Summary
Receptor tyrosine kinases (RTKs) control stem cell maintenance vs. differentiation decisions. Casitas B-lineage lymphoma (CBL) family ubiquitin ligases are negative regulators of RTKs, but their stem cell regulatory roles remain unclear. Here, we show that Lgr5+ intestinal stem cell (ISC)-specific inducible Cbl-knockout (KO) on a Cblb null mouse background (iDKO) induced rapid loss of the Lgr5Hi ISCs with transient expansion of the Lgr5Lo transit-amplifying population. LacZ-based lineage tracing revealed increased ISC commitment toward enterocyte and goblet cell fate at the expense of Paneth cells. Functionally, Cbl/Cblb iDKO impaired the recovery from radiation-induced intestinal epithelial injury. In vitro, Cbl/Cblb iDKO led to inability to maintain intestinal organoids. Single-cell RNA sequencing in organoids identified Akt-mTOR (mammalian target of rapamycin) pathway hyperactivation upon iDKO, and pharmacological Akt-mTOR axis inhibition rescued the iDKO defects. Our results demonstrate a requirement for Cbl/Cblb in the maintenance of ISCs by fine-tuning the Akt-mTOR axis to balance stem cell maintenance vs. commitment to differentiation.
Subject areas: Natural sciences, Biological sciences, Biochemistry, Molecular biology, Cell biology, Stem cells research
Graphical abstract

Highlights
-
•
Cbl/Cblb deletion in Lgr5+ intestinal stem cells (ISCs) depletes the ISC pool
-
•
Cbl/Cblb-KO ISCs produce more goblet cells/enterocytes and less Paneth cells
-
•
Cbl/Cblb-KO in ISCs impaired intestinal epithelial recovery from radiation injury
-
•
Cbl/Cblb maintain ISCs by negatively regulating the Akt-mTOR pathway
Natural sciences; Biological sciences; Biochemistry; Molecular biology; Cell biology; Stem cells research
Introduction
Receptor tyrosine kinases (RTKs) and their peptide growth factor ligands function as versatile regulators of stem cell maintenance and cell fate decisions during embryonic development as well as in adult tissues.1,2,3 Thus, mechanisms that positively or negatively regulate RTK function have a strong potential to impinge on stem cell homeostasis and fate decisions. Unraveling these regulatory mechanisms at the organismic and molecular levels is imperative to fully understand the biological underpinnings of organ and tissue development, homeostasis, and regeneration as well as how disease affects these processes.
Biochemical and cell biological studies have established the Cbl (Casitas B-lineage lymphoma) family of ubiquitin ligases (Cbl, Cbl-b, and Cbl-c) as activation-dependent negative regulators of RTKs and non-RTKs linked to cell surface receptors.4 Cbl proteins possess canonical tyrosine kinase-binding (TKB) and RING finger domains separated by a conserved linker, regions required for their ubiquitin ligase activity, with Cbl and Cbl-b including an extended C-terminal region with proline-rich sequences, tyrosine phosphorylation sites, and a ubiquitin-associated domain, a region lacking in Cbl-c. Furthermore, Cbl and Cbl-b are expressed in overlapping patterns across many tissues while Cblc mRNA expression is primarily localized to epithelial tissues.4,5 While all three Cbl-family proteins function as negative regulators of tyrosine kinases in vitro, only Cbl and Cbl-b have emerged as key regulators of physiological functions in vivo.4 Cblc-null mice show no overt phenotypes, and a protein product of the Cblc gene has not been categorically identified.5 Cbl-null mice show impairment of spermatogenesis and thymic development along with mildly hypercellular hematopoietic organs6,7,8 and exhibit a lean phenotype due to hyperactive insulin receptor signaling.9 Cblb-null mice are developmentally normal, but their immune cells are hyper-responsive to antigenic stimulation thus evoking autoimmunity.10,11 Cblb-null mice are also resistant to disuse muscle atrophy due to hyperactive IGF1R signaling.12 Thus, Cbl and Cbl-b are clearly physiologically relevant as negative regulators of RTK signaling.
While Cbl or Cblb null mice have a relatively normal lifespan under pathogen-free laboratory conditions, combined germline Cbl and Cblb gene deletion is embryonic lethal, supporting their redundant roles during embryonic development. This redundancy is further demonstrated by a rapidly lethal inflammatory disease upon combined Cbl and Cblb deletion in T cells13,14 and a lethal myeloproliferative disease upon combined deletion in hematopoietic stem cells (HSCs).15,16,17 These latter studies in HSCs showed that Cbl proteins are required to regulate physiological levels of HSC proliferation, and their deletion led to reduced HSC quiescence and impaired ability to reconstitute hematopoiesis upon bone marrow transplantation.16 In vitro studies using short hairpin RNA (shRNA) depletion have shown a role for Cbl and Cbl-b in maintaining asymmetric neural stem cell division.18 Recently, we observed that mammary epithelium-specific Cbl/Cblb double deletion led to substantial retardation of the postnatal mammary gland development, with depletion of mammary stem cells (MaSCs).19 Contrary to the impact of Cbl/Cblb double deletion in HSCs, no apparent hyperproliferation of intermediate progenitors or accumulation of progeny cells was observed in the double knockout (KO) (DKO) mammary glands. Furthermore, the fate of Cbl/Cblb-null MaSCs remained unclear primarily due to lack of clear-cut systems to track the stem-progenitor-mature cell hierarchy in the mammary gland.19 Given the well-established stem-progeny cell relationships in the intestinal epithelium, we have undertaken a comprehensive examination of the role of Cbl and Cblb in regulating ISC homeostasis.
The intestinal epithelial cell monolayer is a major environment-organism interface and is responsible for critical functions of digestion and absorption of food while maintaining a barrier against microbes. The intestinal epithelium is organized to ensure its maintenance throughout adult life in the face of rapid turnover of the mature, functional epithelial cells that line the villi in the small intestine. It is estimated that the entire epithelium is renewed every 3–5 days.20 The homeostatic maintenance of such a rapidly renewing tissue requires a well-regulated program to rapidly generate mature epithelial cell types from intestinal stem cells (ISCs) located at the base of the crypts. These ISCs divide daily to form rapidly cycling progenitor cells to expand the number of progeny cells. The progenitors further migrate along the crypt-villus axis as they adopt specific differentiated fates of secretory, absorptive, or other specialized cell types. Thus, the intestinal epithelial stem cell program provides an elegant system to explore mechanisms that control adult epithelial stem cell maintenance in the context of epithelia, an area of great interest since epithelial malignancies account for most of human cancers.21
Recent advances have elucidated the general hierarchy of ISCs and how they help maintain the epithelium under homeostasis or following extrinsic insults that lead to rapid loss of epithelial integrity or that eliminate rapidly dividing stem and progenitor cell pools.22,23,24,25 It is now well-accepted that crypt-base columnar cells (CBCs) are rapidly dividing stem cells that maintain the epithelium under homeostatic conditions.26 A less abundant and relatively slowly dividing ISC population located at the +4 position with respect to crypt base is thought to serve as a reserve pool that is rapidly recruited to replenish the more actively cycling CBC-ISCs when the latter are eliminated, such as upon gamma radiation exposure.22,23,24 Additional studies suggest that the slowly dividing ISCs, commonly identified as label-retaining cells, may differentiate into secretory cells under homeostatic conditions.24 Other studies point to the plasticity of these populations, with interconversion between the rapidly dividing and the slowly cycling cells.22,23,24 There is considerable overlap in the markers that have been used to define the various ISC populations, but the wingless-related integration site (WNT) target gene product Lgr5 (leucine-rich repeat-containing G protein-coupled receptor 5) is generally accepted as a marker of the majority of long-lived but rapidly cycling ISCs.26,27 Lgr5 is a multi-pass cell surface receptor for the R-spondin family of growth factors that enhances WNT signaling, a pathway required for ISC proliferation and colorectal oncogenesis.28,29 Aside from WNT signaling, other signaling pathways, including those mediated by Notch, BMP (bone morphogenetic proteins), and growth factor RTKs, are known to function as key regulators of ISCs and intestinal epithelial maintenance.30,31
Substantial evidence supports the prominent roles of RTKs and/or their ligands in intestinal development, maintenance, and disease. Newborn epidermal growth factor receptor (EGFR)-null mice, on a genetic background that allows live pups to be born, display gross defects in the intestinal mucosa along with reduced cell proliferation.32,33 The ISC niche role of the Paneth cell requires secretion of EGFR ligands by these cells.34 Application of epidermal growth factor (EGF) to intestinal mucosa of whole animals enhances epithelial cell proliferation.35 The importance of EGFR-mediated signals in maintaining a proliferating pool of ISCs is further established by recent studies of intestinal epithelial organoid cultures in which inhibition of EGFR activation induced the Lgr5+ ISCs to enter quiescence.36 RTKs of the EphB family, fibroblast growth factor receptor (FGFR), and insulin-like growth factor receptors (IGFRs) have also been shown to play crucial roles during gut morphogenesis, mucosal proliferation, crypt survival, and differentiation and positioning of cells along the crypt-villus axis.37,38,39,40,41 Lrig1, a negative regulator of the ErbB family and other RTKs,42,43 was shown to be enriched within the crypt compartment, and Lrig1-null mice display hyper-proliferation of intestinal epithelium and duodenal adenomas as early as 3 months of age.42 On the other hand, loss of Shp2, a positive regulator of RTK signaling, was shown to cause expansion of Paneth cells along with an increase in the Lgr5+ stem cells due to an imbalance of mitogen-activated protein kinase (MAPK) and Wnt pathways.44 Additionally, altered differentiation and loss of stemness have been reported upon overexpression of FGF10.40 These studies support the potentially dichotomous roles of RTK signaling in the maintenance of intestinal epithelial homeostasis.
Currently, nothing is known about the physiological roles of Cbl-family proteins in the regulation of ISCs. Previous studies have suggested that Lrig1 may function in part through its interaction with Cbl to promote RTK ubiquitination and degradation.45 Thus, the dramatic intestinal epithelial hyperproliferation and duodenal adenoma formation phenotypes in Lrig1-null mice42 supported the possibility that Cbl and Cbl-b may be involved in the regulation of ISCs. Here, we use Cbl and Cblb gene deletion in the Lgr5+ ISC compartment to provide evidence that Cbl and Cbl-b are redundantly required to maintain Lgr5+ cycling ISCs and that Cbl/Cbl-b deletion leads to rapid loss of ISCs through enhanced Akt-mTOR-dependent differentiation of ISCs into progeny cells followed by unequal lineage differentiation. Our studies therefore reveal a novel and previously unanticipated requirement for a tyrosine kinase-negative regulatory mechanism provided by Cbl and Cbl-b in the maintenance of cycling ISCs.
Results
CBL and CBL-B expression is enriched in intestinal crypts compared to villi
Since the roles of Cbl-family proteins in the intestine have not been examined previously, we first assessed the expression pattern of Cbl and Cbl-b in the intestine (Figures 1A–1E). Immunohistochemical (IHC) staining of formalin-fixed and paraffin-embedded intestinal tissue sections from 6-week-old wild-type (WT) C57Bl/6 mice revealed higher Cbl expression in crypts with lower signal in the villus area; lack of any crypt and villus epithelium staining of the Cbl−/− mouse intestinal sections established the specificity of Cbl staining (Figure 1A). IHC for Cbl-b showed a similar expression pattern (Figure 1B). To confirm these findings, we analyzed the lysates of crypt and villus fractions of intestinal mucosal tissue by western blotting (WB) (Figure 1C). As expected, the levels of phospho-histone3 (pH3) and Cyclin D1, markers of crypt-associated cell proliferation,46,47 were higher in the crypt fraction whereas Villin-148,49 was enriched in the villus fraction (Figure 1C). Notably, WB confirmed the crypt vs. villus enrichment of Cbl and Cbl-b. The crypt enrichment of Cbl and Cbl-b expression is reminiscent of that of the RTK-negative regulator Lrig1 previously shown to partially function through Cbl.42,43,45
Figure 1.
Preferential expression of Cbl and Cbl-b proteins in the crypt regions of the intestine
(A and B) Immunohistochemical (IHC) staining for Cbl (A) and Cbl-b (B) on formalin-fixed paraffin-embedded (FFPE) tissue sections from 6-weeks-old mice reveals their expression in a gradient from crypt to villus in WT animals; lack of epithelial staining in sections from Cbl−/− or Cblb−/− knockout mice demonstrates the specificity of antibody staining.
(C) Crypt-villus fractionation of intestinal mucosa followed by immunoblotting confirmed the crypt enrichment of Cbl and Cbl-b expression. Blotting for crypt (pH3 and Cyclin D1)- and villus (Villin-1)-associated markers confirmed the purity of villus and crypt fractions. Hsc-70 served as a loading control.
(D) Immunoblot analysis of crypt and villus fractions prepared from age- and gender-matched WT, Cbl−/−, and Cblb−/− mice. Within the crypt compartment, deletion of Cblb resulted in an increase in the expression of Cbl while Cbl-b expression was unchanged upon deletion of Cbl. The reverse was seen in the villus compartment, with an increase in Cbl-b expression upon Cbl deletion whereas Cbl expression was unchanged in Cblb−/− animals. Scale bars in A and B are 50 μm; data are representative of 3 experiments.
(E and F) Altered intestinal epithelial cell proliferation and differentiation in Cbl-/- mice. (E) WT and Cbl−/− mice were injected with BrdU 4 h prior to euthanasia and FFPE sections of intestines were stained with anti-BrdU antibody (Red) and DAPI (Blue); scale bar = 20 μm. (F) Quantification of the number of BrdU+ cells per crypt shows an increased proportion of cells in S phase of the cell cycle in Cbl null mice.
(G and I) Muc2 staining of tissue sections from age-matched WT, Cbl−/−, and Cblb−/− mice revealed a significant increase in the number of goblet cells (Crypt-villus unit) in both Cbl−/− and Cblb−/− mice (H and J) and a significant reduction in the Paneth cell (Lysozyme+ cells in crypt regions) number in Cbl−/− mice and Cblb−/− (H), scale bar = 50 μm. Data are presented as mean ± SEM of 3 independent experiments from counting 50 crypt-villus units for goblet cells and 50 crypts for Paneth cells. Statistical analysis used the student’s two-tailed t test; ∗∗∗; p ≤ 0.001.
(K and L) Chromogranin A staining (Red), showed no significant difference in the number of enteroendocrine cells between WT and Cbl−/− mice; scale bar = 50 μm. Data are presented as mean ± SEM of 4 independent experiments. Statistical analysis used the student’s two-tailed t test; ns, p ≤ 0.05, ∗; p ≤ 0.01, ∗∗.
WB of crypt and villus fractions of WT vs. Cbl−/− or Cblb−/− single-KO mice revealed comparable Cbl-b levels in WT and Cbl−/− crypts, but a marked elevation of Cbl levels in Cblb−/− crypt fractions as well as an increase in Cbl-b levels in the Cbl−/− villi fractions (Figure 1D). The overlapping crypt-enriched Cbl and Cbl-b expression and compensatory changes in their expression upon deletion of the other gene supported the likelihood of redundant Cbl/Cbl-b roles in the intestinal epithelium, as we observed during hematopoiesis.15,16 Due to lack of authenticated antibodies against Cbl-c, we examined the Cbl-c expression indirectly using the Cblc−/− mice with a LacZ reporter driven by Cblc promoter.5 Co-staining with X-gal (substrate for β-galactosidase) and Ki67 (to demarcate the crypt compartment) showed that Cblc (LacZ) was predominantly localized to villi and non-overlapping with the Ki67+ crypts (Figure S1A). Given the predominant Cbl/Cbl-b expression in crypts and Cbl-c expression in differentiated villus cells, our further studies focused on the roles of Cbl and Cbl-b in the context of ISCs.
