Abstract
Retrovirus Gag precursor (PrGag) proteins direct the assembly of roughly spherical immature virus particles, while after proteolytic processing events, the Gag capsid (CA) and nucleocapsid (NC) domains condense on viral RNAs to form mature retrovirus core structures. To investigate the process of retroviral morphogenesis, we examined the properties of histidine-tagged (His-tagged) Moloney murine leukemia (M-MuLV) capsid plus nucleocapsid (CANC) (His-MoCANC) proteins in vitro. The His-MoCANC proteins bound RNA, possessed nucleic acid-annealing activities, and assembled into strand, circle (or sphere), and tube forms in the presence of RNA. Image analysis of electron micrographs revealed that tubes were formed by cage-like lattices of CANC proteins surrounding at least two different types of protein-free cage holes. By virtue of a His tag association with nickel-chelating lipids, His-MoCANC proteins also assembled into planar sheets on lipid monolayers, mimicking the membrane-associated immature PrGag protein forms. Membrane-bound His-MoCANC proteins organized into two-dimensional (2D) cage-like lattices that were closely related to the tube forms, and in the presence of both nickel-chelating lipids and RNAs, 2D lattice forms appeared similar to lattices assembled in the absence of RNA. Our observations are consistent with a M-MuLV morphogenesis model in which proteolytic processing of membrane-bound Gag proteins permits CA and NC domains to rearrange from an immature spherical structure to a condensed mature form while maintaining local protein-protein contacts.
Retrovirus particles adopt two major morphological forms (13, 42, 49). Immature virions show an electron-dense layer immediately inside the lipid bilayer envelopes of roughly spherical particles. Mature virions show central, electron-dense cone-shaped, tube-shaped, or spherical ribonucleoprotein structures. The major determinants for mammalian retrovirus particle formation are the retroviral gag genes, which encode precursor Gag (PrGag) proteins (13, 42). While cellularly expressed PrGag proteins are capable of directing the assembly of immature virus particles, proteolytic processing events are necessary for the formation of mature virions (13, 42). In the case of Moloney murine leukemia virus (M-MuLV), PrGag processing yields the mature Gag proteins matrix (MA), p12, capsid (CA), and nucleocapsid (NC). For M-MuLV, the major PrGag protein interaction (I) domain essential for virus particle assembly (42) appears to be composed of a C-terminal portion of CA plus a portion of NC. Like human immunodeficiency virus type 1 (HIV-1), deletion of the amino-terminal MA domain has proven compatible with M-MuLV particle assembly (4, 31, 37, 47), as long as an amino-terminal membrane-anchoring (M) sequence (42) is retained. Similarly, studies have shown that p12-deleted M-MuLV gag genes can direct the assembly of virus-like particles (4, 27, 50). By comparison, investigations have shown that an intact NC domain is essential for efficient M-MuLV particle assembly (4, 27), while major M-MuLV CA deletions compatible with virus formation have yet to be identified.
Relatively little is known about the structures of avian or mammalian retrovirus particles. Nevertheless, electron microscopy (EM) studies on intact and partially disrupted immature HIV (21, 23, 35, 36) and M-MuLV (49) have been informative. In particular, analysis of immature HIV particles expressed by baculovirus vectors showed that PrGag proteins appear to form cage-like lattices on membranes, where protein areas surround protein-free cage holes, spaced at 67- to 70-Å distances (35, 36). Radial-distribution analyses of similar immature HIV particles showed that PrGag proteins are arranged as 15- to 20-nm cylinders, aligned perpendicular to particle surfaces (21). In immature M-MuLV particles, PrGag proteins also were observed as radially arranged cylinders in which the CA and NC domain lengths were approximately 100 and 25 Å, respectively (49). Although M-MuLV particles did not show icosahedral symmetry, some immature virions displayed hexagonal diffraction patterns, and circularly averaged Fourier transforms yielded peaks corresponding to real space distances of 67 ± 11 and 45 ± 10 Å (49).
Because direct examination of retrovirus particles has proven difficult, several researchers have taken the approach of studying structures assembled from retroviral components in vitro (4, 5, 8–10, 22, 24, 46, 53). Initially, Campbell and Vogt demonstrated that proteins composed of the HIV-1 or avian Rous sarcoma virus (RSV) capsid plus nucleocapsid (CANC) proteins assembled in vitro into circular sheets and roughly spherical structures (9). In the presence of RNA, CANC proteins were induced to form 30- to 60-nm-diameter tubes of up to 1 μm long, implying a direct role for RNA in the retrovirus assembly process (9). Subsequent studies supported the observation that CANC plus RNA incubations resulted in the formation of tube- or cone-shaped structures (8, 10, 22) and demonstrated that amino-terminally extended CANC proteins preferentially formed spherical rather than tube-like structures (8, 10). While these investigations suggested that NC and RNA are essential to the assembly process, prior work showed that purified HIV-1 CA proteins were capable of forming higher-order structures in vitro (18). Furthermore, recent studies demonstrated that HIV-1 CA proteins are capable of tube or cylinder formation and that N-terminal extensions of CA proteins also fostered sphere rather than tube formation (24, 46). Although in vitro assembly of tubes from HIV-1 CA required higher protein concentrations than did CANC-plus-RNA tube assembly incubations (24, 46), these results proved that RNA and NC domains were not essential requirements for in vitro tube formation of retroviral Gag proteins.
The in vitro tube and sphere assembly studies have yielded new insights in the retrovirus assembly field. However, they have not addressed the role of membranes in retrovirus particle assembly. This is of interest because HIV, M-MuLV, RSV, and a number of other avian and mammalian retroviruses assemble on membranes (42). Our approach to the in vitro analysis of retrovirus Gag protein interactions has been to assemble N-terminally histidine-tagged (His-tagged) Gag proteins onto membrane monolayers containing nickel-chelating lipids (4, 5, 53). Thus, we have shown that His-tagged HIV-1 CA proteins assemble on lipid monolayers in a fashion similar to PrGag proteins in immature HIV-1 particles (5, 35, 36). His-tagged M-MuLV CA (His-MoCA) proteins were demonstrated to assemble onto membranes in hexamer-hexamer cages (4), which showed notable similarities to immature M-MuLV particles (49). In the present study, we have examined the assembly properties of MuLV His-MoCANC proteins which possess RNA binding and annealing activities. In the presence of RNA, the His-MoCANC proteins assembled helical tubes 68 ± 8 nm in diameter, while in the presence of membranes, the proteins formed two-dimensional (2D) crystals comparable to those of His-MoCA. Remarkably, the His-MoCANC tube and 2D crystal structures were similar to each other, to His-MoCA 2D crystals, and to immature M-MuLV particles. In particular, tube helix lattice vectors corresponded to 1,0 and 1,1 reflections from His-MoCA (4, 50) and His-MoCANC 2D crystal diffraction patterns and to the two predominant Fourier spacings from immature M-MuLV diffraction patterns (49). Both CANC RNA tube and membrane-bound assemblies were composed of protein cage networks incorporating two different types of cage holes, apparently coordinated by hexamers of subunits. Our results suggest that similar CA and NC interactions are utilized in immature and mature M-MuLV particle assemblies.
MATERIALS AND METHODS
Materials.
Egg phosphatidylcholine (PC) was purchased from Avanti Polar Lipids. 1,2-Di-O-hexadecyl-sn-glycero-3-(1′-(2"-R-hydroxy-3′-N-(5-amino-1-carboxypentyl)iminodiacetic acid) propyl ester (DHGN), which was prepared by D. Thompson, and 1,2-dioleoyl-sn-glycero-3-((N-(5-amino-1-carboxypentyl)iminodiacetic acid) succinimide) (DOGS) (Avanti; kindly supplied by L. Wilson-Kubalek [48]) were charged with nickel as described previously (4). Lacey and carbon EM grids (300 mesh) were from Ted Pella. Water was filtered using a MilliQ purification system.
RNA expression constructs and RNA preparation.
The related plasmids pMMwt, pMMΔCD, pMM2ΔPsi, pPsi4Pinawt, and pPsi4PinaΔPsi were modified from previously described constructs (34) by using common cloning techniques (32). Plasmid pMMwt has the M-MuLV region from nucleotides (nt) 1 to 731 directly downstream of a T7 transcription promoter. Plasmid pMMΔCD is derived from pMMwt but has deletions corresponding to M-MuLV nt 310 to 352 plus 355 to 376. Plasmid pMM2ΔPsi is similar to pMMwt but carries two direct repeats of M-MuLV nt 1 to 212 in place of nt 1-731. The 6,700-bp pPsi4Pinawt plasmid contains the M-MuLV region from nt 1 to 731 directly downstream of a T7 transcription promoter, followed by the neo gene and a complete M-MuLV 3′ long terminal repeat sequence. Plasmid pPsi4PinaΔPsi is similar to pPsi4Pinawt but with the sequence from nt 212 to 731 deleted. Another plasmid, pKpngagRR, contains M-MuLV nt 563 (EcoRI linked) to 2856 (KpnI site) in a pGEM3 (Promega) backbone, while pGEM4 was from Promega. For transcriptions, plasmids pMMwt, pMMΔCD, and pMM2ΔPsi were digested with BamHI, pPsi4Pinawt and pPsi4PinaΔPsi were cleaved by SspI, pKpngagRR was cleaved with XhoI, while pGEM4 was cleaved with EcoRI. These digests yielded the following approximate predicted RNA sizes: pMMwt, 730 nt; pMMΔCD, 680 nt; pMM2ΔPsi, 420 nt; pPsi4Pinawt, 4200 nt; pPsi4PinaΔPsi, 3,980 nt; pKpngagRR, 1,010 nt; pGEM4, 60 nt. Endonucleases were removed by phenol-chloroform extraction, and DNAs were ethanol precipitated and resuspended in water treated with 0.05% (vol/vol) diethylpyrocarbonate. Radioactive RNAs were prepared using [32P]GTP (Amersham) and purified by denaturing polyacrylamide gel electrophoresis (5% polyacrylamide) as described previously (26). Unlabeled RNA transcriptions were performed by standard procedures, and aliquots were fractionated by denaturing agarose gel electrophoresis (3) and then analyzed after capillary blotting with 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) onto nylon sheets, UV cross-linking of the RNA to the nylon, and visualization by 20 to 60 s of staining with 0.03% methylene blue in 300 mM sodium acetate (pH 5.2)–5 mM EDTA.
