Abstract
We determined nucleotide sequences of the VP1 and 2AB genes and portions of the 2C and 3D genes of two evolving poliovirus lineages: circulating wild viruses of T geotype and Sabin vaccine-derived isolates from an immunodeficient patient. Different regions of the viral RNA were found to evolve nonsynchronously, and the rate of evolution of the 2AB region in the vaccine-derived population was not constant throughout its history. Synonymous replacements occurred not completely randomly, suggesting the need for conservation of certain rare codons (possibly to control translation elongation) and the existence of unidentified constraints in the viral RNA structure. Nevertheless the major contribution to the evolution of the two lineages came from linear accumulation of synonymous substitutions. Therefore, in agreement with current theories of viral evolution, we suggest that the majority of the mutations in both lineages were fixed as a result of successive sampling, from the heterogeneous populations, of random portions containing predominantly neutral and possibly adverse mutations. As a result of such a mode of evolution, the virus fitness may be maintained at a more or less constant level or may decrease unless more-fit variants are stochastically generated. The proposed unifying model of natural poliovirus evolution has important implications for the epidemiology of poliomyelitis.
Analysis of polioviruses isolated either during a given outbreak of poliomyelitis (30, 35, 51) or from sequential fecal samples from infected individuals (29, 31, 34) has revealed rapid changes in the nucleotide sequence of the viral 7.5-kb RNA. Oligonucleotide fingerprinting (51) and genome sequencing (30, 34) suggested that, during the epidemic, the nucleotide substitutions ranged from 1 to 2% per year. The molecular basis of such genetic instability, common to all RNA viruses, resides in a high error rate of the viral RNA-dependent RNA polymerases, estimated to be, for poliovirus, on the order of 10−4 to 10−5 substitutions per base per replication (12, 70, 71), and the absence of proofreading mechanisms for the correction of the errors made. The combination of these properties results in a high heterogeneity (the “quasispecies” nature) of all populations of the virus (8, 10, 11, 63).
Less understood, however, are the rules governing the fixation of specific mutations upon passages of a viral population. Generally speaking, a mutation may decrease or increase the level of virus fitness for a particular ecological niche or leave it unchanged. Nucleotide substitutions associated with changes in the “sense” of codons (nonsynonymous mutations) are less likely to be neutral than substitutions resulting in no change of the codon meaning (synonymous mutations). Fixation of mutations conferring a selective advantage is readily understood in the framework of positive Darwinian selection. Adverse mutations may be eliminated by negative selection.
The likelihood of fixation of a mutation depends not only on the associated changes in virus fitness but, to a large extent, on the sizes of propagated populations (48, 52–54). Consecutive passages involving small viral populations, the situation very typical of the natural poliovirus infection, may well result in the accumulation of neutral or even debilitating mutations (5, 7, 13, 18), the phenomenon known as the Muller's ratchet (5, 6, 8). Thus, the consecutive changes in viral lineages are not necessarily adaptive but may be due to random sampling. There are reasons to believe that the majority of generated mutations may in fact be neutral or nearly so (33). Since the same appears to be true of the fixed mutations (60), the rate of nucleotide changes should be approximately constant in time (33). This concept of a “molecular clock” was suggested to be valid for the evolution of RNA viruses (17, 24, 69) and is not infrequently used for estimations of the viral “age” (31, 65, 73).
Here, we undertook an analysis of genomic changes in two evolving poliovirus populations. One was represented by a lineage of wild-type (wt) viruses of T geotype (40) in circulation in the territory of the former Soviet Union (FSU) during 1991 to 1995. The other consisted of the strains successively isolated from an immunodeficient patient (IDP) with paralytic poliomyelitis (31). The obtained sequences revealed basic similarities in the character of accumulation of mutations in the two lineages, even though some quantitative differences were also evident. Different regions of the viral genome have evolved at nonidentical rates, and these rates might have changed during the history of a population. Nevertheless, the major contribution to the evolving populations came from the linear accumulation of predominantly synonymous substitutions. From the perspective of current theories of viral evolution (8, 24), the results obtained suggest a unifying model for the evolution of circulating poliovirus on the one hand and for virus growing in the gut of an individual on the other. This model has important implications for the epidemiology of poliomyelitis.
MATERIALS AND METHODS
Virus isolation and typing.
Two sets of polioviruses of serotype 1 were analyzed (Table 1; Fig. 1). One contained 24 wt strains isolated in the territory of the FSU, China, and Pakistan from 1991 to 1995. Of these, 19 strains belonged to the T geotype (40). The other set included five vaccine-related strains (GenBank accession numbers for VP1 sequences, AF083933 to AF083937) obtained from an IDP with paralytic poliomyelitis from Missouri in 1981 (31).
TABLE 1.
Poliovirus isolates studied
| Straina | Geographic location | Geotypeb | Date of sampling (da/mo/yr, mo/yr or yr) |
|---|---|---|---|
| PV1/2677USA81,c day 23 major | Missouri, USAe | 23/08/81 | |
| PV1/2677USA81,c day 48 major | Missouri, USA | 17/09/81 | |
| PV1/3038USA81,c day 126 major | Missouri, USA | 04/12/81 | |
| PV1/3038USA82,c day 158 major | Missouri, USA | 05/01/82 | |
| PV1/3038USA82,c day 200 major | Missouri, USA | 16/02/82 | |
| PV1/169AZB59 | Baku, Azerbaijan | 11/59 | |
| PV1/7TAJ91d | Kurgan-Tyube, Tajikistan | T | 02/08/91 |
| PV1/4TAJ91d | Kurgan-Tyube, Tajikistan | T | 03/08/91 |
| PV1/5341UKR92d | Ukraine | T | 11/92 |
| PV1/5794UZB94d | Kashkadaryinsk region, Uzbekistan | T | 11/07/94 |
| PV1/6011TAJ94d | Tajikistan | T | 17/11/94 |
| PV1/6064CHE95d | Chechnya | T | 14/06/95 |
| PV1/6484CHE95d | Grozny region, Chechnya | T | 17/08/95 |
| PV1/6486CHE95d | Grozny region, Chechnya | T | 22/08/95 |
| PV1/6490CHE95d | Grozny region, Chechnya | T | 08/95 |
| PV1/6405ING95d | Ingush Republic | T | 28/09/95 |
| PV1/6427ING95d | Nasyr-Kort, Ingush Republic | T | 14/10/95 |
| PV1/827GEO85 | Tbilisi, Georgia | T | 05/02/85 |
| PV1/832GEO85 | Tbilisi, Georgia | T | 12/11/85 |
| PV1/434MOL91 | Kishinev, Moldova | T | 29/08/91 |
| PV1/6070CHN94 | China | T | 1994 |
| PV1/5990TAJ94 | Shaartuz, Tajikistan | T | 24/09/94 |
| PV1/6013TAJ94 | Kolkhozabad, Tajikistan | T | 17/11/94 |
| PV1/6430PAK95 | Pakistan | T | 1995 |
| PV1/6433PAK95 | Pakistan | T | 1995 |
| PV1/422RUS91 | Inza, Russia | A | 17/06/91 |
| PV1/919GEO90 | Zugdidi, Georgia | G | 03/12/90 |
| PV1/5937RUS94 | Moscow, Russia | G | 03/06/94 |
| PV1/717RUS94 | Moscow region, Russia | G | 12/08/94 |
Each strain designation consists of the serotype, number given at isolation, and place and year of the corresponding polio case.
As shown in reference 40.
IDP lineage.
