C3D) a decrease in biomass and a rise in optical density in a flow-cell's effluent, was an indication of biofilm dispersion [32]. This declined as the biofilm dispersion process continued till day 7, where on polystyrene it began to increase (OD = 0.034, day 6; OD = 0.041, day 7) as new cells are growing and preparing for a new cycle (Fig. 1D). In fact, when dispersion takes place, the majority of cells will lyse, and the remaining cells will become free cells with distinguished traits capable of forming a new biofilm. The biofilm may disperse totally, and thus all the architecture will be destroyed completely or partially, which will allow imbedded, alive cells, even in dormant stages, to sense condition changes and may reestablish a new biofilm at the original site of formation. The fluctuation in OD was similar on both surfaces, with a delay on borosilicate for a new reformation of the biofilm. This was proved by the obtained OD of the planktonic bacteria at day 1 (OD = 0) (Fig. 1C), whereas on borosilicate, not all planktonic cells formed a biofilm as the recorded OD was higher (0.013) (Fig. 1A). Thus, microbial cell attachment to surfaces is influenced by a multitude of variables, such as surface electrostatic charges, van der Waals attraction, gravitational forces, and Brownian movement [33]. This dissimilarity was confirmed by applying ANOVA, which revealed a significant difference between the two surfaces in terms of biofilm formation during the same period (p < 0.05). The differences that account for this delay originate from the type of culturing surface, where polystyrene (hydrophobic) had a higher affinity for cells' attachment [34,35]. As illustrated in the study done by Oliveira et al. four polymeric materials, including silicone rubber, that are often used in indwelling medical devices were tested for S. epidermidis adhesion. All substances were regarded as hydrophobic (with different degrees of hydrophobicity), and the quantity of adhered cells was linearly associated with an increase in hydrophobicity [36]. Therefore, the comparison between biofilm formation on glass and plastic by applying ANOVA reveals that there is no difference between the two surfaces in terms of biofilm formation during the same period except in primary attachment (p = 0.037 < 0.05).
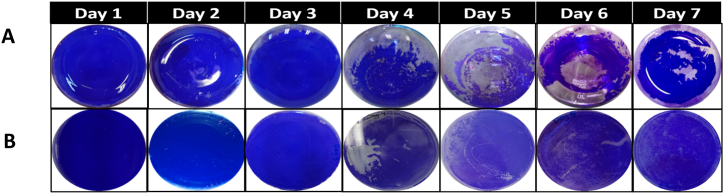
Colorimetric assay using Crystal violet. With respect to the crystal violet assay, the direct observation of the biofilm after fixation and staining with crystal violet (0.1 %) showed a more obvious illustration in terms of dispersion that occurred after biofilm maturation. The stained area begins to decrease on day 4, showing a clear detachment of the biofilm on different supports (borosilicate glass, polystyrene) (Fig. 2A, B). After six days of incubation, the coverage area and intensity of the violet color increased this was visualized at day 7 on both surfaces indicating a neo-formation of the biofilm. Crystal violet stains both bacteria and the surrounding biofilm matrix by binding negatively charged molecules [37]. Gram-positive bacteria have a thick peptidoglycan coating on their cell walls, which the crystal violet dye adheres to, coloring them violet under a light microscope. Consequently, it offers the benefit of allowing for quick monitoring of biofilm development on physical supports. However, it does not provide a quantitative evaluation of the stained surface. Hence, the results of the physical methods used were correlated, suggesting that detachment occurred on day 4.
Fig. 2.
Biofilm of S. epidermidis after fixation by heat and crystal violet (blue color) staining at different periods of biofilm life cycle on two surfaces. Borosilicate Pyrex (A) and Polystyrene (B). The stain was added for 10 min to the biofilm then rinsed with distilled water. The remained blue stain reveals the biofilm that will be visualized by the naked eye. Detachment was noticeable on day 4 (decrease in blue color stain) on both supports and biofilm regeneration on day 7 (re-increase in the stained area in terms of blue color). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
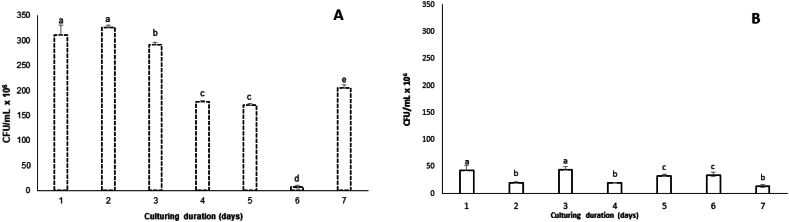
Viable Count (CFU). According to colony count, an increase in biofilm population (Fig. 3A) was followed by a decrease in planktonic population in terms of CFU/mL on day 1 (Fig. 3B). This indicates the switch of planktonic cells to biofilm mode. The biofilm viable cells decline from 291.6 × 10 6 CFU/mL (day 3) to 177.6 × 106 CFU/mL (day 4). As mentioned so far, dispersion is possibly the cause, viable cells begin to increase at day 7 (205 × 106 CFU/mL), which assures the emergence of a new population and verifies the hypothesis of new biofilm. The CFU measurement accounts for culturable cells in the biofilm, not the total number of alive cells. Viable but not culturable cells (vbnc), in addition to dormant cells, are also a portion of the biofilm-embedded population that may be a cause for a new formation after signaling an activation message [38,39].