Protein expression vectors.
The M-MuLV CANC proteins were expressed as His-tagged proteins in Escherichia coli strain BL21 (DE3)/pLys (Novagen) from the bacterial expression plasmids pET15B-MoCANC or pET15B- MoCANCexact. Both plasmids contain a M-MuLV CANC region cassette inserted into the BamHI site of pET15B (Novagen). Plasmid pET15B-MoCANC contains the M-MuLV CANC region from viral nt 1261 to 2189 (310 residues), and when expressed in bacteria, the His-tagged MoCANC protein has an amino-terminal sequence leader of MGSSH HHHHH SSGLV APRGS HMLGD and a carboxy-terminal tail of ADPAA NKARK EAELA AATAEQ. Plasmid pET15B-MoCANCexact contains the M-MuLV CANC region from viral nt 1261 to 2234 (325 residues), and when expressed in bacteria, the His-tagged protein has an amino-terminal sequence leader of MGSSH HHHHH SSGLV APRGS HMLGD. Note also that the chemically synthesized 56-residue M-MuLV NC peptide for annealing assays was prepared by Macromolecular Resources (Colorado State University), high-pressure liquid chromatography purified to >90% pure, and stored at −80°C in 1-mg/ml aliquots in storage buffer (25 mM HEPES [pH 6.5], 25 mM NaCl, 0.1 mM ZnCl2, and 0.1 mM β-mercaptoethanol in nanopure water) (17) prior to use.
Protein expression and purification.
Cells of E. coli strain BL21(DE3)/pLys S containing pET15B-MoCANC (or pET15B-MoCANCexact) were grown at 37°C in Luria broth plus 15 μg of chloramphenicol per ml and 50 μg of ampicillin per ml to an optical density at 600 nm of 0.7. Protein expression then was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.5 mM and incubation for 3 h at room temperature. The proteins were purified and stored as described previously (3, 4).
DNA-annealing assays.
DNA-annealing assays were modified from the method of Tsuchihashi and Brown (43). Sense strand DNA oligonucleotide 28-mers (0.2 to 2 ng), end labeled to 104 to 105 cpm/ng, were heated to 90°C in 20 μl of 30 mM Tris (pH 7.5)–60 mM NaCl–1 mM MgCl2–5 mM dithiothreitol (DTT) plus 30 ng of 18-mer antisense strand DNA and 0.3 to 3 ng of antisense strand 28-mer DNA. After being heated, the mixtures were cooled on ice and supplemented with either 2.5 μl of buffer or 2.5 μl of 0.1 to 1.0 mg of protein per ml. Samples then were incubated for 15 min at 42°C and then for 15 min at 25°C, after which nucleic acids were separated from proteins by addition of 2.5 μl of 10% sodium dodecyl sulfate (SDS) and phenol-chloroform extraction. Extracted nucleic acids from protein-minus and protein-plus incubations were fractionated by electrophoresis in parallel with 28-mer single-stranded probes (28s), double-stranded (28-mer plus 18-mer (28s+18as) hybrids, and double-stranded 28-mer plus 28-mer (28s+28as) hybrids. Electrophoresis was performed with 15% polyacrylamide gels (30:0.8, acrylamide/bisacrylamide ratio) and 1× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA [pH 8.2]) for 2 to 2.5 h at 500 V/15.7 cm (30 mA/15.7 cm) and 4°C. After electrophoresis, the products were detected by autoradiography on X-ray film with intensifying screens. For our studies, probe 1 (28s) was AAAATCTCTC TCTCT CTCTC TCTCA AAA, and its antisense 18-mer (18as) and 28-mer (28as) counterparts were TTTTG AGAGA GAGAG AGA and TTTTG AGAGA GAGAG AGAGA GAGAT TTT, respectively. Probe 2 was the 28as counterpart of probe 1; probe 1 was the 28-mer partner of probe 2; while the 18-mer partner of probe 2 was composed of the 5′ 18 nt of probe 1.
RNA mobility shift assays.
Reaction mixtures (30 μl) containing 0 to 600 nM His-tagged protein, E. coli tRNA, or poly(A) (0.5 μg, 1.43 nmol of nucleotides; 48 μM nucleotides) and 20,000 dpm (3 nM nucleotides) of radiolabeled RNAs were incubated for 30 min at room temperature in 50 mM morpholinepropanesulfonic acid (MOPS) (pH 7.8)–100 mM NaCl–2 mM DTT, with or without 2 mM MgCl2, 10 μM ZnCl2, and 5% glycerol. Afterwards, 12 μl of loading buffer (25% glycerol, 0.5× TBE, 0.2% [wt/vol] bromophenol blue) was added, and the samples were fractionated by nondenaturing polyacrylamide gel electrophoresis (5% polyacrylamide) with 10% glycerol–0.5× TBE at 0 to 4°C (60 V for 4 h) and autoradiographed. For studies with different concentrations of RNA, 10-μl volumes containing 6 μg of protein (12 μM) were incubated with 0 to 246 μM unlabeled RNA plus 2 × 104 cpm of radiolabeled RNA as a tracer. After 2-h incubations at 30°C, samples were electrophoresed and autoradiographed as described above. For quantitation, autoradiographs were scanned and analyzed by densitometry using the NIH Image 1.60 software package.
In vitro assembly of RNA-protein complexes.
Drops (10 μl) containing 2.5 to 15 μM His-MoCANC or His-MoCANCexact plus 30 to 180 μM nucleotides in vitro-transcribed RNA in MED (25 mM MOPS [pH 7.8], 1 mM EDTA, 100 mM NaCl, 1 mM DTT) with or without 10 to 100 μM ZnCl2 were incubated for 2 to 24 h at 0 or 30°C on depression well slides (Carlson Scientific Inc., product 101206) that had been placed in Parafilm-sealed 150-mm plastic petri plates containing filter paper wetted with 2.5 ml distilled water. After the incubations, complexes were transferred to uncoated or polylysine-coated ultrathin carbon grids (Pella product 1822-F, Formvar removed) by placing the grids on top of the drop surfaces for 1 min. Grids with adherent particles were placed on 100-μl water drops for 30 s for washing, wicked from one side, placed for 1 min on 60-μl drops of freshly filtered 1.33% uranyl acetate, wicked again, and air dried before being viewed.
Array formation on lipid monolayers.
Lipid monolayer 2D crystallization incubations followed previously established protocols (4, 5, 15, 30, 45) using nickel-chelating lipids (4, 5, 30, 41, 48). Crystalline arrays were transferred onto lacy carbon grids (Pella, product 01883) by placing grids on top of the drops for 10 s. Samples then were processed by placing grids on top of 100-μl water drops for 30 s, wicking from the side, staining for 45 s on 50 μl of 1.3% uranyl acetate (freshly diluted and filtered), blotting, and air drying.
Analysis of RNA binding abilities of monolayer-bound His-MoCANC arrays.
His-tagged proteins (2 μl, 1 to 10 pmol) and radiolabeled RNAs (4 μl; 1 to 10 pmol of nucleotides; specific activity, 5 × 104 to 10 × 104 cpm/pmol of nucleotides) were incubated in lipid monolayer incubations as described above. Arrays were transferred onto lacy carbon grids, after which the materials adhering to the grids were extracted with 50 μl of IPB buffer (20 mM Tris HCl buffer [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate). In small-scale experiments, samples were directly subjected to radioactive counting. In large-scale studies, 10% of each mixture was used to quantify radioactive RNA and the remainder was subjected to SDS-polyacrylamide gel electrophoresis. After electrophoresis, the gels were electroblotted onto nitrocellulose filters and Gag proteins were immunodetected using the mouse anti-CA antibody Hy187, which was visualized by using an alkaline phosphatase-conjugated anti-mouse antibody, followed by a color reaction (26).
Transmission electron microscopy and image digitization.