T geotype lineage.
USA, United States.
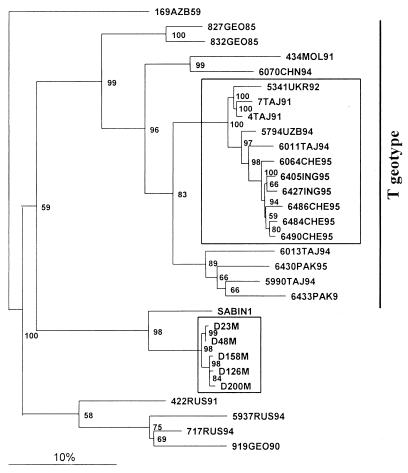
FIG. 1.
The VP1/2ABC′ maximum-likelihood bootstrap consensus tree (PUZZLE, version 4.0) rooted on PV1/169AZB59 showing the relationships of poliovirus type 1 wild and vaccine strains. The scale corresponds to 10% nucleotide divergence. The isolates belonging to the T geotype and IDP lineages are boxed.
Virus isolation from stool samples was done by standard methods (72). wt isolates were passaged once on monolayers of RD or HEp-2 cell lines and were typed in microneutralization tests with type-specific sera. The isolation and typing of vaccine-related viruses were described earlier (31).
Reverse transcription, PCR, and sequencing of poliovirus RNAs.
RNA was extracted from cell lysates with Ultraspec 3 (Biotexc) and was reverse transcribed using random hexamer primers (Boehringer Ingelheim) with avian myeloblastosis virus reverse transcriptase (Promega). DNA copies of selected genomic portions were amplified by PCR with appropriate primers (the primer sequences are available upon request), and the PCR products were purified with the QIAquick system (Qiagen) and were directly sequenced either automatically (Applied Biosystems) or manually by using the Sequenase kit, version 2.0 (U.S. Biochemicals). The following regions were analyzed in all the 29 strains: the complete VP1 (906 nucleotides [nt]; nt 2500 to 3408), 2A (447 nt; nt 3409 to 3855), and 2B (291 nt; nt 3856 to 4146) genes as well as a portion of the 2C gene (51 nt; nt 4147 to 4197; called 2C′). A portion of the 3D gene (792 nt; nt 6601 to 7392; called 3D′) in PV1/169AZB59, -/7TAJ91, -/5794UZB94, -/6484CHE95, -/6486CHE95, -/6490CHE95, -/6405ING95, -/422RUS91, and -/919GEO90 was analyzed. The coordinates are given according to the consensus numbering system of Toyoda et al. (68).
The sequences of the following regions have been previously reported: the VP1/2A junction (150 nt; nt 3319 to 3368) of PV1/827GEO85, -/832GEO85, -/919GEO90, -/434MOL91, -/422RUS91, -/4TAJ91, and -/7TAJ91 (40) as well as the VP1 gene of the IDP strains (31).
Comparative analysis of nucleotide sequences.
Multiple alignment of the determined sequences was performed by the program CLUSTAL W, version 1.74 (67). Phylogenetic analysis was carried out by the PHYLIP, version 3.5c, program package (20), PUZZLE, version 4.0 (64), and METREE (58). Analysis under maximum-likelihood optimality criteria was done with empirical base frequencies and the HKY85 model of substitution (25). The following algorithms have also been used for testing the consistency of branching: maximum parsimony (16), neighbor joining (59), minimum evolution (57), and the unweighted average-linkage method of clustering. Evolutionary distances were estimated using the two-parameter method of Kimura (32) and the Tamura-Nei model (66). The reliability of clustering was tested by 10,000 iterations in the quartet-puzzling method (PUZZLE, version 4.0) and by 1,000 bootstrapping replicates in other methods mentioned above.
To assess the degrees of nonsynonymous and synonymous nucleotide divergence separately, the estimation of pairwise genetic distances was performed according to Li's algorithm (38). A site is considered fourfold degenerate if all possible changes at the site are synonymous; a site is twofold degenerate if one of the three possible changes is synonymous; at a nondegenerate site, all possible changes are nonsynonymous. The following parameters were calculated: the percentage of mutated synonymous sites among all synonymous sites (Ks), the percentage of mutated nonsynonymous sites among all nonsynonymous sites (Ka), the percentage of mutated sites among all sites (Kt), the overall number of mutations (Nt), the number of synonymous differences (Ns), and the number of nonsynonymous differences (Na) (50). The rate of mutation fixation was calculated by regression analysis.
Statistical analysis of rare-codon conservation.
A codon was defined as rare if its abundance was no more than 30% of that of its most abundant synonym in the poliovirus codon usage table. Five families of codons (encoding Ala, Arg, Pro, Ser, and Thr) contain codons meeting this criterion. The same codons were also scarce in humans. To eliminate the bias due to the close relatedness of some of the strains in our data set, the 6 most-diverged VP1 and 2AB sequences were selected out of 29 sequences available. Upon pairwise comparisons, these sequences exhibited divergences higher than 14 and 18%, respectively. The following strains were used in the analysis of both regions: PV1/169AZB59, -/7TAJ91, -/832GEO85, -/5937RUS94, and -/Day 200; in addition, the strains PV1/6070CHN94 and -/6013TAJ94 were used for the VP1 and 2AB sets, respectively. It was assumed that all of the sites mutated independently and that the mutation rate was uniform across the region in question. The ratios of the respective rare codons to all their synonyms in the whole poliovirus (Sabin 1) RNA were calculated. The Bernoulli formula was used to determine the probability of the occurrence of a certain number of rare codons (1 to 6) in the positions of the six aligned sequences that corresponded to each of the above amino acid families. The positions having significant deviations (P < 0.05) from random codon usage were considered anomalously conserved.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper are available from the GenBank nucleotide sequence database under accession no. AF233098 to AF233222.
RESULTS
General characterization of the two poliovirus populations.
Two evolving poliovirus populations of serotype 1 were studied (Table 1). One corresponded to 11 wt strains isolated from poliomyelitis patients in the territory of the FSU in 1991 to 1995 and constituted a distinct cluster of the T geotype (Fig. 1). The other set of viruses was represented by five vaccine-related strains successively isolated from an IDP with paralytic poliomyelitis (31). The primary structures of the 150-nt VP1/2A region of the RNA of most of these T geotype isolates and of the VP1 genes of the vaccine-derived viruses have been determined previously (31, 40). Here, the sequenced portions were extended to include the entire 1,694-nt VP1/2ABC′ region (nt 2500 to 4197) of all of the viruses and a 792-nt portion of the 3D gene (nt 6601 to 7392) of all IDP isolates and five T geotype strains.
The selected cluster of T geotype strains was considered to represent a single lineage since they appeared to have a recent common ancestor, a strain imported to the FSU from Pakistan in 1991 (49). Indeed, the earliest T geotype strain of this lineage (PV1/7TAJ91) shared 148 out of 150 nt of the VP1/2A junction region with Pakistani strain PV1/18643PAK91 isolated the same year. The strains of this group were closely related to each other, having 92.3 to 98.8% nucleotide identity in the entire VP1/2AB region. All of them but one demonstrated a close relatedness in the 3D′ region as well (97.9 to 99% identity). The 3D′ region of the deviating strain, PV1/7TAJ91, differed from those of the other T geotype strains by 12.8 to 13.4%. It seems that a recombination event involving the 3′-terminal part of the genome took place in the history of the T geotype lineage. The 3D′ sequence of PV1/7TAJ91 was excluded from the subsequent comparative analysis.
