Abstract
The repertoire of functional CD4+ T lymphocytes in human immunodeficiency virus type 1-infected individuals remains poorly understood. To explore this issue, we have examined the clonality of CD4+ T cells in simian immunodeficiency virus (SIV)-infected macaques by assessing T-cell receptor complementarity-determining region 3 (CDR3) profiles and sequences. A dominance of CD4+ T cells expressing particular CDR3 sequences was identified within certain Vβ-expressing peripheral blood lymphocyte subpopulations in the infected monkeys. Studies were then done to explore whether these dominant CD4+ T cells represented expanded antigen-specific cell subpopulations or residual cells remaining in the course of virus-induced CD4+ T-cell depletion. Sequence analysis revealed that these selected CDR3-bearing CD4+ T-cell clones emerged soon after infection and dominated the CD4+ T-cell repertoire for up to 14 months. Moreover, inoculation of chronically infected macaques with autologous SIV-infected cell lines to transiently increase plasma viral loads in the monkeys resulted in the dominance of these selected CDR3-bearing CD4+ T cells. Both the temporal association of the detection of these clonal cell populations with infection and the dominance of these cell populations following superinfection with SIV suggest that these cells may be SIV specific. Finally, the inoculation of staphylococcal enterotoxin B superantigen into SIV-infected macaques uncovered a polyclonal background underlying the few dominant CDR3-bearing CD4+ T cells, demonstrating that expandable polyclonal CD4+ T-cell subpopulations persist in these animals. These results support the notions that a chronic AIDS virus infection can induce clonal expansion, in addition to depletion of CD4+ T cells, and that some of these clones may be SIV specific.
CD4+ T cells are likely to play an important role in maintaining the human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-lymphocyte and humoral immune responses in HIV-1-infected individuals. Interleukin-2 (IL-2) produced by CD4+ T cells has been shown in vitro to enhance CD8+ T-lymphocyte-mediated suppression of HIV-1 replication (16). CD4+ T-cell loss has been shown to be associated with waning anti-HIV-1 antibody responses in infected individuals (2). Moreover, potent HIV-1-specific proliferative CD4+ T-cell responses are associated with the control of viremia in a small group of infected humans whose viremia is controlled in the absence of antiretroviral treatment (26). Yet, virus-specific CD4+ T-cell responses have proven difficult to characterize in most HIV-1-infected individuals. Although proliferative responses of HIV-1-specific CD4+ T cells can be detected in peripheral blood lymphocytes (PBLs) of some HIV-1-infected humans (1, 18, 28, 29), the magnitude and frequency of detectable CD4+ T-cell proliferation are low in the majority of chronically infected individuals (18, 28). Determination of the extent to which the difficulty in detecting virus-specific CD4+ T cells in vitro is attributable to the suppressive nature of an AIDS virus infection or the virus-induced deletion of reactive CD4+ memory T cells is important.
In vivo studies assessing molecular aspects of T-cell receptor (TCR) repertoires will be an important contribution to our understanding of HIV-1-specific CD4+ T cells in virus-infected individuals. While a rapid turnover of CD4+ T cells is observed during HIV-1 infections (21, 27), little is known about the evolution of HIV-1-specific CD4+ T cells in infected humans. It is generally thought that the CD4+ T cells activated through TCR signaling are susceptible to a productive viral infection and virus-induced death. This activation-dependent viral infection and cell death has been seen in vitro in CD4+ T-cell infections (11, 13). However, it is not clear to what extent HIV-1 infections can drive an expansion rather than simply a depletion of viral-antigen-specific CD4+ T cells in infected individuals. Although a dominance of CD4+ T cell clones has been identified at single points in time during clinical progression to AIDS in HIV-1-infected humans (10, 12, 20), longitudinal studies of these clonally dominant CD4+ T cells have not formally been done. It has been argued that such a dominance of CD4+ T cells in advanced infection may be driven by opportunistic pathogens rather than HIV-1. It has also been argued that dominant CD4+ T-cell clones identified during chronic infection may represent selected Vβ-expressing lymphocyte subpopulations remaining following virus-induced polyclonal lymphocyte deletion (14). Further studies are needed to characterize the dynamics of HIV-1-specific CD4+ T-cell population changes in infected individuals.