Fig. 3.
The quantification of viable bacteria in biofilm (A) and planktonic cells (B) of S. epidermidis CIP 444 during seven days after inoculation with an initial concentration 1 × 106 CFU/mL. The number of CFUs was counted to assess the time of biofilm dispersion. Biofilm samples were sonicated and diluted before plating. The formula: number of colonies x dilution factor/volume of the culture plate was used to calculate the concentration of living bacteria in CFU/mL. Bars labeled by different letters on the graph are significantly different from each other in terms of viable count (CFU) during the incubation period (p < 0.05) based on one-way ANOVA and Duncan's multiple range test. Error bars indicate standard deviations of the means.
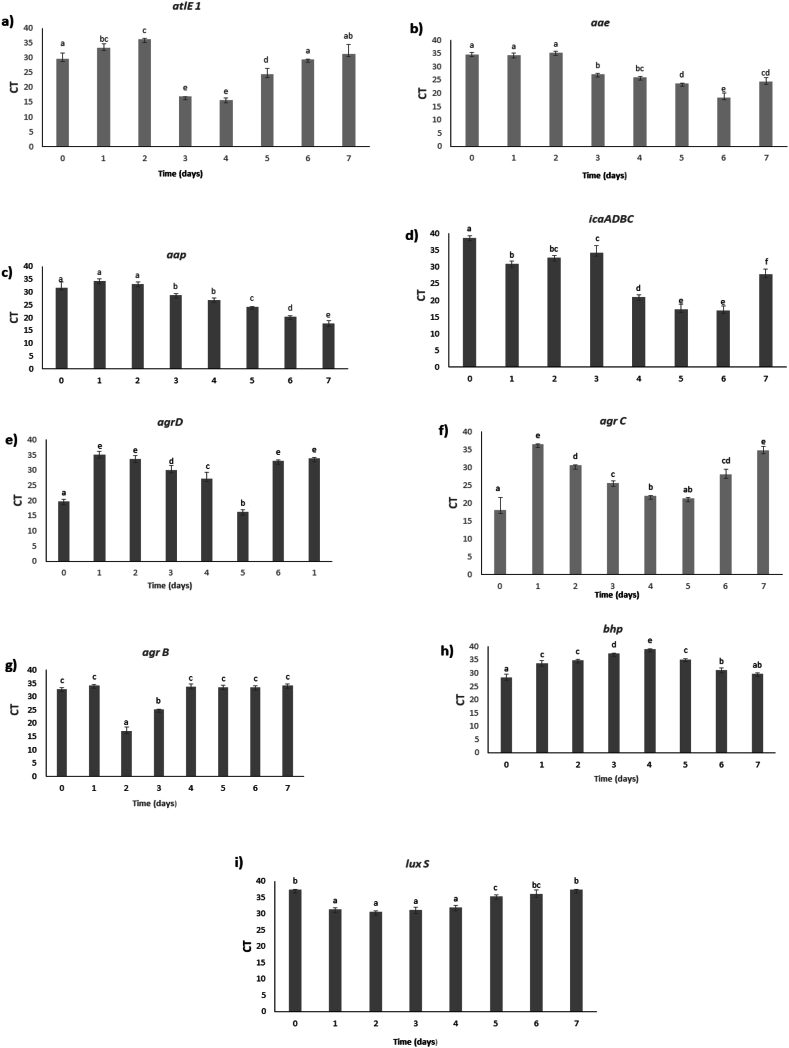
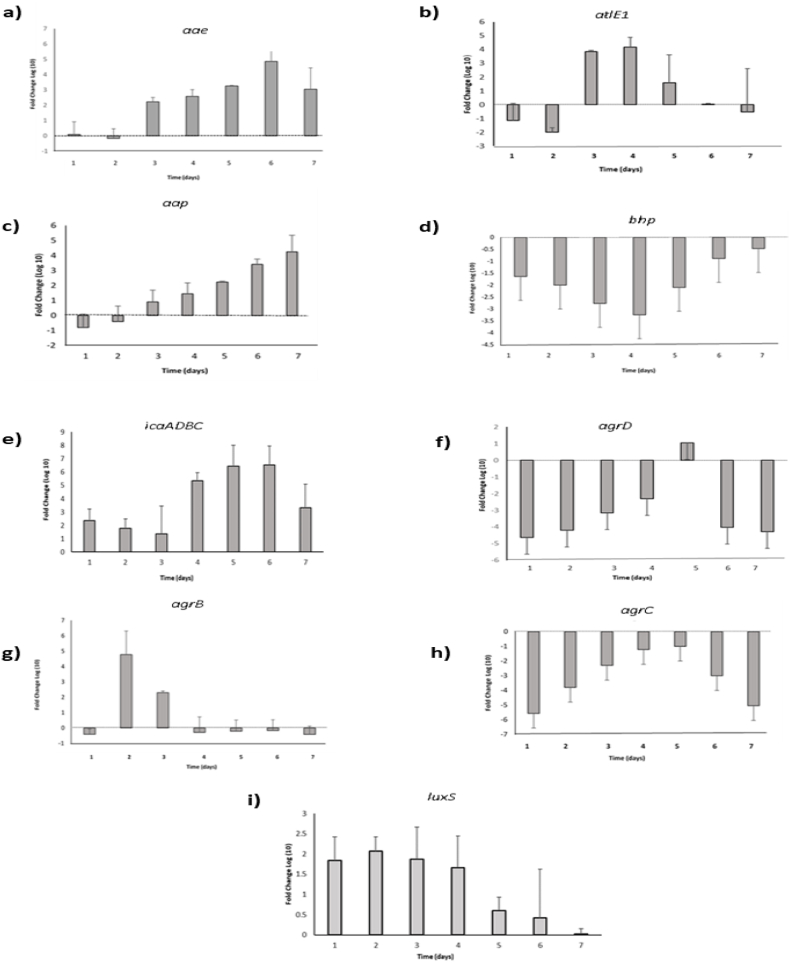
Real-time PCR. Over time, changes in gene expression levels were observed (Fig. 4a–i), qPCR results showed a high expression of atlE1 (days 3 and 4) (Fig. 4a) and aae genes (days 4,5, and 6) (Fig. 4b). Genetic data shows that the S. epidermidis autolysin atlE1 promotes bacterial binding to unmodified polystyrene, as illustrated by the defect in the S. epidermidis O-47 atlE::Tn917 transposon mutant, which was unable to cling to plastic surfaces [7]. Significant changes in cell surface hydrophobicity are induced by atlE1 expression [40]. Also, it is clear that atlE1 plays a crucial role in S. epidermidis biofilm production that is mediated by eDNA [41,42]. Besides, aae represents a novel member of the staphylococcal autolysin/adhesin family that is enrolled in the colonization process. A study revealed that aae possesses bacteriolytic activity and adhesive properties when expressed in E. coli [43]. Hence, the expression of these two autolysins strongly support our hypothesis of new biofilm formation after dispersion, allowing cells to re-attach to the surface and move back to the sessile mode. Yet, we can't predict which type of cells inside the biofilm are expressing these autolysins and promoting a novel formation. In addition, high expression of aap was detected on days 6 and 7 (Fig. 4c). It is known that aap has a role in the accumulation of cells to construct the 3D structure of the biofilm [44]. When fully expressed, aap aids in the initial adhesion of bacterial cells (e.g., abiotic surface or host protein), promotes intercellular accumulation and biofilm formation, in addition, contributes