Altered intestinal epithelial cell proliferation and differentiation in Cbl−/− mice
Given the intestinal crypt-enriched expression of Cbl and Cbl-b (Figures 1A–1D), we first examined the status of cell proliferation and differentiation in the intestinal epithelium of 6-week-old Cbl−/− or Cblb−/− mice compared to age-matched WT controls. Assessment of bromodeoxyuridine (BrdU) incorporation demonstrated an increase in cell proliferation in the Cbl−/− mouse intestinal crypts compared to WT mice, suggesting increased stem/progenitor cell proliferation (Figures 1E and 1F). On the other hand, immunofluorescence staining for pH3 revealed no significant difference in proliferating crypt epithelial cells between Cblb−/− and WT mice (Figures S1B and S1C). Hematoxylin and eosin (H&E)-stained sections demonstrated an intact overall intestinal epithelial architecture in Cbl−/− and Cblb−/− mice, but with increased abundance of goblet cells (Figure S1D). IHC staining for Mucin 2 (Muc2) or lysozyme (Lyz) to directly visualize goblet and Paneth cells, respectively,34,50,51 revealed a moderate or mild goblet cell hyperplasia in Cbl−/− or Cblb−/− mouse intestines, respectively (Figures 1G and 1I), and a reduction in Paneth cell numbers in Cbl−/− and Cblb−/− compared to WT mice (Figures 1H and 1J). Chromogranin A staining for neuroendocrine cells52 showed no significant differences between Cbl−/− and WT mice (Figures 1K and 1L). Collectively, the increased crypt epithelial cell proliferation in Cbl−/− mice and the altered numbers of goblet and Paneth cells in Cbl−/− and Cblb−/− mice suggested a functionally significant role of Cbl and Cbl-b in ISC proliferation and differentiation, with a more prominent role for Cbl.
Inducible Lgr5+ ISC-specific Cbl KO on a Cblb−/− background leads to expansion of progenitors with shrinkage of the stem cell compartment
Given the results presented earlier, we generated an ISC-specific gene deletion model to directly examine the role of Cbl and Cbl-b in the Lgr5+ ISCs, which are required for homeostatic maintenance of the intestinal epithelium and known to self-renew while cycling. To circumvent the embryonic lethality of constitutive DKO of Cbl and Cblb in mice,13 we crossed the Cblflox/flox; Cblb−/− mice previously used for Cbl/Cblb DKO in HSCs15,16,17 or MaSCs19 with mice harboring the Lgr5-EGFP-IRES-creERT2 and R26-LSL-LacZ reporter knockin alleles26 to generate the Cblflox/flox; Cblb−/−; Lgr5-EGFP-IRES-creERT2; Rosa26-LacZ mice. In these mice, Lgr5+ ISC-specific inducible DKO (iDKO) of Cbl and Cblb can be achieved upon tamoxifen (TAM)-induced Cre activation and deletion of floxed-Cbl alleles, and lineage tracing of double KO ISCs can be monitored by X-Gal staining. Lgr5 promoter-driven GFP expression also allowed us to track the fate of Lgr5+ ISCs based on the levels of their GFP expression.
Following three consecutive daily intraperitoneal injections of TAM, mouse intestines were analyzed on day 5 or 10 (Figure 2A). Quantitative real-time PCR (real-time qPCR) of isolated GFP+ intestinal epithelial cell mRNA confirmed the floxed-Cbl deletion at both time points (Figures S2A, 2B, and 2C). Consistent with the specificity of Lgr5-cre expression,26 loss of Cbl expression was seen in GFP-high (GFP Hi) Lgr5+ stem cells and their GFP-low (GFP Lo) daughter cells but not in GFP-negative (GFP neg) cells, the latter representing the non-ISC population and ISCs in which Lgr5-cre was not expressed due to well-established mosaic Lgr5-cre expression.26 To assess the impact of iDKO on Lgr5+ ISC maintenance, we quantified the GFP Hi (stem) and GFP Lo (progenitor) intestinal epithelial cell populations in control vs. iDKO mice by flow cytometry 5- or 10-days after initiating TAM induction (Figures 2D–2G; gating strategy outlined in Figure S2A). To appreciate the abundance of GFP Hi vs. Lo population in the iDKO mice, we analyzed the percentage of GFP+ cells in parental Lgr5-EGFP-IRES-creERT2 mice and observed 25% of total epithelial cells. Further gating with high-GFP and low-GFP population showed 30% high-GFP vs. 70% low-GFP population (Figure S2B). The iDKO mice exhibited a significant reduction in the percentage of GFP Hi ISCs and a concomitant increase in the percentage of GFP Lo progenitor cells at both 5- and 10-day time points (Figures 2E and 2G). A similar trend was seen when absolute cell numbers were quantified (Figures S2C and S2D). qPCR for stemness-associated genes Lgr5 and Olfm4 confirmed the gating strategy and demonstrated reduced Lgr5 and Olfm4 expression in the GFP-Hi ISC compartment of iDKO compared to control mice (Figures S2E and S2F). These results suggested that concurrent Cbl and Cblb deletion leads to a rapid shrinkage of the Lgr5+ stem cell compartment, with a concomitant expansion of the transit-amplifying (TA) compartment.
Figure 2.
Expansion of the intestinal epithelial progenitor compartment at the expense of stem cells upon inducible Cbl/Cblb double KO in Lgr5+ stem cells
(A) Schematic showing the experimental plan followed. Cblflox/flox; Cblb−/−; Lgr5-creERT2; R26-LacZ (iDKO) mice received tamoxifen (TAM) on days 0, 1 and 2 to induce conditional deletion of floxed Cbl alleles in Lgr5+ cells on a Cblb null background.
(B and C) Deletion of Cbl was assessed using real-time PCR of FACS-sorted GFP-high and GFP-low vs. GFP-negative epithelial cells at day 5 and day 10 of induction.
(D–G) Flow cytometric analysis of live epithelial cells showed a significant expansion of Lgr5-GFP Lo (progenitor) cells and a concomitant decrease in Lgr5-GFP Hi (stem cells) at 5 days (D and E) and 10 days (F and G) after TAM induction in Cbl/Cblb iDKO vs. control mice. Data are represented as mean ± SEM of 4 independent experiments. Statistical analysis using student’s two-tailed t test is shown; ns, p ≤ 0.05, ∗; p ≤ 0.01, ∗∗.
Cbl/Cblb iDKO mice exhibit increased cell proliferation in intestinal crypts
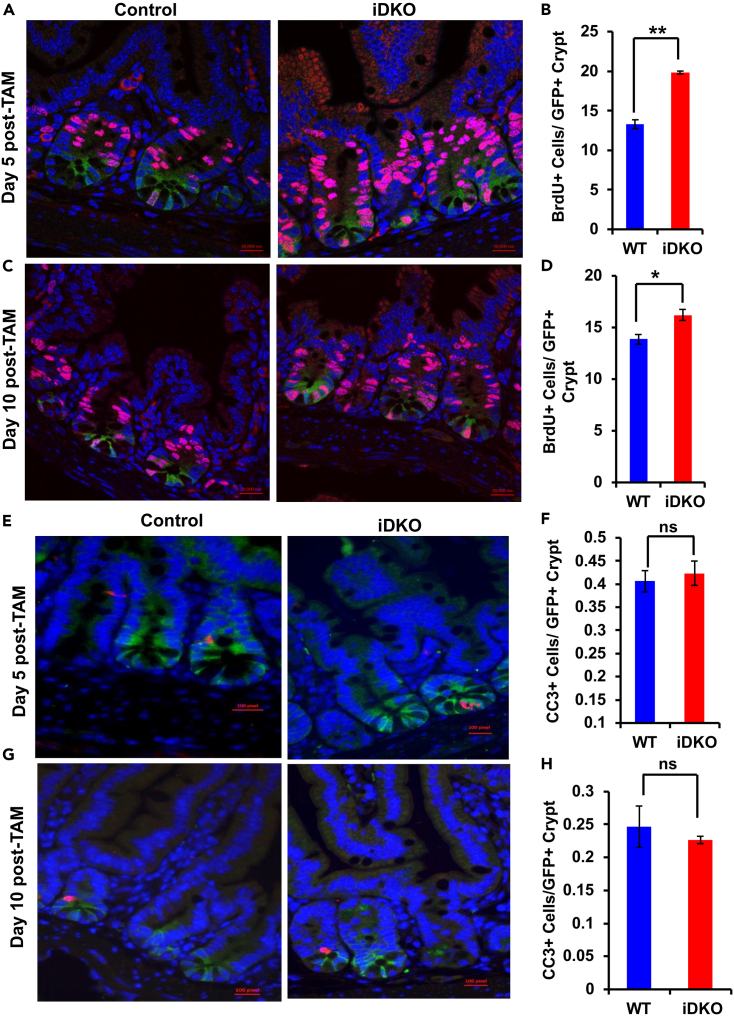
As we observed a significant depletion of Lgr5+ stem cells upon Cbl/Cblb iDKO, we examined the status of crypt cell proliferation in control vs. iDKO mice by quantifying BrdU-incorporating cells specifically in GFP+ crypts, representing those in which Lgr5-cre was active.26 The iDKO GFP+ crypts showed a significant increase in BrdU+, proliferating cells compared to those in control animals at day 5 of TAM induction (Figures 3A and 3B), with a similar albeit less-pronounced trend at day 10 (Figures 3C and 3D). Immunostaining for cleaved caspase 3 did not show any differences in the frequency of apoptotic cells in GFP+ crypts of iDKO vs. control mice (Figures 3E–3H); a higher relative number of apoptotic cells in both genotypes at day 5 relative to day 10 of TAM treatment is consistent with the reported transient toxicity of TAM on proliferating cells53 (Figures 3F and 3H). Thus, concurrent loss of Cbl and Cbl-b in Lgr5+ ISCs is associated with increased cell proliferation.
Figure 3.
Increased crypt cell proliferation with no change in apoptosis upon Lgr5+ cell-specific Cbl/Cblb iDKO in the intestine
(A–D) Control and Cbl/Cblb iDKO mice were injected with BrdU 4 h prior to euthanasia and FFPE sections of intestines were stained for BrdU (Red), GFP (Green) and DAPI (Blue). Quantification of BrdU+ cells within GFP+ crypts showed a significant increase in the fraction of iDKO stem/progenitor cells in S phase of the cell cycle at day 5 (A and B) and day 10 (C and D) of initiating TAM treatment.
(E–H) Cleaved caspase 3 (Red) co-immunostaining with GFP (Green) and DAPI (Blue) revealed no significant difference in the number of dead cells between control and iDKO mice at day 5 (E and F) or day 10 (G and H) after initiating TAM treatment. Analyses at each time point were repeated three times. Data are presented as mean ± SEM with statistical analysis using student’s two-tailed t test; ns, p ≤ 0.05, ∗; p ≤ 0.01, ∗∗. Scale bar, 20 mm.
Loss of Cbl/Cbl-b promotes commitment to differentiation
As the progenitor cells arising from Lgr5+ ISCs are known to proliferate faster,26 and we observed an expansion of the GFP Lo (Lgr5-low) progenitors with a reduction in the GFP Hi (Lgr5+) stem cells upon iDKO (Figure 2), the increased crypt cell proliferation without any change in apoptosis (Figure 3) suggested an increased commitment of the iDKO Lgr5+ ISCs into differentiated progeny. To assess if this is the case, we performed lineage tracing of the Lgr5+ stem cells in iDKO and control mice by tracking their LacZ+ progeny after TAM induction.26 At day 5 and 10 time points, we observed more LacZ+ cells in the crypts and villi with a significant increase in the percentage of partially or completely blue crypts in iDKO compared to control mice (Figures 4A–4C); this trend persisted at day 10 after induction, albeit only the increase in the percentage of fully blue crypts remained significant (Figures 4B and 4D). Notably, the number of crypts with a single blue cell was significantly reduced in iDKO mice at day 10 of TAM induction (Figure 4D); such single LacZ+ cells within crypts likely represent quiescent Lgr5+ cells, possibly those that become secretory progenitors without further division.22,24 The quiescent progenitor pool is known to express high levels of Dll1 (Delta-like ligand 1) and Ngn3 (Neurogenin 3).23,54 qPCR analysis revealed a marked alteration of these markers within the sub-fractions of Lgr5+ cells in iDKO mice, with an elevated Dll1 and Ngn3 expression in the GFP Hi (Lgr5-Hi ISCs) fraction and a decrease in their expression in the GFP Lo (Lgr5-low progenitor) fraction (Figures 4E and 4F).
Figure 4.
Increased commitment to differentiation upon Lgr5+ cell-specific Cbl/Cblb iDKO in the intestine
(A and B) X-Gal staining of intestinal sections at 5 (A) and 10 (B) days post-TAM induction to trace the fate of Lgr5+ ISCs shows an increase in blue progeny in Cbl/Cblb iDKO mice at both time points as compared to control mice; scale bar = 1,000 μm.
(C and D) Quantitative analysis of the staining shown in A & B was performed by counting 2,000 crypts per genotype and assessing the number of crypts with a single blue cell or with fully or partially blue crypts. An increase in the percentage of full and partial blue crypts was observed together with a reduction in the percentage of crypts with single blue cells.
(E and F) Real-time PCR analysis shows a mis-localization of the quiescent secretory progenitor markers Delta-like 1 (Dll1) (E) and Neurogenin 3 (Ngn3) (F) in GFP Hi and GFP Lo populations in Cbl/Cblb iDKO vs. control mice.
(G and H) Enumeration of the number of LacZ+ crypts undergoing fission shows an increase in iDKO mice at 10 days post-TAM induction as compared to control mice.
(I) LacZ staining at 4 months post-TAM induction showed comparable staining between control and Cbl/Cblb iDKO mice with no signs of hyperplasia in the latter; scale bar = 400 μm. Analyses at each time point were repeated four times. Data are presented as mean ± SEM with statistics using student’s two-tailed t test. ns, p ≤ 0.05, ∗; p ≤ 0.01, ∗∗.
Many LacZ+ crypts in iDKO mice appeared as clusters, suggesting increased crypt fission. Enumerating the % of LacZ+ crypts undergoing fission indeed revealed this to be the case in the iDKO mice (Figures 4G and 4H). Notably, analysis at 4 months of TAM induction revealed no differences in LacZ staining of crypts in iDKO vs. WT mice, and we observed normal architecture throughout the gastrointestinal tract with no signs of hyperplasia (Figure 4I), in sharp contrast to adenomas observed in Lrig1-null mouse intestine.42
Given the increased Dll1 expression in the Lgr5-Hi ISC population, we further assessed the secretory cell fate of the iDKO ISC progeny by combined PAS (Periodic acid-Schiff) X-gal staining.23 The iDKO mice showed a significant increase in the number of LacZ+/PAS+ goblet cells and a concomitant reduction in the number of LacZ+/PAS+ Paneth cells compared to control mice (Figures S3A and S3B). LacZ and chromogranin A co-staining revealed no significant difference in the number of enteroendocrine cells in iDKO vs. control mice (Figures S3C and S3D). Staining of Muc2 and Lyz to visualize goblet and Paneth cells, respectively, confirmed the significant increase in goblet cell numbers (Figures S4A and S4B) and a decrease in Paneth cell numbers (Figures S4C and S4D) of iDKO compared to control villi and crypts. IHC analysis of Cbl and Cbl-b expression confirmed the expected deletion of Cbl and Cbl-b in iDKO intestines compared to control intestines (Figures S4E and S4F).
Overall, these results support the conclusion that concurrent loss of Cbl and Cbl-b proteins commits the Lgr5+ ISCs and their rapidly dividing progeny toward differentiation, with skewing of the secretory cell fate toward goblet cells.