We have recently initiated studies of CD4+ T-cell repertoires in simian immunodeficiency virus (SIV) SIVmac-infected rhesus monkeys (3, 4, 30). Our previous studies using PCR-based quantitation of Vβ family expression did not demonstrate a consistent expansion or deletion of selected Vβ family-expressing CD4+ PBL subpopulations in genetically unselected SIVmac-infected monkeys (3). This observation suggests that mechanisms other than superantigen-mediated depletion contribute to the CD4+ T-lymphocyte decline in SIVmac-infected monkeys. Nevertheless, it still remains possible that clonal expansion or depletion within individual Vβ family-expressing CD4+ T-lymphocyte subpopulations occurs as a result of persistent antigen stimulation in these virus-infected individuals. We reasoned that prospective studies of macaque TCR complementarity-determining region 3 (CDR3) profiles during SIV infections might provide a useful setting in which to test the hypothesis that an AIDS virus can drive a prolonged CD4+ T-cell clonal response. We therefore utilized CDR3 profile and sequence analyses to evaluate the dynamics of macaque CD4+ T-cell repertoire changes during persistent SIV infections. We report that SIV infection of macaques can result in prolonged periods of clonal dominance of CDR3-restricted CD4+ T cells despite the decline of CD4+ PBL counts.
MATERIALS AND METHODS
Animals and viruses.
Five rhesus monkeys (Macaca mulatta), 3 to 5 years old, were used in these studies. These animals were maintained in accordance with the guidelines of the Committee on Animals for Harvard Medical School and the Guide for the Care and Use of Laboratory Animals (National Academy Press, Washington, D.C.). The rhesus monkeys were inoculated intravenously with SIVmac251 as described previously (3). Three pig-tailed macaques (M. nemestrina) were inoculated intravenously with the pathogenic SIV FGb as described previously (22).
Superinfection with SIV-infected CD4+ T cells.
Rhesus macaques infected with SIVmac for 2 years were subjected to superinfection by using autologous SIV-infected CD4+ T-cell lines. PBLs and lymph node cells obtained from the macaques were depleted of CD8+ T cells by using immunomagnetic beads as previously described (3). The lymphocytes depleted of CD8+ T cells were stimulated for 3 days in culture in the presence of RPMI 1640 medium containing concanavalin A (5 μg/ml). The concanavalin A-stimulated cells were then infected with the SIVmac251 isolate (109 copies of SIV RNA in the supernatant) and expanded in IL-2-containing medium. The infected cells were harvested from the culture on day 12 and inoculated into chronically SIVmac-infected monkeys. Each macaque was inoculated intravenously with 3 × 107 CD4+ T cells (109 copies of SIV RNA in 106 cells).
SEB inoculation.
SIVmac-infected monkeys were injected intramuscularly with staphylococcal enterotoxin B (SEB; Toxin Technology, Sarasota, Fla.) at a dose of 0.3 μg/kg (18). Following SEB inoculation, blood was drawn for flow-cytometric analysis of Vβ expansion and isolation of CD4+ PBLs as previously described (18).
Isolation and fractionation of lymphocyte populations in blood.
PBLs were isolated from EDTA-anticoagulated blood of the monkeys by Ficoll-diatrizoate gradient centrifugation. CD4+ lymphocytes were purified by using monoclonal anti-CD4 antibody-conjugated Dynabeads (Dynal, Inc., Great Neck, N.Y.) as described previously (3). PBLs were incubated with these immunomagnetic beads for 30 min at room temperature and then selected in two cycles with a magnetic particle concentrator. The cells isolated by this method were more than 97% CD4+ lymphocytes.
Monoclonal antibodies and flow-cytometric analysis.
The following anti-human monoclonal antibodies that cross-react with corresponding macaque antigens were used: phycoerythrin (PE)-conjugated anti-rhesus monkey CD3 (FN18; Biosource, Camarillo, Calif.), PE-conjugated anti-human CD4 (Ortho Diagnostic Systems, Raritan, N.J.), and PE-conjugated anti-human CD8 (Dako Corporation, Carpinteria, Calif.). Whole-blood staining was employed according to the instructions accompanying the immunolysis kit (ImmunoPrep; Coulter Corp., Hialeah, Fla.). Two-color flow-cytometric analyses were performed on a Coulter XL flow cytometer. Lymphocytes were gated by forward- and side-scatter characteristics, and up to 10,000 gated cells were analyzed.
mRNA extraction and cDNA synthesis.
mRNA was extracted from purified CD4+ lymphocytes, using guanidinium thiocyanate and oligo(dT) spin columns (mRNA extraction kit; Pharmacia, Piscataway, N.J.). The first-strand cDNA was synthesized in a 20-μl final volume at 42°C for 1 h, using 0.2 to 1 μg of mRNA, 1 μg of random hexanucleotides, and 5 U of reverse transcriptase (Promega, Madison, Wis.). The samples were heated for 5 min at 95°C to terminate the reaction.