to S. epidermidis virulence [44]. Furthermore, aap from S. epidermidis drives amyloid fiber formation, [45]. Amyloids are highly aggregative proteins that form organized, self-templating fibers that play a role in the stability of biofilms. The production of the amyloid by aap is driven by the release of eDNA [46] from lysed bacterial cells that is maintained through the bacterial cell wall hydrolase autolysin gene atlE as mentioned so far. Therefore, this explains the early expression of atlE1 gene (days 3 and 4) followed by aae and aap genes Figs. (4a, 4b and 4c). Expression of all four icaADBC genes, is necessary for the production of a functionally active PIA molecule [47]. In our study, the transcript level of icaADBC increased over time (day 4, day 5, and day 6) (Fig. 4d). This expression indicates the progressive production of the biofilm matrix components, thus preparing for new establishment. icaADBC was not expressed in the first three days of the biofilm cycle since at this stage the biofilm is already mature. In addition, agr genes were expressed mainly in the period between days 3 and 5, Figs. (4e, 4f, and 4g). Evidently, the agr quorum sensing (QS) system regulates the secretion of proteases in S. epidermidis and S. aureus [48,49]. Therefore, by triggering the release of extracellular proteases, the addition of AIP to preexisting biofilms may reactivate agr and aid in biofilm detachment [30]. These findings are synchronized with the expression of agr system genes between days 3 and 5, when dispersion of the old biofilm is taking place which implies a dual process occurring at this stage. Furthermore, in an animal model of device-associated infection, luxS, another QS system of S. epidermidis, restricts biofilm development and pathogenicity. In our case, luxS had a low expression during the entire experiment and to date, the exact function of luxS is still unknown, maybe narrowing the study period will provide us with more information regarding this complicated system. As for luxS, bhp expression was also low on all days, as it is reported in the absence of PIA, bhp promotes biofilm development and accumulation [50] (Fig. 4h and i). Since S. epidermidis CIP 444 has a PIA-dependent biofilm [51], it appears that there is no direct role for bhp in biofilm formation. Due to its presence, bhp may have another function, or its expression may be limited to more stressful conditions.
