Abstract
Simian immunodeficiency virus (SIV) infection of the rhesus macaque is currently the best animal model for AIDS vaccine development. One limitation of this model, however, has been the small number of cytotoxic T-lymphocyte (CTL) epitopes and restricting major histocompatibility complex (MHC) class I molecules available for investigating virus-specific CTL responses. To identify new MHC class I-restricted CTL epitopes, we infected five members of a family of MHC-defined rhesus macaques intravenously with SIV. Five new CTL epitopes bound by four different MHC class I molecules were defined. These included two Env epitopes bound by Mamu-A*11 and -B*03 and three Nef epitopes bound by Mamu-B*03, -B*04, and -B*17. All four restricting MHC class I molecules were encoded on only two haplotypes (b or c). Interestingly, resistance to disease progression within this family appeared to be associated with the inheritance of one or both of these MHC class I haplotypes. Two individuals that inherited haplotypes b and c separately survived for 299 and 511 days, respectively, while another individual that inherited both haplotypes survived for 889 days. In contrast, two MHC class I-identical individuals that did not inherit either haplotype rapidly progressed to disease (survived <80 days). Since all five offspring were identical at their Mamu-DRB loci, MHC class II differences are unlikely to account for their patterns of disease progression. These results double the number of SIV CTL epitopes defined in rhesus macaques and provide evidence that allelic differences at the MHC class I loci may influence rates of disease progression among AIDS virus-infected individuals.
Products of the highly polymorphic major histocompatibility complex (MHC) class I loci determine how many viral peptides will be presented by a given individual for cytotoxic T-lymphocyte (CTL) recognition (48). Since all epitopes are not equal with respect to their MHC class I binding affinity, T-cell receptor recognition, or the degree of sequence variation the virus can tolerate, MHC class I polymorphism may introduce considerable variation in the quality of virus-specific CTL responses between individuals. Thus, allelic differences at the MHC class I loci probably contribute to the variable patterns of disease progression observed among human immunodeficiency virus (HIV)-infected people. In support of this hypothesis, statistical analyses have revealed associations between certain HLA class I alleles and time until AIDS onset (7, 26). Furthermore, those HLA molecules associated with slower courses of disease progression were generally predicted to bind more HIV epitopes than those associated with more rapid disease progression (22, 31). Yet, despite these associations, there is little direct experimental evidence demonstrating that MHC class I differences can influence survival time following HIV infection. This likely reflects the difficulties inherent in studying human subjects, which include uncontrolled sources, doses, and routes of infection; the inaccuracies of estimating dates of infection from patient histories; and, more recently, the influence of antiretroviral drug therapies. Additionally, it has been difficult to separate the effects of allelic differences at the MHC class I loci from those of the class II loci, largely because of the exceptional polymorphism of the MHC class II DRB genes.
To control for the many variables associated with infection and host immunogenetics in humans, we infected five MHC class I-disparate, Mamu-DRB-identical offspring from a family of MHC-defined rhesus macaques with a well-defined stock of simian immunodeficiency virus (SIV) to explore the relationship between MHC class I differences and disease progression.
MATERIALS AND METHODS
SIV infection.
Rhesus macaques were infected intravenously with 40 50% tissue culture infectious doses of a heterogeneous virus stock originally derived from SIVmac251 (11, 33, 43). Virus dilutions were prepared in 1 ml of sterile saline and injected into the saphenous vein of anesthetized animals at a rate of 1 ml per min. SIV-infected animals were cared for according to an experimental protocol approved by the University of Wisconsin Research Animal Resource Committee.
Viral loads, CD4 counts, and antibody titers.
Plasma SIV titers were monitored at regular time points postinfection (p.i.) by using the infectious centers assay. CEM×174 cells (2 × 105) were cultured with decreasing volumes of plasma (200, 100, 50, 10, and 2 μl) in duplicate six-well plates and scored for syncytium formation over a 4-week period. Infectious doses per milliliter of plasma were calculated by dividing 1,000 μl by the volume of initial plasma that resulted in a positive viral culture. Plasma viral loads were also determined by branched DNA analysis (9). CD4+ T-lymphocyte counts were monitored by flow cytometry. Peripheral blood lymphocytes (PBLs) were isolated from blood drawn in EDTA tubes, fixed in 1% paraformaldehyde, stained with anti-CD4-phocyerythrin conjugate (M220020; Antigenix, Franklin Square, N.Y.), and run on a Beckton-Dickinson FACS Caliber. Antibody responses were detected by using the HIV-2 enzyme immunoassay (EIA) kit (GeneticSystems, Chaska, Minn.) at plasma dilutions of 1:400 with a GeneticSystems LP400 enzyme-linked immunosorbent assay (ELISA) plate reader.
Molecular analysis of the MHC class I and II loci.
MHC class I alleles were cloned and sequenced from cDNA libraries constructed from animals D and B as described previously (5, 6). MHC class II DRB cDNAs were separated by denaturing gradient gel electrophoresis and sequenced directly as described by Knapp et al. (25). For the Mamu-DP and -DQ loci, the polymorphic second exon was amplified from genomic DNA extracted from 5 × 106 B-lymphoblastoid cell lines (B-LCLs) by using the QIAmp Blood kit (Qiagen, Chatsworth, Calif.) as previously described (23, 32, 40, 41). PCR primer pairs included GH98 and GH99 for the Mamu-DPA1 locus, DPB SalI and DPB XbaI for the Mamu-DPB-1 locus, GH26 and GH27 for the Mamu-DQA1 locus, 89–445 and GH27 for the Mamu-DQA2 locus, and 5′ DQβ SalI and 3′ DQB XbaI for the Mamu-DQB1 locus. PCR products were cloned into the M13 vectors tg130 and tg131 and sequenced.
CTL cultures and assays.
CTL cultures were established from fresh or frozen PBL samples. PBLs were stimulated 1:1 with 5 × 106 paraformaldehyde-fixed, autologous B-LCLs infected overnight with vaccinia virus constructs expressing the SIVmac251 Gag, Pol, Env, or SIVmac239 Nef proteins provided by Therion Biologics Corporation (Cambridge, Mass.). Half of the medium was replaced after 2 days with R10 supplemented with 20 U of recombinant interleukin-2 (rIL-2) provided by Hoffman-LaRoche (Nutley, N.J.). After 7 days, viable cells were isolated on a Ficoll-Hypaque gradient and restimulated with fixed, vaccinia virus-infected B-LCLs. CTLs were then expanded in the presence of rIL-2 and tested for CTL activity after 13 days in culture. CTL cultures from animal D were prepared from PBLs isolated 55, 83, 139, 342, 398, 426, 566, 622, 643, and 776 days p.i. For animal C, CTL cultures were derived from PBLs isolated 245, 273, and 299 days p.i., and for animal A, CTL cultures were derived from PBLs isolated 245, 273, and 397 days p.i.