TCR Vβ family expression.
PCR-based quantitation of Vβ family expression was undertaken as previously described (3). Briefly, cDNA isolated from each lymphocyte sample was aliquoted into 25 tubes, each containing a sense Vβ family-specific primer and an antisense Cβ primer. As an internal control, each reaction tube also contained a pair of primers that amplified a 105-bp fragment of the constant region of the macaque TCR α chain. The radiolabeled PCR products were electrophoresed through a 5% polyacrylamide gel, dried, and exposed to X-ray film. The radioactivity in the separated Vβ-Cβ and Cα-Cα bands was measured with an Ambis 100 (Ambis, San Diego, Calif.). Using this assay system, we assessed Vβ family expression in CD4+ T cells obtained from monkeys 315 and 320. In these experiments, we were unable to identify a significant expansion or deletion at the level of Vβ family expression. This finding was consistent with our previous observation suggesting the relative conservation of a given Vβ family among genetically unrelated monkeys (3).
TCR β-chain CDR3 length analysis.
CDR3 profiles were analyzed by both autoradiogram display of CDR3 lengths and Genescan-based spectrotyping (10, 12, 30). For autoradiogram display, cDNAs were amplified by PCR for expression of 24 Vβ families, using individual Vβ-specific primers and a Cβ-specific primer as described previously (3, 4, 17, 30). The second round of PCR was performed with nested Vβ primers and a Cβ primer, designed as described previously (30). The internal Cβ primer was labeled at its 5′ end with 32P. The first-round PCR products were amplified for 15 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. The amplified TCR β chains bearing various CDR3 lengths were visualized as a series of radiolabeled bands 3 bases apart on a 6% polyacrylamide sequencing gel. Reactions demonstrating selected Vβ families that exhibited a change in CDR3 length were repeated at least once by the same method, followed by quantitation by the Genescan method. For the spectrotyping analysis of CDR3 profiles, the first-round PCR products were amplified in a second-round PCR using individual nested Vβ primers and the internal Cβ primer coupled with the Fam fluorophore (Applied Biosystems, Foster City, Calif.). The 15-cycle PCR was performed in a 15-μl volume with 0.2 μM each primer under the conditions described above for autoradiographic analysis. One microliter of each reaction product was mixed with deionized formamide and a ROCK-500 size standard and then electrophoresed on a 5% acrylamide gel on a model 377 DNA sequencer (Applied Biosystems). Size and fluorescence intensity were analyzed by using the Genescan software. Experiments performed with samples obtained at three different times from four normal monkeys indicated that these CDR3 length analyses were highly reproducible and Vβ specific. Further cloning and sequencing in conjunction with the CDR3 length display allowed for the prediction of CDR3 lengths. These lengths were expressed as predicted numbers of amino acids. As controls for SIV-induced changes in CDR3 profiles, normal macaques were inoculated with virus-free culture supernatant from CEM174 cells, the cells that were used to expand the SIVmac virus. We saw no significant changes in CDR3 profiles during the ≤5 weeks of follow-up after the inoculation with the supernatant (30).
Molecular cloning and sequencing of selected TCR CDR3-bearing cDNA.
Cloning of TCR CDR3-bearing cDNA was done by a PCR-based technique (6, 18). Briefly, dominant CDR3 length-bearing cDNA, determined by CDR3 length display studies (see Fig. 3B) (17, 30), was cut out of the gel and recovered by boiling at 100°C followed by ethanol precipitation. The recovered DNA was then amplified by PCR using corresponding Vβ-specific primers containing EcoRI restriction sites and a Cβ primer containing an XbaI restriction site (22, 23). cDNAs generated from CD4+ PBL and lymph node samples obtained from the monkeys before SIV infection were included as controls. PCR was performed for 30 cycles as previously described (17, 30). To minimize PCR-generated misincorporation, Pfu DNA polymerase was used in the PCRs. The PCR products were digested with EcoRI and XbaI and ligated into the plasmid pSP65 (Promega) for cloning and sequencing. At least 20 clones were analyzed for each cDNA sample. The frequencies of the clonotypic sequences were determined based on the percentage of each clone in the total clones representing the same Vβ family.