Fig. 4.
The expression level of the genes (agrD, agrC, agrB, bhp, atlE1, aae, aap, icaADBC and luxS) were assessed by real-time PCR reactions. The expression levels of all tested genes were normalized using the housekeeping gene 16SrRNA. Results are presented as threshold cycles (Ct) values of genes expressed in biofilm cells of S. epidermidis CIP 444 (a–i) during 7 days (day 1 to day 7), in addition to planktonic (day 0). Ct values ≥ 30 were chosen as very low or no expression. Columns with different annotation letters (a,b,c.) indicate days with a significant difference in gene expression with a p-value <0.05 based on Duncan's multiple range test.
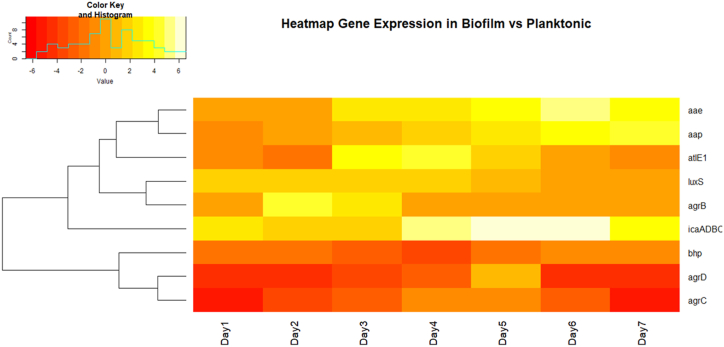
Fold Change relative to Planktonic 2-ΔΔCt. The transcripts from the biofilm cultures were normalized by the reference gene (16SrRNA), and compared to the planktonic ones by calculating the fold change (2 -ΔΔCt) (Fig. 5a- 5i). A common method of visualizing gene expression data is to display it as a heat map combined with clustering methods which group genes together based on the similarity of their gene expression pattern this can be useful for identifying genes that are commonly regulated in a particular condition. Some of the genes in biofilm were significantly increased and others were significantly decreased when compared to planktonic over the seven-day study (Fig. 6). The columns in the heat map indicate the comparison between the studied genes in biofilm at different days of incubation and rows represent the expression of these genes relative to planktonic cells. The colors of heatmap indicate scaled expression of genes (folds log10), the color value ranging from red to yellow while red indicates downregulation and yellow indicates up regulation. According to the obtained results, in comparison with planktonic transcript's level atlE1 and aae that function as adhesins increase at the early stage that promote biofilm formation on abiotic or biotic surfaces, and this was found in our study, as they increase about 4 folds compared to planktonic free cells which was also, clearly illustrated in the heat map (orange-yellow color). It must be mentioned that cells in each step may exhibit different expression of genes even at the same time as a variety of cellular modes are present, and this is also dependent on their position and how it is influenced by the microenvironment inside the biofilm. So free cells will typically have a low expression of adhesins in addition to the genes required for accumulation and maturation. The expression of ica operon genes and aap later in the biofilm cycle explained that fact. icaADBC and aap genes exhibit an upsurge on days 6 and 7 by 6.5 and 4.2 positive folds (Fig. 5a and e), respectively with a yellow-white range on heat map indicating very high expression.
Fig. 5.
Fold change 2-ΔΔct expressed in Log 10 for biofilm genes (a–i) after normalization with reference gene16srRNA compared to planktonic sample during seven days. Where ΔΔCt = ΔCt (sample) -ΔCt (control), the control is the planktonic sample. Some genes in biofilm were enhanced while others were significantly decreased as compared to planktonic. Fold change (log10) = 0 indicate same expression of the gene in biofilm and planktonic sample. Folds >0 indicate positive increase in the expression relative to the control, folds <0 indicate negative expression (downregulation) of the gene relative to control. The assay was performed with at least three independent sample. Analysis of variance was not performed; error bars indicate standard deviation of the means.
Fig. 6.