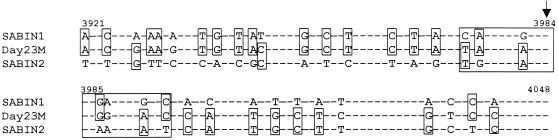
The isolates from the IDP exhibited a serotype 1 specificity and were closely related to the Sabin 1 virus in the primary structure of the VP1 gene (31). However, more-extensive sequencing demonstrated that at least a portion of their 2B and 2C genes was derived from the Sabin 2 genome. The crossover region appeared to map to a short sequence between positions 3973 and 3996, exhibiting similar degrees of relatedness to Sabin 1 and Sabin 2 genomes (Fig. 2). The 583-nt 2AB sequence upstream of the arbitrarily assumed crossover point (in the middle of the crossover region, that is, between nt 3983 and 3984) showed 7.0 and 17.9% divergences from the Sabin 1 and Sabin 2 sequences, respectively, and for the VP1 gene, the corresponding values were 9.7 and 34.1%. On the other hand, corresponding values were 20.5 and 10.4% for the remaining 155-nt portion of the 2B gene and the 51-nt 5′-terminal portion of the 2C gene, respectively. The 3D′ region showed again a closer similarity to the Sabin 1 sequence (4.7%) than to the Sabin 2 sequence (11.7%). We concluded that the IDP isolates were represented by double (type 1/type 2/type 1) recombinants. The portions of the 2B gene upstream and downstream of the assumed crossover point were denoted 2B1 and 2B2, respectively.
FIG. 2.
The crossover region within the 2B gene of the strain Day23M. Dashes, identical bases in all three sequences; framed segment (nt 3973 to 3996), hypothetical recombination region; arrow, beginning of the 2B2 region. Identical nucleotides are boxed.
Time course of genomic evolution of the poliovirus populations.
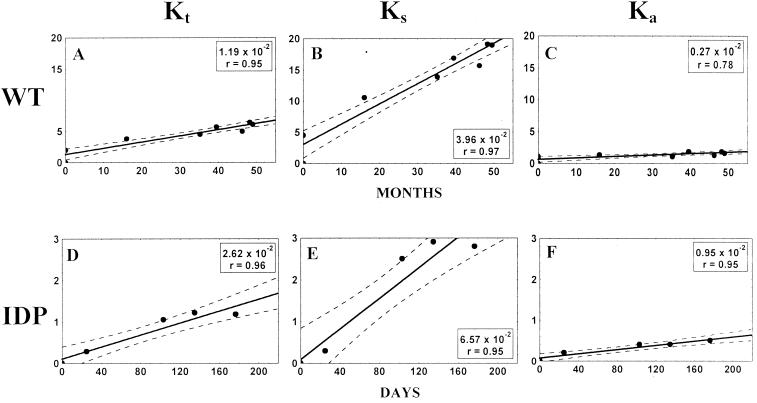
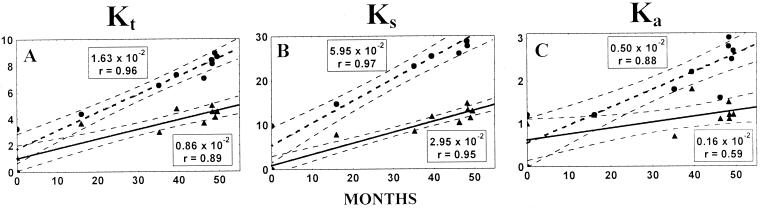
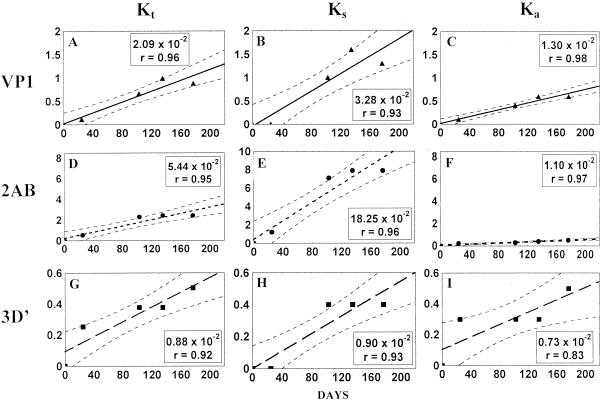
The total number of accumulated mutations in the sequenced portion of the genomes was plotted against time. The time course of mutation accumulation could be well approximated by straight lines (the regression coefficient, r, was ≥0.95) corresponding to the rates of 1.19 × 10−2 and 2.62 × 10−2 substitutions/site/year for the T geotype and IDP lineages, respectively (Fig. 3A and D).
FIG. 3.
Mutation fixation rates in the sequenced regions of 11 wt (VP1/2ABC′) and 5 IDP (VP1/2ABC′/3D′) isolates in substitutions per site per year (r, correlation coefficient) for all (Kt), synonymous (Ks), and nonsynonymous (Ka) sites. y axis, percentage of mutated sites. The position of the first strain in each of the lineages corresponds to x = 0 and y = 0. Dashed lines, 95% confidence intervals.
The majority of the detected mutations (∼75% in both populations) were synonymous substitutions. The linear (r ≥ 0.95) rates of accumulation of synonymous substitutions per synonymous site per year were 3.96 × 10−2 and 6.57 × 10−2 for the T geotype and IDP lineages, respectively (Fig. 3B and E). The corresponding parameters for nonsynonymous substitutions (per nonsynonymous site per year) were significantly lower, 0.27 × 10−2 and 0.95 × 10−2, respectively (Fig. 3C and F).
The preponderance of synonymous substitutions among the fixed mutations and the linear character of mutation fixation were consistent with the notion that random sampling was a major factor contributing to the evolution of both the viral populations studied.
Nonuniform rates of evolution of different parts of the viral genome.
The character of mutations accumulated was analyzed for the three genomic regions (VP1, 2AB, and 3D′) separately (Fig. 4 and 5). The total and synonymous mutations accumulated linearly with time in all the studied regions of both lineages (with r varying between 0.89 and 0.97). However, in both sets of viruses, different RNA regions varied significantly in their relative mutabilities. The 2AB region evolved 2.5 to 5.5 times faster than the VP1 gene, and again a major contribution to this difference came from synonymous substitutions, with the relevant rate for the 2AB region in the IDP strains being exceptionally high, 18.25 × 10−2 (Fig. 5E). No significant difference between the rates of evolution of the 2A and 2B genes studied separately was noted (not shown).
FIG. 4.
Mutation fixation rates in the T geotype (11 isolates) lineage in all (Kt), synonymous (Ks), and nonsynonymous (Ka) sites. Solid and boldface dashed lines, data for VP1 and 2AB regions, respectively. For other details, see the legend to Fig. 3.
FIG. 5.
Mutation fixation rates in the IDP lineage (five isolates) in all (Kt), synonymous (Ks), and nonsynonymous (Ka) sites in the VP1, 2AB, and 3D′ regions. For other details, see the legend to Fig. 3.
The rate of evolution of the portion of 3D gene in the IDP lineage was found to be the lowest among the regions investigated, being more than two- and threefold lower than the rates of accumulation of total and synonymous substitutions, respectively, in the VP1 gene (Fig. 5G and H). The lowest mutability of the 3D′ region was also noted for the T lineage sequences (data not shown), but due to the insufficient number of 3D′ sequences available, a quantitative comparison with the other regions in this case seemed unwarranted.