Electron microscopy was performed on the Portland VA hospital JEOL JEM1200EX instrument, operated at 100 kV, or on the OHSU Philips CM120/Biotwin instrument, operated at 100 kV. Low-dose photography was performed at ambient temperature using Kodak SO163 film. Digitization of areas on negatives was accomplished with an Optronics DEI-470 charge-coupled device camera mounted on a Fisher Stereomaster dissecting microscope equipped with a 0.5× objective lens, using Scion image 1.57 digitization software. As necessary, digitized TIFF images were converted to pgm images using xv and pgm images were converted to MRC img format using pgm_fimg and fimg_flip (to correct image flipping) programs. In some cases, MRC img files were subsequently converted to SUPRIM image files (11) using the SUPRIM program frommrc. For figures, images were reconverted to TIFF files with xv or with the SGI capture and imgview functions.
Image analysis of RNA-protein complexes and tubes.
For morphological analysis of RNA-protein complexes, random areas were photographed, scanned, and analyzed as TIFF or MRC img files (14), using xv, Adobe Photoshop, or boxmrc (14) tools for distance measurements. For RNA-protein tube analysis, 75 tube segments were boxed with boxFilament, Fourier transformed using ICE (28), and viewed as power spectra using SPECTRA (40). Helix unit vectors (33) were determined empirically using the SPECTRA refine lattice option, and vector distances and angles were obtained from the SPECTRA information header box. All 75 diffraction patterns could be defined by unit vectors of 1/(63.8 ± 1.5 Å) and 1/(37.2 ± 1.2 Å), separated by an angle of 22.6 ± 4.4°, although in 33 helices these appeared to correspond to the 3 and 5 layer lines while in 42 helices they appeared to be the 4 and 7 layer lines. For Fourier filtration of tube images, helix images were converted to SUPRIM format (11), padded, transformed, and masked by hand from the power spectrum using the irsel command. Masking was performed to include reflections corresponding either to both helix surfaces or to one helix surface, based on the unit vectors described above. Masks were applied to Fourier transforms using the maskex and mfilter commands, after which filtered tube images were obtained by backtransformation using the SUPRIM dift command. A magnified portion of the Fourier-filtered helix image was obtained by image resizing with Adobe Photoshop.
2D crystallographic analysis.
Crystalline areas on electron micrographs were scanned, digitized, and converted to MRC img format images as described above. Using the ICE and SPECTRA versions of the MRC image analysis packages (2, 14, 28, 29, 40, 44), real-space images were Fourier transformed and diffraction patterns were indexed by hand. Lattices were refined and unbent using the MRC-derived programs MMBOX and UNBEND (2, 29, 40), and unit cell dimensions were obtained from SPECTRA information headers. To correct for contrast transfer function (CTF) variations, amplitude and phase (aph) files were CTF corrected using the ICE-implemented versions of CTF-determine and CTF-apply with the Weiner filter option. Space group fits were calculated using CTF-corrected aph files in conjunction with the MRC ALLSPACE program (14), and ALLSPACE phase origin search output data were used to direct the phase origin centering of His-MoCANC aph file f2x012342a, which was applied with the program aph_phshift. The unbent, CTF-corrected, centered f2x012342a aph file was used as the reference for aph file merging. Merging to 20 Å was performed using iq≤5 reflections (14), with origtiltb for the p6 merge and origtiltc for the p1 merge. Note that for p1 merges, each aph file was reindexed six times to correspond to the six possible hexagonal lattice vector pairs, and the best merging unit cell index for aph file was used in the final p1 merge. After merging, iq≤5 amplitudes and phases for each reflection from origtilt output files were vector averaged to obtain merged, averaged aph files. The quality of these data (R factor) was obtained by calculating the ratio (Q factor [39]) of the amplitude vector sum by the scalar sum of amplitudes, where the value expected for random data is 1 divided by the square root of the number of vectors added (38). To obtain averaged 2D projections of His-MoCANC crystals, vector-averaged, merged amplitude and phase values were assigned to symmetry-related reflections in a p1 unit cell and the resulting aph files were backtransformed using the ICE programs (28) create_tnf, fftrans, and ice_skew. Backtransformed images derived from aph files including data out to the 3,0 reflections, yielding a practical resolution limit of 25.7Å.
RESULTS
Characterization of MoCANC proteins.
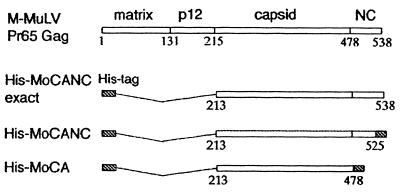
The recombinant His-tagged MoCANC proteins His-MoCANC and His-MoCANCexact were produced from bacterial expression vector plasmids in E. coli. As shown in Fig. 1, His-MoCANCexact encodes a 40-kDa protein composed of the entire CA and NC domains plus an amino-terminal sequence of 25 residues containing a histidine tag. His-MoCANC encodes a 41-kDa protein similar to His-MoCANCexact but which has 21 vector-encoded residues in place of the 11 C-terminal residues of the NC domain (Fig. 1). In parallel with the CANC proteins, we also expressed His-MoCA, a previously described M-MuLV His-tagged CA protein (4). The proteins were purified under nondenaturing conditions by using two rounds of nitrilotriacetic acid-nickel chromatography (4, 5). Based on stain and immunoblot analysis of electrophoretically separated proteins, the protein purities were >90%, with low levels of contaminants or degradation products.
FIG. 1.
Schematic representation of M-MuLV Gag proteins. The M-MuLV Pr65gag precursor protein consists of four domains, matrix (MA), p12, CA, and NC, which are cleaved by the viral protease during maturation. The gag gene contains 538 codons, and the N-terminal methionine of the precursor polypeptide is replaced with a myristyl group during or shortly after synthesis. The modified protein, His-MoCANCexact, expresses the polypeptide CANC regions corresponding to gag codons 213 to 538 and an amino-terminal polyhistidine tag of MGSSH HHHHH SSGLV APRGS HMLGD. His-MoCANC is composed of the His tag, the polypeptide CANC region corresponding to codons 213 to 523, and a carboxy-terminal tail, ADPAA NKARK EAELA AATAEQ. His-MoCA contains an N-terminal polyhistidine tag, the region corresponding to M-MuLV gag codons 213 to 478 and a C-terminal tail of ADPAA NKARK EAELA AATAEQ.
Although retroviral Gag proteins have no known enzymatic activities, the NC domains should bind RNA (1, 6, 7, 9, 12, 16, 42). Consequently, we tested the abilities of His-tagged proteins to bind RNAs. One approach was to employ RNA mobility shift assays (6, 12). To do so, radioactive riboprobes (3 nM labeled nucleotide concentration) were mixed with increasing concentrations of protein in the presence of 48 μM (total nucleotide concentration) RNA competitor. After being subjected to binding incubations at room temperature, samples were analyzed following polyacrylamide gel electrophoresis (5% polyacrylamide) and autoradiography. Not surprisingly, while His-MoCA did not appear to bind RNAs, 200 to 600 nM His-MoCANC and His-MoCANCexact proteins bound nonspecifically to RNA (data not shown). Employing the mobility shift technique with increasing RNA input concentrations, it was possible to obtain a rough indication of the stoichiometry of His-MoCANC binding to RNA. This was accomplished by performing mobility shift assays with 12 μM His-MoCANC plus 0 to 246 μM probe RNA and densitometrically quantitating bound RNA levels. Using this method, maximal binding of 12 μM His-MoCANC was achieved with 50 to 100 μM (total nucleotides) input RNA (data not shown), corresponding to a ratio of 4 to 8 nt of RNA bound per protein monomer, in agreement with previous reports (9, 16).
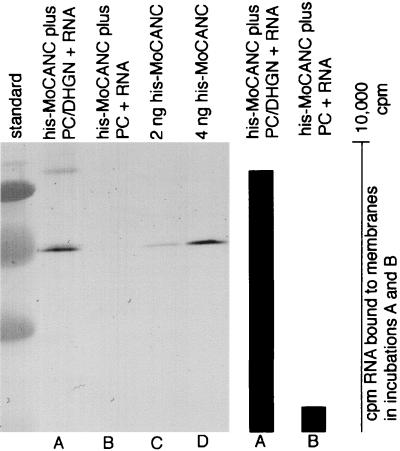
Because the His-MoCANC and His-MoCANCexact proteins retained their His tags, it was possible to assay RNA binding in a different fashion. Specifically, based on the fact that proteins retained His tags, the binding of proteins and protein-bound RNAs to nickel-chelating (and His tag-binding) lipid monolayers could be monitored. Thus, combinations of unlabeled proteins and radiolabeled RNAs were incubated beneath lipid monolayers consisting of PC and the nickel-chelating lipid DHGN (4, 5). After the incubations, monolayers plus bound materials were lifted onto lacy EM grids, washed, eluted, and assayed for levels of bound radioactive RNA. The results, shown in Table 1, demonstrate that His-MoCANC and His-MoCANCexact proteins bound more RNA than did protein-minus reactions while the control His-MoCA protein RNA binding levels were equal to those of the negative control (Table 1). Binding did not require viral encapsidation signals, as demonstrated by the fact that pGEM4, pMM2ΔPsi, and pMMΔCD RNAs were as suitable substrates as the pMMwt viral RNA substrate was. Furthermore, in agreement with previous work (16, 20), the NC domains on Gag proteins were capable of binding RNAs as short as 60 nt (pGEM RNA) but were unable to bind free nucleotide (GTP). That RNA binding to monolayers was mediated by a protein His tag-DHGN interaction was verified in a pair of larger-scale incubations. In particular, His-MoCANC proteins plus labeled RNAs were incubated in large-scale assays beneath monolayers of PC alone or PC plus DHGN. After incubations, the bound and eluted materials were monitored for RNA levels by measuring radioactive counts and for Gag protein levels by immunoblotting. As illustrated in Fig. 2, when DHGN was excluded from lipid mixes, no protein (lane B) or RNA (bar graph B) binding to monolayers was observed.