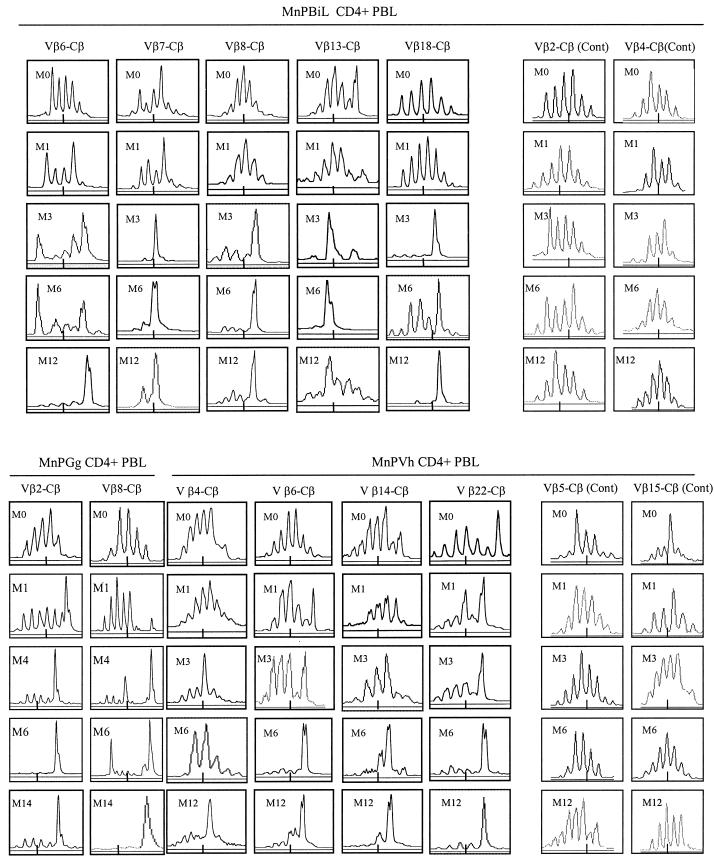
FIG. 3.
Histograms from spectrotyping (A) and autoradiographic (B) analyses of CDR3 lengths employed by selected Vβ-expressing PBL subpopulations in SIVmac-infected rhesus macaques. Spectrotyping data are displayed as described in the legend to Fig. 2. For autoradiographic analysis of CDR3 lengths, 32P-incorporated CDR3 lengths were separated for display in a 5% sequencing gel as previously described (4, 30).
RESULTS
Dominance of selected CDR3-bearing CD4+ T cells was identified in PBLs of SIV-infected monkeys whose CD4+ PBL counts were decreased.
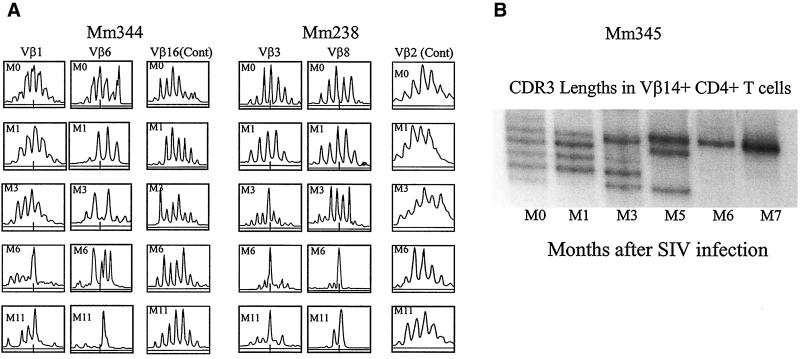
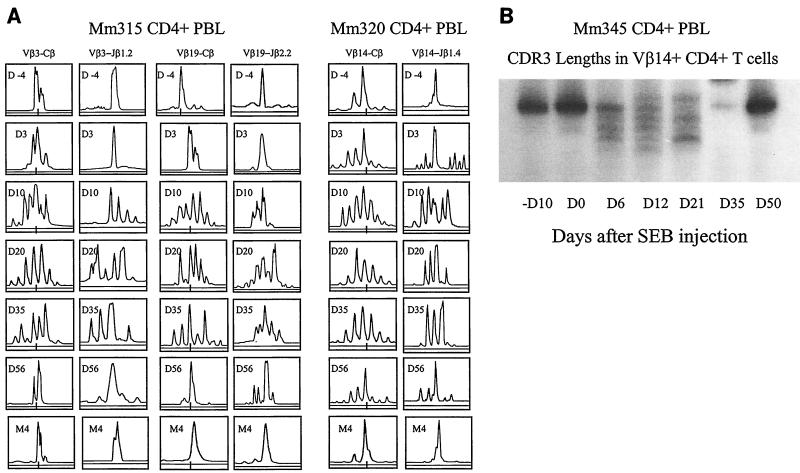
While CD4+ T cells are destroyed by HIV-1 and SIV during the course of an infection, it is possible that some CD4+ T cells may expand clonally in response to these viruses. To explore this possibility, six macaques were infected with two pathogenic SIV isolates, SIV FGb (22) and SIVmac251 (6). The statuses of these SIV-infected macaques were investigated prospectively by examining their virus loads and CD4+ PBL counts. In addition, CD4+ T cells were purified and assessed for alterations in the CDR3 profiles of each of the 24 Vβ gene families. These macaques exhibited a marked decline in their CD4+ PBL counts 2 to 5 months after SIV infection (Fig. 1). Associated with this decline in CD4+ PBL counts were changes in CDR3 profiles in some Vβ family-expressing CD4+ PBL subpopulations (Fig. 2). CDR3 length changes were evident only in selected Vβ gene families in the SIVmac-infected monkey, with the majority of Vβ families showing no evolution (Fig. 2 and data not shown). Similarly, a change in CDR3 profiles from multiple lengths to one- or two-length dominance was identified in certain Vβ families in the SIV FGb-infected macaques (Fig. 2 and data not shown). This dominance of selected CDR3 length-bearing