SIV-specific CTL activity was assessed by using standard 51Cr release assays. Herpesvirus papio-immortalized B-LCLs were labeled with 75 μCi of sodium [51Cr]chromate, and either infected overnight with vaccinia virus recombinants (multiplicity of infection [MOI] of 4:1) or pulsed with peptides (5 μg) for 1 h on the day of the assay. B-LCL targets were washed and plated at 5 × 103 cells per well in round-bottom 96-well plates. CTL effectors were added at different effector/target ratios and incubated for 5 h. Spontaneous and maximal chromium release was determined by incubating target cells in medium alone and with 5% Triton X-100, respectively. CTL activity was calculated from the counts per minute present in harvested supernatants by the formula: % specific release = [(experimental release − spontaneous release)/(maximal release − spontaneous release)] × 100.
CTL epitope mapping.
CTL epitopes of the SIV Env and Nef proteins were mapped with sets of peptides synthesized according to the predicted amino acid sequences of SIVmac251. Autologous B-LCLs pulsed for 1 h with pools of overlapping 20-mers (5 μg each) were first tested as CTL targets in 51Cr release assays. In subsequent assays, individual 20-mers were tested from positive pools followed by overlapping 9-mers spanning each positive 20-mer. To define the ends of each CTL epitope, additional peptides with amino acid additions or deletions at the N and C termini (8- to 10-mers) were tested for CTL recognition over a range of concentrations.
1-D IEF.
MHC class I molecules were analyzed by one-dimensional isoelectric focusing (1-D IEF) as previously described (46).
Creation of stable MHC class I transfectants.
Stable transfectants expressing rhesus macaque MHC class I molecules were created in the HLA class I-deficient human B-cell line 721.221. Full-length cDNAs encoding Mamu-A*03, -A*04, -A*11, -A*12, -B*02, -B*03, -B*04, -B*17, and -B*29 were subcloned into the expression vector pKG5 by using the restriction sites XhoI and HindIII and were electroporated into 721.221 cells (39). Log-phase cells (7.5 × 106) were suspended in 250 μl of medium with 25 μg of DNA in a chilled 0.4-cm cuvette and electroporated at 200 V and 950 μF. After cooling for 1 min on ice, the cells were warmed to room temperature, diluted to 50 ml in medium, and seeded to 24-well plates in 1-ml aliquots. The cells were allowed to recover for 3 days before beginning selection in medium containing 650 μg of Geneticin per ml (Gibco-BRL). Geneticin-resistant clones were screened for MHC class I surface expression by flow cytometry after staining with fluorescein isothiocyanate-conjugated W6/32 antibody (Sigma, St. Louis, Mo.). The electroporation and selection medium consisted of RPMI supplemented with 10% defined supplemented calf serum (Hyclone, Logan, Utah), 5% fetal bovine serum, l-glutamine, penicillin (100 IU/ml), and streptomycin (100 μg/ml).
RESULTS
Viral load and disease progression in five SIV-infected family members.
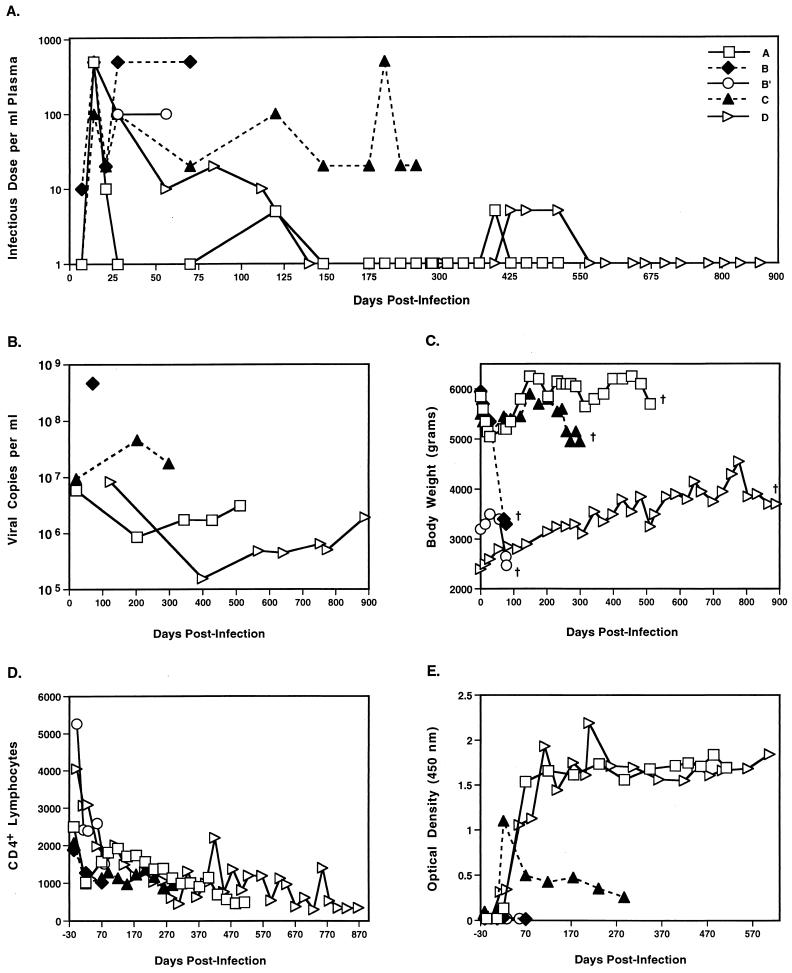
Five offspring of an extended family of rhesus macaques were infected intravenously with the same dose of SIV. All five animals exhibited similar peak titers of primary viremia between 7 and 21 days after infection. However, after the acute phase, their courses of infection differed greatly (Fig. 1). The two siblings B and B′ were unable to control their viral loads and rapidly progressed to AIDS, with symptoms including wasting, diarrhea, lymphadenopathy, and maculopapular rashes. The kinetics of their decline were remarkably similar, despite differences in age and weight at the time of infection. In contrast, their sister, animal A, showed a much slower course of disease progression. Animal A resolved her primary viremia by day 28, and with the exception of two later time points, maintained plasma virus titers below the limits of detection by the infectious center assay. Furthermore, she remained asymptomatic for well over a year after infection despite steadily declining CD4+ T-cell counts. Animal D showed an even slower course of disease progression, maintained low plasma virus loads, and died 889 days after infection. The course of disease exhibited by animal C was intermediate to her rapid- and slow-progressing half-siblings, since she survived for 299 days before developing AIDS-associated wasting and maintained moderate plasma virus titers in the range of 10 to 100 infective doses per ml throughout infection. While the two rapid progressors did not mount detectable antibody responses, the slow progressors mounted strong antibody responses.
FIG. 1.
Viral loads and disease progression in five SIV-infected rhesus macaques. (A) Plasma SIV titers were determined by culturing serial dilutions of plasma with CEM×174 cells and scoring for syncytium formation over a period of 4 weeks. (B) SIV loads in plasma as determined by branched DNA analysis (Chiron). (C) Body weight changes during the course of SIV infection. All five of the macaques died or were euthanized as a result of their infections on the day indicated by †. (D) CD4+ T-cell counts per μl of blood as determined by flow cytometry. (E) SIV-specific antibody responses determined at 1:400 plasma dilutions with the HIV-2 EIA kit (GeneticSystems). Antibody titers for animal D were not assessed after day 600.