TABLE 1.
RNA binding abilities of His-MoCANC bound to membranesa
| RNA typeb | Amt of RNA (cpm) bound to proteins assembled on monolayersc
|
|||
|---|---|---|---|---|
| MoCA | MoCANC | MoCANCexact | No protein | |
| pGEM4 | 654 | 2,755 | NDd | 432 |
| pMM2ΔPsi | 102 | 1,948 | 2,104 | ND |
| pMMΔCD | 558 | 4,861 | 4,020 | 668 |
| pMMwt | 623 | 3,181 | 2,210 | ND |
| GTP | 360 | 497 | ND | 399 |
Drop incubations (10 μl) containing proteins (10 pmol) and 1 pmol (nucleotide concentration) of the indicated radiolabeled RNAs or GTP (specific activity, 2.2 × 105 cpm/pmol of nucleotide; except GEM4 RNA, which was 8.8 × 104 cpm/pmol of nucleotide) were overlaid with 1 μl of lipid monolayers composed of PC and Ni2+-DHGN, as described in Materials and Methods. After overnight incubations, monolayers with adherent proteins and RNAs were transferred to EM grids, which were washed, and bound radioactive materials were counted.
Approximate RNA sizes were as follows: pGEM4, 60 nt; pMM2ΔPsi, 420 nt; pMMΔCD, 680 nt; pMMwt, 730 nt.
Values are listed as cpm bound.
ND, not done.
FIG. 2.
RNA binding activity of membrane-bound His-MoCANC proteins. Drops (10 μl), each containing His-MoCANC (4 pmol) and radiolabeled 58-nt RNA (3 pmol of total nucleotides) were overlaid with a lipid monolayer composed of PC and Ni2+-DHGN (lane A), or PC alone (lane B). After overnight incubations at 30°C, monolayers with adherent proteins and RNA were transferred to EM grids, which were stripped to remove bound material. Aliquots (90%) of the mixtures were subjected to SDS-polyacrylamide gel electrophoresis (lanes A and B), along with a size standard (unlabeled left lane) and 2 ng (lane C) or 4 ng (lane D) of purified His-MoCANC protein standards. After electrophoresis and electroblotting, Gag proteins were immunodetected with the mouse anti-CA antibody Hy187, which was revealed by using an alkaline phosphatase-conjugated anti-mouse antibody followed by a color reaction. On the right-hand side of the figure, the bar graph indicates the corrected levels of RNA bound to the protein-membrane samples in lanes A and B. These levels were obtained by radioactive counting of 10% aliquots of the bound material and correspond to 9,100 cpm (A) and 950 cpm (B) of RNA.
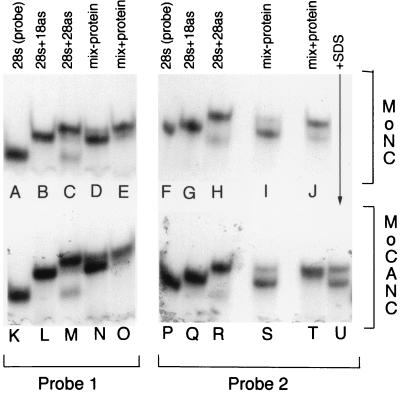
In addition to their abilities to bind nucleic acids, retroviral NC proteins possess annealing activities (13, 19, 43). Thus, it was of interest to test the annealing activities of our His-tagged CANC proteins. These experiments were performed using two different radiolabeled 28-mer oligonucleotide (28s) probes (Fig. 3, lanes A, F, K, and P). As expected, when the probes were annealed with 18-mer antisense (18as) oligonucleotides, their electrophoretic mobilities decreased (lanes B, G, L, and Q), while annealing to full-length 28-mer antisense (28as) oligonucleotides generated even lower-mobility complexes (lanes C, H, M, and R). Incubations at 42°C of mixtures containing probe, limiting amounts of 28as oligonucleotides (0.3 ng), and an excess of 18as oligonucleotides (30 ng) resulted in the preferential formation of probe plus 18as (28s+18as) hybrids (lanes D, I, N, and S). These results were anticipated since formation of the 28s+18as hybrids was kinetically favored over formation of the more stable 28s+28as complexes (19, 43). In contrast, addition of chemically synthesized M-MuLV NC protein to reaction mixtures shifted the balance of products toward 28s+28as hybrids (lanes E and J), consistent with the ability of NC to foster the annealing of thermodynamically stable duplexes. In a similar fashion, we found that purified His-MoCANC facilitated 28s+28as hybrid annealing (lanes O and T). Because His-MoCANCexact also fostered annealing whereas bovine serum albumin and His-MoCA proteins did not display such activity at fivefold-higher concentrations (data not shown) and because the activity of His-MoCANC was impaired by the addition of SDS as a denaturant (lane U), our results indicate that the NC domains of CANC proteins possess both nucleic acid binding and annealing activities. Indeed, equivalent annealing activities were observed with 375 ng (9 pmol) of His-MoCANC and 250 ng (36 pmol) of NC. Although we do not have a measure for the fractions of native His-MoCANC and NC proteins in our preparations, these results suggest that their nucleic acid-annealing activities may be roughly comparable.
FIG. 3.
DNA-annealing assays. DNA-annealing assays employed 28-mer (28s) probe 1 (lanes A to E and K to O) or probe 2 (lanes F to J and P to U), described in Materials and Methods. Lanes A, F, K, and P show autoradiographed probes (0.3 ng) after fractionation on a nondenaturing 15% polyacrylamide gel. Lanes B, G, L, and Q show the migration of probes after hybridization with 10 ng of their 18-mer antisense (18as) counterparts, while lanes C, H, M, and R show the migration of probes hybridized with 0.3 ng of the 28-mer antisense (28as) counterparts. The remaining lanes (D, E, I, J, N, O, and S to U) all contained mixes of probe (0.3 ng), 18as (30 ng), and 28as (0.3 ng) oligonucleotides, incubated in the absence of protein (lanes D, I, N, and S) or in the presence of 2,500 ng of chemically synthesized M-MuLV NC (lanes E and J), or 375 ng of His-MoCANC protein (lanes O, T, and U). Note that protein was added along with SDS to a final concentration of 0.1% in lane U and that 10-fold reduction of M-MuLV NC (250 ng) appeared to have the same annealing activity as the amounts used in lanes E and J.
In vitro assembly of CANC proteins in the presence of RNA.
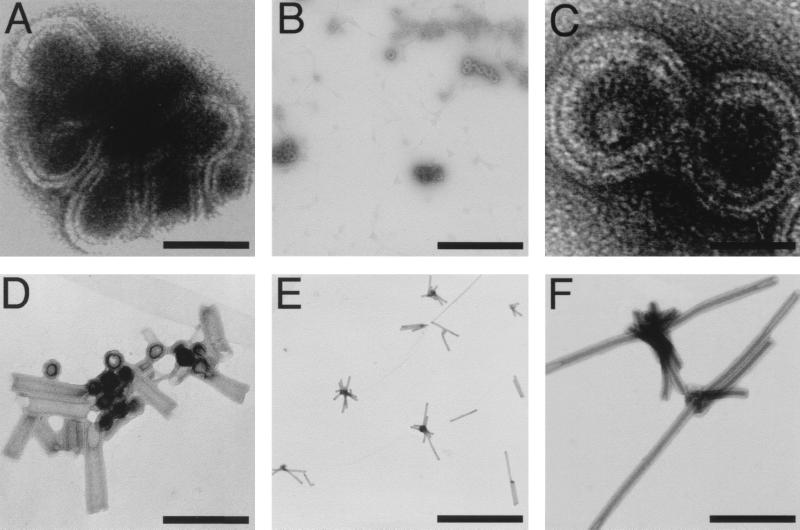
The results described above demonstrated that MoCANC proteins possessed nucleic acid binding and annealing activities, consistent with the notion that they might be able to form ribonucleoprotein complexes (8–10, 22). To examine the ultrastructure of such complexes, CANC proteins were incubated with in vitro-transcribed RNAs, adhered to EM grids, negatively stained, and viewed at a magnification of ×5,000 to ×100,000. As shown in Fig. 4, a number of different morphologies, from strands (Fig. 4A) to circles (Fig. 4B and C) to tubes (Fig. 4D to F), were seen. It is important to emphasize that while the terms “strands” and “circles” denote the appearance of forms in 2D projections, the actual 3D structures represented by these forms may be sheets or spheres. Another point is that tube morphologies did not appear to depend on a specific type or size of RNA but were observed only with fresh, high-purity preparations of RNAs and His-MoCANC or His-MoCANCexact proteins in 16-h incubations at 4°C with protein-to-nucleotide ratios of 1:5 to 1:10. In contrast, strand and circle forms were observed with most of our protein and RNA preparations at different temperatures (4 to 30°C), after different incubation times (2 to 16 h), and with variable protein-to-nucleotide ratios (1:1 to 1:20). Despite these differences, common features were observed between circle and tube morphologies, suggesting that the forms were related. In particular, the apparent wall thicknesses of the circle and tube forms were nearly identical (172 and 174 Å, respectively [Table 2]). Additionally, the diameters of circles (737 ± 69 Å) and tubes (686 ± 81 Å) were in the same range, and circle or tube diameters and tube lengths did not appear to vary with RNA input lengths (Table 2). These results suggest that the circle forms either may be intermediates in the process of tube assembly or may represent spheres with diameters corresponding to tube diameters.