Cbl/Cblb iDKO mice exhibit delayed recovery from radiation-induced intestinal mucosal injury
Rapid replacement of intestinal epithelial cells shed/lost under normal homeostasis or following mucosal insults, such as exposure to radiation, is critical to maintain an intact epithelial barrier. Radiation treatment induces a rapid loss of cycling Lgr5+ stem and progenitor compartments in the gut with regeneration by re-entry of the quiescent Lgr5+ secretory progenitors into the cell cycle and their rapid expansion to re-establish epithelial homeostasis.23,24 Thus, we reasoned that the impact of the premature differentiation of rapidly dividing Lgr5+ ISCs and their progeny in iDKO mice would be more discernible in a post-radiation recovery setting despite mosaic gene deletion directed by the Lgr5-cre.26 To avoid concurrent bone marrow dysfunction from whole-body irradiation commonly used to induce the intestinal mucosal injury,55 we used focused beam abdominal radiation, known to reduce the hematopoietic toxicity and to effectively target the gut epithelium with 100% recovery through re-establishment of the intestinal epithelium by the quiescent pool of gut stem cells in normal mice.56
We tested irradiation using 8, 10, 12, or 14 Gy and found 14 Gy to be optimal at depleting ISCs as reported but without lethality, as expected.25,57 Groups of control and iDKO mice were subjected to 14Gy of focused beam abdominal radiation and TAM treatment as indicated (Figure 5A), and mice were weighed daily to monitor weight loss, which provides an accurate surrogate indicator of induced mucosal injury in this model.56 While control mice began to recover their weight by day 5 post-radiation and reached their pre-radiation weight by day 9 (data not shown), iDKO mice lagged significantly (Figure S5A). H&E staining of intestinal sections harvested on day 7 showed blunted villi and several crypt-less regions in iDKO mice compared to substantially better-preserved histology in control mice (Figure 5B). Ki67 staining revealed a significant reduction in the number of surviving crypts (defined as a crypt with at least 5 adjacent Ki67+ cells25) in iDKO as compared to control mice (Figures S5B and S5C). Fluorescence-activated cell sorting (FACS) analysis at day 5 after radiation demonstrated a reduction in the percentage as well as absolute number of Lgr5-Hi cells and a mild increase in the percentage and number of Lgr5-Lo cells in iDKO compared to control mice (Figures 5C–5E, S5D, and S5E).
Figure 5.
Cbl/Cblb iDKO mice exhibit delayed recovery from abdominal radiation injury
Control and Cbl/Cblb iDKO mice were exposed to CT-based conformal X-ray radiation (14 Gy) focused on the abdomen to assess the ability of intestinal stem/progenitor cells to re-establish epithelial homeostasis post-damage.
(A) The schematic shows the experimental plan followed.
(B) Histological analysis of H&E-stained sections of different regions of intestine on day 7 post-injury showed hyperproliferating crypts with fission and repopulation of villi in the control mice while Cbl/Cblb iDKO mice showed bare, crypt-less patches and blunted villi with little recovery; representative pictures are shown; scale bar, 200 μm.
(C–E) The inability of Cbl/Cblb iDKO mice to regenerate injured intestinal epithelium is associated with a precipitous drop in the Lgr5-Hi stem cell population (C and D) and a mild increase in the Lgr5-Lo progenitors (C and E) analyzed at d5 post-injury. Analysis at each time point was repeated three times. Data are presented as mean ± SEM with statistics using student’s two-tailed t test; ns, p ≤ 0.05, ∗; p ≤ 0.01, ∗∗.
Cbl/Cblb iDKO impairs crypt stem cell self-renewal by hyperactivating the Akt-mTOR pathway
Organoid cultures initiated from isolated intestinal epithelial crypts or purified Lgr5+ stem cells provide a facile system to dissect mechanisms of ISC self-renewal.34,58 Given the mosaic expression of Lgr5-Cre, we examined the crypt epithelium-derived organoids isolated from Cblf/f; Cblbf/f (double-floxed or FF control) or Cblf/f; Cblbf/f; R26-creERT+ (FF/creERT) mice19 and cultured in the presence of 4-hydroxy tamoxifen (4-OH-TAM) for the indicated time points. Of note, in this system both Cbl and Cblb are deleted only after 4-OH-TAM induction. The FF control organoids showed a marked increase in crypt (seen as evaginations) and villus domain formation over the 72-h observation period, and H&E staining at the end showed a uniform epithelial layer surrounding the more amorphous interior (Figures 6A and 6B upper panels), as expected.58,59 In contrast, the FF/creERT organoids cultured in 4-OH-TAM showed an increase in crypt domain formation at 24 and 48 h time points, but these receded rapidly by 72 h, with the H&E staining showing marked disintegration with no clear epithelial layer (Figures 6A and 6B lower panels). Quantification of the crypt domains confirmed these results (Figure 6C). Upon replating, the TAM-treated control organoids re-created the original organoid architecture with intact crypt and villus domains; in contrast, no intact organoid structures could be seen upon replating of TAM-treated FF/creERT organoids (Figure 6D). The expected Cbl and Cbl-b deletion was confirmed upon 48 h and 72 h of 4-OH-TAM treatment of FF/creERT organoids (Figure 6E). Thus, Cbl and Cbl-b are required for intestinal epithelial homeostasis in vitro.
Figure 6.
Conditional Cbl/Cblb deletion impairs self-renewal in intestinal crypt organoid cultures by upregulating the Akt-mTOR pathway
Equal number of crypts isolated from Cblflox/flox; Cblbflox/flox (FF Control) and Cblflox/flox; Cblbflox/flox; R26creERT2 (FF/creERT) mice were plated in 100% Matrigel in the presence of growth factors to form organoids. Once formed, organoids were replated at a 1:4 split ratio and treatment with 400 nM 4-OH TAM was initiated after 24 h to induce Cbl/Cblb deletion.
(A) Bright-field imaging showed that while FF control mouse intestinal organoids exhibited a steady increase in budding (crypt domains) over time (up to 72 h of observation), the FF/creERT mouse organoids showed increased budding until 48 h after 4-OH-TAM induction but rapidly lost crypt domains by 72 h post-induction; scale bar = 40 μm.
(B) Representative H&E-stained images of FF control and FF/creERT organoids confirm the loss of morphological features in the latter; scale bar = 50 μm.
(C) Ten distinct organoids per genotype were followed up to 72 h and change in the number of buds (crypt domains) at each time point relative to time 0 was quantified.
(D) FF Control and FF/creERT organoids grown in 4-OH-TAM for 72 h were re-passaged and imaged after 48 h. Note the lack of organoid structures with intact morphology in FF/creERT organoid cultures, supporting a loss of self-renewal contrary to growth and intact morphology of control organoids; scale bar = 400 μm.
(E) Immunoblotting of organoid lysates at different time points confirmed effective Cbl/Cblb deletion by 48 and 72 h time points in FF/creERT organoids. HSC-70, loading control. FF/creERT organoids derived from two independent female mice (2F and 3F) were cultured in the presence (iDKO) or absence (control) of 400 nM 4-hydroxy-TAM for 72 h. Single cells isolated from the organoids were processed through the GemCode Single Cell Platform (10X Genomics) to perform single-cell RNA-seq and analyzed using Seurat.
(F) tSNE map of combined controls and combined Cbl/Cblb iDKO mouse organoid cells. Cells are grouped into seven clusters based on transcriptome profiles and are colored accordingly. The cell types were assigned based on gene expression profile of stem cells (Lgr5, Ascl2, Axin2, Olfm4, Gkn3), transit-amplifying (TA) cells (Mki67, Cdk4, Mcm5, Mcm6, Pcna), enterocytes (Alpi, Apoa1, Apoa4, Fabp1), Paneth cells (Lyz1, Defa17, Defa22, Defa24, Ang4), enteroendocrine cells (Chga, Chgb, Tac1, Tph1, Neurog3), goblet cells (Muc2, Tff3, Agr2) and tuft cells (Dclk1, Trpm5, Gfi1b).
(G) Heatmap shows the expression levels of top cluster-specific genes in each cluster. Yellow represents the highest expression while purple represents low or no expression.
(H) Histogram depicting the percentage of cells in each cluster.
(I) Validation of Cbl and Cblb deletion in the 2F and 3F organoids by qPCR.
(J) Gene set enrichment analysis (GSEA) of each cell type between control and iDKO organoids shows upregulation the of PI3K-Akt-mTOR signaling pathway upon Cbl/Cblb iDKO in stem cells, goblet cells, enteroendocrine cells, Paneth cells, and enterocytes. NES (normalized enrichment score), FDR (false discovery rate), and p values are indicated in the GSEA plots.
(K) To validate the alterations in PI3K-mTOR signaling upon Cbl/Cblb iDKO, organoid lysates were collected at different time points of culture in the presence of 4-OH-TAM and analyzed by immunoblotting for EGFR, p-EGFR, Akt, p-Akt, S6 and p-S6. Clear upregulation of p-AKT/mTOR pathway components (p-Akt and p-S6) is seen and pEGFR upregulation is observed only after 72 h of 4-OH-Tam induction. Hsc-70 served as a loading control. Densitometries of Cbl, Cbl-b, p-EGFR/EGFR, p-Akt/Akt and p-S6/S6 after normalizing to loading control in comparison to FF control sample in each time point are indicated on the top of the immunoblots.
(L) Intestinal tissue sections from Control and Cbl/Cblb iDKO mice 10 days after initiating TAM treatment were stained with antibodies against p-S6 to assess the impact of Cbl/Cblb deletion. Sections stained with secondary antibody alone were used as a negative control. p-S6 levels (red in color) were sharply increased in iDKO as compared to control sections; scale bar = 20 μm. Quantified data are presented as mean ± SEM of three independent experiments with statistics using student’s two-tailed t test. ns, not significant; p ≤ 0.05, ∗; p ≤ 0.01, ∗∗; p ≤ 0.001, ∗∗∗.
To gain unbiased mechanistic insights into the nature of molecular alterations caused by Cbl/Cblb iDKO, we carried out single-cell RNA sequencing (RNA-seq) analysis of cells isolated from untreated (control) vs. 4-OH-T-treated (Cbl/Cblb iDKO) organoid cultures from FF/creERT mice. Expression of cell type-specific index genes in the transcriptomic profiles identified seven cell clusters corresponding to stem cells (Lgr5, Ascl2, Axin2, Olfm4, Gkn3), TA cells (Mki67, Cdk4, Mcm5, Mcm6, Pcna), enterocytes (Alpi, Apoa1, Apoa4, Fabp1), Paneth cells (Lyz1, Defa17, Defa22, Defa24, Ang4), enteroendocrine cells (Chga, Chgb, Tac1, Tph1, Neurog3), goblet cells (Muc2, Tff3, Agr2), and tuft cells (Dclk1, Trpm5, Gfi1b) in both control and iDKO organoids (Figures 6F–6H), consistent with the major cell types observed in previous single-cell RNA-seq analyses of intestinal epithelial organoids.60,61 qPCR analyses verified the loss of Cbl and Cblb gene expression in iDKO relative to control organoids (Figure 6I). Notably, Cbl/Cblb deletion was observed in all (not only Lgr5+) cells in intestinal organoids with crypt domain expansion in DKO. We did not observe any decrease in the Lgr5+ compartment, and in fact the ISC and TA compartments exhibited an increase (Figures 6F and 6H). We could not use the RNA-seq results as a validation of the loss of ISCs we observed upon in vivo Lgr5-Cre-mediated Cbl deletion for an Lgr5 compartment-selective DKO (Figures 6F and 6H). We reason that the expansion of stem cells and TA cell pools in DKO organoids revealed by single-cell RNA-seq experiments reflects our capturing the expansion phase of DKO organoids. Given the concurrent deletion of Cbl and Cblb in all cell types in the organoid model, we performed gene set enrichment analysis (GSEA) of the single-cell RNA-seq results between controls and DKOs across all identified cell types (Table S3). The signaling pathways shared across cell types were prioritized for further analyses for the potential signaling mechanism.
GSEAs identified an upregulation of the PI3-kinase/AKT/mTOR pathway in multiple cell compartments in iDKO organoids, including stem cells, goblet cells, enterocytes, and enteroendocrine cells (Figure 6J). Notably, Notch pathway, well-known to play an important role in regulating intestinal epithelial homeostasis,62,63,64 showed a downregulation in DKO TA cells, while this pathway was upregulated in DKO enterocytes and enteroendocrine cells (Figure S6). We therefore focused on the PI3K-AKT-mTOR signaling as a potential mechanism by which loss of Cbl/Cbl-b alters the intestinal epithelial homeostasis. WB analysis of FF control vs. FF/creERT organoid lysates confirmed a nearly complete loss of Cbl and Cbl-b expression by 72 h after adding TAM (Figure 6K). Immunoblotting showed an upregulation of p-Akt levels in FF/creERT organoids compared to controls at 24 and 48 h that remained sustained at 72 h of TAM treatment, and a similar pattern was seen with p-S6 (Figure 6K). EGFR being an upstream of PI3K/AKT signaling and a direct target of Cbl proteins, total and activation status was assessed by immunoblotting and revealed upregulation of p-EGFR at 72 h of Tam treatment in FF/creERT organoid lysates (Figure 6K). IHC staining of EGFR showed upregulation in single Cbl and Cblb KO and Cbl/Cblb iDKO compared to WT mice (Figure S7). Immunostaining of tissue sections from TAM-induced control vs. Cbl/Cblb iDKO mice showed that p-S6 levels were markedly increased in iDKO crypts compared to crypts in control mice (Figure 6L). Collectively, these results supported the conclusion that the AKT/mTOR pathway is hyperactivated upon Cbl/Cblb iDKO in ISCs, consistent with the AKT-mTOR pathway hyperactivation upon Cbl/Cbl-b deletion in other systems16,19,65,66 and the importance of the Akt/mTOR pathway downstream of RTKs in intestinal epithelial homeostasis.
To assess the role of the hyperactive AKT/mTOR pathway in the observed defective intestinal epithelial maintenance upon Cbl/Cblb iDKO, we treated FF/creERT organoids with 4-OH-TAM in the absence or presence of specific inhibitors of AKT (MK-2206; 250 nM) and/or mTOR (rapamycin; 10 nM). The dual-fluorescent reporter of Cre activation (red before and green after Cre activation67) in the organoids revealed uniform activation of Cre upon 4-OH-TAM treatment (Figure 7A). Compared to 4-OH-TAM-treated organoids without inhibitors, which showed the expected disruption of the organoid structure and reduced crypt domain number by day 3, MK-2206 treatment led to a small but statistically insignificant preservation of the crypt domains and epithelial integrity (Figures 7A and 7B). Notably, treatment with rapamycin or rapamycin plus MK-2206 led to a marked rescue of the organoid disruption induced by Cbl/Cblb deletion (Figures 7A and 7B). Of note, treatment with either inhibitor led to some reduction in crypt domain formation at the 24 h time point (Figure 7B); however, this effect was independent of Cbl/Cblb deletion and also seen with control organoids (Figures S8A and S8B). Replating of FF/creERT organoids cultured under the more effective inhibitor conditions for 72 h showed a substantial retention of organoid structures when analyzed 48 h after replating (Figure 7C). GFP reporter expression ruled out the organoids seen after replating arising from cells in which Cre activation had not occurred. Western blot analysis revealed the expected upregulation of p-Akt and p-S6 in FF/creERT organoids cultured with 4-OH-TAM alone (Figure 7D). The levels of p-Akt and p-S6 phosphorylation were drastically reduced in organoids treated with rapamycin or rapamycin plus MK-2206 (Figure 7D). Collectively, the sustained in vitro and in vivo Akt/mTOR pathway activation, together with the reversal of the disruption of Cbl/Cblb iDKO organoid architecture upon Akt/mTOR pathway inhibition, supports the conclusion that negative regulation of the Akt-mTOR pathway by Cbl and Cbl-b is required to maintain the ISCs.