The rates of fixation of nonsynonymous substitutions were in general significantly lower and less regular. For the T geotype, the relevant values for 2AB appeared to be higher than those for VP1 (Fig. 4C), but the reverse was true of the IDP population (Fig. 5C and F). The accumulation of nonsynonymous mutations in the VP1 genes of the former population deviated from linearity (r = 0.59) (Fig. 4C).
Character of mutation fixation.
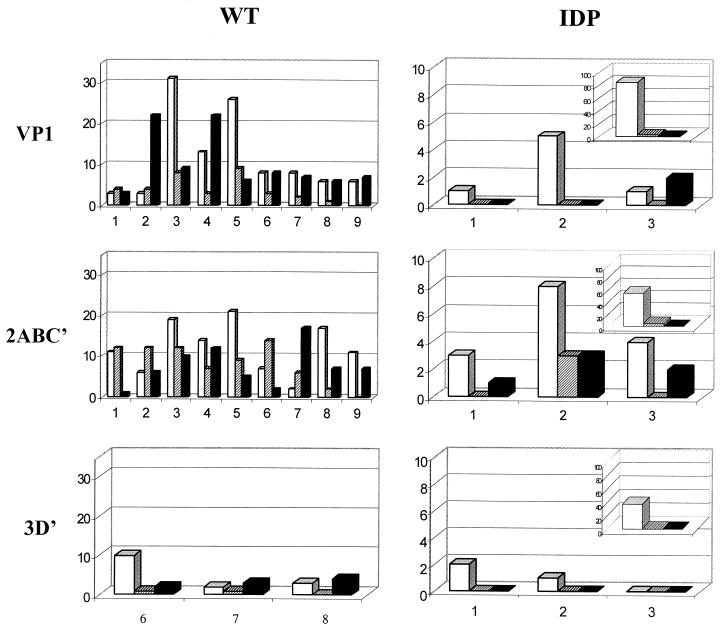
In the vaccine-derived viruses, a significant proportion of mutations, once fixed, continued to be present in the genomes of all of the subsequent isolates. This was true of the mutations accumulated both before and after the isolation of the first strain (Fig. 6). Such a character of mutation accumulation was consistent with the assumption that the successive isolates of the IDP lineage were related to each other in the parent-progeny fashion. Such a direct relationship was less evident for the T geotype populations, where a significant proportion of mutations present in a given isolate was absent from some or all subsequent isolates (Fig. 6).
FIG. 6.
Characterization of mutation fixation in the wt and IDP lineages. y axis, numbers of mutations that once appeared, became fixed in all (open bars) or at least some (hatched bars) of the subsequent isolates, as well as numbers of solitary mutations (black bars). Insets in the IDP panels, comparison of the first isolate (day 23) to the vaccine sequences. For the wt lineage, the numbers on the x axis correspond to the following isolates: 1, PV1/4TAJ91 (compared to PV1/7TAJ91); 2, PV1/5341UKR92; 3, PV1/5794UZB94; 4, PV1/6011TAJ94; 5, PV1/6064CHE95; 6, PV1/6484CHE95; 7, PV1/6486CHE95; 8, PV1/6490CHE95; and 9, PV1/6405ING95; for the IDP lineage, the numbers correspond to the following isolates: 1, day 48 (compared to day 23); 2, day 126; 3, day 158. The last isolates in the lineages are omitted.
Remarkably, about half of the detected replacements in the VP1 and 2AB1 regions of the IDP viruses resulted in the return to the Sabin 1 sequence, and such reversions were more or less evenly distributed over these regions (no reversions were observed in 3D′ out of a total of three replacements). A smaller but significant (22%) proportion of reversions to Sabin 2 in the 2B2 region was noted.
Codon usage and the nonuniform evolution of the viral genome.
A possible explanation of different rates of fixation of synonymous mutations along the viral genome may be related to the codon usage bias. Different synonymous codons are known to be used with different frequencies in different organisms, different tissues, and even different genes, contributing thereby to the translational control of gene expression (61, 74). It can be argued that highly preferred codons are favorable for efficient translation and may therefore be less prone to be replaced during evolution. Consequently, the genomic regions with an abundance of such codons might be expected to undergo nucleotide replacements relatively less frequently. No significant heterogeneity in codon usage among the genomic regions studied could be found (not shown). From this analysis, however, a statistically significant conservation of rarely used codons at certain positions of the VP1, but not the 2AB, region was noticed (Fig. 7A). In this analysis, not only the true conservation of a rare codon but also the cases in which a rare codon was replaced by another synonymous rare codon were taken into account. At certain positions, the consecutive conserved rare codons were clustered (Fig. 7A). In the vicinity of 9 out of the 15 conserved rare codons (at a distance of 1 to 3 codons) additional rare codons were also present in at least 75% of the 29 aligned VP1 sequences (not shown).
FIG. 7.
Conserved rare codons of the VP1 gene. (A) Sites with anomalous conservation of the rare codons in the six most divergent sequences of the type 1 poliovirus. The codons are numbered from the start of the VP1 gene. The rare codons are underlined. The number of each of the individual codons in a given position of the six sequences is shown in parentheses. (B) VP1 secondary structure wire plot (Protein Data Bank; www.biochem.ucl.ac.uk/bsm/pdbsum/2plv/main.html). The amino acids corresponding to the conserved rare codons in panel A are underlined and in boldface. Straight arrows, beta structural elements; curved arrows, alpha helices. The amino acids are numbered from the start of the VP1 gene.
It should be noted, however, that the number of conserved rare codons in the VP1 gene was too small to account for the observed difference in the rates of synonymous replacements in the VP1 and 2AB regions.
Does the RNA secondary structure contribute to the nonuniform evolution of the viral genome?
It could be hypothesized that local minima in the accumulation of synonymous substitutions were due to constraints imposed by the necessity to maintain specific high-order RNA structures. An analysis of this problem was hampered by the dynamic rather than static nature of the RNA structural organization. RNA regions with relatively stable secondary structure elements are expected to possess a low value of the so-called pairing number (P-num), a quantitative measure of the propensity of a certain base to pair with alternative partners in a collection of suboptimal folds (28, 56). We calculated that the proportions of bases with low P-num values (<100) for the VP1 and 2AB regions of the Mahoney genome were quite similar, being 91 out of 906 nt (10%) and 62 out of 738 nt (8.4%), respectively. This fact failed to support the notion that the two RNA regions differed from one another with regard to possessing strictly conserved secondary structures. No such support was obtained from attempts to derive a consensus secondary structure by comparative sequence analysis of these regions either. However, a higher percentage (17.7% or 140 out of 792 nt) of nucleotides with low P-num values was found in the 3D′ region. This observation was indicative of substantial structural constraints, which may be partially responsible for lowering the mutation fixation rate in the sequenced portion of the polymerase gene. The close relatedness of the 13 sequences available did not enable us to search for a consensus secondary structure of the 3D′ region.
Deviation from the molecular clock mode of evolution.