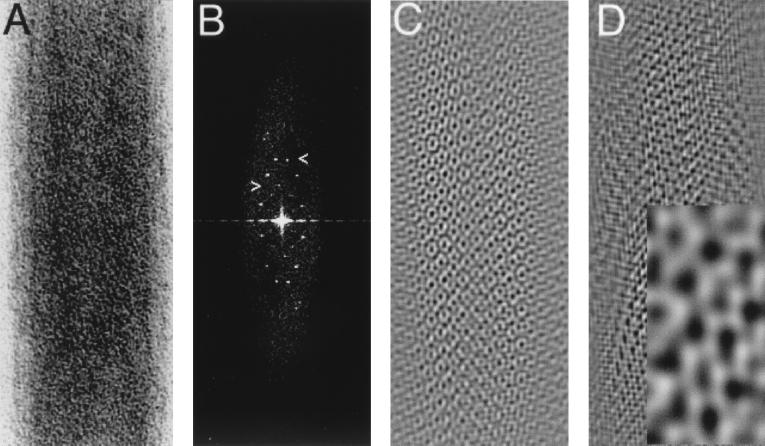
FIG. 4.
In vitro assembly of protein-RNA complexes. His-MoCANC-plus-RNA complexes were assembled, transferred to EM grids, stained, and photographed as described in Materials and Methods and are shown with protein areas appearing white. Samples contained 12 μM His-MoCANC under the following conditions: (A) RNA pPsi4Pinawt (4,200 nt; 180 μM total nucleotide concentration), 16-h incubation at 30°C, photographed at ×27,500 (bar, 87 nm); (B) RNA pPsi4PinaΔPsi (3,980 nt; 180 μM total nucleotide concentration), 16-h incubation at 30°C, photographed at ×27,500 (bar, 924 nm); (C) RNA pPsi4PinaΔPsi (3,980 nt; 180 μM total nucleotide concentration), 16-h incubation at 30°C, photographed at ×52,000 (bar, 46 nm); (D) RNA pMMΔCD (680 nt; 78 μM total nucleotide concentration), 16-h incubation at 4°C, photographed at ×60,000 (bar, 318 nm); (E) RNA pMMΔCD (680 nt; 78 μM total nucleotide concentration), 16-h incubation at 4°C, photographed at ×11,000 (bar, 2,309 nm); (F) RNA pMMΔCD (680 nt; 78 μM total nucleotide concentration), 16-h incubation at 4°C, photographed at ×52,000 (bar, 488 nm).
TABLE 2.
Morphologies of CANC plus RNA particlesa
| Characteristicb | Circles | Tubes |
|---|---|---|
| Frequency (per 100 μm2) | 31–1,471 | 0.6–60 |
| Diameter (Å) | 737 ± 69 (n = 79) | 686 ± 81 (n = 86) |
| Wall thickness (Å) | 172 ± 19 (n = 103) | 174 ± 22 (n = 91) |
| Length (nm) | NAc | 873 ± 453 (n = 118) |
| RNA size (nt) | 424–4,200 | 678–4,200 |
| No. of samples (grids) | 8 | 6 |
Fourteen EM grids, representing separate RNA-plus-His-MoCANC or His-MoCANCexact incubations performed as described in Materials and Methods, were examined for the presence of circle and tube morphological forms. Electron micrographs were obtained at magnifications of ×5,000 to ×75,000 and scanned for analysis.
Frequencies of morphological forms derive from areas of approximately 125 μm2 for circle forms and 300 μm2 for tube forms. Diameters, wall thicknesses, and lengths (for tube forms) were averaged from scanned images, where n indicates the number of individual circles or tubes contributing to the value. Note that the term “circle” is used here to describe the appearance of structures in 2D projections and that the actual 3D morphology of circles may be a spherical form. Note also that since morphological forms did not appear to vary with protein or RNA type, compiled values include results from incubations with His-MoCANC and His-MoCANCexact and with in vitro-transcribed RNAs of 424 to 4200 nt.
NA, not applicable.
Because some of our negatively stained MoCANC-plus-RNA incubations gave apparently regular tube forms, it was of interest to determine whether they showed helical symmetry. Thus, digitized images of straight filament sections from 75 His-MoCANC-plus-RNA tubes were boxed, as in Fig. 5A, after which diffraction patterns were calculated and displayed as power spectra. As illustrated in Fig. 5B, power spectra showed reflections on regularly spaced layer lines, as expected of helical objects. (Note that on some layer lines of some spectra, pairs of reflections were not of equal intensity.) Interestingly, reflections from all diffraction patterns fell on lattice points defined by unit vectors from the origin to layer lines at about 1/66 and 1/38 Å (Fig. 5B, arrowheads). Indeed, reciprocal space lattice dimensions as defined by unit cell vectors for all 75 tubes were consistent (1/a* = 63.8 ± 1.5 Å, 1/b* = 37.2 ± 1.2Å, γ* = 22.6 ± 4.4° [Fig. 5 legend]). As discussed below, ribonucleoprotein tube lattices have similarities to lattices of MoCANC proteins assembled on membranes, suggesting a similarity of the two structural forms.
FIG. 5.
Tubes formed by His-MoCANC plus RNA. (A) His-MoCANC-plus-RNA tube segment. Shown is a section of a His-MoCANC-plus-RNA tube (tube 353-3) with protein areas appearing white. The apparent tube diameter (width) is 774 Å. (B) Power spectrum of the His-MoCANC-plus-RNA tube. The tube segment from panel A was Fourier transformed and is displayed as a power spectrum with the same dimensions as in the original image. The right-pointing arrowhead indicates a layer line at 1/65.7Å, and the left-side reflection on this line defines a lattice vector of 1/64.5Å. The left-pointing arrowhead indicates a layer line at 1/38.6Å, and the right-side reflection on this line defines a lattice vector of 1/37.8Å. (Both lattice vectors were derived after correction for the diffraction pattern rectangularity.) All His-MoCANC-plus-RNA tube diffraction patterns that could be indexed used lattice vectors corresponding to those shown (n = 75), although with some diffraction patterns these corresponded to the 3 and 5 layer lines (n = 33) while in others they corresponded to the 4 and 7 layer lines (n = 42). The average reciprocal space lattice dimensions for measured diffraction patterns were 1/a* = 63.8 ± 1.5 Å, 1/b* = 37.2 ± 1.2 Å, γ* = 22.6 ± 4.4° (n = 35). (C) Filtered image of the His-MoCANC-plus-RNA tube. A Fourier-filtered image of panel A was generated by masking reflections from panel B and applying this mask to the Fourier transform prior to backtransformation. As in panel A, protein areas appear white. (D) Image of one side of the His-MoCANC-plus-RNA tube. A Fourier-filtered image of one side of the tube segment in panel A was generated by masking the subset of reflections from panel B defined by the indicated lattice vectors, followed by filtration and backtransformation. The inset shows a magnification in which the inset width corresponds to 106 Å. As in panel C, protein areas appear white. Note that since the handedness of the tubes has not been defined, it is unclear whether the image represents the proximal or distal surfaces of the tube.
Unfortunately, while the diffraction patterns of negatively stained His-MoCANC-plus-RNA tubes demonstrated helical symmetry, evidence indicated that the helices were somewhat flattened, as exemplified by the well-defined, nonstreaky appearance of the reflections (Fig. 5B). Thus, the tube images were intractable for normal helical reconstruction analysis. Nevertheless, it was possible to perform simple Fourier filtration steps to improve the signal-to-noise ratio of individual tube images. To do so, diffraction patterns were masked to include only reflections (removing noise, which does not lie on helix lattice points) and then backtransformed to yield filtered images. As shown in Fig. 5C, tubes were composed of an array of protein units surrounding protein-free zones. In some areas, the protein-free zones appeared to be coordinated by six protein units, although they were difficult to resolve. Because the image in Fig. 5C was derived from a mask in which all diffraction pattern reflections were included, it depicts the entire ribonucleoprotein tube (upper and lower surfaces of the helix). To obtain a filtered image of only one side of the tube, only reflections representing one side of the tube were included in a backtransformation. The resultant filtered image (Fig. 5D) represents one side of the tube, although because the helical handedness is not known, we are uncertain whether Fig. 5D portrays the upper or lower tube surface. Despite this, some features were evident. In particular, at least two types of protein-free zones were apparent (Fig. 5D, inset), and, as with Fig. 5C, some of the zones appeared to be coordinated by hexamer units. The distances between nearest-neighbor protein-free zones were 34.0 ± 2.6 Å, while the distances between the non-nearest-neighbor protein-free zones were 63.8 ± 9.0 Å. These spacings are slightly smaller but are reminiscent of cage hole-to-hole spacings previously observed with 2D crystals of MoCA proteins assembled on lipid monolayers and bilayers (4, 52). As discussed below, the MoCANC protein arrangement in ribonucleoprotein tubes also is comparable to that observed in CANC membrane-bound arrays, indicating that Gag protein interactions are conserved in the two forms.