Figure 7.
p-Akt/mTOR pathway inhibition rescues defective maintenance of Cbl/Cblb iDKO intestinal crypt organoids
(A) Organoids established from FF/creERT mice carrying the mT/mG dual fluorescent reporter to document gene deletion were treated with 400 nM 4-OH-TAM, without (4-OH-TAM only control) or with Akt inhibitor MK-2206 (250 nM), mTOR inhibitor Rapamycin (10 nM) or their indicated combinations (5 nM Rapamycin plus 125 nM MK-2206 or 10 nM Rapamycin plus 250 nM MK-2206) and imaged at various time points over a 72 h period. While 4-OH-TAM alone treated organoid structures showed the expected disruption of architecture by 72 h, inhibition of Akt, mTOR or their combination rescued the organoid morphology with robust budding. Fluorescent imaging showed successful activation of Cre following TAM treatment under all conditions (conversion from uniform red to uniform green).
(B) Quantification of crypt domains showed that Rapamycin by itself or in combination with MK-2206 significantly improved the crypt budding in comparison to TAM alone at 72 h; significant reduction in crypt budding was seen at 24 h, while no differences were seen at 48 h.
(C) After 72 h of organoid culture treatments, the 4-OH-TAM treated, 10 nM Rapamycin+4-OH-TAM treated and 10 nM Rapamycin+250nM MK-2206+4-OH-TAM treated organoids (the latter two showing the best rescue) were replated in complete ADF medium and imaged at 48 h after replating. The organoids previously treated with Rapamycin or Rapamycin plus MK-2206 showed robust budding while such structures were absent in secondary cultures of 4-OH-TAM alone treated organoids.
(D) Western blot analysis of FF control organoids and FF/creERT organoids at 72 h of treatment showed the expected downregulation of p-Akt, p-S6 or p-S6 + p-Akt levels with 4-OH-TAM+MK-2206, 4-OH-TAM+ Rapamycin or 4-OH-TAM+MK-2206+Rapamycin treatment, respectively, as compared to 4-OH-TAM alone treatment in both FF Control and FF/creERT organoids. Densitometries of p-Akt/Akt and p-S6/S6 after normalizing to loading control in comparison to 4-OH-TAM treated FF control sample are indicated on the top of p-Akt and p-S6 bands. Quantified data are presented as mean ± SEM of three independent experiments with statistics using student’s two-tailed t test. ns, p ≤ 0.05, ∗; p ≤ 0.01, ∗∗.
Discussion
In addition to serving as a critical environment-organism interface with essential digestive and absorptive functions, the intestinal epithelium provides an elegant system to unravel gene functions critical for the maintenance and regeneration of epithelial tissues. Tyrosine kinase signaling linked to growth factor receptors represents one component of signaling mechanisms that orchestrate ISC maintenance, proliferation, and differentiation.30,31 Cbl-family ubiquitin ligases are activation-dependent negative regulators of tyrosine kinases, but their roles in intestinal epithelial stem cell homeostasis are unknown. Here, using unique genetic deletion approaches, we establish a novel and redundant requirement for two Cbl-family members, Cbl and Cbl-b, in the maintenance of Lgr5+ ISCs and in their commitment to differentiate along various mature lineages. We also demonstrate the importance of negative regulation of the Akt-mTOR pathway by Cbl and Cbl-b as a critical mechanistic underpinning for their role in ISC regulation.
While in vitro studies of CBL family proteins have defined their key potential roles as negative regulators of both receptor and non-RTKs, their in vivo physiologic roles have been primarily defined in the context of immune and hematopoietic cell compartments,4,15,16,17,68,69,70,71,72 with only sporadic studies of other physiological pathways and organ/tissue systems despite a pervasive role of tyrosine kinase signaling in physiological systems.32,33,34,35,37,38,39,40,41 Notably, studies of the roles of Cbl proteins in epithelial biology are distinctly lacking except for our recent findings that Cbl and Cbl-b are vital for the postnatal development of mouse mammary gland, with their combined deletion leading to reduced ductal branching and defects in MaSC self-renewal.19 Given the counterintuitive nature of these findings in the face of a linkage of hyperactive tyrosine kinase signaling to breast and other epithelial cancers,73,74,75,76,77 and a meager understanding of mammary epithelial stem cell maintenance and commitment into mature cell types or the signaling pathways involved,78,79 the broader significance of Cbl proteins in epithelial stem cell maintenance has remained unclear. Thus, the current studies establishing a requirement for Cbl and Cbl-b in the maintenance of intestinal epithelial stem cells represent a significant step forward in our understanding of the biology of Cbl proteins, while expanding our understanding of the nature and importance of negative regulation of tyrosine kinase signaling in intestinal epithelial stem cell homeostasis.
Since no prior studies have systematically examined the expression or function of Cbl and Cbl-b proteins in the intestinal epithelium, we first assessed their expression pattern in the intestinal epithelium. We show that Cbl and Cbl-b are expressed throughout the length of the intestinal epithelial crypt-villus axis but are enriched in the crypt (Figures 1A–1D), which pointed to their potential role in regulating the intestinal stem/progenitor cell compartment well-established to reside within crypts.26 Their overlapping expression pattern also suggested their potentially redundant functional roles in the intestine, reminiscent of their redundancy in regulating hematopoietic15,16 and mammary19 stem cells. Our findings of compensatory upregulation of Cbl expression in the crypts of Cblb-null mice and of Cbl-b in the villi of Cbl-null mice supported such redundant roles. In contrast to Cbl and Cbl-b, the expression of Cbl-c, measured indirectly using a LacZ reporter incorporated into the Cblc gene in Cblc-null mice, was confined to villi (Figure S1A). The lack of a compensatory increase in Cbl-b expression in the crypts of Cbl-null mice suggested a potentially more significant role for Cbl. Indeed, Cbl-null mice showed increased cell-cycle entry within the crypt, goblet cell hyperplasia, and reduction in the number of Paneth cells, while Cblb-null mice showed a moderate increase in goblet cell numbers and significant decreased Paneth cells (Figures 1G–1I, S1B, and S1C).
Since whole-body Cbl/Cblb DKO mice are embryonic lethal,13 we utilized the Lgr5+ ISC-specific inducible DKO of Cbl and Cblb19 to explore the redundant functional roles of Cbl and Cbl-b in ISCs. We note that the iDKO designation is used for simplicity to denote that the DKO state is only induced in a specific cell type upon TAM-inducible Cbl deletion, but the studies utilize a model with a germline Cbl-b deletion. The rationale for the use of such a model for cell-specific DKO studies is well established by prior studies and reflects the lack of any detectable phenotypes in germline Cbl-b KO mice10,11 but profound cell type-specific phenotypes when Cbl is concurrently deleted in a cell-selective manner13,15,16,19,80,81 while germline Cbl and Cbl-b DKO is embryonic lethal.13 The Cbl/Cblb iDKO led to a rapid and marked drop in the size of the Lgr5+ ISC compartment (Figures 2D–2G and S2C–S2F), with a concomitant increase in the Lgr5-Lo population, suggesting that Cbl and Cbl-b negatively regulate the commitment of ISCs into TA progenitors, known to be Lgr5-Lo.27,36 Supporting this conclusion, the iDKO mouse intestinal crypts exhibit increased cell proliferation, with no alterations in the rate of apoptosis (Figures 3A–3H); a similar but less-pronounced crypt hyper-proliferation was seen in Cbl−/− mice (Figures 1E and 1F). Furthermore, Cbl/Cblb iDKO was associated with crypt fission (Figures 4G and 4H), a process that can be fueled by increased progenitor cell numbers.20,82 Thus, our results support a role for Cbl and Cbl-b in the maintenance of a self-renewing pool of Lgr5+ ISCs by controlling the rate at which they commit to differentiation vs. self-renewal. Despite the hyper-proliferative crypt phenotype of Cbl/Cblb iDKO mice, however, a long-term hyperplasia of the epithelium or adenoma formation was never seen, a phenotype quite distinct from that seen in mice with deletion of the RTK-negative regulator Lrig1, which induced epithelial hyperplasia and adenomas,42 even though Lrig1 is thought to function partly by facilitating RTK targeting by Cbl.45 Notably, although Lrig1 is expressed in Lgr5+ ISCs, its expression in the intestinal epithelium appears to be broader.43 The phenotype of Lrig1 deletion specifically in Lgr5+ cells has not been determined.
The lack of hyperplasia or adenomas upon Cbl/Cblb deletion in Lgr5+ ISCs, despite an immediate crypt hyper-proliferation phenotype, is likely to arise from multiple factors. Lineage tracing showed a rapid transition of Lgr5+ cells with iDKO to enterocytes and goblet cells, and goblet cell numbers were increased in Cbl or Cblb single-KO mice and much more so in iDKO mice (Figures 4A–4D, 1G–1J, S3A, S3B, and S4A–S4D). BrdU labeling showed proliferation zones to be limited to crypts (Figures 3A and 3C), suggesting that the expanded TA population must rapidly exit the cell cycle as the cells mature into differentiated cell types. We contend that the expansion of the TA Lgr5-Low population occurs at the expense of the Lgr5-Hi compartment, which eventually eliminates all hyperproliferating iDKO cells. Supporting that notion, lineage tracing showed a decrease in the proportion of crypts with single blue (iDKO) cells (Figures 4A–4D), which are thought to represent a population of quiescent Lgr5+ cells that directly adopt a secretory cell fate, primarily into Paneth cells.23,24 Indeed, the numbers of Paneth cells were reduced in iDKO crypts (Figures S4C and S4B). A reduction in Paneth cell numbers could in turn further reduce the ISC numbers due to deficient niche function.34,83,84,85 The quiescent and cycling Lgr5+ pools are known to be in equilibrium22; apparently, the iDKO quiescent ISCs may be defective in resuming cycling Lgr5+ stem cell fate and instead differentiate into progeny cells, thereby disrupting the quiescent-cycling ISC equilibrium. Interestingly, it has been previously shown that signaling downstream of RTKs can tilt the balance among different secretory cell types.39,40,44 A decrease in the quiescent Lgr5+ iDKO ISCs is consistent with the loss of quiescence in HSCs upon Cbl/Cblb deletion, but the shrinkage of the Lgr5-Hi cycling ISCs contrasts with an expansion of cycling HSCs.16 Whether this reflects a significant loss of Paneth cell niche in iDKO mice, rapid maturation of quiescent Lgr5+ cells along alternate fates (such as goblet cells or enterocytes), or a defective interconversion between the two different states of ISCs will require further studies.
It is noteworthy that the impact of Cbl/Cblb iDKO on mature intestinal epithelial cell types was not uniform, with a remarkable increase in goblet cells and reduction in Paneth cells providing a sharp contrast. Molecular marker analyses also suggested an increase in enterocytes with no change in enteroendocrine cells. These findings support the idea that Cbl/Cbl-b-mediated regulation of ISC signaling is a determinant of differential fates of ISCs along various lineages. Consistent with altered fates of iDKO ISCs, the expression of markers of quiescent secretory progenitors, Dll1 and Ngn3, was elevated in Lgr5-Hi cells while it was reduced in Lgr5-Lo cells of iDKO mice (Figures 4E and 4F). High expression of Dll1, a ligand for Notch receptors, is a characteristic of +5 crypt cells, immediate descendants of Lgr5 ISCs that adopt a secretory cell fate.24,86 These cells are also thought to stochastically turn off Notch signaling and exit the cell cycle, further promoting their secretory cell fate. Notably, inhibition of Notch signaling is known to reduce the Lgr5-Hi cell population and the expression of stemness-associated genes such as Lgr5 and Olfm4.62,63 Ectopic expression of Notch in the Ngn3+ enteroendocrine progenitor cells, on the other hand, has been shown to drive differentiation toward enterocyte and goblet cell lineages.64 Thus, it is possible that iDKO Lgr5-Lo cells experience greater-than-usual Notch signals propelling their expansion and further differentiation along enterocyte and goblet cell lineages. Further studies are needed to understand the basis of the differential impact of Cbl/Cblb iDKO on mature intestinal epithelial cell populations.
While Cbl/Cblb iDKO led to a rapid loss of Lgr5+ ISCs, no overt effects on mice were observed. One plausible reason for a mild phenotype with a lack of overt homeostatic imbalance in the composition of the intestinal epithelium over time is the replacement by ISCs without gene deletion since Lgr5-Cre expression is mosaic.26 We reason that mosaic Lgr5-Cre-dependent DKO combined with a loss-of-function phenotype of Cbl/Cblb DKO (elimination of the cells in which Cbl and Cbl-b co-deletion has occurred) further adds to the mild phenotype.
To experimentally reveal a potential functional impact of the loss of Lgr5+ cells induced by Cbl/Cblb iDKO, we used a radiation injury model as recovery from such injury involves a rapid conversion of quiescent ISCs to cycling Lgr5+ ISCs.22,23,24,87 For these studies, we used focused beam radiation to the abdomen to spare hematopoietic compartments to reduce the toxicity. Indeed, the Cbl/Cblb iDKO following focused beam abdominal radiation showed significantly slower recovery and slower re-establishment of the stem/progenitor compartments in the intestine (Figures 5 and S5).
Since many growth factors and their cognate receptors have been linked to the maintenance of ISCs30,31 and signaling downstream of these could be impacted by Cbl/Cblb iDKO, we used unbiased single-cell RNA-seq analysis of control vs. iDKO intestinal organoids to discern the likely signaling pathways since Lgr5+ ISCs are known to divide, self-renew, and differentiate into mature cell types in organoid culture.58 Using a fully inducible DKO model (with TAM-inducible creERT and floxed Cbl/Cblb), our analyses revealed upregulation of the Akt/mTOR signaling pathway in iDKO ISCs and other cell compartments (Figure 6J), findings that were confirmed by elevated pAkt and p-S6 levels in iDKO organoids (Figure 6K) and an increase in p-S6 staining in ISCs of iDKO mice (Figure 6L). The likelihood of the Akt-mTOR pathway being mechanistically important for ISC regulation by Cbl/Cbl-b is further strengthened by prior studies showing this axis to be a major target of Cbl/Cbl-b-imposed negative regulation in various systems16,65,66 and our recent implication of the hyperactive Akt-mTOR pathway in MaSC depletion induced by loss of Cbl/Cbl-b.19 Reversal of the loss of crypt domain-forming ability upon Cbl/Cblb iDKO by inhibiting Akt and/or mTOR in the organoid system (Figure 7) further supports the conclusion that negative regulation of Akt-mTOR signaling by Cbl/Cbl-b is required to maintain functional ISCs.
We note that single-cell RNA-seq did not show any depletion of the ISC compartment of the Cbl/Cbl-b DKO organoids in contrast to our findings in vivo. We surmise that this may reflect the concurrent TAM-inducible Cbl/Cbl-b deletion in all cellular compartments of organoids vs. selective Lgr5+ compartmental DKO in vivo and reflects our capturing the expansion phase of DKO organoids (Figures 6F and 6H). Regardless of this discrepancy, we reason the mechanistic insights obtained from this experimental model are valid. Consistent with this argument, intestinal sections of the iDKO model used for our in vivo analyses confirmed the upregulation of p-S6 (Figure 6L) levels in DKO vs. control mice, similar to what we observed using organoid models (Figure 6K). Furthermore, consistent with higher total and p-EGFR levels in DKO vs. control organoids in vitro (Figure 6K), IHC analysis showed higher EGFR staining in iDKO intestinal sections relative to control mice (Figure S7). Clearly, coordinate in vivo and organoid studies using Lgr5+ ISC compartment directed fully inducible Cbl/Cblb deletion will be necessary to fully validate the linkage of Cbl/Cbl-b to regulation of the Akt/mTOR axis in ISCs we describe here.