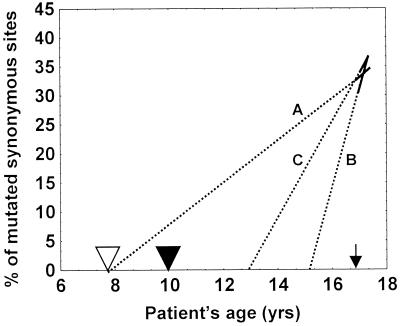
As shown above, the accumulation of total, synonymous, and nonsynonymous mutations appeared to linearly depend on time in all the RNA regions studied with only one exception (nonlinearity of nonsynonymous substitutions in the T geotype VP1 gene). However, a more serious deviation from linearity was revealed by an analysis of mutation rates throughout the whole history of the Sabin vaccine-derived population. Indeed, the rate of synonymous substitutions in 2AB, during the time interval studied here, was approximately fivefold higher than that in VP1. On the other hand, the proportions of synonymous substitutions (Ks values) in the VP1 and 2AB1 regions of the earliest IDP isolate, compared to the corresponding values for the Sabin progenitor virus, were similar, 30.4 and 27.9, respectively, suggesting that these regions evolved at comparable rates. This discrepancy became clearly evident when the apparent times of divergence of the relevant IDP lineage genes from the Sabin vaccine were calculated by extrapolations using the estimated evolution rates taken from Fig. 5B and E. For the synonymous nucleotide replacements in the VP1 region (Fig. 8A), this time (9.6 years) corresponded reasonably well to the time (6.9 years) elapsed between the last oral polio vaccine vaccination and the onset of paralysis (31). However, the 2AB1 region (Fig. 8B) appeared to require only one-fourth of the time (∼ 2.5 years) to achieve the observed divergence from the progenitor. Possible reasons for this paradox are discussed below.
FIG. 8.
Extrapolation of evolution rates of synonymous sites calculated for the IDP lineage for VP1 (A), 2AB1 (B), and combined VP1/2AB1/3D′ (C) regions to zero divergence with Sabin 1. The solid upper portions of the lines correspond to the data taken from Fig. 5B and E and 3E. Open arrowhead, extrapolated age of initial infection as calculated by Kew et al. (31); solid arrowhead, time of the last vaccination with oral polio vaccine; arrow, time of the paralysis onset.
DISCUSSION
Basic mechanisms of poliovirus evolution.
The rate and character of fixation of mutations generated by an error-prone viral RNA polymerase are controlled by positive selection of more-fit variants, negative selection of variants with decreased fitness, and accumulation of neutral or nearly neutral (33) replacements through random sampling of viral exemplars from the “swarm” of genomic variants (7, 9, 11, 13, 14, 18). Admittedly, the discussion of the nature of evolutionary changes would have been more factual if accompanying alterations in the level of viral fitness were known (8). Unfortunately, no adequate animal model to assess the relative fitness of polioviruses in their natural niche, the human gut, is available. It seems likely, however, that all three of the above-mentioned mechanisms contributed, to a certain extent, to the changes registered in this study. Although the exact contribution of each of them cannot as yet be defined, the preponderance of synonymous replacements and the constant rates of mutation fixation suggest a major role for random sampling as the mechanism of neutral evolution. This appeared to be true of both poliovirus populations studied here, even though they differed from one another in several respects: (i) the starting populations were represented by wt (presumably relatively well adapted) and attenuated (less-fit) viruses; (ii) the wt lineage was interrupted by human-to-human transfers, whereas the other one corresponded to a continuously growing population in a single host; and (iii) accordingly, the wt isolates were hardly related to each other in the direct ancestor-progeny fashion, whereas this was almost certainly true of the IDP isolates.
The accumulation of a significant proportion of synonymous substitutions in the IDP population requires some specification. Too little is known about the mode of poliovirus reproduction in the human gut to explain how consecutive random sampling from a heterogeneous viral population can be achieved in this case. A relatively low content of infectious virus in the feces (on the order 3.0 to 6.5 log10 50% tissue culture infective doses/g [45]) makes it likely that either relatively few susceptible cells are available at each given moment or only a tiny minority of susceptible cells are effectively infected. If so, virus reproduction in the gut may be to some extent likened to the propagation of a virus by consecutive small-population passages, resulting in the accumulation of neutral and adverse mutations (7, 18). In line with this reasoning, polioviruses inducing neurological disease are believed to represent just stochastic variants of the heterogeneous population present in the gut (23). It is worthwhile to note that Muller's ratchet can be enabled only if the initial poliovirus variants are sufficiently optimized for their particular ecological niche, the gut, through adaptive selection. Previous studies indicated that selection against the attenuating mutations in the Sabin viruses did indeed occur at the initial steps of infection (15).
Note that the rates of evolution of the populations studied here fit well values, on the order of 10−2 to 10−3 substitution/site/year, observed for other picornaviruses (65, 69, 73) and other RNA-containing viruses (12, 24, 26).
Nonuniform rates of evolution of different genomic regions and different genomes.
A nonuniform rate of mutation accumulation along the viral RNA is not unique for poliovirus. The bias in nonsynonymous mutation fixation in different genomic regions of different RNA viruses has been observed (24, 27) and can readily be explained by differences in constraints imposed on the structures of different viral proteins. More interesting, in the context of the present study, were unequal rates of accumulation of synonymous substitutions.
The nature of the selective pressure on synonymous sites is not clear. A higher rate of mutation fixation in 2AB than in VP1 and 3D′ may be caused by the additional contribution of either positive selection directed to 2AB or negative selection directed to VP1 and 3D′. The slowing down of the evolution of a genomic region could be due to the need for conservation of certain sequences. In our case, these sequences were represented predominantly by synonymous codons. Hypothetically, conservation of certain rare codons could be important to preserve an optimal translation elongation rate and hence the proper folding of nascent proteins (1, 36, 37, 55, 74). Statistically significant conservation of rare codons at certain positions of the VP1 gene (but not in the 2AB region) may be interpreted as indirect evidence for this hypothesis. Remarkably, the conserved loci with one or more rare codons tend to be positioned close to the borders of structured elements of VP1 (Fig. 7B), again consistent with their hypothetical role in proper folding. However, the number of such sites is too small to account for the difference between the evolutionary rates of VP1 and 2AB nucleotide sequences.
In addition to other mechanisms, conservation of some synonymous codons could be caused by the necessity to maintain the RNA structure as such. A relatively low variability in the subterminal coding regions of the hepatitis G virus RNA was explained by postulating the participation of these regions in the formation of structured replicative cis elements (62). Similar reasons may explain the extraordinary evolutionary stability of the 3D′ region. The internal coding regions of picornavirus RNAs are also known to harbor cis-acting replicative signals, usually in the form of hairpin elements (41, 43). Such signals may well be present in the sequenced region of the poliovirus genome. However, they hardly occupy extended RNA regions and therefore could not make a significant contribution to the observed difference in the rates of evolution of VP1 and 2AB. On the other hand, the importance of RNA secondary structure may be related not only to certain conserved domains but also to the rule “fold as you please, but fold you must” (2). Implementation of this rule may result in negative selection of certain synonymous substitutions and hence in retardation of the evolutionary rate.
On the other hand, a relatively high rate of mutation fixation in the 2AB region could be caused by positive selection of certain substitutions (see below).
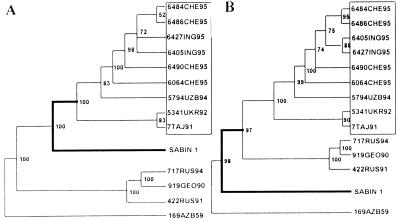
Regardless of the mechanism, local differences in the mutation fixation rates may result in the ambiguity of dendrograms illustrating the relatedness of the viral strains. Indeed, the dendrograms of type 1 viruses based on the VP1 and 2AB sequences are topologically different with regard to the position of the Sabin 1 strain (Fig. 9).
FIG. 9.