MHC typing of family members.
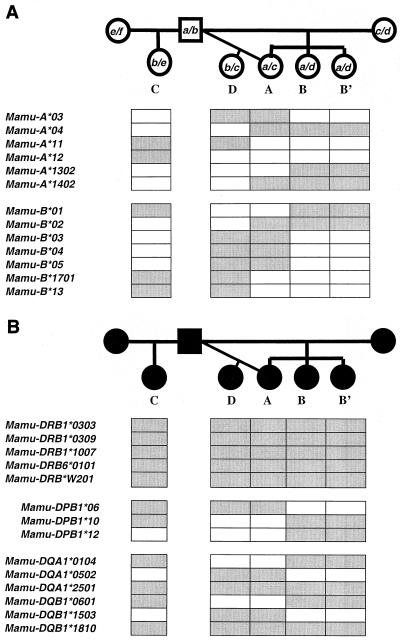
To explore the possibility that there was a relationship between the MHC of these macaques and their variable courses of disease progression, we carried out an analysis of the MHC class I molecules expressed by five family members (data not shown). Surprisingly, animals B and B′ were MHC class I identical (as defined by identical 1-D IEF focusing patterns) and shared one MHC class I haplotype with animal A. To more rigorously define the MHC class I haplotypes within this family, we carried out a more comprehensive molecular analysis of the MHC class I and class II loci of each family member (Fig. 2). A haplotype diagram is shown in Fig. 2, detailing a number of the class I alleles identified by cDNA library screening. For simplicity, only a few alleles, including all of those shown to restrict the CTL epitopes identified in this article, are included. A more comprehensive analysis of haplotype organization in the rhesus macaque is discussed elsewhere (H. Horton et al., unpublished data). The two rapid progressors, B and B′, were confirmed to be MHC class I identical and to share an MHC class I haplotype (haplotype a) with their slow-progressing sister, animal A (Fig. 2A). Additional MHC class I haplotypes were also shared between the intermediate and slow progressors. Animals A and D both inherited haplotype c, and animals C and D both inherited haplotype b (Fig. 2A). Thus, each of the five SIV-infected offspring in this family shared at least one MHC class I haplotype with another sibling. Interestingly, these macaques expressed multiple MHC class I alleles (i.e., animal D has five Mamu-B alleles). This is likely due to duplication of both the Mamu-A and -B loci seen in rhesus monkeys (6).
FIG. 2.
Simplified diagram depicting the inheritance of certain MHC class I (A) and class II (B) alleles in an extended family of rhesus macaques. The MHC class I and class II alleles present in each member of an extended family of rhesus macaques were analyzed by molecular methods. The MHC class I alleles segregated with six different haplotypes designated a to f. The shaded boxes below the pedigree indicate the MHC class I alleles and class II alleles present in each individual.
Remarkably, analysis of the MHC class II genes revealed that all five offspring had identical Mamu-DRB loci, thereby eliminating the most polymorphic MHC class II loci as factors contributing to the variable courses of disease progression within this family (Fig. 2B). Furthermore, while there were differences at the Mamu-DQ and -DP loci among the five offspring, there was no clear segregation of MHC class II haplotypes between rapid and slow progressors. The intermediate progressor (C) and the two rapid progressors (B and B′) shared the same set of Mamu-DQ alleles and one Mamu-DPB1 allele (Fig. 2B). Additionally, the slow progressors (A and D) shared Mamu-DQ alleles with the rapid progressors. Thus, the MHC class II alleles present in the rapid progressors differed from their siblings by only a single DPB1 allele. Mamu-DPB1*06 was present in animals A, C, and D, while Mamu-DPB1*12 was present in animals B and B′ (Fig. 2B).
Multiple CTL epitopes of the SIV Env and Nef proteins were mapped in two SIV-infected rhesus macaques.
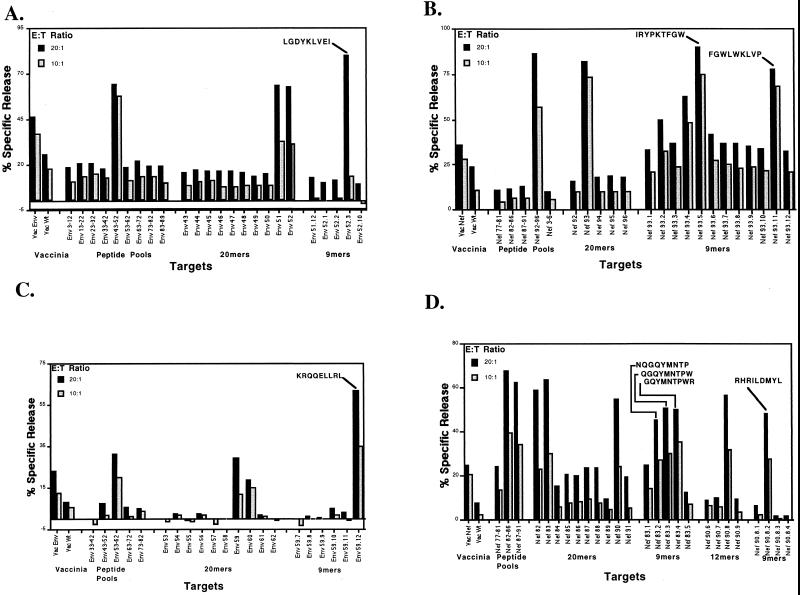
To investigate the hypothesis that MHC class I-restricted CTL responses were responsible for the differences in disease progression among these Mamu-DRB-identical siblings, we first tested for CTL responses to the SIV Gag, Pol, Env, and Nef proteins in each of the five offspring. SIV-specific CTL responses against Env and Nef proteins were detected only in the MHC class I-disparate individuals, A and C. Additionally, animal D responded only to Env and Nef in vaccinia virus enzyme-linked immunospot (ELISPOT) assays and did not exhibit responses to Gag or Pol (data not shown). Epitope mapping studies using sets of overlapping peptides revealed that animals A and C each recognized one Env and two Nef CTL epitopes (Fig. 3).
FIG. 3.
Mapping of SIV Env (A and C) and Nef (B and D) CTL epitopes in macaques C (A and B) and A (C and D). A single Env epitope and two Nef CTL epitopes were mapped in both animals C and A. Autologous B-LCL targets were pulsed with peptides synthesized according to the predicted amino acid sequences of the Env and Nef proteins of SIVmac251 and used as targets in 5-h chromium release assays. Each panel summarizes three separate assays in which peptide pools were used in the first experiment, individual 20-mers were used in the second experiment, and 9-mers were used in the final experiment. Target cells infected with wild-type vaccinia virus (Vac Wt) and either an Env- or Nef-expressing recombinant vaccinia virus (Vac Env or Vac Nef) were included as controls in the first assay. E:T, effector/target ratio.