We posit that Cbl/Cbl-b provide a redundant but necessary negative regulatory mechanism to fine-tune the level of Akt-mTOR signaling in Lgr5+ ISCs to favor self-renewal with orderly differentiation, while hyperactive Akt-mTOR signaling leads to rapid transition of ISCs into progeny at the cost of stem cells and aberrant lineage differentiation. The mTOR pathway is a major regulator of proliferation and growth during development as well as in adult tissues in mammals88 and plays its vital roles by integrating diverse signals including nutrient status, growth factor signals, and redox/mitochondrial stress to control cellular metabolism. As such, mTOR signaling has emerged as critical in the control of self-renewal and differentiation in many stem cell systems, including the intestine.89 Overexpression of dominant negative TSC2, the negative regulator of mTOR, was shown to induce crypt proliferation but reduce goblet and Paneth cells by activating the Notch pathway.90 On the other hand, inducible mTOR deletion in the intestine led to replacement of mTOR-deficient ISCs by WT ISCs while constitutive mTOR deletion led to altered differentiation, including blunted villi, loss of alkaline phosphatase, and loss of goblet and Paneth cells.91 In addition, mTOR-deleted crypts failed to form organoids in vitro. Interestingly, calorie restriction was shown to inhibit the mTOR pathway in Paneth cells and enhance their niche function to support ISC self-renewal, but exposure of both Paneth and Lgr5+ stem cells to rapamycin enhanced organoid formation ability of the latter in vitro.83 PTEN (phosphatase and tensin homolog), a negative regulator of PI3-kinase-Akt signaling, has been linked to the maintenance of a quiescent ISCs92 and progenitors.93 Loss of YY1, an effector of mTOR, has been shown to cause exhaustion of Lgr5+ stem cells.94 It is likely that the various reported results represent the distinct outcomes of mTOR signaling in different intestinal epithelial cell and ISC compartments. It is also noteworthy that initial elevation of proliferation and subsequent loss of ISCs upon Cbl/Cblb DKO observed in vivo (Figures 3A, 4C, and 4D) as well as in vitro (Figure 6C), together with apparently enhanced differentiation to certain mature lineages (such as goblet cells) (Figures S3A, S3B, S4A, and S4B), suggest a potential role for Cbl and Cbl-b in protecting ISCs against exhaustion, a possibility consistent with our previous demonstration of HSC exhaustion16 and MaSC depletion19 upon Cbl/Cblb DKO in these lineages and the roles of the AKT/mTOR pathway in maintaining ISC characteristics and quiescence alluded to earlier discussion.83,91,92,94 Further analyses will be needed to determine the mechanisms by which DKO ISCs are depleted, including the relative importance of their aberrant/rapid differentiation vs. regulated forms of cell death. We also note that our current understanding of Cbl/Cbl-b as tyrosine-kinase-directed ubiquitin ligases suggests that their negative regulatory role in Akt-mTOR signaling in ISCs reflects their activation downstream of tyrosine kinase-coupled receptors.4 Whether Cbl/Cbl-b might also regulate the Akt-mTOR axis in response to nutritional cues remains to be determined. Future studies of perturbations in Akt-mTOR signaling pathways specifically in Lgr5+ ISCs with or without concurrent Cbl/Cblb deletion should further elucidate the importance of Cbl/Cbl-b regulation of the Akt-mTOR pathway in ISC maintenance.
Pertinent to our findings, previous studies reported that Cbl can serve as a negative regulator of beta-catenin through direct binding and ubiquitination.95,96 Subsequently, the same investigators reported an inverse correlation between Cbl expression and nuclear beta-catenin levels in colorectal tumors, with increased proliferation and tumorigenic ability of colorectal tumor cell lines with downregulation of CBL expression.97,98 Furthermore, haploinsufficiency of Cbl in mice enhanced the atypical hyperplasia and adenocarcinoma numbers in APCΔ14/+ mice but had no effect on its own.99 While we have not directly examined the WNT signaling pathway, we consider it unlikely that the aberrations of ISCs and mature intestinal epithelial cell lineages in Cbl/Cblb iDKO mouse model reflect a potential increase in beta-catenin activity since Lgr5+ ISC-specific increased WNT/beta-catenin pathway activation upon Apc deletion has been demonstrated to be tumorigenic.28
Finally, it is notable that aberrant tyrosine kinase and Akt-mTOR signaling are linked to oncogenesis, including in colorectal cancer,100,101 yet somatic mutations of Cbl/Cbl-b found in myeloid leukemias102,103,104,105 are distinctly rare in colorectal and other epithelial malignancies. Future studies to determine if the requirement for Cbl/Cbl-b for the maintenance of stem cell compartments in normal epithelial tissues, such as the intestine (here) and mammary glands,19 extends to cancer stem cells that drive these malignancies28,106,107,108 will therefore be of great interest.
Limitations of the study
While the conclusions of this study that Cbl and Cbl-b ubiquitin ligases function as negative regulators of Akt-mTOR signaling to maintain the Lgr5+ stem cell compartment during homeostasis and regeneration from injury are based on multiple complementary approaches, these conclusions should be viewed together with the following limitations. Our studies used mice with whole-body Cblb KO to generate Lgr5+ compartment-specific iDKO; future studies with fully conditional Lgr5-specific Cbl/Cblb KO are needed to exclude impact of the global Cblb KO. The known mosaic expression of Lgr5-Cre restricted the iDKO to a subpopulation of ISCs, which likely contributes to generally mild and transient phenotypes in the absence of intestinal injury. The organoid model studies used the creERT2 system thus leading to Cbl/Cblb iDKO in all cell types; thus, the mechanistic findings in this model must be verified using ISC-specific KO in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Monoclonal anti-CBL | BD Biosciences | Cat# 610442; RRID: AB_397816 |
| Mouse Monoclonal anti-CBL-B for IHC | Abcam | Cat# ab54362; RRID: AB_879652 |
| Rat Monoclonal anti-Ki67 | eBioscience™ | Cat# 14-5698-82 |
| Rabbit Polyclonal anti-Chromogranin A | Abcam | Cat# ab15160; RRID: AB_301704 |
| Rabbit Polyclonal anti-MUC2 | Invitrogen | Cat# PA5-21329 |
| Rabbit Monoclonal anti-Lysozyme | Abcam | Cat# ab108508; RRID: AB_10861277 |
| Rabbit Monoclonal anti-EGFR | Cell Signaling Technology | Cat#4267; RRID: AB_2895042 |
| Mouse Monoclonal anti-BrdU | Developmental Studies Hybridoma Bank, Iowa | Cat# G3G4 |
| Rabbit Monoclonal anti-Cleaved Caspase 3 | Cell Signaling Technology | Cat#9664; RRID: AB_2070042 |
| Rabbit Polyclonal anti-Phospho Histone 3 (Ser10) | Cell Signaling Technology | Cat#9701; RRID: AB_331535 |
| Rabbit Monoclonal anti-GFP | Cell Signaling Technology | Cat#2956; RRID: AB_1196615 |
| Chicken Polyclonal anti-GFP | Aveslabs | Cat#GFP1010 |
| Rabbit Monoclonal anti-Phospho S6 Ribosomal Protein (Ser235/236) for IF | Cell Signaling Technology | Cat#4858; RRID: AB_916156 |
| Rabbit Monoclonal anti-CBL-B for WB | Cell Signaling Technology | Cat#9498; RRID: AB_2797707 |
| Mouse Monoclonal anti-HSC-70 | Santa Cruz | Cat#7298; RRID: AB_627761 |
| Rabbit Polyclonal anti-Phospho EGFR (Y1068) | Cell Signaling Technology | Cat#2234; RRID: AB_331701 |
| Rabbit Monoclonal anti-EGFR | Cell Signaling Technology | Cat#4267; RRID: AB_2895042 |
| Rabbit Monoclonal anti-Phospho AKT (Ser473) | Cell Signaling Technology | Cat#4060; RRID: AB_2315049 |
| Rabbit Monoclonal anti-AKT | Cell Signaling Technology | Cat#4691; RRID: AB_915783 |
| Rabbit Monoclonal anti-Phospho S6 Ribosomal Protein (Ser240/244) for WB | Cell Signaling Technology | Cat#5364; RRID: AB_10694233 |
| Rabbit Monoclonal anti-S6 Ribosomal Protein | Cell Signaling Technology | Cat#2217; RRID: AB_331355 |
| Rabbit Polyclonal anti-Villin-1 | Cell Signaling Technology | Cat#2369; RRID: AB_2215958 |
| Rat Monoclonal anti-CD45 (30-F11), Alexa Fluor™ 700 | BioLegend | Cat#103128 |
| Chemicals, peptides, and recombinant proteins | ||
| Isoflurane | Piramal Healthcare | Cat# NDC66794-017-25 |
| Tamoxifen for In vivo injections | MP Biomedicals | Cat# ICN15673880 |
| Sunflower oil | Sigma Aldrich | Cat# S5007 |
| Matrigel | Corning | Cat# 356231 |
| 4-hydroxy tamoxifen for In vitro | Sigma Aldrich | Cat #H7904 |
| Rapamycin | Sigma Aldrich | Cat#R0395 |
| Akt-inhibitor, MK2206 2HCl | Selleckchem.com | Cat#s1078 |
| Formalin solution, neutral buffered, 10% | Sigma Aldrich | Cat# HT501128 |
| Antigen unmasking solution | Vector Laboratories | Cat#H-3300-250 |
| Tween 20 | BioRad | Cat#1610781 |
| Goat serum | Sigma Aldrich | Cat#G9023 |
| 3,3′-diaminobenzidine, DAB | Vector Laboratories | Cat#SK-4100 |
| Nuclear fast red counter stain | Vector Laboratories | Cat#H3403-500 |
| Anti-rabbit secondary antibody conjugated to a dextran-labeled polymer and HRP | DakoCytomation | Cat#K4003 |
| ROCK inhibitor, Y-27632 dihydrochloride | TOCRIS | Cat#1254 |
| TrypLE™ Express Enzyme (1X), no phenol red | Gibco, Thermo Fisher Scientific | Cat#12604013 |
| N-acetylcysteine | Sigma Aldrich | Cat#A7250 |
| DNase-I | Stem Cell Technologies | Cat#07900 |
| N-2 Supplement (100X) | Gibco, Thermo Fisher Scientific | Cat#17502048 |
| B-27™ Supplement (50X), minus vitamin A | Gibco, Thermo Fisher Scientific | Cat#12587010 |
| GlutaMAX™-I Supplement | Gibco, Thermo Fisher Scientific | Cat#35050061 |
| Propidium iodide solution | BioLegend | Cat#421301 |
| Recombinant Murine EGF | PeproTech | Cat#31509 |
| Recombinant Murine Noggin | PeproTech | Cat#25038 |
| Recombinant Murine Rspondin-1 | PeproTech | Cat#31532 |
| Recombinant Murine Wnt3a | PeproTech | Cat#31520 |
| Critical commercial assays | ||
| Mouse on Mouse (MOM) kit, Mouse Ig Blocking Reagent | Vector Laboratories | Cat#PK-2200 |
| The ABC amplification IHC kit | Vector Laboratories | Cat#PK-6100 |
| RNAqueous®-Micro Total RNA Isolation Kit | Thermo Fisher Scientific | Cat # AM1931 |
| QuantiTect Reverse Transcription kit (50) | QIAGEN | Cat#205311 |
| QuantiTect SYBR® Green PCR Kit (200) | QIAGEN | Cat#204143 |
| ECL Western Blotting Substrate Kit | Thermo Fisher Scientific | Cat#32209 |
| The Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23227 |
| Deposited data | ||
| Single Cell RNAseq Raw and analyzed data | This paper | GEO: GSE2361863 |
| Experimental models: Cell lines | ||
| HEK293T cells overexpressing recombinant R-spondin1 | a gift from Dr. Mark R. Frey, Keck School of Medicine, University of Southern California | NA |
| L Wnt3a Cell line | ATCC | Cat#CRL-2647 |
| Experimental models: Organisms/strains | ||
| Mouse: Lgr5-EGFP-IRES-creERT2 knock-in | Jackson Laboratory | Stock No: 008875 |
| Mouse: Rosa26-LacZ reporter | Jackson Laboratory | Stock No: 003474 |
| Mouse: Cblflox/flox; Cblb−/−l; Lgr5-creERT2; R26-LacZ (iDKO) | Mohapatra et al.19 | N/A |
| Mouse: Cbl−/− | Gu et al.7 | N/A |
| Mouse: Cblb−/− | Chiang et al.10 | N/A |
| Mouse: Cblflox/flox; Cblbflox/flox (FF Control) | Goetz et al.14 | N/A |
| Mouse: Rosa26-CreERT2 | Jackson Laboratory | Stock No: 008463 |
| Mouse: mT/mG-Reporter | Jackson Laboratory | Stock No: 007676 |
| Mouse: Cblflox/flox; Cblbflox/flox; R26CreERT2; mT-mG (FF CreRT) | Mohapatra et al.19 | N/A |
| Oligonucleotides | ||
| Primers for PCR-based genotyping of various alleles listed in Table S1 | This paper | N/A |
| Real time qPCR primers used to detect mRNA levels are listed in Table S2 | This paper | N/A |
| Software and algorithms | ||
| ImageJ software | NIH | https://imagej.net/ij/ |
| FACS DIVA flow cytometry analysis | BD | https://www.bdbiosciences.com/en-us/products/software/instrument-software/bd-facsdiva-software |
| Seurat for t-SNE clustering of genes in single cell RNAseq with R package | Butler et al.110 | https://github.com/satijalab/seurat |
| GraphPad Prism for statistical Analysis | GraphPad Prism 10.0.3 | https://www.graphpad.com/ |
| BioRender for making graphical abstract | BioRender | https://app.biorender.com/illustrations/6516ed04964eb05e9a6e30f7 |
Resource availability
Lead contact
Data are available by request to the lead contact. The correspondence should be addressed to Hamid Band (hband@unmc.edu).
Materials availability
This study did not generate any new mouse model or unique reagent. The mouse models used in this study are already published and available upon request to the lead contact.
Data and code availability
-
•
Single cell RNAseq data used in this publication have been deposited in NCBI’s Gene Expression Omnibus and are publicly accessible through GEO series accession number GSE236186.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Animals
The source of mice used in this study is provided in the key resources table. All mouse strains were maintained on a C57BL/6J background under specific pathogen-free conditions. The Institutional Animal Care and Use Committee (IACUC) of UNMC approved all the mice experiments (IACUC Protocol#19-042-05-FC and # 17-137-00-FC).
Cell lines
HEK293T cells overexpressing recombinant R-spondin1 was a gift from Dr. Mark R. Frey, Keck School of Medicine, University of Southern California and L cells overexpressing Wnt-3a was purchased from ATCC. R-spondin secreting HEK293T cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS (Fetal Bovine Serum) and 1% penicillin/streptomycin. L cells secreting Wnt-3a were cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin and 0.4 mg/mL G-418. Plated cells were incubated at 37oC in 5% CO2 tissue culture incubators.
Conditional media collected from the above-mentioned cell lines were used for organoid culture.
Method details
Mouse strains
The Lgr5-EGFP-IRES-creERT2 knock-in (Lgr5tm1(cre/ERT2)Cle; Stock No: 008875) and Rosa26-LacZ reporter (B6.129S4-Gt(ROSA)26Sortm1Sor/J; Stock No: 003474; aka R26R) mice were purchased from The Jackson Laboratory and interbred; this genotype served as a control. These mice were further bred with Cblflox/flox; Cblb−/− mice15 to obtain Cblflox/flox; Cblb−/−l; Lgr5-creERT2; R26-LacZ (iDKO) experimental mice for tamoxifen-inducible intestinal stem cell specific deletion of Cbl on a background of germline Cbl-b deletion. The Cbl−/−7 and Cblb−/−10 mice used to examine the function of CBL and CBL-B individually have been reported previously. Age and gender matched wild-type (WT) C57BL/6J mice served as controls. Cblflox/flox; Cblbflox/flox (Conditional Cbl/Cblb double floxed (FF control) mice generated in our lab14 were crossed to Rosa26-CreERT2 (B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J; Stock No: 008463; aka R26-CreERT2; The Jackson Laboratory) and mT/mG-Reporter mice (B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J; Stock No: 007676; aka mT/mG, mTmG; The Jackson Laboratory) to obtain the Cblflox/flox; Cblbflox/flox; R26CreERT2; mT-mG model used to isolate intestinal crypts for organoid culture studies. All mouse strains were maintained on a C57BL/6J background under specific pathogen-free conditions and handled in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of UNMC. All mice used in this study were euthanized by first anesthetizing using isoflurane (Piramal Healthcare, Catalog no. NDC66794-017-25) followed by cervical dislocation to confirm death. Mice were genotyped using tail DNA PCR (primer sequences in Table S1).