Different topologies of the maximum-likelihood bootstrap consensus trees (PUZZLE, version 4.0) of the poliovirus type 1 wild and vaccine (Sabin 1) strains (rooted on PV1/169AZB59) when based on the VP1 (A) and 2AB (B) sequences. For the sake of brevity, a selected set of strains was used; a comparison of the complete trees also demonstrated the same difference in topology (not shown). Strains of the T geotype lineage are boxed. PV1/422RUS91 belongs to geotype A; PV1/919GEO90 and PV1/717RUS94 correspond to geotype G (40).
Disturbances in the operation of molecular clocks.
Although the accumulation of mutations is generally linear, deviation from the molecular clock is possible. Mutations within the 2AB region of the IDP lineage appeared to accumulate nonlinearly with time, the rate of evolution being apparently higher during the last period of the population history.
Although the nonlinearity of the viral evolutionary rates has been reported previously (17, 69), the underlying mechanism(s) remains obscure. The nonlinearity of 2AB evolution could hardly be explained by changes in the fidelity of the viral RNA polymerase or in the number of viral generations per time unit because of the local nature of the anomaly. One may speculate that the apparent increase in the rate of 2AB evolution was caused by the prior acquisition of an adverse mutation(s) within or outside this region. If so, the enhanced mutability of 2AB might reflect the necessity to mutually readjust different portions of the genome and thus was largely adaptive, even though it involved predominantly synonymous substitutions. For example, one may imagine that a recombination event destroyed (or destabilized) an extended RNA secondary structure element in the 2AB region, causing a concomitant drop of the fitness level. The selection pressure tended to restore the function, which could be achieved by stepwise accumulation of numerous slightly advantageous (nearly neutral) mutations.
On the other hand, the nonlinearity of 2AB evolution could be only apparent. For example, recombination with a bona fide Sabin strain could have occurred ∼2.5 years before the isolation of the first IDP strain. As a result, the IDP lineage could have acquired an unaltered segment of the 2AB region which evolved rapidly but with a constant rate afterwards. An apparent deviation from linearity might be encountered if the time interval of observations is too long. Indeed, due to a high mutation rate, the probability of back mutations also becomes high, resulting in illusory “slowing down” of the accumulation of detectable mutations.
Given the real or apparent deviations from the molecular clock, one should be cautious in the determination of the viral age by back-extrapolating the rates of mutation fixation. The longer is the sequence used for the estimation of the mutation fixation rate, the more reliable should be the results of such extrapolations. However, even combining the sequences of the VP1, 2AB1, and 3D′ regions for such calculations appeared to result in an underestimated time of divergence of the IDP lineage from the Sabin vaccine (3.8 years) (Fig. 8C).
A hypothetical scenario of viral evolution in the immunodeficient host.
The events that occurred in the IDP host between the vaccination and the isolation of the first virus are not documented. Indirect evidence suggests that the lineage studied might have become predominant not long before the first isolate (PV1/2677USA81) was obtained. First, the relevant fecal sample contained two variants, one of which, the minor variant (m), subsequently disappeared (31), suggesting that the major variant (M) exhibited better fitness. This selective advantage of M should have likely precluded the long-term coexistence of the two variants in a single host. Second, the paralytic disorder that developed in the IDP after several years of asymptomatic virus carriage was most likely due to a newly arisen more pathogenic variant.
Thus, the evolution of the Sabin virus in the immunodeficient host might occur according to the following hypothetical scenario. The vaccine virus evolved as a quasispecies, and a predecessor of the M variant diverged from the predominantly m population relatively shortly before the onset of paralysis (31) and existed as a minor component. Then a stochastic increase in fitness (9, 14, 19, 52) and hence in neurovirulence occurred in the M lineage due to either a mutation(s) (not necessarily in the sequenced region) or recombination (21, 22, 39). The newly emerged M variant eventually outcompeted the then-predominant m variant and caused paralytic poliomyelitis.
Implications for epidemiology of poliomyelitis.
We propose a unifying model of evolution of both wt poliovirus upon its natural circulation and of vaccine-derived virus replicating in the human gut. According to this model, evolution proceeds essentially through consecutive sampling of small portions of a viral population, ensuring predominant fixation of neutral and possibly adverse mutations. As a result the virus fitness is expected to be maintained at a more or less constant level or to decrease, except for the early stage after the immunization, when loss of some attenuating mutations (47) may result in an enhanced fitness. The possible accumulation of adverse mutations may serve to self-limit viral reproduction in the human gut and during person-to-person circulation and may contribute to the relatively low pathogenicity of poliovirus, which is known to cause overt disease only in a tiny minority of infected nonimmune subjects (44). The fitness decrease may be particularly conspicuous for attenuated viruses, leading to the very low level of their transmissibility. Some of the deleterious mutations may be eliminated by negative selection and by intratypic or intertypic recombination (4, 21, 22, 39, 46). Concurrently, the positive selection of rarely arisen more fit and more neurovirulent variants may facilitate the development of vaccine-associated poliomyelitis in the case of vaccine-derived variants and outbreaks of poliomyelitis in the case of circulating wt viruses.
By the time this work was submitted, two papers on prolonged poliovirus replication in IDP were published (3, 42). Although the authors have not addressed the main issues studied here, a comparison of some of their and our data is of interest. The mutation fixation rates calculated in these studies for the entire poliovirus genome were comparable to that found here for all studied genomic regions combined. Also, the divergences from Sabin 1 of the VP1 and 2AB regions in the last isolates found by Bellmunt et al. (3) and those in our lineages were quite similar, (9.2 versus 9.9% and 9.3 versus 8.1%, respectively). In spite of this, the difference in nucleotide sequences of the isolates from these two IDPs amounted to 14.8 and 13.2% for the VP1 and 2AB regions, respectively. Such a striking divergence may be interpreted as additional evidence for the largely neutral (nondirected) rather than adaptive character of the evolution of vaccine viruses in IDP hosts. It may be also added that when the VP1 region sequence of the most diverged isolate from the study by Bellmunt et al. (3) was added to the analysis of the rare codons, the anomaly of the conservation of all of the rare codons determined here remained statistically significant.
ACKNOWLEDGMENTS
We thank A. V. Alekseevsky, S. A. Spirin, and E. A. Korotkova for useful suggestions on statistical analysis and Mick N. Mulders for making the VP1/2A sequence of 18643PAK91 strain available. We also express our gratitude to the virologists from NIS countries for providing us with the poliovirus strains and O. E. Ivanova from the M. P. Chumakov Institute of Poliomyelitis and Viral Encephalitides for the propagation and typing work.
Financial support for a short-term visit of E.V.G. to the Centers for Disease Control and Prevention, Atlanta, Ga., is gratefully acknowledged. This work was supported by EU grants Copernicus and INTAS as well as by the Russian Foundation for Basic Research. V.I.A. is a Soros Professor.