CD4+ T cells persisted in the PBLs of the SIV-infected macaques, in some animals as long as 14 months, although the disappearance of other dominant CDR3-bearing cells was seen over time in those same Vβ families (Fig. 2 and 3 and data not shown). These results, therefore, provide evidence that the persistent infection of two macaque species with different SIV isolates results in the dominance of selected CDR3-bearing CD4+ T cells.
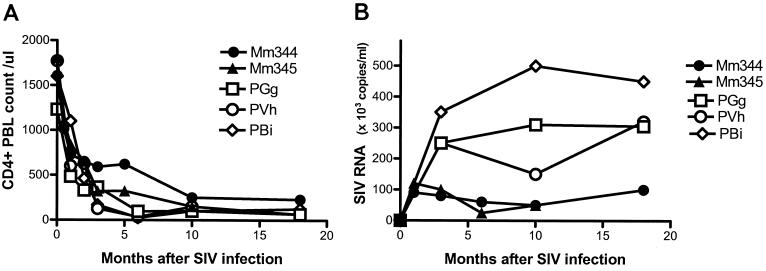
FIG. 1.
Clinical sequelae of SIV infections of macaques evaluated in this study. (A) Marked declines in CD4+ PBL counts were evident in SIVmac-infected rhesus (Mm344 and Mm345) and SIV FGb-infected pig-tailed macaques (PGg, PVh, and PBi). (B) Persistently detectable levels of plasma SIV RNA were present in SIV-infected macaques.
FIG. 2.
Pathogenic SIV infections were associated with the appearance of dominant CDR3-bearing CD4+ T cells in certain Vβ-expressing CD4+ PBL subpopulations. Shown are histograms for spectrotyping analysis of CDR3 lengths employed by selected Vβ-expressing PBL subpopulations of SIV FGb-infected pig-tailed macaques. Monkey designations are noted at the top. Vβ families are indicated, with different months (M) following SIV infection shown at the upper left corners of the histograms. Fragment length in nucleotides is on the x axis, and fluorescence intensity is on the y axis. The numbers of nucleotides in different CDR3 lengths were determined in control experiments (see Materials and Methods) and are expressed as predicted numbers of amino acids. The short line at the bottom of each histogram represents the length of a CDR3 molecule with 10 amino acids. Shown are only those histograms of Vβ families in which a significant change in CDR3 was observed after SIV infection. Histograms of two families that do not exhibit consistent changes in CDR3 are also shown as controls (Cont).