Definition of optimal CTL epitopes.
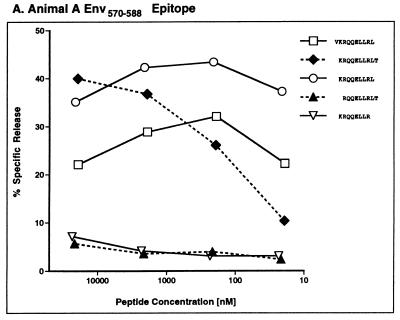
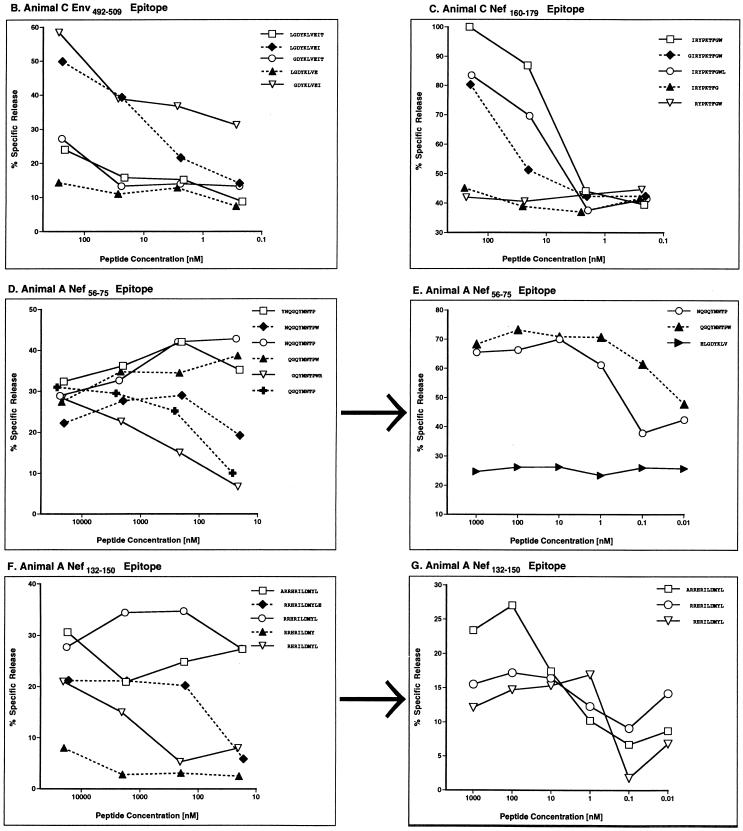
To identify the optimal peptide for each CTL epitope, additional amino acids were added or subtracted from the N and C termini of each nonamer and tested for CTL recognition over a range of peptide concentrations (Fig. 4). Unfortunately, we could not define the ends of one of the Nef epitopes recognized by animal C (FGWLWKLVP), since we were unable to reproduce CTL responses to this peptide from frozen PBL samples. CTL assays also failed to unambiguously define the N termini of two Nef epitopes (Fig. 4D to G). However, for these epitopes, live-cell binding assays identified QGQYMNTPW and ARRHRILDMYL as the optimal peptides for MHC class I binding (12).
FIG. 4.
Fine mapping of SIV Env and Nef CTL epitopes. Additional peptides were synthesized with residues added or subtracted from the N and C termini for five of the 9-mers that were mapped in animals C and A. Serial 10-fold dilutions of these variants were pulsed onto self B-LCL targets and tested for CTL recognition at a 20:1 effector/target ratio in a 5-h chromium release assay. We were unable to resolve the ends of two of the Nef epitopes in these experiments (D to G). However, additional live-cell binding assays indicate that QGQYMNTPW (D and E) and ARRHRILDMYL (F and G) represent the optimal-length peptides for each of these epitopes (12).
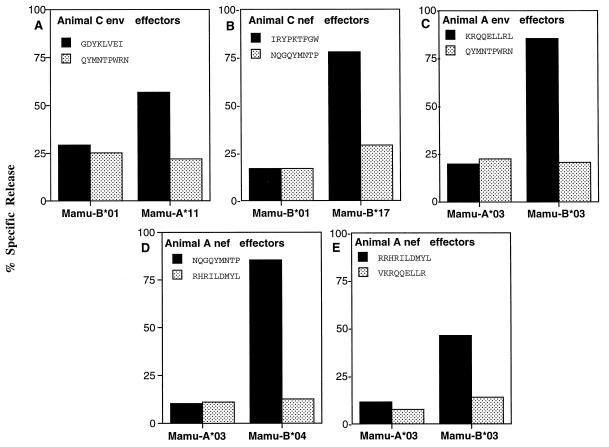
Five different CTL epitopes are bound by four rhesus MHC class I molecules.
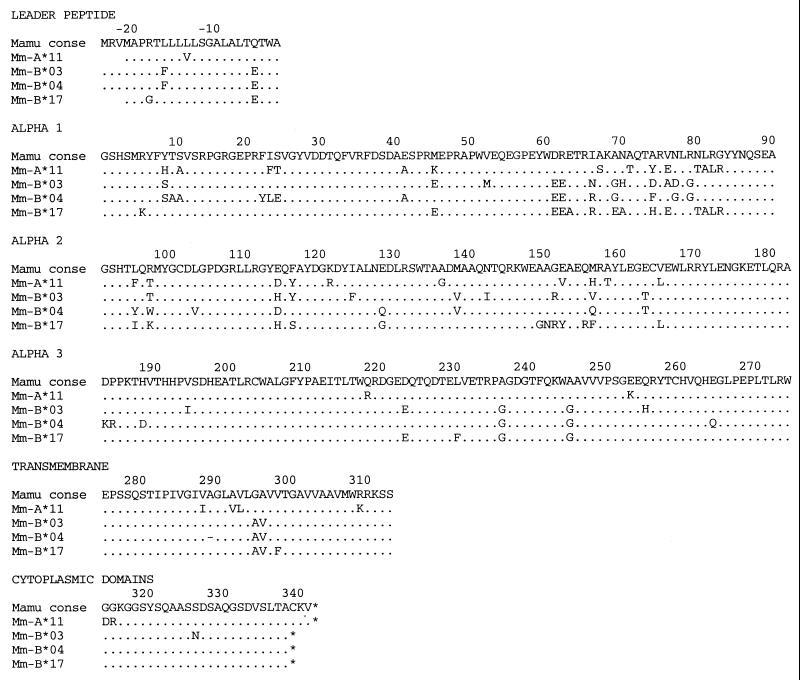
Full-length MHC class I cDNAs from animals A and C were cloned into an expression vector and transfected into the HLA class I-deficient cell line 721.221 (39) to create a panel of stable transfectants, each expressing a single rhesus MHC class I molecule (Fig. 5). CTL recognition of these transfectants pulsed with epitope and control peptides revealed the restricting MHC class I molecules for all five defined CTL epitopes (Fig. 6 and Table 1). Surprisingly, all three of the CTL epitopes mapped in animal A were bound by Mamu-B*03 or -B*04 encoded on the maternal c haplotype, and both CTL epitopes mapped in animal C were bound by Mamu-A*11 and -B*17 encoded on the paternal b haplotype.