Heat map or clustering technique, which place genes in groups based on how similarly their gene expression patterns behave, a typical approach of displaying gene expression data can help discover the genes that are often regulated in a given circumstance, showing the expression patterns of 9 genes in biofilm cells compared to planktonic cells over a period of seven-days. The columns in the heat map indicate the comparison between the studied genes in biofilm at different days of incubation and the rows represent the expression of these genes relative to planktonic cell. The colors indicate scaled expression of genes (folds log10). Yellow and red represent up-and downregulated gene expression respectively color density indicating levels of fold change was displayed. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
agrC transcripts were higher in the planktonic bacteria except for days 4 and 5 where the relative expression was similar in both cultures (Fig. 5h). Whereas agrD transcript level was double in the biofilm culture compared to the planktonic at day 4 (Fig. 5f), while agrB accounts for a 4.7-fold increase on day 2 than the planktonic culture (Fig. 5g). bhp had a low expression in the planktonic cells in contrast with the dispersed biofilm cells at days 4 and 5, where it had no expression (Fig. 5d). This result indicates that in this strain, bhp has no direct role in self-detachment. Many pathogenic bacteria with luxS mutants exhibit reduced pathogenicity, in contrast, the S. epidermidis luxS mutant has higher virulence in a model of catheter-associated infection. Most likely, the enhanced PIA production and intensified biofilm formation contribute to the higher pathogenicity [52]. In our findings, the expression of luxS was low and similar in planktonic and biofilm cells except for the period after day 5, when a lower expression was detected, and possibly this further decrease promoted new biofilm formation and the slight increase in its expression in the first three days may indicate its contribution in the downregulation of icaADBC expression to prevent biofilm formation (Fig. 5i). In addition, the expression of luxS was significantly inversely proportional to bhp and bhp was downregulated when compared to the planktonic (red color in heat map). bhp in S. epidermidis isolates serve as early attachment elements in the development of biofilms [53] however, its low expression in the biofilm over the studied period with a slight increase on days 6 and 7 raise a question about its role in virulence or early biofilm formation, further work will be needed to investigate more about its expression in both planktonic and biofilm cells. Despite the idea that dispersed cells become planktonic and then return to the bulk, it was demonstrated that dispersed P. aeruginosa cells are different from both biofilm and planktonic cell, and surpassed their planktonic counterparts in their ability to penetrate and kill macrophages [54].
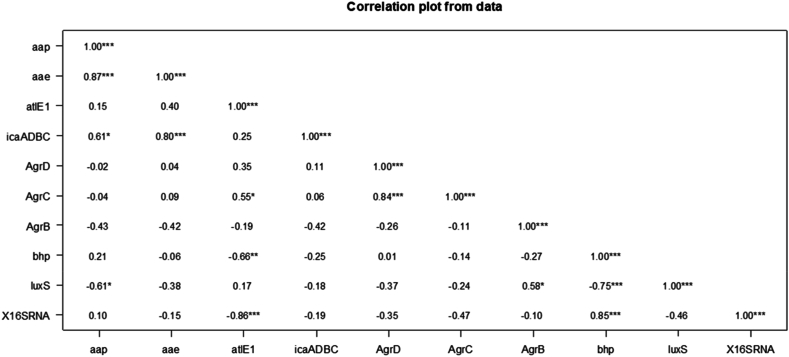
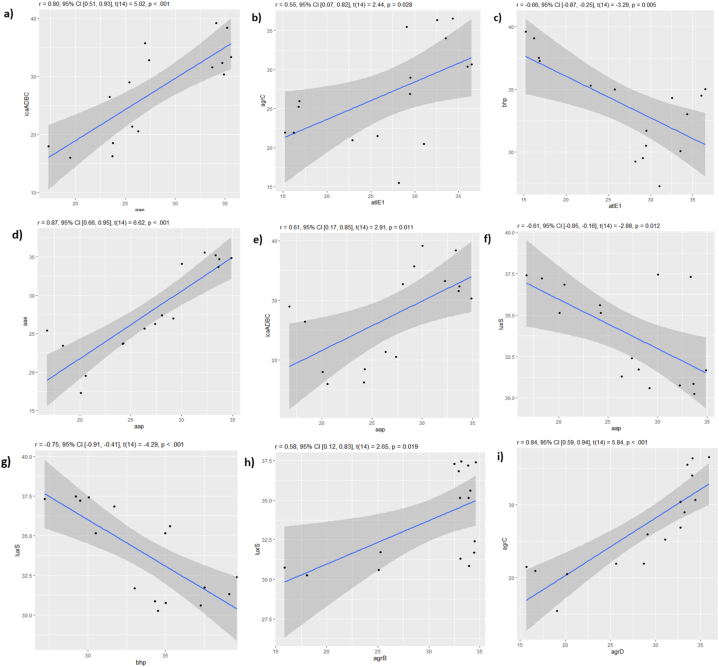
The Pearson test (Fig. 7) showed a strong correlation between agrD and agrC with r = 0.84, aap and aae genes, (r = 0.87) and between aae, icaADBC (r = 0.80). Whereas a moderate correlation was found between aap and icaADBC (r = 0.61), atlE1 and agrC (r = 0.55), agrB and luxS (r = 0.58). aap showed a moderate correlation inversely proportional with luxS (r = −0.61). While bhp revealed a moderate inversely proportional correlation with atlE1 (r = −0.66) and a strong correlation varying inversely with luxS (r = −0.75) and no correlation with other genes. All mentioned correlations were significance with a p-value <0.05 (Fig. 8a–i).
Fig. 7.