Selected CDR3-bearing CD4+ T cells exhibited a monoclonal or oligoclonal dominance during chronic SIV infection of macaques.
We then assessed the CDR3 sequences employed by the dominant CDR3-bearing CD4+ T cells in chronically SIVmac-infected monkeys. Since the dominance of selected CDR3-bearing CD4+ T cells was most readily appreciated in monkeys with particularly low CD4+ PBL counts (data not shown), longitudinal molecular analyses of single CDR3 length-bearing TCR β-chain cDNA corresponding to the dominant CDR3 were undertaken (17, 30) throughout the clinical course of the SIV infections. Interestingly, monoclonal or oligoclonal sequences were observed in the PCR-derived DNA clones isolated from Vβ cDNA bearing the selected CDR3 lengths, suggesting that selected CDR3-bearing CD4+ T cells in the infected monkeys had undergone clonal expansion (Fig. 4). In accordance with the results revealed by these CDR3 profile analyses, the dominance of these clonal sequences was consistently identified in those Vβ family-expressing CD4+ PBL subpopulations of the monkeys for up to 14 months (Fig. 2 to 4). In fact, dominance of these clones during early infection was confirmed by a frequency analysis of the clones bearing the same CDR3 lengths (Fig. 4). These molecular analyses suggest that SIV infections are temporarily associated with the dominance of selected CDR3-bearing CD4+ T-cell clones in chronically infected macaques.
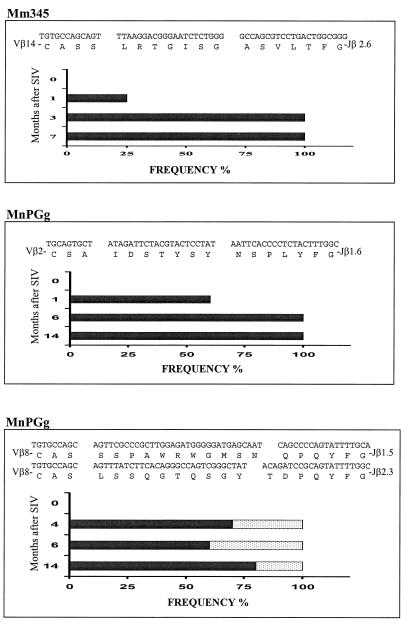
FIG. 4.
Sequence and frequency analyses showed that SIV infections were associated with the prolonged clonal dominance of selected CDR3-bearing CD4+ T cells within certain Vβ-expressing CD4+ PBL subpopulations. cDNA was prepared from purified CD4+ PBLs obtained from SIV-infected macaques. The selected CDR3-bearing clones were generated from the dominant CDR3 bands and the corresponding bands revealed by the autoradiogram CDR3 display. The dominant nucleotide and amino acid sequences of the CDR3-restricted clones are shown at the top of each panel, with their frequencies (x axis) in different months after SIV infections (y axis) displayed in the graphics. The frequency of clonotypic sequences is expressed as the percentage of that clone among the total clones bearing the same CDR3 lengths (17, 30). At least 20 clones were analyzed for each cDNA sample.
Inoculation of SIV-infected macaques with autologous SIV-infected CD4+ T cells induced or enhanced the dominance of selected CDR3-bearing CD4+ T-cell clones.