REFERENCES
- 1.Adzhubei A A, Adzhubei I A, Krasheninnikov I A, Neidle S. Non-random usage of ‘degenerate’ codons is related to protein three-dimensional structure. FEBS Lett. 1996;399:78–82. doi: 10.1016/s0014-5793(96)01287-2. [DOI] [PubMed] [Google Scholar]
- 2.Arora R, Priano C, Jacobson A B, Mills D R. Cis-acting elements within an RNA coliphage genome: fold as you please, but fold you must!! J Mol Biol. 1996;258:433–446. doi: 10.1006/jmbi.1996.0260. [DOI] [PubMed] [Google Scholar]
- 3.Bellmunt A, May G, Zell R, Pring-Akerblom P, Verhagen W, Heim A. Evolution of poliovirus type 1 during 5.5 years of prolonged enteral replication in an immunodeficient patient. Virology. 1999;265:178–184. doi: 10.1006/viro.1999.0003. [DOI] [PubMed] [Google Scholar]
- 4.Cammack N, Phillips A, Dunn G, Patel V, Minor P D. Intertypic genomic rearrangements of poliovirus strains in vaccinees. Virology. 1988;167:507–514. [PubMed] [Google Scholar]
- 5.Chao L. Fitness of RNA virus decreased by Muller's ratchet. Nature. 1990;348:454–455. doi: 10.1038/348454a0. [DOI] [PubMed] [Google Scholar]
- 6.Chao L. Evolution of sex and the molecular clock in RNA viruses. Gene. 1997;205:301–308. doi: 10.1016/s0378-1119(97)00405-8. [DOI] [PubMed] [Google Scholar]
- 7.Clarke D K, Duarte E A, Moya A, Elena S F, Domingo E, Holland J. Genetic bottlenecks and population passages cause profound fitness differences in RNA viruses. J Virol. 1993;67:222–228. doi: 10.1128/jvi.67.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingo E, Martinez-Salas E, Sobrino F, de la Torre J C, Portela A, Ortin J, Lopez-Golindez C, Perez-Brena P, Villanueva N, Najera R, VandePol S, Steinhauer D, DePolo N, Holland J J. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene. 1985;40:1–8. doi: 10.1016/0378-1119(85)90017-4. [DOI] [PubMed] [Google Scholar]
- 9.Domingo E, Holland J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 10.Domingo E, Escarmis C, Sevilla N, Baranowski E. Population dynamics in the evolution of RNA viruses. Adv Exp Med Biol. 1998;440:721–727. doi: 10.1007/978-1-4615-5331-1_93. [DOI] [PubMed] [Google Scholar]
- 11.Domingo E, Escarmis C, Menendez-Arias L, Holland J J. Viral quasispecies and fitness variations. In: Domingo E, Webster R, Holland J J, editors. Origin and evolution of viruses. San Diego, Calif: Academic Press; 1999. pp. 141–161. [Google Scholar]
- 12.Drake J W, Holland J J. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. 1999;96:13910–13913. doi: 10.1073/pnas.96.24.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duarte E, Clarke D, Moya A, Domingo E, Holland J J. Rapid fitness losses in mammalian RNA virus clones due to Muller's ratchet. Proc Natl Acad Sci USA. 1992;89:6015–6060. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duarte E A, Novella I S, Ledesma S, Clarke D K, Moya A, Elena S F, Domingo E, Holland J J. Subclonal components of consensus fitness in an RNA virus clone. J Virol. 1994;68:4295–4301. doi: 10.1128/jvi.68.7.4295-4301.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn G, Begg N T, Cammack N, Minor P. Virus excretion and mutation by infants following primary vaccination with live oral poliovaccine from two sources. J Med Virol. 1990;32:92–95. doi: 10.1002/jmv.1890320205. [DOI] [PubMed] [Google Scholar]
- 16.Eck R V, Dayhoff M O. Atlas of protein sequence and structure. Silver Spring, Md: National Biomedical Research Foundation; 1966. [Google Scholar]
- 17.Elena S F, Gonzalez-Candelas F, Moya A. Does the VP1 gene of foot-and-mouth disease virus behave as a molecular clock? J Mol Evol. 1992;35:223–229. doi: 10.1007/BF00178598. [DOI] [PubMed] [Google Scholar]
- 18.Escarmis C, Davila M, Charpentier N, Bracho A, Moya A, Domingo E. Genetic lesions associated with Muller's ratchet in an RNA virus. J Mol Biol. 1996;264:255–267. doi: 10.1006/jmbi.1996.0639. [DOI] [PubMed] [Google Scholar]
- 19.Escarmis C, Davila M, Domingo E. Multiple molecular pathways for fitness recovery of an RNA virus debilitated by operation of Muller's ratchet. J Mol Biol. 1999;285:495–505. doi: 10.1006/jmbi.1998.2366. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. PHYLIP: phylogeny inference package, version 3.52c. Seattle, Wash: University of Washington; 1993. [Google Scholar]
- 21.Furione M, Guillot S, Otelea D, Balanant J, Candrea A, Crainic R. Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology. 1993;196:199–208. doi: 10.1006/viro.1993.1468. [DOI] [PubMed] [Google Scholar]
- 22.Georgescu M M, Delpeyroux F, Tardy-Panit M, Balanant J, Combiescu M, Combiescu A A, Guillot S, Crainic R. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J Virol. 1994;68:8089–8101. doi: 10.1128/jvi.68.12.8089-8101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgescu M M, Balanant J, Ozden S, Crainic R. Random selection: a model for poliovirus infection of the central nervous system. J Gen Virol. 1997;78:1819–1828. doi: 10.1099/0022-1317-78-8-1819. [DOI] [PubMed] [Google Scholar]
- 24.Gojobori T, Moriyama E N, Kimura M. Molecular clock of viral evolution, and the neutral theory. Proc Natl Acad Sci USA. 1990;87:10015–10018. doi: 10.1073/pnas.87.24.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa M, Kishino H, Yano K. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 26.Imazeki F, Omata M, Ohto M. Heterogeneity and evolution rates of delta virus RNA sequences. J Virol. 1990;64:5594–5599. doi: 10.1128/jvi.64.11.5594-5599.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito T, Gorman O, Kawaoka Y, Bean W, Webster R G. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kew O M, Nottay B K, Hatch M H, Nakano J H, Obijeski J F. Multiple genetic changes can occur in the oral poliovaccines upon replication in humans. J Gen Virol. 1981;56:337–347. doi: 10.1099/0022-1317-56-2-337. [DOI] [PubMed] [Google Scholar]
- 30.Kew O M, Mulders M N, Lipskaya G Y, da Silva E E, Pallansch M A. Molecular epidemiology of polioviruses. Semin Virol. 1995;6:401–414. [Google Scholar]
- 31.Kew O M, Sutter R W, Nottay B K, McDonough M J, Prevots D R, Quick L, Pallansch M A. Prolonged replication of a type 1 vaccine-derived poliovirus in an immunodeficient patient. J Clin Microbiol. 1998;36:2893–2899. doi: 10.1128/jcm.36.10.2893-2899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 33.Kimura M. The neutral theory of molecular evolution. Cambridge, United Kingdom: Cambridge University Press; 1983. [Google Scholar]
- 34.Kinnunen L, Huovilainen A, Poyry T, Hovi T. Rapid molecular evolution of wild type 3 poliovirus during infection in individual hosts. J Gen Virol. 1990;71:317–324. doi: 10.1099/0022-1317-71-2-317. [DOI] [PubMed] [Google Scholar]
- 35.Kinnunen L, Poyry T, Hovi T. Genetic diversity and rapid evolution of poliovirus in human hosts. Curr Top Microbiol Immunol. 1992;176:49–61. doi: 10.1007/978-3-642-77011-1_4. [DOI] [PubMed] [Google Scholar]
- 36.Kolb V A, Makeyev E V, Spirin A S. Folding of firefly luciferase during translation in a cell-free system. EMBO J. 1994;13:3631–3637. doi: 10.1002/j.1460-2075.1994.tb06670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolb V A, Makeyev E V, Kommer A, Spirin A S. Cotranslational folding of proteins. Biochem Cell Biol. 1995;73:1217–1220. doi: 10.1139/o95-131. [DOI] [PubMed] [Google Scholar]