FIG. 5.
Predicted amino acid sequences of four new rhesus macaque MHC class I molecules used to bind CTL epitopes derived from SIV. Identity with the consensus (conse) MHC class I sequence is indicated by a dot.
FIG. 6.
Restricting MHC class I molecules for five CTL epitopes of the SIV Env (A and C) and Nef (B, D, and E) proteins. Stable transfectants expressing macaque MHC class I molecules were created by electroporation of 721.221 cells (39) with full-length MHC class I cDNAs from animals A (C to E) and C (A and B) cloned into the expression vector pKG5. Transfectants were pulsed with the three epitopes mapped in animal A and two of the epitopes mapped in animal C (solid bars) and tested for CTL recognition at a 20:1 effector/target ratio. Transfectants were also pulsed with irrelevant peptides to control for nonspecific killing (stippled bars). Mamu-A*11 presented an Env epitope (A) and Mamu-B*17 presented a Nef epitope (B) for recognition by CTLs from animal C. Mamu-B*03 and -B*04 presented Env and Nef epitopes for recognition by CTLs from animal A (C to E).
TABLE 1.
MHC class I haplotypes, CTL epitopes, and disease progression in a family of SIV-infected macaques
| Macaque (class I haplotype) | CTL epitope (allele) | Survival time (days) | Disease progression |
|---|---|---|---|
| C (b/e) | GDYKLVEIenv497–504 (Mamu-A*11) | 299 | Intermediate |
| IRYPKTFGWnef165–173 (Mamu-B*17) | |||
| FGWLWKLVPnef171–179 | |||
| D (b/c) | KRQQELLRLenv575–583 (Mamu-B*03) | 889 | Slow |
| QGQYMNTPnef61–69 (Mamu-B*04) | |||
| ARRHRILDMYLnef136–146 (Mamu-B*03) | |||
| GDYKLVEIenv497–504 (Mamu-A*11) | |||
| IRYPKTFGWnef165–173 (Mamu-B*17) | |||
| A (a/c) | KRQQELLRLenv575–583 (Mamu-B*03) | 511 | Slow |
| QGQYMNTPnef61–69 (Mamu-B*04) | |||
| ARRHRILDMYLnef136–146 (Mamu-B*03) | |||
| B (a/d) | Not detected | 77 | Rapid |
| B′ (a/d) | Not detected | 78 | Rapid |
Analysis of the CTL responses against the SIV Gag, Pol, Env, and Nef proteins in animals B′ and D.
Given the rapid onset of disease in animals B and B′, we investigated whether one of these animals (B′) ever had an SIV-specific CTL response. Macaques B′ and D were tested for CTL responses against the SIV Gag, Pol, Env, and Nef proteins at 2 and 8 weeks p.i. With the exception of a transient response against the Env protein at 2 weeks, we were unable to detect a CTL response to any of these four viral antigens in animal B′ (data not shown). However, significant CTL responses against the SIV Env and Nef proteins were detected in animal D by 8 weeks p.i. Since animal D shared the c haplotype with animal A and the b haplotype with animal C, we next asked if her CTLs could recognize the same epitopes as both of these individuals. CTL cultures were derived from animal D by peptide stimulation and tested for the ability to recognize self targets pulsed with each of the five CTL epitopes of animals A and C. These CTL cultures recognized all three of the epitopes mapped in animal A and two of the epitopes mapped in animal C (data not shown). Thus, animal D recognized at least five different SIV Env and Nef CTL epitopes.
CTL epitope variation.
To determine whether the CTL responses of animals A, C, and D exerted selective pressure on SIV replication in vivo, we sequenced the CTL epitope coding regions of the plasma virus population for each individual at selected time points throughout infection. Statistical analysis of the nonsynonymous (amino acid replacement) versus synonymous (silent) nucleotide substitution rate in regions of env and nef coding for restricted CTL epitopes versus nonrestricted epitopes or flanking sequence revealed clear evidence of CTL selection (15). Furthermore, by the time animals A, C, and D died or were euthanized as a result of their infections, all of their CTL epitopes had accumulated amino acid substitutions that diminished MHC class I binding, CTL recognition, or both (15). Thus, the selection of escape variants within the CTL epitopes recognized by animals A, C, and D provides additional evidence for the importance of MHC class I-restricted CTL responses in controlling the infections of these individuals. Moreover, it may also explain why their CTL responses ultimately failed to prevent disease progression.
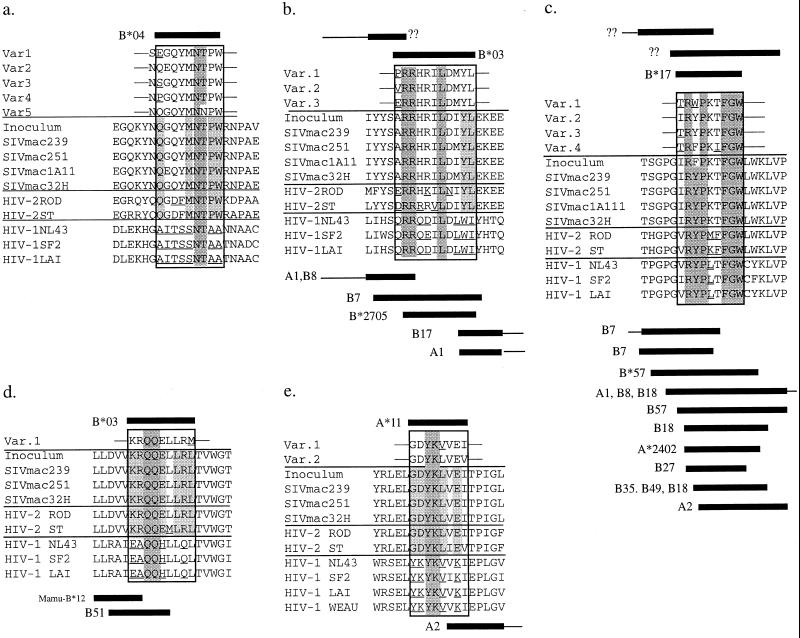
Each of our newly defined CTL epitopes appears to be derived from regions of the Env and Nef proteins that are relatively well conserved between SIV, HIV-1, and HIV-2 (Fig. 7). Few of the CTL escape variants resulted in changes of highly conserved residues. Only the Mamu-B*03 Env and a Mamu-B*04 Nef epitope variant (Var5) altered residues normally shared between SIV and HIV-1. Additionally, four of the five new rhesus SIV CTL epitopes overlapped with known HIV CTL epitopes in humans (Fig. 7). Most notably, the Mamu-B*03 and -B*17 Nef epitopes overlapped precisely with HLA-B27-restricted epitopes. Since HLA-B27 preferentially binds peptides with arginine at position 2, this observation is consistent with peptide binding experiments indicating that arginine at the second position is an anchor residue for binding of these epitopes to Mamu-B*03 and -B*17 (12).