Assembly of His-MoCANC proteins on lipid membranes.
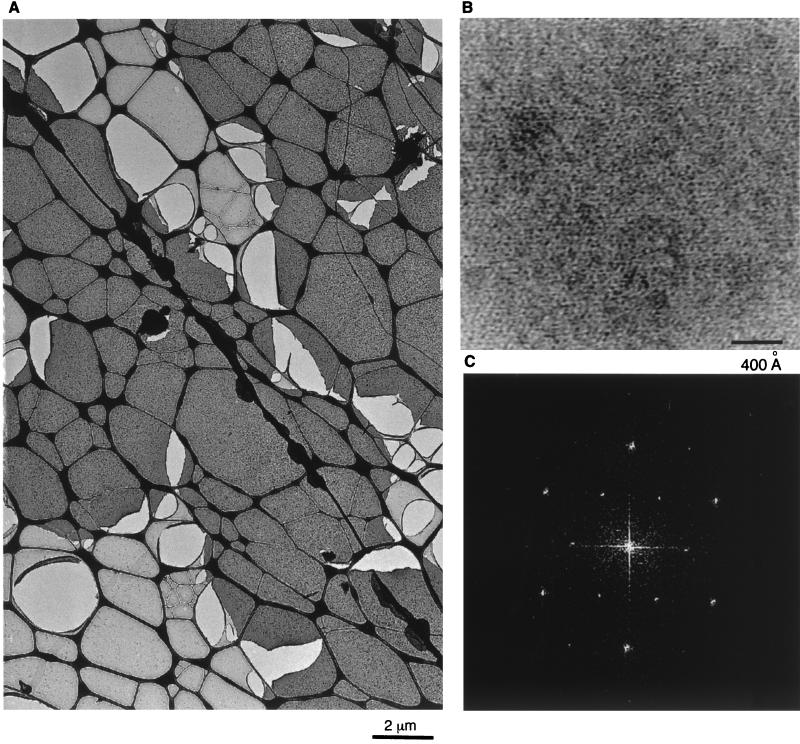
Because M-MuLV Gag proteins naturally assemble on the plasma membranes of infected cells (13), we wished to examine how His-MoCANC proteins assembled at membranes in vitro. To do so and to mimic the membrane-anchoring function of the amino-terminal Gag protein myristate moiety (31, 38), His-CANC proteins were assembled on lipid monolayers containing engineered nickel-chelating lipids (4, 5, 30, 41). Using this approach, we found that His-MoCANC array formation occurred under similar conditions to those for His-MoCA (4). Monolayers with associated proteins were transferred to lacy EM grids, which were washed, stained, and viewed. As shown in Fig. 6A, His-MoCANC proteins formed extensive assemblies on membrane monolayers, in which crystalline arrays could be identified at higher magnification (Fig. 6B). Diffraction patterns from such areas (Fig. 6C) appeared either hexagonal or orthorhombic, with systematic absences. When indexed hexagonally, the averaged real space unit cell for His-MoCANC crystals was a = b = 77.2 ± 2.6 Å, γ = 118.6 ± 2.3° (Table 3), similar to that of His-MoCANCexact (a = b = 76.5 ± 1.4 Å; γ = 120.4 ± 0.9° [Table 3]) and that of His-MoCANC plus RNA (a = b = 77.0 ± 1.5 Å; γ = 120.1 ± 0.0° [Table 3]). Not surprisingly, when data from diffraction patterns were subjected to analysis for space group fitting, best fits (lowest-phase residuals) were obtained for primitive (p1), p2, trigonal (p3), and hexagonal (p6) space groups (Table 3).
FIG. 6.
Assembly of His-MoCANC proteins on a lipid membrane. His-MoCANC proteins were incubated beneath PC-plus-Ni2+-DHGN monolayers overnight at 30°C, after which the membranes with bound proteins were lifted onto lacy EM grids, washed, and stained with uranyl acetate. (A) Stained protein arrays appear as homogenous dark-staining layers which cover the lacy carbon grid support (thick dark lines). Also visible are membrane areas devoid of protein (light grey) and lighter lacy holes indicative of broken membranes. Bar, 2 μm. (B) Shown is a high-magnification image of His-MoCANC proteins arrayed on a lipid monolayer, in which protein areas appear white. Bar, 400 Å. (C) The diffraction pattern of a His-MoCANC protein array was calculated as described in Materials and Methods and is displayed as a power spectrum. The pattern can be indexed in either hexagonal or orthorhombic fashion. The six innermost reflections correspond to the 1,0; 0,1; −1,1; −1,0; 0,−1 and 1,−1 reflections for a γ* = 60° unit cell and correspond to real-space unit cell edges of 77.2 Å.
TABLE 3.
Characteristics of membrane-bound arraysa
| Unit cell dimensions |
| His-MoCANC |
| a = b = 77.2 ± 2.6 Å |
| γ = 118.6 ± 2.3° |
| His-MoCANCexact |
| a = b = 76.5 ± 1.4 Å |
| γ = 120.4 ± 0.9° |
| His-MoCANC plus RNA |
| a = b = 77.0 ± 1.5 Å |
| γ = 120.1 ± 0° |
| Space group determination |
| His-MoCANC phase residuals |
| p1 = 25.4 ± 3.5° |
| p2 = 25.8 ± 6.0° |
| p3 = 16.1 ± 5.9° |
| p6 = 18.1 ± 5.6° |
| Merge statistics |
| His-MoCANC |
| p1 phase residual = 13.6 ± 4.1° |
| p1 R factor = 0.96 ± 0.04 |
| p6 phase residual = 16.5 ± 6.7° |
| p6 R factor = 0.86 ± 0.18 |
| His-MoCANCexact |
| p1 phase residual = 18.6 ± 3.8° |
| His-MoCANC plus RNA |
| p1 phase residual = 34.0 ± 5.4° |
2D His-MoCANC arrays were formed on PC plus Ni+2-DHGN monolayers and stained, photographed, and digitized as described in Materials and Methods. Ten His-MoCANC images were Fourier transformed and viewed as power spectra to permit indexing of diffraction patterns, from which real-space unit cell dimensions are derived. For space group determinations, CTF-corrected, unbent aph files from indexed transforms were used in conjunction with the MRC program ALLSPACE to obtain averaged phase residuals to 20-Å resolution, where 0° represents perfect agreement with a designated space group while a 90° value is indicative of random agreement. Note that the average numbers of comparisons used for p1, p2, p3, and p6 calculations were 23, 12, 17, and 47, respectively, and no other space groups gave phase residuals of less than 30°. Merging of aph files to 20-Å resolution was performed with the program origtiltc for the p1 merge and origtiltb for the p6 merge. Average phase residuals for both merges derive from an average of 11 comparisons per aph file. Average R factors represent vector-averaged amplitude values divided by the corresponding scalar sum of amplitudes for each reflection. R-factor values for random data depend on the number of vectors averaged and correspond to values of 0.41 and 0.23 for the p1 and p6 merges, respectively. Note that while His-MoCANCexact and His-MoCANC+RNA arrays generally gave lower-quality crystals than did His-MoCANC, seven His-MoCANCexact and two His-MoCANC+RNA images also were examined. The His-MoCANCexact images yielded a unit cell size of a = b = 76.5 ± 1.4 Å, γ = 120.4 ± 0.9°, and gave an average phase residual of 18.6 ± 3.8° in a p1 merge to His-MoCANC images. The His-MoCANC+RNA images gave cell parameters of a = b = 77.0 ± 1.5 Å, γ = 120.1 ± 0°, and merged with His-MoCANC files to a phase residual of 34.0 ± 5.4°.