- 38.Li W H, Wu C I, Luo C C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 39.Lipskaya G Y, Muzychenko A R, Kutitova O K, Maslova S V, Equestre M, Drozdov S G, Bercoff R P, Agol V I. Frequent isolation of intertypic poliovirus recombinants with serotype 2 specificity from vaccine-associated polio cases. J Med Virol. 1991;35:290–296. doi: 10.1002/jmv.1890350415. [DOI] [PubMed] [Google Scholar]
- 40.Lipskaya G Y, Chervonskaya E A, Belova G I, Maslova S V, Kutateladze T N, Drozdov S G, Mulders M, Pallansch M A, Kew O M, Agol V I. Geographical genotypes (geotypes) of poliovirus case isolates from the former Soviet Union: relatedness to other known poliovirus genotypes. J Gen Virol. 1995;76:1687–1699. doi: 10.1099/0022-1317-76-7-1687. [DOI] [PubMed] [Google Scholar]
- 41.Lobert P E, Escriou N, Ruelle J, Michiels T. A coding RNA sequence acts as a replication signal in cardioviruses. Proc Natl Acad Sci USA. 1999;96:11560–11565. doi: 10.1073/pnas.96.20.11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin J, Dunn G, Hull R, Patel V, Minor P. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J Virol. 2000;74:3001–3010. doi: 10.1128/jvi.74.7.3001-3010.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKnight K L, Lemon S M. Capsid coding sequence is required for efficient replication of human rhinovirus 14 RNA. J Virol. 1996;70:1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melnick J L. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 45.Melnick J L, Rennick V. Infectivity titers of enterovirus as found in human stools. J Med Virol. 1980;5:205–220. doi: 10.1002/jmv.1890050305. [DOI] [PubMed] [Google Scholar]
- 46.Minor P D, John A, Ferguson M, Icenogle J P. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J Gen Virol. 1986;67:693–706. doi: 10.1099/0022-1317-67-4-693. [DOI] [PubMed] [Google Scholar]
- 47.Minor P D, Dunn G. The effect of sequences in the 5′ non-coding region on the replication of polioviruses in the human gut. J Gen Virol. 1988;69:1091–1096. doi: 10.1099/0022-1317-69-5-1091. [DOI] [PubMed] [Google Scholar]
- 48.Miralles R, Gerrish P J, Moya A, Elena S F. Clonal interference and the evolution of RNA viruses. Science. 1999;285:1745–1747. doi: 10.1126/science.285.5434.1745. [DOI] [PubMed] [Google Scholar]
- 49.Mulders M N, Lipskaya G Y, van der Avoort H G A M, Koopmans M P G, Kew O M, van Loon A M. Molecular epidemiology of wild poliovirus type 1 in Europe, the Middle East and the Indian subcontinent. J Infect Dis. 1995;171:1399–1405. doi: 10.1093/infdis/171.6.1399. [DOI] [PubMed] [Google Scholar]
- 50.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 51.Nottay B K, Kew O M, Hatch M H, Heyward J T, Obijeski J F. Molecular variation of type 1 vaccine-related and wild poliovirus during replication in humans. Virology. 1981;108:405–423. doi: 10.1016/0042-6822(81)90448-7. [DOI] [PubMed] [Google Scholar]
- 52.Novella I S, Duarte E A, Elena S F, Moya A, Domingo E, Holland J J. Exponential increases of RNA virus fitness during large population transmissions. Proc Natl Acad Sci USA. 1995;92:5841–5844. doi: 10.1073/pnas.92.13.5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novella I S, Elena S F, Moya A, Domingo E, Holland J J. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novella I S, Quer J, Domingo E, Holland J J. Exponential fitness gains of RNA virus populations are limited by bottleneck effects. J Virol. 1999;73:1668–1671. doi: 10.1128/jvi.73.2.1668-1671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oresic M, Shalloway D. Specific correlations between relative synonymous codon usage and protein secondary structure. J Mol Biol. 1998;281:31–48. doi: 10.1006/jmbi.1998.1921. [DOI] [PubMed] [Google Scholar]
- 56.Palmenberg A C, Sgro J-Y. Topological organization of picornaviral genomes: statistical prediction of RNA structural signals. Semin Virol. 1997;8:231–241. [Google Scholar]
- 57.Rzhetsky A, Nei M. Theoretical foundation of the minimum evolution method of phylogenetic inference. Mol Biol Evol. 1993;10:1073–1095. doi: 10.1093/oxfordjournals.molbev.a040056. [DOI] [PubMed] [Google Scholar]
- 58.Rzhetsky A, Nei M. METREE: program package for inferring and testing minimum evolution trees. Comput Appl Biosci. 1994;10:409–412. doi: 10.1093/bioinformatics/10.4.409. [DOI] [PubMed] [Google Scholar]
- 59.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 60.Sala M, Wain-Hobson S. Drift and conservatism in RNA virus evolution: are they adapting or merely changing? In: Domingo E, Webster R, Holland J J, editors. Origin and evolution of viruses. San Diego, Calif: Academic Press; 1999. pp. 115–140. [Google Scholar]
- 61.Sharp P M, Matassi G. Codon usage and genome evolution. Curr Opin Genet Dev. 1994;4:851–860. doi: 10.1016/0959-437x(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 62.Simmonds P, Smith D B. Structural constraints on RNA virus evolution. J Virol. 1999;73:5787–5794. doi: 10.1128/jvi.73.7.5787-5794.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steinhauer D A, Holland J J. Rapid evolution of RNA viruses. Annu Rev Microbiol. 1987;41:409–433. doi: 10.1146/annurev.mi.41.100187.002205. [DOI] [PubMed] [Google Scholar]
- 64.Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 65.Takeda N, Tanimura M, Miyamura K. Molecular evolution of the major capsid protein VP1 of enterovirus 70. J Virol. 1994;68:854–862. doi: 10.1128/jvi.68.2.854-862.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 67.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toyoda H, Kohara M, Kataoka Y, Suganuma T, Omata T, Imura N, Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes. Implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984;174:561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- 69.Villaverde A, Martinez M A, Sobrino F, Dopazo J, Moya A, Domingo E. Fixation of mutations at the VP1 gene of foot-and-mouth disease virus. Can quasispecies define a transient molecular clock? Gene. 1991;103:147–153. doi: 10.1016/0378-1119(91)90267-f. [DOI] [PubMed] [Google Scholar]
- 70.Ward C D, Stokes M A, Flanegan J B. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J Virol. 1988;62:558–562. doi: 10.1128/jvi.62.2.558-562.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward C D, Flanegan J B. Determination of the poliovirus RNA polymerase error frequency at eight sites in the viral genome. J Virol. 1992;66:3784–3793. doi: 10.1128/jvi.66.6.3784-3793.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.World Health Organization. Manual for the virologic investigation of poliomyelitis. WHO/EPI/GEN/97.1. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 73.Zhang G, Haydon D T, Knowles N J, McCauley J W. Molecular evolution of swine vesicular disease virus. J Gen Virol. 1999;80:639–651. doi: 10.1099/0022-1317-80-3-639. [DOI] [PubMed] [Google Scholar]
- 74.Zhou J, Liu W J, Peng S W, Sun X Y, Frazer I. Papillomavirus capsid protein expression level depends on the match between codon usage and tRNA availability. J Virol. 1999;73:4972–4982. doi: 10.1128/jvi.73.6.4972-4982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]