FIG. 7.
Alignment of SIVmac, HIV-2, and HIV-1 (clade B) amino acid sequences in regions encoding the newly identified SIV Env and Nef CTL epitopes. Residues conserved between the HIV-1, HIV-2, and SIV are shaded dark gray. Residues conserved only between HIV-2 and SIV are shaded light gray. Boxes above the alignments represent previously identified SIV CTL epitopes and their restricting class I alleles (if known). Boxes below the alignment represent previously identified HIV CTL epitopes and their restricting class I alleles. Residues that are not present in the SIVmac isolates are underlined. (a) Nef56–75, (b) Nef132–150, (c) Nef160–179, (d) Env570–588, (e) Env492–509. Var., variant.
DISCUSSION
To date, only four MHC class I molecules that restrict virus-specific CTL responses in SIV- or simian/human immunodeficiency virus-infected rhesus macaques have been identified (1, 2, 14, 16, 30, 44, 45). These molecules bind a total of eight different CTL epitopes (five from SIV and three from HIV Env), five of which are bound by the same molecule, Mamu-A*01. Thus, the analysis of virus-specific CTL responses in SIV-infected rhesus macaques has been limited to a small number of epitopes and has relied predominantly on Mamu-A*01-positive individuals. The five new CTL epitopes and four new restricting MHC class I molecules described here double the number of CTL epitopes and MHC class I types available for CTL studies in the SIV-rhesus macaque model. Furthermore, we have also described the first MHC class I-restricted CTL epitopes in Nef in SIV-infected rhesus macaques. These three epitopes may be particularly useful for evaluating the significance of Nef as a CTL target, given that Nef is well expressed early in each cycle of infection and may be unusually immunogenic (17). Tetramers can now be synthesized for each of these five new peptide-MHC class I pairs (3, 28), enabling more quantitative and sensitive analysis of SIV-specific CTLs after vaccination or infection (10). Preliminary evidence also suggests that two of these new MHC class I alleles (Mamu-A*11 and -B*17) are at appreciable frequencies in rhesus macaques of Indian descent. This should significantly increase the value of the rhesus macaque as an animal model for HIV infection and AIDS vaccine development.
Given the large number of variables associated with HIV infection, it has been difficult to assess the relative contribution of the MHC class I alleles to disease progression. Molecular analysis of the rhesus macaque MHC has facilitated the development of a genetically defined animal model to address this issue. The ability to contain SIV replication and resist progression to AIDS followed the segregation of the MHC class I haplotypes among five Mamu-DRB-identical offspring in an extended family of rhesus macaques. Interestingly, the MHC class I haplotypes of the intermediate and slow progressors encoded molecules that bound multiple CTL epitopes derived from the SIV Env and Nef proteins. In animal A, all three CTL epitopes were restricted by two molecules, Mamu-B*03 and -B*04, encoded on her maternal c haplotype. Similarly, both of the CTL epitopes recognized by animal C were restricted by Mamu-A*11 and -B*17 encoded on her paternal b haplotype. These two macaques survived for 511 and 299 days, respectively, thereby exhibiting slow and intermediate courses of disease progression. Another family member, animal D, which inherited both the b and the c haplotypes, recognized five different CTL epitopes and survived for 889 days after infection. In contrast, the MHC class I-identical siblings, animals B and B′ (haplotypes a/d), failed to mount consistently detectable CTL responses and rapidly developed AIDS-associated wasting within 80 days after infection. Although we have studied only a single family of macaques, these observations suggest that the inheritance of certain MHC class I alleles may influence the course of disease progression in SIV-infected macaques.
Allelic differences at the MHC class II loci are less likely to account for the variable pathology of the SIV-infected offspring of this family, since all five SIV-infected individuals had identical Mamu-DRB loci. While there were several differences at the MHC class II DQ and DP loci of these individuals, the only unique difference between the MHC class II alleles of the rapid and slow or intermediate progressors was a single Mamu-DPB1 allele. Although products of the MHC class II DP loci have been shown to present peptides in humans (8, 13, 18, 19, 27), the vast majority of MHC class II-restricted T-cell responses involve products of the more polymorphic HLA-DR and -DQ loci. Therefore, it is unlikely that a single difference at the Mamu-DPB1 locus would account for the dramatic differences in disease progression observed within this family. However, further experiments are needed to more carefully evaluate the influence of the MHC class II loci on progression to AIDS.
The emergence of escape mutations in the CTL epitopes recognized by the intermediate and slow progressors provided additional evidence that the MHC class I-restricted CTL responses of these animals contributed to their resistance to disease progression (15). By the time animals A, C, and D died or were euthanized due to AIDS, amino acid replacements had accumulated within all 10 of the CTL epitopes recognized by these individuals. Furthermore, most of the new epitope variants greatly reduced or eliminated CTL recognition and/or MHC class I binding. The selection of CTL escape variants suggests that the CTL responses of the intermediate and slow progressors exerted considerable selective pressure on virus replication in vivo and thus provides further support for the hypothesis that certain MHC class I alleles may confer resistance to disease progression.
The relationship between certain MHC class I molecules and longer survival times following SIV infection in our family study implicates the MHC class I genes as determinants of disease progression in this family of macaques. These results therefore support the hypothesis that allelic differences at the MHC class I loci are important determinants of time until AIDS onset among HIV-infected people (7). Interestingly, a recent report describes the association of rapid disease progression with homozygosity and with HLA-B*35 and -Cw*04 in particular. This association of HLA-B*35 and rapid progression to disease has also been reported in other studies (20, 24, 36). Interestingly, however, this molecule can bind many different HIV-derived CTL epitopes (21, 35, 38, 42). It is, therefore, not entirely clear why HLA-B*35 should predispose individuals to rapid progression. The use of MHC-defined macaques in future experiments should provide valuable insights to resolve the role of cellular immune responses in protecting against AIDS virus infection.
ACKNOWLEDGMENTS
We thank Lettie Smith for help in preparing the manuscript and Bob Becker for help with illustration. We thank Joan Scheffler for initially identifying this family of macaques.
This work was supported by grants from the National Institutes of Health (AI32426, AI42641, AI41913, and RR00167 to D.I.W.; AI15486 to R.D.). D.I.W. is an Elizabeth Glaser Scientist.