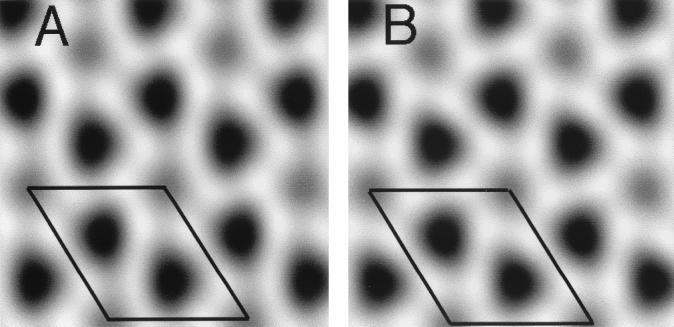
To obtain 2D projection reconstructions from images of MoCANC proteins assembled on lipid monolayers, amplitude and phase data from the reflections of 10 His-MoCANC diffraction patterns were merged, assuming either hexagonal (p6) or no (p1) symmetry, and backtransformed to yield Fourier-filtered projection images (Fig. 7). The p1 (Fig. 7A) and p6 (Fig. 7B) reconstructions gave roughly equivalent merges based on phase residuals and R factors (Table 3), and backtransformed images appeared similar. In particular, each projection (Fig. 7) shows a membrane-bound cage of proteins surrounding at least two different types of protein-free cage holes. In projection, the cage holes appeared to be coordinated by six protein units while distances between nearest-neighbor holes were 46.2 ± 2.5 Å and spacings between the less dark cage holes corresponded to the unit cell distances (77.2 Å). These measurements were similar to those seen with His-MoCA proteins assembled on lipid monolayers (4) and bilayers (53). They also were typical of reconstructions from membrane-bound His-MoCANCexact and His-MoCANC-plus-RNA arrays, observations which are supported by the relatively low phase residuals from merges (Table 3). Furthermore, there was a marked similarity between the membrane-bound (Fig. 7) and RNA-bound (Fig. 5) cages formed by the MoCANC proteins. These similarities are discussed below.
FIG. 7.
Projection reconstructions of membrane-bound His-MoCANC arrays. Ten images of lipid monolayer-bound His-MoCANC arrays were scanned, boxed, Fourier transformed, indexed, CTF corrected, and merged as described in Materials and Methods. Amplitude and phase values for each reflection were vector averaged, after which the averaged values were used in backtransformations to obtain projection reconstructions. Merges were performed assuming no symmetry constraints (p1) (A) or assuming hexagonal (p6) symmetry (B). Images show protein regions as white and protein-free regions as dark. The entire backtransformation grey scale was used in each rendering, and reflections to 25.7 Å were used in reconstructions. The outlined areas indicate unit cells of a = b = 77.2 Å and γ = 120°, and merge statistics are provided in Table 3 and Materials and Methods.
DISCUSSION
We have investigated aspects of retrovirus assembly by in vitro characterization of the properties of M-MuLV Gag proteins composed of the Gag capsid (CA) plus nucleocapsid (NC) domains. The MoCANC proteins (His-MoCANC and His-MoCANCexact) appear to constitute the major PrGag interaction (I: 41) domains (4, 26) and were supplied with amino-terminal His tags to facilitate purification. Additionally, we have demonstrated that His-tagged Gag proteins, used in conjunction with nickel-chelating lipids, can mimic the membrane-anchoring function of the Gag matrix domain, permitting the examination of membrane-bound Gag protein assemblies (4, 5, 52).
In contrast to a His-tagged CA domain expressed in the absence of NC (His-MoCA [4, 53]), MoCANC proteins showed RNA binding and annealing activities (Table 1; Fig. 2 and 3). Our estimates that CANC proteins bind 4 to 8 nt per monomer are consistent with previous studies on other NC domains (9, 16, 20). However, while others have achieved specific in vitro binding of encapsidation signal-containing RNAs by NC-containing polypeptides (12), we were unable to demonstrate specific binding to viral RNA fragments (Table 1). Thus, either our binding conditions were not appropriately stringent or binding specificity requires another viral or nonviral component (1).
Following previously described methods (8–10, 22, 24, 46), His-CANC and His-CANCexact proteins could be induced to assemble ribonucleoprotein strand, circle (or spherical), and tube forms. That circle and tube forms were related was implied by the fact that they had similar diameters and wall thicknesses (Table 2). Thus, circles may constitute intermediates in tube assembly or may represent the final products of an alternate spherical assembly pathway (10, 24, 25, 46). For either tube or circle assembly, it is intriguing that diameters and lengths appeared independent of input RNA lengths (Table 2). Indeed, we observed relatively constant circle and tube diameters (60 to 80 nm) and variable tube lengths (0.1 to 5 μm) regardless of the RNA included in the incubations. These results suggest that CANC protein-protein interactions have determined the diameters of circles and tubes, while tube lengths were determined by as yet undefined factors. Our observations also leave unclear the role of RNA in strand, circle, and tube assembly. Although RNA was required for assembly of these forms, it simply may have been needed to initiate the process or it may have been an essential building block, with multiple pieces of RNA contributing to the final structures.
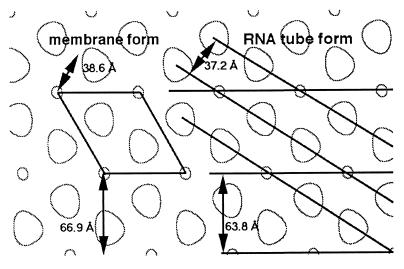
The tubes generated by MoCANC-plus-RNA incubations yielded diffraction patterns consistent with helical symmetry (Fig. 5B). Unfortunately, the helices appeared somewhat flattened, making them unsuitable for conventional helical reconstruction analysis. Nevertheless, Fourier-filtered images showed that tubes were composed of cage-like lattices of proteins and protein-free zones (Fig. 5C and D). The tube lattices were remarkably similar to lattices formed by His-MoCANC (Fig. 7), His-MoCANCexact (Table 3), His-MoCANC plus RNA (Table 3), and His-MoCA (4, 52) assembled on membranes. Specifically, at least two types of protein-free cage holes, apparently coordinated by hexamer units, have been observed in all of the above incubations. The average tube hole-to-hole distances were shorter than the corresponding distances from monolayers, but this distance disparity is likely to be due in part to some residual tube curvature versus relatively flat monolayer arrays. Comparison of reciprocal space unit vectors showed that tube vectors of a* = 1/63.8 Å, b* = 1/37.2 Å, and γ* = 22.6° corresponded well to the distances to 1,0 (1/66.9 Å) and 1,1 (1/38.6 Å) 2D crystal reflections and the angle (29.6°) between them. Mapping of these vectors onto a cartoon of the real-space monolayer-bound lattice of the His-CANC and His-CANCexact proteins (Fig. 8) underscores the close relationship between the MoCANC RNA-bound and membrane-bound forms. Furthermore, the distances between nearest-neighbor cage holes (46 Å) and between ribonucleoprotein a helix or membrane 1,0 planes (63 to 67 Å) closely correspond to the major Fourier spacings of immature M-MuLV particles (45 and 67 Å [49]); these results strongly suggest that protein associations in in vitro assemblies accurately reflect those made in vivo.
FIG. 8.
Comparison of His-MoCANC membrane-bound and RNA tube forms. Real-space His-MoCANC membrane-bound and RNA tube forms are compared on an edge-enhanced representation of a 2D crystal reconstruction, where small circles and larger off-circles indicate protein-free cage holes. On the left is shown a 2D crystal unit cell, along with the distances between the 1,0 (66.9 Å) and 1,1 (38.6 Å) planes (not corrected for γ = 120°). On the right are shown the basic helices of the RNA tube form, where a and b helices are separated by 63.8 and 37.2 Å, respectively. The tube form can be generated from the 2D net by rotating the 2D ab plane slightly and rolling the net into a tube.
From our observations, a number of conclusions concerning the assembly of M-MuLV Gag proteins can be made. One conclusion is that M-MuLV NC domains do not grossly alter the arrangement of membrane-bound Gag proteins, even in the presence of RNA. This supports the idea that at low resolution, membrane-bound CA proteins faithfully mimic the membrane organization of PrGag proteins (4, 49). Our results also imply that in the presence of membrane and RNA binding partners, membrane-Gag interactions preclude the establishment of ribonucleoprotein circle and rod forms. However, in the absence of membrane-Gag interactions, M-MuLV CA and NC domains appear to be free to assemble ribonucleoprotein structures, even in the presence of CA amino-terminal extensions, which inhibit the formation of rod structures by HIV-1 capsid proteins (24, 46). Our observations are consistent with a C-type retroviral morphogenesis model in which proteolytic processing of membrane-bound Gag proteins permits CA and NC domains to rearrange from an immature spherical structure to a condensed mature form while maintaining local protein-protein contacts. Such a model is not without its caveats. For instance, it presumes that in vitro His-tag/Ni2+-DHGN interactions reflect natural PrGag-membrane interactions and defines local protein-protein contacts as those which are detectable at our current level of resolution in 2D projections. In this regard, it should be noted that although current 2D reconstructions suggest that hexamer units coordinate protein-free zones in our crystals and tubes, the precise arrangements of Gag proteins in these structures will not be known until 3D structures are available. Also unknown is the mechanism by which retroviral morphogenesis might occur. Conceivably, morphogenesis might be mediated by rearrangements of putative Gag protein spirals (9), such as those observed in strands and circles (Fig. 4). However, determination of the details of this or alternative models will require further in vivo and in vitro investigations.
ACKNOWLEDGMENTS
This research was supported by a grant from the National Institutes of Health (5R01 GM 52914) to E.B. G.Z. received fellowship support from the Association pour la Recherche sur le Cancer (ARC) and the Human Frontier Science Program. The OHSU Philips CM120/Biotwin transmission electron microscope was obtained with grant support from the National Center for Research Resources (1 S10 RR12935).
We are grateful to Haoyu Qian and Zac Love for help in RNA preparations and to Jessica Willey, Josh Seeds, Brian Arvidson, Keith Mayo, and Doug Huseby for helpful advice and discussions.