Tamoxifen-induced cre activation
For in vivo deletion of floxed Cbl alleles, Tamoxifen free base (MP Biomedicals, Catalog no. ICN15673880) was resuspended in sunflower oil (Sigma Aldrich, Catalog no. S5007) at a concentration of 10 mg/mL. At 6 weeks of age, gender matched Lgr5-EGFP-IRES-creERT2; R26-LacZ (WT-Control) and Cblflox/flox; Cblb−/−l; Lgr5-creERT2; R26-LacZ (iDKO) mice were given an intra-peritoneal injection of 1mg tamoxifen for 3 consecutive days to activate inducible Cre recombinase. Animals were analyzed 3 days after the last injection (5 days after the first injection), 8 days after the last injection (10 days after the first) or 4 months after the last injection. For in vitro activation of Cre, crypts were isolated from FF control and FFcreERT mice and equal numbers were plated in Matrigel (Corning, catalog# 356231) under conditions as described34 to obtain organoids by day 7. The organoids thus generated were re-plated at 1:4 split ratio and exposed to 400 nM 4-hydroxy tamoxifen (Sigma; Catalog #H7904) in medium to achieve the deletion of Cbl and Cblb. For rescue experiments, FF control and FFcreERT organoids were treated with Rapamycin (Sigma, Catalog#R0395) and Akt-inhibitor, MK2206 2HCl (Selleckchem.com, Catalog#s1078) alongside 4-OH-tamoxifen.
Tissue harvest and histological analysis
Mouse small intestine was carefully excised from the peritoneal cavity by first snipping at the ileo-cecal junction and pulling away to disentangle the intestine from the mesh of mesentery and finally cutting at the pyloric end of stomach. The intestine was immediately placed in a Petri dish, and gently flushed with cold phosphate buffered saline (PBS) to remove fecal matter using a 30 mL syringe and a blunt ended needle. Once cleaned, intestine was placed on a blotting sheet, cut open longitudinally, Swiss-rolled and placed in formalin (Sigma Aldrich, Catalog # HT501128) for overnight fixation at room temperature. Fixed tissue was embedded in paraffin and sectioned to 5 μM thickness.
Immunofluorescence (IF)
Tissue sections were deparaffinized and rehydrated followed by antigen retrieval in sodium citrate antigen unmasking solution (Vector Laboratories; catalog #H-3300-250) by heating in a microwave oven for 20 min. Slides were then washed in PBS and blocked for 1h in 20 mm Tris-Buffered Saline with 0.1% Tween 20 (BioRad, catalog #1610781) (TBS-T) containing 5% goat serum (Sigma Aldrich, G9023) followed by overnight incubation at 4°C with optimized concentrations of primary antibodies [(BrdU, Developmental Studies Hybridoma Bank, Iowa, G3G4,1:25), Chromogranin A (Abcam, Catalog No.15160, 1:200), Cleaved Caspase 3 (CST, Cat# 9664, 1:200), phospho-histone 3 (Ser10) (Cat# 9701, CST, 1:200), GFP (CST, Cat# 2956, 1:200), GFP (Aves Labs, Chicken, Cat#GFP-1010, 1:2000), phospho-S6 (CST, Cat# 4858,dilution 1:200). Tissue sections were then washed thrice in PBS for 5 min each, followed by incubation with fluorescence-tagged secondary antibodies (Molecular Probes Alexa series, all 1:400 in PBS) for 1h at room temperature, washed 3X with PBS and mounted with Vectashield and DAPI to stain nuclei (Vector Laboratories).
Immunohistochemistry (IHC)
Tissue sections were deparaffinized and rehydrated followed by antigen retrieval in sodium citrate antigen unmasking solution (Vector Laboratories) using microwaving for 20 min. Slides were then washed in PBS and incubated in 5% H2O2 for 30 min at room temperature to inactivate endogenous peroxidase. The slides were washed with PBS and incubated with the following blocking solutions for 1 h at room temperature: Mouse Ig Blocking Reagent in Mouse on Mouse (MOM) kit from Vector Laboratories (PK-2200) for staining with antibodies against Cbl (mouse monoclonal, anti-Cbl, BD Bioscience, 1:400) and Cbl-b (mouse monoclonal, anti-Cbl-b, Abcam, 1:100) and 5% goat serum (Sigma Aldrich, G9023) for Chromogranin A (Cat#15160, Abcam,1:200) and Ki67 (Rat monoclonal, eBioscience, 145698-82, 1:200). Slides were washed twice with PBS and incubated with primary antibodies indicated above at 4°C overnight. The next day, slides were washed thrice with PBS for 5 min each and incubated with species-matched biotinylated secondary antibodies for 30 min. The ABC amplification IHC kit (Vector Laboratories, PK-6100) was used as per vendor’s instructions to amplify the signal of Cbl and Cbl-b staining. Slides were washed twice, and peroxidase-based signals were developed using DAB (3,3′-diaminobenzidine, Vector Laboratories, SK-4100) as substrate. The slides were counterstained with hematoxylin, dehydrated and mounted in automated counter-stainer (Sakura Tissue-tek Prisma) at the UNMC Tissue Science Core Facility. Muc2 (rabbit polyclonal, Invitrogen#PA5-21329, 1:500), Lysozyme (rabbit monoclonal, Abcam, ab108508, 1:400) and EGFR (rabbit monoclonal, CST#4267, 1:500) IHC staining were performed with minor modifications. The blocking was performed using 5% goat serum (Sigma Aldrich, G9023) in PBS after peroxidase blocking. The sections were then stained with primary antibodies overnight at 4oC. After rinsing in TBST, the sections were incubated for 15 min with an anti-rabbit secondary antibody conjugated to a dextran-labeled polymer and HRP (DakoCytomation K4003), followed by incubation in DAB solution (3,3′-diaminobenzidine, Vector Laboratories, SK-4100) for 5 min. Counterstaining and mounting was carried out as above.
β-Galactosidase (LacZ) staining
The β-galactosidase (LacZ) staining for lineage tracing was performed in intestinal sections of mice with a lacZ reporter, as described.26 Briefly, freshly dissected intestinal tissue was flushed clean with cold PBS and fixed in glutaraldehyde fixative (1% formaldehyde, 0.2% glutaraldehyde, 0.02% NP-40 in PBS) for 2 h at 4°C. Tissue was washed twice with PBS and incubated in equilibration buffer (2 mM MgCl2, 0.02% NP-40, 0.01% sodium deoxycholate in PBS) for 30 min at room temperature. Subsequently, the tissue was incubated in LacZ substrate [5mM K3Fe(CN)6, 5mM K4Fe(CN)6, 2mM MgCl2, 0.02% NP-40, 0.1% sodium deoxycholate, 1 mg/mL X-gal in PBS] for 4 h at 37°C, protected from light. Once the blue color developed, tissue was washed twice with PBS, Swiss-rolled, and incubated in 4% paraformaldehyde at 4°C overnight. The fixative was then removed, and tissue was washed with PBS and dehydrated in alcohol gradient (70%, 96% and 100% for 1 h each) and paraffin embedded. 5 μM tissue sections were cut, paraffin cleared and rehydrated, followed by either counterstaining with nuclear fast red (Vector Labs), PAS staining or IHC as indicated above.
Intestinal epithelial stem cell (ISC) isolation and flow cytometric analysis
A modification of the Intestinal Stem Cell Consortium protocol109 was used to obtain stem cell-enriched single cell suspensions of intestinal epithelium. Briefly, freshly dissected intestinal tissue was flushed clean with ice-cold wash buffer (100 μg/mL penicillin/streptomycin, 10mM HEPES, 2mM Glutamax in HBSS) and cut open longitudinally to expose the lumen. A glass coverslip was used to scrape off the villi and the tissue chopped into 5 mm long pieces. The chopped pieces were then washed several times with cold wash buffer by vigorously pipetting until the supernatant was almost clear. After this, the tissue was placed in dissociation reagent #1 [15 mM EDTA, 1.5 mM DTT, 10 μM ROCK inhibitor (Y27632, TOCRIS), 100 μg/mL penicillin/streptomycin, 10mM HEPES, 2mM Glutamax in HBSS] and rocked slowly at 4°C for 30 min. Supernatant was removed and replaced with fresh cold wash buffer and the tubes were shaken vigorously. The tissue pieces were allowed to settle down, and the supernatant was passed through 70 μM cell strainers and centrifuged at 1200 rpm for 5 min at 4°C. The pellet was resuspended in 10 mL wash buffer and centrifuged at 600 rpm for 2 min at 4°C. The cell pellet obtained was resuspended in dissociation reagent # 2 [TrypLE Express (Thermo Scientific), 10μM ROCK inhibitor (Y27632, TOCRIS), 0.5 mM N-acetylcysteine (Sigma, Catalog#A7250), and 200 μg/mL DNase-I (Stem Cell Technologies, Catalog#07900) in Intestinal Epithelial Stem Cell Media (ISC Media) [Advanced DMEM/F12 supplemented with 1XN2 (Gibco, Catalog#17502048), 1XB27 without vitamin A (Gibco, Catalog#12587010), 10 mM HEPES, 2mM Glutamax (Gibco, Catalog#35050061),100 μg/mL penicillin/streptomycin] and incubated for 8 min at 37°C, with intermittent pipetting to reduced cell clumping. Equal volume of neutralization buffer (10% FBS in ISC Media) was added, and the cell suspension was passed through 20 μM cell strainers. The cells were then centrifuged at 1000 rpm for 5 min at 4°C. The supernatant was discarded, and cells were resuspended in 1 mL of cold ISC medium and cell numbers counted to assess the yield. The isolated cells were stained with Alexa 700 conjugated anti-CD45 antibody (BioLegend, Catalog No. 103128, 0.25 μg per 106 cells) at 4°C for 30 min, washed twice and propidium iodide (BioLegend, Catalog No. 421301, 5 μg per 106 cells) added before FACS analysis on a BD LSR2 instrument. FACS data were analyzed using BD FACS DIVA software.
RNA isolation, cDNA synthesis and quantitative PCR analysis
Cell analyzed by flow cytometry were directly sorted into 500 μL of RNA lysis buffer of the RNAqueous-Micro Total RNA Isolation Kit from Thermo Scientific (Catalog # AM1931). Instructions from the kit were followed to isolate the RNA. The RNA yield was assessed using a Nanodrop instrument and cDNA was synthesized using a kit from QIAGEN (QuantiTect Reverse Transcription kit, catalog # 205311). QuantiTect SYBR Green PCR Kit was used to perform real-time PCR analysis on a Bio-Rad CFX-96 Thermo Cycler. Sequences of primers (obtained from Sigma-Aldrich) are listed in Table S2. Relative gene expression was calculated according to 2ΔΔCt method.
Droplet-based scRNA-seq
Single cell gene expression profiling was performed as per manufacturer’s protocol (10X genomics, Pleasanton, CA) by the UNMC Genomics core facility. Briefly, intestinal crypts were isolated from two 10-week-old Cblflox/flox; Cbl-bflox/flox; R26CreERT2 mice and grown into intestinal organoids using our established protocol. The organoids were replated and grown in presence or absence of 4-hydroxy-tamoxifen (400 nM) for 3 days to delete Cbl and Cblb. Organoids were dissociated into single cells. The single cells were checked for viability and processed through the GemCode Single Cell Platform. An input of 7,000 cells was added to each channel of a chip with a recovery rate of 1,200 cells on average. The cells were then partitioned into Gel Beads in Emulsion (GEMs) in the GemCode instrument, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, shearing and 5′ adaptor and sample index attachment. Libraries were sequenced on an Illumina NextSeq 500 in the Genomic Core facility, UNMC. Demultiplexing, alignment to the transcriptome and UMI-collapsing were performed through 10X genomics recommended cell ranger standard pipeline (Cell Ranger toolkit, version 3.0.1) by the Bioinformatics and Systems Biology Core at UNMC. Seurat was used to discard low quality cells with aberrant gene expression, normalization, identification of highly variable features, scaling and linear dimensionality reduction. Subsequently clustering the cells was carried out by t-distributed stochastic neighbor embedding (t-SNE) method.110 Cluster markers were then identified in Seurat by inspecting differentially expressed features among the clusters.61 Signaling pathways were analyzed using Gene Set Enrichment Analysis (GSEA). The gene set enrichment score was calculated using the Bioconductor GSVA package and the plots were generated using R.
Lysate preparation and western blot
Freshly-excised, cleaned and chopped fragments of intestine were incubated in 2 mM EDTA in cold PBS at 4°C for 30 min with vigorous shaking to detach the intestinal epithelium from the remaining tissue. Once the epithelial layer was separated, the supernatant containing the detached epithelium was pelleted and homogenized in RIPA lysis buffer containing protease and phosphatase inhibitors. Intestinal organoid lysates were also prepared in the same lysis buffer. The lysate proteins were quantified using the BCA kit from Thermo Scientific. 40 μg of lysate protein aliquots were resolved on 8% SDS–PAGE and transferred to PVDF membranes, which were then blocked with 5% BSA in TBS-T and incubated overnight at 4°C with specific antibodies diluted in TBS-T. The following antibodies were procured from commercial sources: Anti-Cbl (Clone 17/c-Cbl, BD Bioscience); anti-Cbl-b (Clone D3C12, Cell Signaling Technology); anti-HSC70 (Clone B6, Santa Cruz Biotechnology Inc. Santa Cruz, CA); phospho EGFR (Y1068) (Cat# 2234, Cell Signaling Technology); EGFR (Cat#4267, Cell Signaling Technology); phospho Akt (Ser473) (Cat# 4060, Cell Signaling Technology); Akt (pan, Cat# 4691, Cell Signaling Technology); phospho S6 (Ser240/244), Cat# 5364, Cell Signaling Technology); Total S6 (5G10, Cat#2217, Cell Signaling Technology), phospho-histone 3 (Ser10) (Cat# 9701, Cell Signaling Technology), and Villin-1 (Cat# 2369, Cell Signaling Technology). Membranes were washed 3X in TBS-T, incubated with HRP-conjugated species-specific secondary antibodies (Zymed Laboratories) and signals were detected with Pierce ECL substrate (Thermo scientific).
Focused beam abdominal radiation
Mouse irradiation was accomplished on the TrueBeam STx linear accelerator (Varian Medical Systems, Palo Alto, CA) in the Radiation Oncology Department at UNMC. Mice were first sedated with Ketamine/Xylazine, restrained to prevent movement to ensure reproducible positioning for radiation and simulated using the simulation CT scanner. The radiation target abdominal region was contoured by an attending radiation oncologist. A specific treatment plan was created for each mouse by the medical physicists, and mice were treated to a dose of 14 Gy photon radiation, deemed optimal based on initial dose titrations (8, 10, 12 or 14 Gray).
Organoid culture
A modification of the organoid protocol established by Sato et al.34 was used to prepare organoid cultures from crypts of Cblflox/flox; Cblbflox/flox; R26CreERT2 mice. Freshly dissected, cleaned and chopped fragments of ileum were incubated in 2 mM EDTA in cold PBS at 4°C for 30 min in a 15-mL Falcon tube. Tissue fragments were allowed to settle down, the supernatant was replaced with wash buffer (described under isolation of ISC) and the tube was shaken by hand for 3 min to release the epithelial layer from the remaining tissue. The supernatant was passed through 70 μm cell strainer and the strainers further rinsed with fresh wash buffer. The crypts were counted under the microscope and aliquots containing 500 crypts was pelleted. The supernatant was removed, and the crypt pellets were resuspended in Matrigel (BD Bioscience, Catalog No. 356237) mixed with growth factors (EGF 50 ng/mL; Noggin 100 ng/mL; R-spondin 500 ng/mL) and plated in wells of 24 well plates (Costar). The plates were kept at 37°C for 15 min to allow the Matrigel to polymerize. 500 μL of ISC medium was added and plates were incubated at 37°C in 5% CO2 tissue culture incubators. On the 4th day after plating, the spent medium was replaced with ISC medium containing EGF, Noggin, R-spondin1 conditioned medium (1:10) and Wnt-3a conditioned medium (1:10). The conditioned media were obtained from late log phase cultures of HEK293T cells overexpressing recombinant R-spondin1 (a gift from Dr. Mark R. Frey, Keck School of Medicine, University of Southern California) or L cells overexpressing Wnt-3a (ATCC). Murine Wnt-3a was purchased from Peprotech (Cat#315-20).