Pearson's Correlation (r) for the studied genes obtained through “R” program, throughout the studied period. Each cell in the table shows the correlation between two specific variables (the vertical and the horizontal). Significant positive correlation r ≥ 0.5, and negatively correlated r ≥ -0.5. (*) refer to degree of correlation.
Fig. 8.
Pearson correlation showing the p-value of the correlated genes and its linear distribution using ‘R’ program. Each square represents the correlation between two genes (a–i), the dots indicate the expression of both genes throughout the studied period from day 0 to day 7. The closer the dots come to forming a straight line when plotted, the higher the correlation between the two variables, or the stronger the relationship. r ≥ ±0.5 are said to be correlated, p-value <0.05 the correlation is significant.
Pearson test showed a significant correlation between the autolysins/adhesins genes expressed on the same period and the icaADBC operon (Fig. 8a) Compared to the planktonic profile, icaADBC showed a great increase in its expression (pale yellow and white in heat map) (Fig. 7).
Finally, it was revealed that genes related to attachment were correlated with the expression of dispersal genes, which explains the fact that two processes are occurring during the same period of the biofilm cycle of S. epidermidis CIP 444. Gene expression results from planktonic and biofilm cells showed interesting differences. Therefore, genetic investigation of biofilm cells particularly during the dispersion phase, will help in understanding the dynamics of microbial biofilms, including different metabolic states of bacteria embedded in the extracellular matrix, time of self-dispersion, characteristics of the dispersed cells and their spatial distribution implemented into a three-dimensional individual-based model of biofilm including the incorporated genes and that can be applied and help in the management and control of infections related to S. epidermidis.
Ethical statements
There are no human or animal experimental work in this study.
Funding
No funding was obtained for this study.
Data availability
Data will be made available on request.
CRediT authorship contribution statement
Suzanne Jonblat: Writing – review & editing, Methodology, Investigation. Falah As-sadi: Software. Andre El Khoury: Writing – review & editing, Supervision, Project administration. Neressa Badr: Methodology. Mireille Kallassy: Writing – review & editing, Supervision, Project administration. Ali Chokr: Writing – review & editing, Visualization, Supervision, Resources, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Our team is grateful to the English Language Center at the Lebanese University, Beirut, Lebanon for the critical reading of the article.
Contributor Information
Suzanne Jonblat, Email: Suzannejonblat@hotmail.com.
Falah As-sadi, Email: falah.assadi@gmail.com.
Andre El Khoury, Email: andre.khoury@usj.edu.lb.
Neressa Badr, Email: neressabadr@outlook.com.
Mireille Kallassy, Email: Mireille.kallassy@usj.edu.lb.
Ali Chokr, Email: alichokr@hotmail.com, alichokr@ul.edu.lb.
References
- 1.Lowy F.D. Staphylococcus aureus infections. N. Engl. J. Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Rupp M.E., Archer G.L. Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 1994:231–243. doi: 10.1093/clinids/19.2.231. [DOI] [PubMed] [Google Scholar]
- 3.Rupp M.E. Clinical characteristics of infections in humans due to Staphylococcus epidermidis. Staphylococcus epidermidis: methods and protocols. 2014:1–16. doi: 10.1007/978-1-62703-736-5_1. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerli W., Trampuz A., Ochsner P.E. Prosthetic-joint infections. N. Engl. J. Med. 2004;351(16):1645–1654. doi: 10.1056/NEJMra040181. [DOI] [PubMed] [Google Scholar]
- 5.Skovdal S.M., Jørgensen N.P., Meyer R.L. JMM profile: Staphylococcus epidermidis. J. Med. Microbiol. 2022;71(10) doi: 10.1099/jmm.0.001597. [DOI] [PubMed] [Google Scholar]
- 6.Otto M. Staphylococcus epidermidis—the'accidental'pathogen. Nat. Rev. Microbiol. 2009;7(8):555–567. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heilmann C., Hussain M., Peters G., Götz F. Evidence for autolysin‐mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 1997;24(5):1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 8.Heilmann C., Thumm G., Chhatwal G.S., Hartleib J., Uekotter A., Peters G. Identifi-cation and characterization of a novel autolysin (Aae) with adhesive properties from Staphy-lococcus epidermidis. Microbiology. 2003;149(10):2769–2778. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- 9.Mack D., Fischer W., Krokotsch A., Leopold K., Hartmann R., Egge H., Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a lin-ear beta-1, 6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 1996;178(1):175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y.Q., Ren S.X., Li H.L., Wang Y.X., Fu G., Yang J.…Wen Y.M. Ge-nome‐based analysis of virulence genes in a non‐biofilm‐forming Staphylococcus epidermidis strain (ATCC 12228) Mol. Microbiol. 