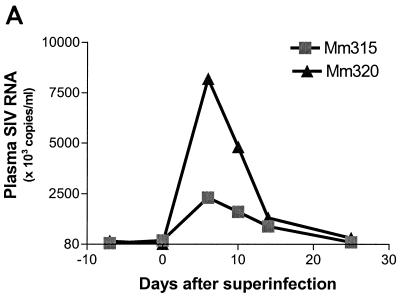
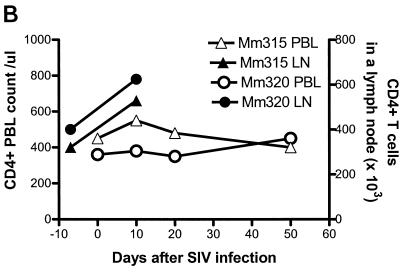
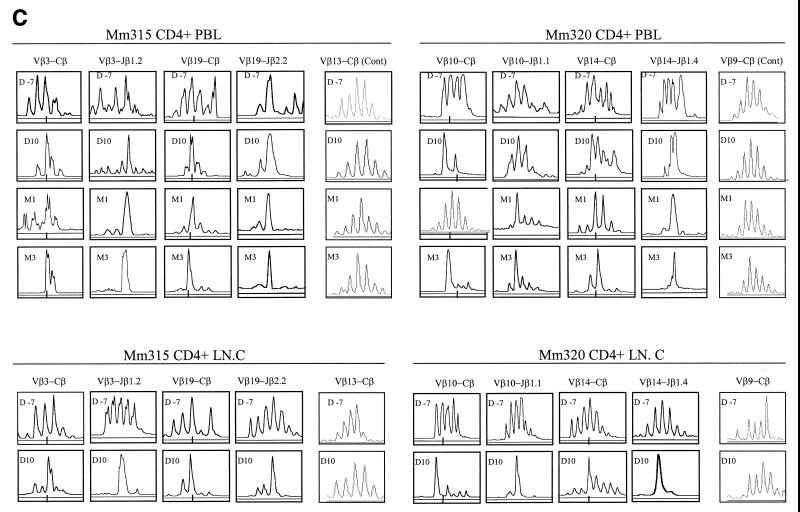
Since the dominance of selected CDR3-bearing CD4+ T cells was most readily appreciated in clinically advanced SIVmac infections, it was important to determine whether the clonal dominance of these CD4+ T cells was associated with SIV rather than with opportunistic pathogens. To address this issue, chronically SIVmac-infected monkeys were superinfected by intravenous inoculation of SIVmac-infected autologous CD4+ T-cell lines and then assessed for changes in both virus loads and CDR3 profiles in each of the 24 Vβ families. The superinfection of two chronically SIVmac-infected monkeys resulted in up to an 80-fold increase in plasma SIV RNA (Fig. 5A). Associated with these changes in viral loads was a transient increase in absolute CD4+ T-cell counts in PBLs and a sampled lymph node (Fig. 5B). Despite these changes, PCR-based TCR repertoire analyses did not reveal a significant expansion of individual Vβ families (there was less than 2.5-fold increase in expression [data not shown]). However, when individual Vβ families were assessed for changes in CDR3 profiles, these transient increases in virus load were shown to be associated with new or an increased magnitude of the dominance of selected CDR3-bearing CD4+ T cells in PBLs and lymph node cells (Fig. 5C). These results strengthen the association between the dominance of selected CDR3-bearing CD4+ T cells and viral antigen in the SIV-infected macaques.
FIG. 5.
Superinfection of two SIVmac-infected macaques with autologous SIV-infected cell lines resulted in an increase in plasma SIV RNA (A), increased numbers of CD4+ T cells (B), and an associated increased dominance of selected CDR3-bearing CD4+ T cells (C). Each macaque was inoculated intravenously with 3 × 107 CD4+ T cells (109 copies of SIV RNA in 106 cells). Plasma RNA quantitation was done by QC-PCR as described elsewhere (30). Absolute numbers of CD4+ PBLs were calculated from data generated by flow cytometry analyses and complete blood counts, whereas those of lymph node CD4+ T cells (LN.C) were derived from flow cytometry data and total lymphocyte counts of a single lymph node (17). The spectrotyping data are displayed as described in the legend to Fig. 2.
Selected CDR3-bearing CD4+ T cells dominated over a polyclonal background in Vβ-expressing T-cell subpopulations during chronic SIV infection.