REFERENCES
- 1.Allen T M, Sidney J, del Guercio M-F, Glickman R L, Lensmeyer G L, Wiebe D A, DeMars R, Pauza C D, Johnson R P, Sette A, Watkins D I. Characterization of the peptide-binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from SIV. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 2.Allen T M, Vogel T U, Fuller D H, Mothe B R, Steffen S, Boyson J E, Shipley T, Fuller J, Hanke T, Sette A, Altman J D, Moss B, McMichael A J, Watkins D I. Induction of AIDS virus-specific CTL activity in fresh, unstimulated PBL from rhesus macaques vaccinated with a DNA prime/MVA boost regimen. J Immunol. 2000;164:4968–4978. doi: 10.4049/jimmunol.164.9.4968. [DOI] [PubMed] [Google Scholar]
- 3.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 4.Borrow P, Lewicki H, Hahn B H, Shaw G M, Oldstone M B A. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J Virol. 1994;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyson J E, Iwanaga K K, Golos T G, Watkins D I. Identification of a novel MHC class I gene, Mamu-AG, expressed in the placenta of a primate with an inactivated G locus. J Immunol. 1997;159:3311–3321. [PubMed] [Google Scholar]
- 6.Boyson J E, Shufflebotham C, Cadavid L F, Urvater J A, Knapp L A, Hughes A L, Watkins D I. The MHC class I genes of the rhesus monkey: different evolutionary histories of MHC class I and II genes in primates. J Immunol. 1996;156:4656–4665. [PubMed] [Google Scholar]
- 7.Carrington M, Nelson G W, Martin M P, Kissner T, Vlahov D, Goedert J J, Kaslow R, Buchbinder S, Hoots K, O'Brien S J. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 8.Celis E, Karr R W. Presentation of an immunodominant T-cell epitope of hepatitis B surface antigen by the HLA-DPw4 molecule. J Virol. 1989;63:747–752. doi: 10.1128/jvi.63.2.747-752.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dailey P J, Zamroud M, Kelso R, Kolberg J, Urdea M. Quantitation of simian immunodeficiency virus (SIV) RNA in plasma of acute and chronically infected macaques using a branched DNA (bDNA) signal amplification assay. J Med Primatol. 1995;24:209. [Google Scholar]
- 10.Doherty P C. The numbers game for virus-specific CD8+ T cells. Science. 1998;280:227. doi: 10.1126/science.280.5361.227. [DOI] [PubMed] [Google Scholar]
- 11.Dykhuizen M, Mitchen J L, Montefiori D C, Thomson J, Acker L, Lardy H, Pauza C D. Determinants of disease in the simian immunodeficiency virus-infected rhesus macaque: characterizing animals with low antibody responses and rapid progression. J Gen Virol. 1998;79:2461–2467. doi: 10.1099/0022-1317-79-10-2461. [DOI] [PubMed] [Google Scholar]
- 12.Dzuris J L, Sidney J, Appella E, Chesnut R W, Watkins D I, Sette A. Conserved MHC class I peptide binding motif between humans and rhesus macaques. J Immunol. 2000;164:283–291. doi: 10.4049/jimmunol.164.1.283. [DOI] [PubMed] [Google Scholar]
- 13.Eckels D D, Lake P, Lamb J R, Johnson A H, Shaw S, Woody J N, Hartzman R J. SB-restricted presentation of influenza and herpes simplex virus antigens to human T-lymphocyte clones. Nature. 1983;301:716–718. doi: 10.1038/301716a0. [DOI] [PubMed] [Google Scholar]
- 14.Egan M A, Kuroda M J, Voss G, Schmitz J E, Charini W A, Lord C I, Forman M A, Letvin N L. Use of major histocompatibility complex class I/peptide/β2M tetramers to quantitate CD8+ cytotoxic T lymphocytes specific for dominant and nondominant viral epitopes in simian-human immunodeficiency virus-infected rhesus monkeys. J Virol. 1999;73:5466–5472. doi: 10.1128/jvi.73.7.5466-5472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans D T, O'Connor D H, Jing P, Allen T M, Horton H, Venham J E, Rudersdorf R A, daSilva J, Pauza C D, Bontrop R E, DeMars R, Hughes A L, Watkins D I. Virus-specific CTL responses select for amino acid variation in SIV Env and Nef. Nat Med. 1999;5:1270–1276. doi: 10.1038/15224. [DOI] [PubMed] [Google Scholar]
- 16.Furchner M, Erickson A L, Allen T, Watkins D I, Sette A, Johnson P R, Walker C M. The simian immunodeficiency virus envelope glycoprotein contains two epitopes presented by the Mamu-A*01 class I molecule. J Virol. 1999;73:8035–8039. doi: 10.1128/jvi.73.10.8035-8039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallimore A, Cranage M, Cook N, Almond N, Bootman J, Rud E, Silvera P, Dennis M, Corcoran T, Stott J, McMichael A, Gotch F. Early suppression of SIV replication by CD8+ nef-specific cytotoxic T cells in vaccinated macaques. Nat Med. 1995;1:1167–1173. doi: 10.1038/nm1195-1167. [DOI] [PubMed] [Google Scholar]
- 18.Henwood J, Loveridge J, Bell J I, Gaston J S. Restricted T cell receptor expression by human T cell clones specific for mycobacterial 65-kDa heat-shock protein: selective in vivo expansion of T cells bearing defined receptors. Eur J Immunol. 1993;23:1256–1265. doi: 10.1002/eji.1830230610. [DOI] [PubMed] [Google Scholar]
- 19.Higgins J A, Lamb J R, Marsh S G, Tonks S, Hayball J D, Rosen-Bronson S, Bodmer J G, O'Hehir R E. Peptide-induced nonresponsiveness of HLA-DP restricted human T cells reactive with Dermatophagoides spp. (house dust mite) J Allergy Clin Immunol. 1992;90:749–756. doi: 10.1016/0091-6749(92)90098-m. [DOI] [PubMed] [Google Scholar]
- 20.Itescu S, Mathur-Wagh U, Skovron M L, Brancato L J, Marmor M, Zeleniuch-Jacquotte A, Winchester R. HLA-B35 is associated with accelerated progression to AIDS. J Acquir Immune Defic Syndr. 1991;5:37–45. [PubMed] [Google Scholar]
- 21.Johnson R P, Trocha A, Buchanan T M, Walker B D. Recognition of a highly conserved region of human immunodeficiency virus type 1 gp120 by an HLA-Cw4-restricted cytotoxic T-lymphocyte clone. J Virol. 1993;67:438–445. doi: 10.1128/jvi.67.1.438-445.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaslow R A, Carrington M, Apple R, Park L, Munoz A, Saah A J, Goedert J J, Winkler C, O'Brien S J, Rinaldo C, Detels R, Blattner W, Phair J, Erlich H, Mann D L. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 23.Kenter M, Otting N, Anholts J, Leunissen J, Jonker M, Bontrop R E. Evolutionary relationships among the primate Mhc-DQA1 and DQA2 alleles. Immunogenetics. 1992;36:71–78. doi: 10.1007/BF00215282. [DOI] [PubMed] [Google Scholar]