REFERENCES
- 1.Bacharach E, Goff S P. Binding of the human immunodeficiency virus type 1 Gag protein to the viral RNA encapsidation signal in the yeast three-hybrid system. J Virol. 1998;72:6944–6949. doi: 10.1128/jvi.72.8.6944-6949.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin J, Henderson R, Beckman E, Zemlin F. Images of purple membrane at 2.8 Å resolution obtained by cryo-electron microscopy. J Mol Biol. 1988;202:585–591. doi: 10.1016/0022-2836(88)90288-4. [DOI] [PubMed] [Google Scholar]
- 3.Barklis E, Lodish H. Regulation of Dictyostelium discoideum mRNAs specific for prespore and prestalk cells. Cell. 1983;32:1139–1148. doi: 10.1016/0092-8674(83)90297-0. [DOI] [PubMed] [Google Scholar]
- 4.Barklis E, McDermott J, Wilkens S, Schabtach E, Schmid M F, Fuller S, Karanjia S, Love Z, Jones R, Rui Y, Zhao X, Thompson D. Structural analysis of membrane-bound retrovirus capsid proteins. EMBO J. 1997;16:1199–1213. doi: 10.1093/emboj/16.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barklis E, McDermott J, Wilkens S, Fuller S, Thompson D. Organization of HIV-1 capsid proteins on a lipid monolayer. J Biol Chem. 1998;273:7177–7180. doi: 10.1074/jbc.273.13.7177. [DOI] [PubMed] [Google Scholar]
- 6.Berkowitz R D, Luban J, Goff S P. Specific binding of human immunodeficiency virus type 1 gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J Virol. 1993;67:7190–7200. doi: 10.1128/jvi.67.12.7190-7200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkowitz R D, Ohagen A, Hoglund S, Goff S P. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69:6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J Virol. 1999;73:2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell S, Vogt V. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J Virol. 1995;69:6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell S, Vogt V. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J Virol. 1997;71:4425–4435. doi: 10.1128/jvi.71.6.4425-4435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carragher B, Whittaker M, Milligan R. Helical processing using PHOELIX. J Struct Biol. 1996;116:107–112. doi: 10.1006/jsbi.1996.0018. [DOI] [PubMed] [Google Scholar]
- 12.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coffin J M, Hughes S H, Varmus H E. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 14.Crowther R, Henderson R, Smith J. MRC image processing programs. J Struct Biol. 1996;116:9–16. doi: 10.1006/jsbi.1996.0003. [DOI] [PubMed] [Google Scholar]
- 15.Darst S, Ahlers M, Meller P, Kubalek E, Blankenburg R, Ribi H, Ringsdorf H, Kornberg R. Two-dimensional crystals of streptavidin on biotinylated macromolecules. Biophys J. 1991;59:387–396. doi: 10.1016/S0006-3495(91)82232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 17.Dememe H, Jullian N, Morellet N, DeRocquigny H, Cornille F, Maigret B, Roques B P. Three-dimensional 1H NMR structure of the nucleocapsid protein Ncp10 of Moloney murine leukemia virus. J Biomol NMR. 1994;4:153–170. doi: 10.1007/BF00175244. [DOI] [PubMed] [Google Scholar]
- 18.Ehrlich L, Agresta B, Carter C. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y, Campbell S, Harvin D, Ehresmann B, Ehresmann C, Rein A. The human immunodeficiency virus type 1 Gag polyprotein has nucleic acid chaperone activity: possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J Virol. 1999;73:4251–4256. doi: 10.1128/jvi.73.5.4251-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher R, Rein A, Fivash M, Urbaneja M, Casas-Finet J, Medaglia M, Henderson L. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller S D, Wilk T, Gowen B E, Krausslich H-G. Cryo-electron microscopy reveals ordered domains in the immature HIV particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 22.Ganser B, Li S, Klishko V, Finch J, Sundquist W. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 23.Gay B, Tournier J, Chazal N, Carriere C, Boulanger P. Morphopoeitic determinants of HIV-1 Gag particles assembled in baculovirus-infected cells. Virology. 1998;242:160–169. doi: 10.1006/viro.1998.9237. [DOI] [PubMed] [Google Scholar]
- 24.Gross I, Hohenberg H, Huckhagel C, Kräusslich H G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gross I, Hohenberg H, Wilk T, Wiegers K, Grattinger M, Muller B, Fuller S, Krausslich H G. A conformation switch controlling HIV-1 morphogenesis. EMBO J. 2000;19:103–113. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen M, Jelinek L, Jones R, Stegeman-Olsen J, Barklis E. Assembly and composition of intracellular particles formed by Moloney murine leukemia virus. J Virol. 1993;67:5163–5174. doi: 10.1128/jvi.67.9.5163-5174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen M, Barklis E. Structural interactions between retroviral Gag proteins examined by cysteine crosslinking. J Virol. 1995;69:1150–1159. doi: 10.1128/jvi.69.2.1150-1159.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardt S, Wang B, Schmid M F. A brief description of I.C.E.: the integrated crystallographic environment. J Struct Biol. 1996;116:68–70. doi: 10.1006/jsbi.1996.0012. [DOI] [PubMed] [Google Scholar]
- 29.Henderson R, Baldwin J, Ceska T, Zemlin F, Beckmann E, Downing K. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 30.Kubalek E, LeGrice S, Brown P. Two-dimensional crystallisation of histidine-tagged HIV-1 reverse transcriptase promoted by a novel nickel-chelating lipid. J Struct Biol. 1995;113:117–123. doi: 10.1006/jsbi.1994.1039. [DOI] [PubMed] [Google Scholar]
- 31.Lee P P, Linial M. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis T, Fritsch E, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 33.Moody M F. Image analysis of electron micrographs. In: Hawkes P W, Valdre U, editors. Biophysical electron microscopy. New York, N.Y: Academic Press, Inc.; 1990. pp. 145–287. [Google Scholar]
- 34.Mougel M, Zhang Y, Barklis E. cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J Virol. 1996;70:4043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nermut M, Hockley D, Jowett J, Jones I, Garreau M, Thomas M. Fullerene-like organization of HIV Gag protein shell in virus-like particles produced by recombinant baculovirus. Virology. 1994;198:288–296. doi: 10.1006/viro.1994.1032. [DOI] [PubMed] [Google Scholar]
- 36.Nermut M, Hockley D, Bron P, Thomas D, Zhang W-H, Jones I. Further evidence for hexagonal organization of HIV Gag protein in prebudding assemblies and immature virus-like particles. J Struct Biol. 1998;123:143–149. doi: 10.1006/jsbi.1998.4024. [DOI] [PubMed] [Google Scholar]
- 37.Reil H, Bukovsky A, Gelderblom H, Gottlinger H. Efficient HIV-1 replication can occur in the absense of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rein A, McClure M R, Rice N R, Luftig R B, Schultz A M. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc Natl Acad Sci USA. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson J, Schmid M F, Morgan D, Chiu W. Three-dimensional structural analysis of tetanus toxin by electron crystallography. J Mol Biol. 1988;200:367–375. doi: 10.1016/0022-2836(88)90247-1. [DOI] [PubMed] [Google Scholar]
- 40.Schmid M, Dargahi R, Tam M. SPECTRA: a system for processing electron images of crystals. Ultramicroscopy. 1993;48:251–264. doi: 10.1016/0304-3991(93)90099-j. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt L, Dietrich C, Tampe R. Synthesis and characterization of chelator-lipids for reversible immobilization of engineered proteins at self-assembled lipid interfaces. J Am Chem Soc. 1994;116:8485–8491. [Google Scholar]
- 42.Swanstrom R, Wills J. Synthesis, assembly and processing of viral proteins. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 43.Tsuchihashi Z, Brown P. DNA strand exchange and selective DNA annealing promoted by the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1994;68:5863–5870. doi: 10.1128/jvi.68.9.5863-5870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unwin P N T, Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol. 1975;94:425–440. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]
- 45.Uzgiris E, Kornberg R. Two-dimensional crystallization technique for imaging macromolecules with application to antigen-antibody-complement complexes. Nature. 1983;301:125–129. doi: 10.1038/301125a0. [DOI] [PubMed] [Google Scholar]
- 46.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C, Zhang Y, McDermott J, Barklis E. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J Virol. 1993;67:7067–7076. doi: 10.1128/jvi.67.12.7067-7076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson-Kubalek E, Brown R, Celia H, Milligan R A. Lipid nanotubes as substrates for helical crystallization of macromolecules. Proc Natl Acad Sci USA. 1998;95:8040–8045. doi: 10.1073/pnas.95.14.8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeager M, Wilson-Kubalek E M, Weiner S G, Brown P O, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryomicroscopy: implication for retroviral assembly mechanisms. Proc Natl Acad Sci USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan B, Li X, Goff S P. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the life cycle. EMBO J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 1995;69:5716–5722. doi: 10.1128/jvi.69.9.5716-5722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuber G, Barklis E. Analysis of retrovirus Gag proteins assembled in vitro on lipid bilayers by EM and AFM. Biophys J. 2000;78:373–384. doi: 10.1016/S0006-3495(00)76600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]