2003;49(6):1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 11.Heilmann C., Schweitzer O., Gerke C., Vanittanakom N., Mack D., Götz F. Molecular basis of intercellular adhesion in the biofilm‐forming Staphylococcus epidermidis. Mol. Microbiol. 1996;20(5):1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 12.Conlon K.M., Humphreys H., O'Gara J.P. icaR encodes a transcriptional repressor in-volved in environmental regulation of ica operon expression and biofilm formation in Staphy-lococcus epidermidis. J. Bacteriol. 2002;184(16):4400–4408. doi: 10.1128/JB.184.16.4400-4408.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateman A., Holden M.T., Yeats C. The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics. 2005;21(8):1301–1303. doi: 10.1093/bioinformatics/bti206. [DOI] [PubMed] [Google Scholar]
- 14.Yao Y., Sturdevant D.E., Otto M. Genomewide analysis of gene expression in Staphy-lococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 2005;191(2):289–298. doi: 10.1086/426945. [DOI] [PubMed] [Google Scholar]
- 15.Waters C.M., Bassler B.L. Quorum sensing: cell-to-cell communication in bacte-ria. Annu. Rev. Cell Dev. Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 16.Kleerebezem M., Quadri L.E., Kuipers O.P., De Vos W.M. Quorum sensing by pep-tide pheromones and two‐component signal‐transduction systems in Gram‐positive bacte-ria. Mol. Microbiol. 1997;24(5):895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 17.Wright I.I.I., Jin R., Novick R.P. Transient interference with staphylococcal quorum sensing blocks abscess formation. Proc. Natl. Acad. Sci. USA. 2005;102(5):1691–1696. doi: 10.1073/pnas.0407661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheung G.Y., Wang R., Khan B.A., Sturdevant D.E., Otto M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus path-ogenesis. Infect. Immun. 2011;79(5):1927–1935. doi: 10.1128/IAI.00046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novick Richard P. [27] genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 20.Ji G., Beavis R.C., Ji G., Beavis R.C., Novick R.P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. USA. 1995;92(26):12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saenz H.L., Augsburger V., Vuong C., Jack R.W., Götz F., Otto M. Inducible ex-pression and cellular location of AgrB, a protein involved in the maturation of the staphylo-coccal quorum-sensing pheromone. Arch. Microbiol. 2000;174:452–455. doi: 10.1007/s002030000223. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., Ji G. Identification of a staphylococcal AgrB segment (s) responsible for group-specific processing of AgrD by gene swapping. J. Bacteriol. 2004;186(20):6706–6713. doi: 10.1128/JB.186.20.6706-6713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lina G., Jarraud S., Ji G., Greenland T., Pedraza A., Etienne J.…Vandenesch F. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol. Microbiol. 1998;28(3):655–662. doi: 10.1046/j.1365-2958.1998.00830.x. [DOI] [PubMed] [Google Scholar]
- 24.Novick R.P., Projan S.J., Kornblum J., Ross H.F., Ji G., Kreiswirth B.…Novick R.P. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aure-us. Mol. Gen. Genet. MGG. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 25.Queck S.Y., Jameson-Lee M., Villaruz A.E., Bach T.H.L., Khan B.A., Sturdevant D.E.…Otto M. RNAIII-independent target gene control by the agr quorum-sensing sys-tem: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell. 2008;32(1):150–158. doi: 10.1016/j.molcel.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Toole G., Kaplan H.B., Kolter R. Biofilm formation as microbial develop-ment. Annu. Rev. Microbiol. 2000;54(1):49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 27.Boles B.R., Horswill A.R. Staphylococcal biofilm disassembly. Trends Microbiol. 2011;19(9):449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu. Rev. Med. 2013;64:175–188. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 29.Cheung G.Y., Joo H.S., Chatterjee S.S., Otto M. Phenol-soluble modulins–critical de-terminants of staphylococcal virulence. FEMS Microbiol. Rev. 2014;38(4):698–719. doi: 10.1111/1574-6976.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boles B.R., Horswill A.R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4(4) doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2021. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ URL. [Google Scholar]
- 32.Barraud N., Kardak B.G., Yepuri N.R., Howlin R.P., Webb J.S., Faust S.N.…Kelso M.J. Cephalosporin‐3′‐diazeniumdiolates: targeted NO‐donor prodrugs for dispersing bacterial biofilms. Angew. Chem. Int. Ed. 2012;51(36):9057–9060. doi: 10.1002/anie.201202414. [DOI] [PubMed] [Google Scholar]
- 33.Van Loosdrecht M.C., Norde W., Lyklema J., Zehnder A.J. Hydrophobic and electro-static parameters in bacterial adhesion: Dedicated to Werner Stumm for his 65 th birth-day. Aquat. Sci. 1990;52:103–114. [Google Scholar]