- 24.Klein M R, Keet I P M, D'Amaro J, Bende R J, Hekman A, Mesman B, Koot M, de Waal L P, Coutinho R A, Miedema F. Associations between HLA frequencies and pathogenic features of human immunodeficiency virus type 1 infection in seroconverters from the Amsterdam cohort of homosexual men. J Infect Dis. 1994;169:1244–1249. doi: 10.1093/infdis/169.6.1244. [DOI] [PubMed] [Google Scholar]
- 25.Knapp L A, Cadavid L F, Eberle M E, Knechtle S J, Bontrop R E, Watkins D I. Identification of new Mamu-DRB alleles using DGGE and direct sequencing. Immunogenetics. 1997;45:171–179. doi: 10.1007/s002510050186. [DOI] [PubMed] [Google Scholar]
- 26.Kroner B L, Goedert J J, Blattner W A, Wilson S E, Carrington M N, Mann D L. Concordance of human leukocyte antigen haplotype-sharing, CD4 decline and AIDS in hemophilic siblings. AIDS. 1995;9:275–280. [PubMed] [Google Scholar]
- 27.Kurane I, Dai L-C, Livingston P G, Reed E, Ennis F A. Definition of an HLA-DPw2-restricted epitope on NS3, recognized by a dengue virus serotype-cross-reactive human CD4+ CD8− cytotoxic T-cell clone. J Virol. 1993;67:6285–6288. doi: 10.1128/jvi.67.10.6285-6288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda M C, Schmitz J E, Barouch D H, Craiu A, Allen T M, Sette A, Watkins D I, Forman M A, Letvin N L. Analysis of gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys by cell staining with a tetrameric MHC class I/peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lifson J D, Nowak M A, Goldstein S, Rossio J L, Kinter A, Vasquez G, Wiltrout T A, Brown C, Schneider D, Wahl L, Lloyd A L, Williams J, Elkins W R, Fauci A S, Hirsch V M. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller M D, Yamamoto H, Hughes A L, Watkins D I, Letvin N L. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J Immunol. 1991;147:320–329. [PubMed] [Google Scholar]
- 31.Nelson G W, Kaslow R, Mann D L. Frequency of HLA allele-specific peptide motifs in HIV-1 proteins correlates with the allele's association with relative rates of disease progression after HIV-1 infection. Proc Natl Acad Sci USA. 1997;94:9802–9807. doi: 10.1073/pnas.94.18.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otting N, Kenter M, van Weeren P, Jonker M, Bontrop R E. Mhc-DQB repertoire variation in hominoid and old world primate species. J Immunol. 1992;149:461–470. [PubMed] [Google Scholar]
- 33.Pauza C D, Emau P, Salvato M S, Trivedi P, MacKenzie D, Malkovsky M, Uno H, Schultz K T. Pathogenesis of SIVmac251 after atraumatic inoculation of the rectal mucosa in rhesus monkeys. J Med Primatol. 1993;22:154–161. [PubMed] [Google Scholar]
- 34.Reimann K A, Tenner-Racz K, Racz P, Montefiori D C, Yasutomi Y, Lin W, Ransil B J, Letvin N L. Immunopathogenic events in acute infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Virol. 1994;68:2362–2370. doi: 10.1128/jvi.68.4.2362-2370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowland-Jones S, et al. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 36.Sahmoud T, Laurian Y, Gazengel C, Sultan Y, Gautreau C, Costagliola D. Progression to AIDS in French haemophiliacs: association with HLA-B35. AIDS. 1992;7:497–500. doi: 10.1097/00002030-199304000-00007. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, Ghrayeb J, Forman M A, Montefiori D C, Rieber E P, Letvin N L, Reimann K A. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 38.Shiga H, Shioda T, Tomiyama H, Yakamiya Y, Oka S, Kimura S, Yamaguchi Y, Gojoubori T, Rammensee H-G, Miwa K, Takiguchi M. Identification of multiple HIV-1 cytotoxic T-cell epitopes presented by human leukocyte antigen B35 molecules. AIDS. 1996;10:1075–1083. [PubMed] [Google Scholar]
- 39.Shimizu Y, Koller B, Geraghty D, Orr H, Shaw S, Kavathas P, DeMars R. Transfer of cloned human class I major histocompatibility complex genes into HLA mutant human lymphoblastoid cells. Mol Cell Biol. 1986;6:1074–1087. doi: 10.1128/mcb.6.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slierendregt B L, Bontrop R E. Current knowledge on the major histocompatibility complex class II region in non-human primates. Eur J Immunogenet. 1994;21:391–402. doi: 10.1111/j.1744-313x.1994.tb00212.x. [DOI] [PubMed] [Google Scholar]
- 41.Slierendregt B L, Hall M, Hart B, Otting N, Anholts J, Verduin W, Claas F, Jonker M, Lanchbury J S, Bontrop R E. Identification of an Mhc− DPB1 allele involved in susceptibility to experimental autoimmune encephalomyelitis in rhesus macaques. Int Immunol. 1995;7:1671–1679. doi: 10.1093/intimm/7.10.1671. [DOI] [PubMed] [Google Scholar]
- 42.Tomiyama H, Miwa K, Shiga H, Moore Y I, Oka S, Iwamoto A, Kaneko Y, Takiguchi M. Evidence of presentation of multiple HIV-1 cytotoxic T lymphocyte epitopes by HLA-B*3501 molecules that are associated with the accelerated progression of AIDS. J Immunol. 1997;158:5026–5034. [PubMed] [Google Scholar]
- 43.Trivedi P, Meyer K K, Streblow D N, Preuninger B L, Schultz K T, Pauza C D. Selective amplification of simian immunodeficiency virus genotypes after intrarectal inoculation of rhesus monkeys. J Virol. 1994;68:7649–7653. doi: 10.1128/jvi.68.11.7649-7653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voss G, Letvin N L. Definition of human immunodeficiency virus type 1 gp120 and gp41 cytotoxic T-lymphocyte epitopes and their restricting major histocompatibility complex class I alleles in simian-human immunodeficiency virus-infected rhesus monkeys. J Virol. 1996;70:7335–7340. doi: 10.1128/jvi.70.10.7335-7340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe N, McAdam S N, Boyson J E, Piekarczyk M S, Yasutomi Y, Watkins D I, Letvin N L. A simian immunodeficiency virus envelope V3 cytotoxic T-lymphocyte epitope in rhesus monkeys and its restricting major histocompatibility complex class I molecule Mamu-A*02. J Virol. 1994;68:6690–6696. doi: 10.1128/jvi.68.10.6690-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watkins D I, Kannagi M, Stone M E, Letvin N L. Major histocompatibility complex class I molecules of nonhuman primates. Eur J Immunol. 1988;18:1425–1432. doi: 10.1002/eji.1830180919. [DOI] [PubMed] [Google Scholar]
- 47.Yasutomi Y, Reimann K A, Lord C I, Miller M D, Letvin N L. Simian immunodeficiency virus-specific CD8+ lymphocyte response in acutely infected rhesus monkeys. J Virol. 1993;67:1707–1711. doi: 10.1128/jvi.67.3.1707-1711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yewdell J W, Bennink J R. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]