Fractionation of mouse crypt and villus
Biochemical fractionation of mouse crypt and villus was carried out as described by Mahmoudi et al.111 Briefly, freshly dissected small intestine of 12-week-old wildtype, Cbl−/− and Cblb−/− mice were cleaned and flushed with ice-cold PBS several times and cut open longitudinally to expose crypts and villi. Small intestine was chopped into small 1–2 cm fragments and incubated in 2mM EDTA in cold Ca2+/Mg2+ free PBS (Chelation buffer) at 4°C for 20 min. Tissue fragments were allowed to be settled down and the supernatant was replaced with fresh chelation buffer and the tube was shaken vigorously for 10 min to release the epithelial layer from the remaining tissue. This process was repeated, and the fractions were passed through 70 μm strainer. The villus structures remain on top of the cell strainer and cells were collected, whereas the flow through was discarded. The process of incubation, shaking and separation through the cell strainer was repeated. The fractions 2–4 was collected for pure villi. The intact crypts remain in flow through after fifth fraction. The purified villi and crypts were washed with cold PBS and protein lysates were prepared.
Quantification and statistical analyses
Quantified results of LacZ stained sections, qPCR, immunohistochemistry, and flow cytometry were compared between groups using the Student’s t-test on GraphPad Prism, and were presented as mean ± SEM, with p ≤ 0.05 deemed as significant. A supplemental excel file (Data S1) with all statistical information related to the figures has been provided.
Acknowledgments
We thank the Band Lab members for their many discussions, Drs. Rakesh Singh and Karen Gould for advice, and the staff of UNMC Core facilities for their invaluable assistance. We thank Dr. Mark R. Frey, Keck School of Medicine, University of Southern California, for providing the R-spondin-1 secreting cell line. This work was supported by the National Institutes of Health (CA105489, CA99163 to H.B.); the Nebraska Department of Health and Human Services LB606 (18123-Y3 to H.B.); a pilot grant from the UNMC Regenerative Medicine Program; a Core Facility usage grant for RNA-seq analysis from the Nebraska Research Initiative; support to the UNMC Confocal, Flow Cytometry, Bioinformatics, Molecular Biology, and other Core facilities from the NCI Cancer Center Support Grant to the Fred & Pamela Buffett Cancer Center (P30CA036727), and the Nebraska Research Initiative. We thank the Dr. Raphael Bonita Fund for partial support of graduate students in the H.B. lab. N.Z., B.C.M., and W.A. received University of Nebraska Medical Center graduate assistantships. B.T.G. was a trainee under the National Cancer Institute Cancer Biology Training Grant T32CA009476.
Author contributions
H.B., V.B., N.Z., and B.C.M. conceived the project design. H.B. and V.B. supervised the project and obtained resources to support the study. N.Z., B.C.M., W.A., and B.T.G. developed methodologies and/or acquired data. P.M. and S.P.T. analyzed the single-cell RNA-seq data. S.W., S.L., and C.L. performed and analyzed focused beam abdominal radiation experiments. H.B., V.B., C.L., J.D.B., S.P.T., A.R.B., and D.F.M. analyzed data. N.Z. and B.C.M. wrote the manuscript, and H.B., V.B., C.L., J.D.B., S.P.T., and D.F.M. edited the manuscript. M.D.S. provided key technical support.
Declaration of interests
H.B. and V.B. received funding from Nimbus Therapeutics for an unrelated project.
Published: May 8, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109912.
Contributor Information
Vimla Band, Email: vband@unmc.edu.
Hamid Band, Email: hband@unmc.edu.
Supplemental information
References
- 1.Mele S., Johnson T.K. Receptor Tyrosine Kinases in Development: Insights from Drosophila. Int. J. Mol. Sci. 2019;21:188. doi: 10.3390/ijms21010188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramachandra C.J.A., Mehta A., Lua C.H., Chitre A., Ja K.P.M.M., Shim W. ErbB Receptor Tyrosine Kinase: A Molecular Switch Between Cardiac and Neuroectoderm Specification in Human Pluripotent Stem Cells. Stem Cell. 2016;34:2461–2470. doi: 10.1002/stem.2420. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi T., Shiraishi A. Stem Cell Signaling Pathways in the Small Intestine. Int. J. Mol. Sci. 2020;21:2032. doi: 10.3390/ijms21062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohapatra B., Ahmad G., Nadeau S., Zutshi N., An W., Scheffe S., Dong L., Feng D., Goetz B., Arya P., et al. Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl-family ubiquitin ligases. Biochim. Biophys. Acta. 2013;1833:122–139. doi: 10.1016/j.bbamcr.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths E.K., Sanchez O., Mill P., Krawczyk C., Hojilla C.V., Rubin E., Nau M.M., Khokha R., Lipkowitz S., Hui C.C., Penninger J.M. Cbl-3-deficient mice exhibit normal epithelial development. Mol. Cell Biol. 2003;23:7708–7718. doi: 10.1128/MCB.23.21.7708-7718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murphy M.A., Schnall R.G., Venter D.J., Barnett L., Bertoncello I., Thien C.B., Langdon W.Y., Bowtell D.D. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol. Cell Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naramura M., Kole H.K., Hu R.J., Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. USA. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Chami N., Ikhlef F., Kaszas K., Yakoub S., Tabone E., Siddeek B., Cunha S., Beaudoin C., Morel L., Benahmed M., Régnier D.C. Androgen-dependent apoptosis in male germ cells is regulated through the proto-oncoprotein Cbl. J. Cell Biol. 2005;171:651–661. doi: 10.1083/jcb.200507076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molero J.C., Jensen T.E., Withers P.C., Couzens M., Herzog H., Thien C.B.F., Langdon W.Y., Walder K., Murphy M.A., Bowtell D.D.L., et al. c-Cbl-deficient mice have reduced adiposity, higher energy expenditure, and improved peripheral insulin action. J. Clin. Invest. 2004;114:1326–1333. doi: 10.1172/JCI21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiang Y.J., Kole H.K., Brown K., Naramura M., Fukuhara S., Hu R.J., Jang I.K., Gutkind J.S., Shevach E., Gu H. Cbl-b regulates the CD28 dependence of T-cell activation. Nature. 2000;403:216–220. doi: 10.1038/35003235. [DOI] [PubMed] [Google Scholar]
- 11.Bachmaier K., Krawczyk C., Kozieradzki I., Kong Y.Y., Sasaki T., Oliveira-dos-Santos A., Mariathasan S., Bouchard D., Wakeham A., Itie A., et al. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature. 2000;403:211–216. doi: 10.1038/35003228. [DOI] [PubMed] [Google Scholar]
- 12.Nakao R., Hirasaka K., Goto J., Ishidoh K., Yamada C., Ohno A., Okumura Y., Nonaka I., Yasutomo K., Baldwin K.M., et al. Ubiquitin ligase Cbl-b is a negative regulator for insulin-like growth factor 1 signaling during muscle atrophy caused by unloading. Mol. Cell Biol. 2009;29:4798–4811. doi: 10.1128/MCB.01347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naramura M., Jang I.K., Kole H., Huang F., Haines D., Gu H. c-Cbl and Cbl-b regulate T cell responsiveness by promoting ligand-induced TCR down-modulation. Nat. Immunol. 2002;3:1192–1199. doi: 10.1038/ni855. [DOI] [PubMed] [Google Scholar]
- 14.Goetz B., An W., Mohapatra B., Zutshi N., Iseka F., Storck M.D., Meza J., Sheinin Y., Band V., Band H. A novel CBL-Bflox/flox mouse model allows tissue-selective fully conditional CBL/CBL-B double-knockout: CD4-Cre mediated CBL/CBL-B deletion occurs in both T-cells and hematopoietic stem cells. Oncotarget. 2016;7:51107–51123. doi: 10.18632/oncotarget.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naramura M., Nandwani N., Gu H., Band V., Band H. Rapidly fatal myeloproliferative disorders in mice with deletion of Casitas B-cell lymphoma (Cbl) and Cbl-b in hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 2010;107:16274–16279. doi: 10.1073/pnas.1007575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An W., Nadeau S.A., Mohapatra B.C., Feng D., Zutshi N., Storck M.D., Arya P., Talmadge J.E., Meza J.L., Band V., Band H. Loss of Cbl and Cbl-b ubiquitin ligases abrogates hematopoietic stem cell quiescence and sensitizes leukemic disease to chemotherapy. Oncotarget. 2015;6:10498–10509. doi: 10.18632/oncotarget.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An W., Mohapatra B.C., Zutshi N., Bielecki T.A., Goez B.T., Luan H., Iseka F., Mushtaq I., Storck M.D., Band V., Band H. VAV1-Cre mediated hematopoietic deletion of CBL and CBL-B leads to JMML-like aggressive early-neonatal myeloproliferative disease. Oncotarget. 2016;7:59006–59016. doi: 10.18632/oncotarget.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferron S.R., Pozo N., Laguna A., Aranda S., Porlan E., Moreno M., Fillat C., de la Luna S., Sánchez P., Arbonés M.L., Fariñas I. Regulated segregation of kinase Dyrk1A during asymmetric neural stem cell division is critical for EGFR-mediated biased signaling. Cell Stem Cell. 2010;7:367–379. doi: 10.1016/j.stem.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 19.Mohapatra B., Zutshi N., An W., Goetz B., Arya P., Bielecki T.A., Mushtaq I., Storck M.D., Meza J.L., Band V., Band H. An essential role of CBL and CBL-B ubiquitin ligases in mammary stem cell maintenance. Development. 2017;144:1072–1086. doi: 10.1242/dev.138164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 21.Andersson-Rolf A., Zilbauer M., Koo B.K., Clevers H. Stem Cells in Repair of Gastrointestinal Epithelia. Physiology. 2017;32:278–289. doi: 10.1152/physiol.00005.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeda N., Jain R., LeBoeuf M.R., Wang Q., Lu M.M., Epstein J.A. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Es J.H., Sato T., van de Wetering M., Lyubimova A., Yee Nee A.N., Gregorieff A., Sasaki N., Zeinstra L., van den Born M., Korving J., et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buczacki S.J.A., Zecchini H.I., Nicholson A.M., Russell R., Vermeulen L., Kemp R., Winton D.J. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 25.Metcalfe C., Kljavin N.M., Ybarra R., de Sauvage F.J. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Barker N., van Es J.H., Kuipers J., Kujala P., van den Born M., Cozijnsen M., Haegebarth A., Korving J., Begthel H., Peters P.J., Clevers H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 27.Muñoz J., Stange D.E., Schepers A.G., van de Wetering M., Koo B.K., Itzkovitz S., Volckmann R., Kung K.S., Koster J., Radulescu S., et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barker N., Ridgway R.A., van Es J.H., van de Wetering M., Begthel H., van den Born M., Danenberg E., Clarke A.R., Sansom O.J., Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 29.Carmon K.S., Gong X., Lin Q., Thomas A., Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 31.Tan S., Barker N. Epithelial stem cells and intestinal cancer. Semin. Cancer Biol. 2015;32:40–53. doi: 10.1016/j.semcancer.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Miettinen P.J., Berger J.E., Meneses J., Phung Y., Pedersen R.A., Werb Z., Derynck R. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- 33.Sibilia M., Wagner E.F. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 34.Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheung Q.C.K., Yuan Z., Dyce P.W., Wu D., DeLange K., Li J. Generation of epidermal growth factor-expressing Lactococcus lactis and its enhancement on intestinal development and growth of early-weaned mice. Am. J. Clin. Nutr. 2009;89:871–879. doi: 10.3945/ajcn.2008.27073. [DOI] [PubMed] [Google Scholar]
- 36.Basak O., Beumer J., Wiebrands K., Seno H., van Oudenaarden A., Clevers H. Induced Quiescence of Lgr5+ Stem Cells in Intestinal Organoids Enables Differentiation of Hormone-Producing Enteroendocrine Cells. Cell Stem Cell. 2017;20:177–190.e4. doi: 10.1016/j.stem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Batlle E., Henderson J.T., Beghtel H., van den Born M.M.W., Sancho E., Huls G., Meeldijk J., Robertson J., van de Wetering M., Pawson T., Clevers H. Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111:251–263. doi: 10.1016/s0092-8674(02)01015-2. [DOI] [PubMed] [Google Scholar]
- 38.Holmberg J., Genander M., Halford M.M., Annerén C., Sondell M., Chumley M.J., Silvany R.E., Henkemeyer M., Frisén J. EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell. 2006;125:1151–1163. doi: 10.1016/j.cell.2006.04.030. [DOI] [PubMed] [Google Scholar]
- 39.Vidrich A., Buzan J.M., Brodrick B., Ilo C., Bradley L., Fendig K.S., Sturgill T., Cohn S.M. Fibroblast growth factor receptor-3 regulates Paneth cell lineage allocation and accrual of epithelial stem cells during murine intestinal development. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;297:G168–G178. doi: 10.1152/ajpgi.90589.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al Alam D., Danopoulos S., Schall K., Sala F.G., Almohazey D., Fernandez G.E., Georgia S., Frey M.R., Ford H.R., Grikscheit T., Bellusci S. Fibroblast growth factor 10 alters the balance between goblet and Paneth cells in the adult mouse small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2015;308:G678–G690. doi: 10.1152/ajpgi.00158.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freier S., Eran M., Reinus C., Ariel I., Faber J., Wilschanski M., Braverman D. Relative expression and localization of the insulin-like growth factor system components in the fetal, child and adult intestine. J. Pediatr. Gastroenterol. Nutr. 2005;40:202–209. doi: 10.1097/00005176-200502000-00023. [DOI] [PubMed] [Google Scholar]
- 42.Powell A.E., Wang Y., Li Y., Poulin E.J., Means A.L., Washington M.K., Higginbotham J.N., Juchheim A., Prasad N., Levy S.E., et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong V.W.Y., Stange D.E., Page M.E., Buczacki S., Wabik A., Itami S., van de Wetering M., Poulsom R., Wright N.A., Trotter M.W.B., et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat. Cell Biol. 2012;14:401–408. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heuberger J., Kosel F., Qi J., Grossmann K.S., Rajewsky K., Birchmeier W. Shp2/MAPK signaling controls goblet/paneth cell fate decisions in the intestine. Proc. Natl. Acad. Sci. USA. 2014;111:3472–3477. doi: 10.1073/pnas.1309342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gur G., Rubin C., Katz M., Amit I., Citri A., Nilsson J., Amariglio N., Henriksson R., Rechavi G., Hedman H., et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Landeghem L., Santoro M.A., Mah A.T., Krebs A.E., Dehmer J.J., McNaughton K.K., Helmrath M.A., Magness S.T., Lund P.K. IGF1 stimulates crypt expansion via differential activation of 2 intestinal stem cell populations. Faseb. J. 2015;29:2828–2842. doi: 10.1096/fj.14-264010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinozaki H., Yang K., Fan K., Edelmann W., Kucherlapati R., Weinstein I.B., Lipkin M. Cyclin D1 expression in the intestinal mucosa and tumors of Apc1638N mice. Anticancer Res. 2003;23:2217–2226. [PubMed] [Google Scholar]
- 48.el Marjou F., Janssen K.P., Chang B.H.J., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., Srinivasan K., Siddiqui M.R., George S.P., Tomar A., Khurana S. A novel role for villin in intestinal epithelial cell survival and homeostasis. J. Biol. Chem. 2008;283:9454–9464. doi: 10.1074/jbc.M707962200. [DOI] [PubMed] [Google Scholar]