- 34.Sousa C., Teixeira P., Oliveira R. International journal of biomateri-als; 2009. Influence of Surface Properties on the Adhesion of Staphylococcus Epidermidis to Acrylic and Silicone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De-la-Pinta I., Cobos M., Ibarretxe J., Montoya E., Eraso E., Guraya T., Quindós G. Effect of biomaterials hydrophobicity and roughness on biofilm development. J. Mater. Sci. Mater. Med. 2019;30:1–11. doi: 10.1007/s10856-019-6281-3. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira R., Azeredo J., Teixeira P., Fonseca A.P. Bioline; 2001. The Role of Hydrophobicity in Bacteri-Al Adhesion. [Google Scholar]
- 37.Merck Safety data sheet. Crystal. Violet. 2017 [Google Scholar]
- 38.Zandri G., Pasquaroli S., Vignaroli C., Talevi S., Manso E., Donelli G., Biavasco F. Detection of viable but non-culturable staphylococci in biofilms from central venous catheters negative on standard microbiological assays. Clin. Microbiol. Infection. 2012;18(7):E259–E261. doi: 10.1111/j.1469-0691.2012.03893.x. [DOI] [PubMed] [Google Scholar]
- 39.Gaio V., Lopes N., Cerca N., França A. codY and pdhA expression is induced in Staphylococcus epidermidis biofilm and planktonic populations with higher proportions of viable but non-culturable cells. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.771666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otto M. Staphylococcus epidermidis pathogenesis. Staphylococcus Epidermidis. Methods and Protocols. 2014:17–31. doi: 10.1007/978-1-62703-736-5_2. [DOI] [PubMed] [Google Scholar]
- 41.Qin Z., Ou Y., Yang L., Zhu Y., Tolker-Nielsen T., Molin S., Qu D. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermid-is. Microbiology. 2007;153(7):2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 42.Christner M., Heinze C., Busch M., Franke G., Hentschke M., Bayard Dühring S.…Rohde H. sarA negatively regulates S taphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis‐dependent release of eDNA. Mol. Microbiol. 2012;86(2):394–410. doi: 10.1111/j.1365-2958.2012.08203.x. [DOI] [PubMed] [Google Scholar]
- 43.Heilmann C., Thumm G., Chhatwal G.S., Hartleib J., Uekotter A., Peters G. Identifi-cation and characterization of a novel autolysin (Aae) with adhesive properties from Staphy-lococcus epidermidis. Microbiology. 2003;149(10):2769–2778. doi: 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- 44.Schaeffer C.R., Woods K.M., Longo G.M., Kiedrowski M.R., Paharik A.E., Büttner H.…Fey P.D. Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect. Immun. 2015;83(1):214–226. doi: 10.1128/IAI.02177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yarawsky A.E., Johns S.L., Schuck P., Herr A.B. The biofilm adhesion protein Aap from Staphylococcus epidermidis forms zinc-dependent amyloid fibers. J. Biol. Chem. 2020;295(14):4411–4427. doi: 10.1074/jbc.RA119.010874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B., Zhan Q., Xiao Y., Xu Y., Zhao H., Rao L.…Yu F. Mupirocin enhances the biofilm formation of Staphylococcus epidermidis in an atlE-dependent man-ner. Int. J. Antimicrob. Agents. 2023;62(4) doi: 10.1016/j.ijantimicag.2023.106904. [DOI] [PubMed] [Google Scholar]
- 47.Gerke C., Kraft A., Sußmuth R., Schweitzer O., Götz F. Characterization of theN-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 1998;273(29):18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 48.Yao Y., Vuong C., Kocianova S., Villaruz A.E., Lai Y., Sturdevant D.E., Otto M. Characterization of the Staphylococcus epidermidis accessory-gene regulator response: quor-um-sensing regulation of resistance to human innate host defense. J. Infect. Dis. 2006;193(6):841–848. doi: 10.1086/500246. [DOI] [PubMed] [Google Scholar]
- 49.Thoendel M., Kavanaugh J.S., Flack C.E., Horswill A.R. Peptide signaling in the staphylococci. Chem. Rev. 2011;111(1):117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tormo M.A., Knecht E., Gotz F., Lasa I., Penades J.R. Bap-dependent biofilm for-mation by pathogenic species of Staphylococcus: evidence of horizontal gene trans-fer? Microbiology. 2005;151(7):2465–2475. doi: 10.1099/mic.0.27865-0. [DOI] [PubMed] [Google Scholar]
- 51.Chokr A., Watier D., Eleaume H., Pangon B., Ghnassia J.C., Mack D., Jabbouri S. Correlation between biofilm formation and production of polysaccharide intercellular adhesin in clinical isolates of coagulase-negative staphylococci. International journal of medical mi-crobiology. 2006;296(6):381–388. doi: 10.1016/j.ijmm.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 52.Xu L., Li H., Vuong C., Vadyvaloo V., Wang J., Yao Y.…Gao Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermid-is. Infect. Immun. 2006;74(1):488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heilmann C., Ziebuhr W., Becker K. Are coagulase-negative staphylococci virulent? Clin. Microbiol. Infection. 2019;25(9):1071–1080. doi: 10.1016/j.cmi.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 54.Chua S.L., Liu Y., Yam J.K.H., Chen Y., Vejborg R.M., Tan B.G.C.…Yang L. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 2014;5(1):4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.