- 50.Gum J.R., Jr., Hicks J.W., Gillespie A.M., Carlson E.J., Kömüves L., Karnik S., Hong J.C., Epstein C.J., Kim Y.S. Goblet cell-specific expression mediated by the MUC2 mucin gene promoter in the intestine of transgenic mice. Am. J. Physiol. 1999;276:G666–G676. doi: 10.1152/ajpgi.1999.276.3.G666. [DOI] [PubMed] [Google Scholar]
- 51.Nyström E.E.L., Martinez-Abad B., Arike L., Birchenough G.M.H., Nonnecke E.B., Castillo P.A., Svensson F., Bevins C.L., Hansson G.C., Johansson M.E.V. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science. 2021;372 doi: 10.1126/science.abb1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gunawardene A.R., Corfe B.M., Staton C.A. Classification and functions of enteroendocrine cells of the lower gastrointestinal tract. Int. J. Exp. Pathol. 2011;92:219–231. doi: 10.1111/j.1365-2613.2011.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Y., Huang Y.F., Kek C., Bulavin D.V. Apoptosis differently affects lineage tracing of Lgr5 and Bmi1 intestinal stem cell populations. Cell Stem Cell. 2013;12:298–303. doi: 10.1016/j.stem.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 54.Schonhoff S.E., Giel-Moloney M., Leiter A.B. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev. Biol. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 55.Hua G., Thin T.H., Feldman R., Haimovitz-Friedman A., Clevers H., Fuks Z., Kolesnick R. Crypt base columnar stem cells in small intestines of mice are radioresistant. Gastroenterology. 2012;143:1266–1276. doi: 10.1053/j.gastro.2012.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Landeghem L., Blue R.E., Dehmer J.J., Henning S.J., Helmrath M.A., Lund P.K. Localized intestinal radiation and liquid diet enhance survival and permit evaluation of long-term intestinal responses to high dose radiation in mice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi C., Lee C., Shin S.W., Kim S.Y., Hong S.N., Park H.C. Comparison of Proton and Photon Beam Irradiation in Radiation-Induced Intestinal Injury Using a Mouse Model. Int. J. Mol. Sci. 2019;20:1894. doi: 10.3390/ijms20081894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 59.Miura S., Suzuki A. Generation of Mouse and Human Organoid-Forming Intestinal Progenitor Cells by Direct Lineage Reprogramming. Cell Stem Cell. 2017;21:456–471.e5. doi: 10.1016/j.stem.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 60.Grün D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N., Clevers H., van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 61.Haber A.L., Biton M., Rogel N., Herbst R.H., Shekhar K., Smillie C., Burgin G., Delorey T.M., Howitt M.R., Katz Y., et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carulli A.J., Keeley T.M., Demitrack E.S., Chung J., Maillard I., Samuelson L.C. Notch receptor regulation of intestinal stem cell homeostasis and crypt regeneration. Dev. Biol. 2015;402:98–108. doi: 10.1016/j.ydbio.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud Å., et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H.J., Kapoor A., Giel-Moloney M., Rindi G., Leiter A.B. Notch signaling differentially regulates the cell fate of early endocrine precursor cells and their maturing descendants in the mouse pancreas and intestine. Dev. Biol. 2012;371:156–169. doi: 10.1016/j.ydbio.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li D., Qu X., Hou K., Zhang Y., Dong Q., Teng Y., Zhang J., Liu Y. PI3K/Akt is involved in bufalin-induced apoptosis in gastric cancer cells. Anti Cancer Drugs. 2009;20:59–64. doi: 10.1097/CAD.0b013e3283160fd6. [DOI] [PubMed] [Google Scholar]
- 66.Li L., Xu L., Qu X., Zhao M., Yu P., Kang J., Liu Y., Hu X. Cbl-regulated Akt and ERK signals are involved in β-elemene-induced cell apoptosis in lung cancer cells. Mol. Med. Rep. 2011;4:1243–1246. doi: 10.3892/mmr.2011.548. [DOI] [PubMed] [Google Scholar]
- 67.Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 68.Rao N., Dodge I., Band H. The Cbl family of ubiquitin ligases: critical negative regulators of tyrosine kinase signaling in the immune system. J. Leukoc. Biol. 2002;71:753–763. [PubMed] [Google Scholar]
- 69.Rathinam C., Thien C.B.F., Langdon W.Y., Gu H., Flavell R.A. The E3 ubiquitin ligase c-Cbl restricts development and functions of hematopoietic stem cells. Genes Dev. 2008;22:992–997. doi: 10.1101/gad.1651408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liyasova M.S., Ma K., Lipkowitz S. Molecular pathways: cbl proteins in tumorigenesis and antitumor immunity-opportunities for cancer treatment. Clin. Cancer Res. 2015;21:1789–1794. doi: 10.1158/1078-0432.CCR-13-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tang R., Langdon W.Y., Zhang J. Regulation of immune responses by E3 ubiquitin ligase Cbl-b. Cell. Immunol. 2019;340 doi: 10.1016/j.cellimm.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X., Gong L., Gu H. Regulation of immune system development and function by Cbl-mediated ubiquitination. Immunol. Rev. 2019;291:123–133. doi: 10.1111/imr.12789. [DOI] [PubMed] [Google Scholar]
- 73.Korkaya H., Paulson A., Iovino F., Wicha M.S. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hynes N.E., Watson C.J. Mammary gland growth factors: roles in normal development and in cancer. Cold Spring Harbor Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Radinsky R., Risin S., Fan D., Dong Z., Bielenberg D., Bucana C.D., Fidler I.J. Level and function of epidermal growth factor receptor predict the metastatic potential of human colon carcinoma cells. Clin. Cancer Res. 1995;1:19–31. [PubMed] [Google Scholar]
- 76.Guo M., Liu S., Lu F. Gefitinib-sensitizing mutations in esophageal carcinoma. N. Engl. J. Med. 2006;354:2193–2194. doi: 10.1056/NEJMc052698. [DOI] [PubMed] [Google Scholar]
- 77.Martinelli E., De Palma R., Orditura M., De Vita F., Ciardiello F. Anti-epidermal growth factor receptor monoclonal antibodies in cancer therapy. Clin. Exp. Immunol. 2009;158:1–9. doi: 10.1111/j.1365-2249.2009.03992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sternlicht M.D. Key stages in mammary gland development: the cues that regulate ductal branching morphogenesis. Breast Cancer Res. 2006;8:201. doi: 10.1186/bcr1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Malhotra G.K., Zhao X., Band H., Band V. Shared signaling pathways in normal and breast cancer stem cells. J. Carcinog. 2011;10:38. doi: 10.4103/1477-3163.91413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kitaura Y., Jang I.K., Wang Y., Han Y.C., Inazu T., Cadera E.J., Schlissel M., Hardy R.R., Gu H. Control of the B cell-intrinsic tolerance programs by ubiquitin ligases Cbl and Cbl-b. Immunity. 2007;26:567–578. doi: 10.1016/j.immuni.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu F., Liu C., Dong Y., Wu W., Xu J., Yan Y., Shao Y., Hao C., Yang Y., Zhang J. Ablation of Cbl-b and c-Cbl in dendritic cells causes spontaneous liver cirrhosis via altering multiple properties of CD103+ cDC1s. Cell Death Dis. 2022;8:142. doi: 10.1038/s41420-022-00953-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dekaney C.M., Fong J.J., Rigby R.J., Lund P.K., Henning S.J., Helmrath M.A. Expansion of intestinal stem cells associated with long-term adaptation following ileocecal resection in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G1013–G1022. doi: 10.1152/ajpgi.00218.2007. [DOI] [PubMed] [Google Scholar]
- 83.Yilmaz Ö.H., Katajisto P., Lamming D.W., Gültekin Y., Bauer-Rowe K.E., Sengupta S., Birsoy K., Dursun A., Yilmaz V.O., Selig M., et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Geiser J., Venken K.J.T., De Lisle R.C., Andrews G.K. A mouse model of acrodermatitis enteropathica: loss of intestine zinc transporter ZIP4 (Slc39a4) disrupts the stem cell niche and intestine integrity. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parry L., Young M., El Marjou F., Clarke A.R. Evidence for a crucial role of paneth cells in mediating the intestinal response to injury. Stem Cell. 2013;31:776–785. doi: 10.1002/stem.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stamataki D., Holder M., Hodgetts C., Jeffery R., Nye E., Spencer-Dene B., Winton D.J., Lewis J. Delta1 expression, cell cycle exit, and commitment to a specific secretory fate coincide within a few hours in the mouse intestinal stem cell system. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan K.S., Chia L.A., Li X., Ootani A., Su J., Lee J.Y., Su N., Luo Y., Heilshorn S.C., Amieva M.R., et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc. Natl. Acad. Sci. USA. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wullschleger S., Loewith R., Hall M.N. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 89.Ochocki J.D., Simon M.C. Nutrient-sensing pathways and metabolic regulation in stem cells. J. Cell Biol. 2013;203:23–33. doi: 10.1083/jcb.201303110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhou Y., Rychahou P., Wang Q., Weiss H.L., Evers B.M. TSC2/mTORC1 signaling controls Paneth and goblet cell differentiation in the intestinal epithelium. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2014.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sampson L.L., Davis A.K., Grogg M.W., Zheng Y. mTOR disruption causes intestinal epithelial cell defects and intestinal atrophy postinjury in mice. Faseb. J. 2016;30:1263–1275. doi: 10.1096/fj.15-278606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He X.C., Yin T., Grindley J.C., Tian Q., Sato T., Tao W.A., Dirisina R., Porter-Westpfahl K.S., Hembree M., Johnson T., et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat. Genet. 2007;39:189–198. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Richmond C.A., Shah M.S., Deary L.T., Trotier D.C., Thomas H., Ambruzs D.M., Jiang L., Whiles B.B., Rickner H.D., Montgomery R.K., et al. Dormant Intestinal Stem Cells Are Regulated by PTEN and Nutritional Status. Cell Rep. 2015;13:2403–2411. doi: 10.1016/j.celrep.2015.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perekatt A.O., Valdez M.J., Davila M., Hoffman A., Bonder E.M., Gao N., Verzi M.P. YY1 is indispensable for Lgr5+ intestinal stem cell renewal. Proc. Natl. Acad. Sci. USA. 2014;111:7695–7700. doi: 10.1073/pnas.1400128111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chitalia V., Shivanna S., Martorell J., Meyer R., Edelman E., Rahimi N. c-Cbl, a ubiquitin E3 ligase that targets active β-catenin: a novel layer of Wnt signaling regulation. J. Biol. Chem. 2013;288:23505–23517. doi: 10.1074/jbc.M113.473801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shivanna S., Harrold I., Shashar M., Meyer R., Kiang C., Francis J., Zhao Q., Feng H., Edelman E.R., Rahimi N., Chitalia V.C. The c-Cbl ubiquitin ligase regulates nuclear β-catenin and angiogenesis by its tyrosine phosphorylation mediated through the Wnt signaling pathway. J. Biol. Chem. 2015;290:12537–12546. doi: 10.1074/jbc.M114.616623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shashar M., Siwak J., Tapan U., Lee S.Y., Meyer R.D., Parrack P., Tan J., Khatami F., Francis J., Zhao Q., et al. c-Cbl mediates the degradation of tumorigenic nuclear β-catenin contributing to the heterogeneity in Wnt activity in colorectal tumors. Oncotarget. 2016;7:71136–71150. doi: 10.18632/oncotarget.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kumaradevan S., Lee S.Y., Richards S., Lyle C., Zhao Q., Tapan U., Jiangliu Y., Ghumman S., Walker J., Belghasem M., et al. c-Cbl Expression Correlates with Human Colorectal Cancer Survival and Its Wnt/β-Catenin Suppressor Function Is Regulated by Tyr371 Phosphorylation. Am. J. Pathol. 2018;188:1921–1933. doi: 10.1016/j.ajpath.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Richards S., Walker J., Nakanishi M., Belghasem M., Lyle C., Arinze N., Napoleon M.A., Ravid J.D., Crossland N., Zhao Q., et al. Haploinsufficiency of Casitas B-Lineage Lymphoma Augments the Progression of Colon Cancer in the Background of Adenomatous Polyposis Coli Inactivation. Am. J. Pathol. 2020;190:602–613. doi: 10.1016/j.ajpath.2019.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zenonos K., Kyprianou K. RAS signaling pathways, mutations and their role in colorectal cancer. World J. Gastrointest. Oncol. 2013;5:97–101. doi: 10.4251/wjgo.v5.i5.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen J., Shao R., Li F., Monteiro M., Liu J.P., Xu Z.P., Gu W. PI3K/Akt/mTOR pathway dual inhibitor BEZ235 suppresses the stemness of colon cancer stem cells. Clin. Exp. Pharmacol. Physiol. 2015;42:1317–1326. doi: 10.1111/1440-1681.12493. [DOI] [PubMed] [Google Scholar]
- 102.Caligiuri M.A., Briesewitz R., Yu J., Wang L., Wei M., Arnoczky K.J., Marburger T.B., Wen J., Perrotti D., Bloomfield C.D., Whitman S.P. Novel c-CBL and CBL-b ubiquitin ligase mutations in human acute myeloid leukemia. Blood. 2007;110:1022–1024. doi: 10.1182/blood-2006-12-061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Muramatsu H., Makishima H., Jankowska A.M., Cazzolli H., O'Keefe C., Yoshida N., Xu Y., Nishio N., Hama A., Yagasaki H., et al. Mutations of an E3 ubiquitin ligase c-Cbl but not TET2 mutations are pathogenic in juvenile myelomonocytic leukemia. Blood. 2010;115:1969–1975. doi: 10.1182/blood-2009-06-226340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kao H.W., Sanada M., Liang D.C., Lai C.L., Lee E.H., Kuo M.C., Lin T.L., Shih Y.S., Wu J.H., Huang C.F., et al. A high occurrence of acquisition and/or expansion of C-CBL mutant clones in the progression of high-risk myelodysplastic syndrome to acute myeloid leukemia. Neoplasia. 2011;13:1035–1042. doi: 10.1593/neo.111192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jawhar M., Schwaab J., Schnittger S., Sotlar K., Horny H.P., Metzgeroth G., Müller N., Schneider S., Naumann N., Walz C., et al. Molecular profiling of myeloid progenitor cells in multi-mutated advanced systemic mastocytosis identifies KIT D816V as a distinct and late event. Leukemia. 2015;29:1115–1122. doi: 10.1038/leu.2015.4. [DOI] [PubMed] [Google Scholar]
- 106.Merlos-Suárez A., Barriga F.M., Jung P., Iglesias M., Céspedes M.V., Rossell D., Sevillano M., Hernando-Momblona X., da Silva-Diz V., Muñoz P., et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 107.Schepers A.G., Snippert H.J., Stange D.E., van den Born M., van Es J.H., van de Wetering M., Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 108.Pece S., Tosoni D., Confalonieri S., Mazzarol G., Vecchi M., Ronzoni S., Bernard L., Viale G., Pelicci P.G., Di Fiore P.P. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 109.Wang F., Scoville D., He X.C., Mahe M.M., Box A., Perry J.M., Smith N.R., Lei N.Y., Davies P.S., Fuller M.K., et al. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology. 2013;145:383–395.e1-21. doi: 10.1053/j.gastro.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mahmoudi T., Li V.S.W., Ng S.S., Taouatas N., Vries R.G.J., Mohammed S., Heck A.J., Clevers H. The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 2009;28:3329–3340. doi: 10.1038/emboj.2009.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single cell RNAseq data used in this publication have been deposited in NCBI’s Gene Expression Omnibus and are publicly accessible through GEO series accession number GSE236186.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.