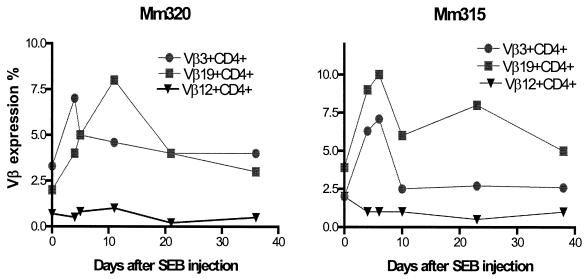
We then examined whether the dominance of selected CDR3-bearing CD4+ T cells represented a clonal expansion from a polyclonal lymphocyte population or, rather, residual clones in a depleted Vβ-expressing CD4+ T-cell subpopulation. We have recently demonstrated that a bacterial superantigen, SEB, can stimulate in vivo polyclonal expansions of reactive Vβ-expressing cell subpopulations in macaques (17). We made use of this observation to examine whether in vivo SEB stimulation could induce a change from mono- or oligoclonal dominance to polyclonal expansion of CD4+ T-cell subpopulations in the SIV-infected macaques. As expected, injection of SIVmac-infected macaques with SEB resulted in a transient expansion of the SEB-reactive Vβ3+ and Vβ19+ cell subpopulations (Fig. 6). Interestingly, this SEB-induced expansion of Vβ3+ and Vβ19+ T cells coincided with a change in the clonal representation in these SEB-reactive cell subpopulations of the SIVmac-infected monkeys. These Vβ-expressing CD4+ T-cell subpopulations underwent a change from the expression of a single dominant CDR3 length to the expression of multiple CDR3 lengths temporally coincident with this SEB-induced expansion (Fig. 7). Following the resolution of these SEB-induced responses, the selected CDR3-bearing CD4+ T cells reemerged as dominant in the Vβ-expressing cell subpopulations. Similarly, sequence analyses of the cDNA from these monkeys showed that the SIVmac-driven clonal dominance in the CD4+ T cells was replaced by a polyclonal representation on day 6 after SEB injection (data not shown), indicating that stimulation by SEB can override SIV-induced clonal dominance and stimulate polyclonal expansion in these T-lymphocyte subpopulations. These SEB-induced changes in clonal representation in the reactive Vβ-expressing subpopulations suggest that a diversity of other expandable clones of CD4+ T lymphocytes remains in chronically SIV-infected macaques.
FIG. 6.
SEB inoculation of SIVmac-infected macaques induced a transient expansion of Vβ3+ and Vβ19+ T cells. Vβ antibody staining of PBLs was done as previously described (21). Macaque Vβ3+, Vβ14+, Vβ18+, and Vβ19+ lymphocyte subpopulations are SEB reactive (21).
FIG. 7.
SEB superantigen inoculation of SIVmac-infected macaques revealed a polyclonal background underlying the dominant selected CDR3-bearing CD4+ T cells in SEB-reactive Vβ families. Transient changes in CDR3 profiles, from a dominant length(s) to multiple CDR3 lengths, occurred after SEB injection. The initial and end time points shown for macaques 315 and 320 were the periods from months 3 through 7 (Fig. 6). The spectrotyping and audioradiograms are displayed as described in the legends to Fig. 2 and 3.
DISCUSSION
In the present studies, we have evaluated virus infection-associated CD4+ T-cell responses in SIV-infected macaques through analyses of TCR CDR3 profiles and sequences. We have shown that dominant CD4+ T-cell clones are seen in SIV-infected macaques following infection. In addition, the relative frequency of these dominant clones increases or new dominant clones emerge in the infected macaques following inoculation with SIV-infected cells. These results suggest that SIV infection can induce the prolonged expansion of virus-specific clonal CD4+ T-cell populations in vivo. These findings also suggest that the dominant Vβ+ CD4+ T-cell clones identified in cross-sectional analyses during the course of HIV-1 infection may represent clonal subpopulations of virus-specific T lymphocytes (10, 12, 20). Nevertheless, we cannot exclude the possibility that changes in CDR3 in selected Vβ families are also driven by antigens derived from opportunistic pathogens replicating in the setting of immunosuppression.
These results also suggest that the dominant CD4+ T-cell clones identified in HIV-1- or SIV-infected individuals are not necessarily residual clones in a depleted pool of T cells. A decrease in the number of clones represented in a given Vβ-expressing T-cell subpopulation might certainly occur as a result of virus-induced declines in total CD4+ T-cell counts. This diminution of background polyclonality would render a clonal expansion more readily detectable. Nevertheless, an SEB superantigen exposure clearly should be capable of revealing a limited polyclonal background underlying an SIV-driven clonal dominance within a selected Vβ-expressing CD4+ T-cell subpopulation. In the present studies, SEB superantigen-induced polyclonal expansions were readily detected in the reactive Vβ-expressing CD4+ T-cell subpopulations that were expanded in response to SIV infection. This finding provides evidence that the clonal dominance of CD4+ T cells in HIV-1 or SIV infections can represent virus-driven expansions rather than residual clones in depleted T-cell subpopulations.
Importantly, these studies suggest that infection-driven CD4+ T cells are detectable during various clinical stages of AIDS virus infections, although the specificity of the clonal expansions observed has not been formally established. In fact, a recent study has demonstrated that HIV-1-specific memory CD4+ T cells can be readily detected by using in vitro peptide stimulation of PBLs followed by intracellular cytokine staining (24). The present studies complement this in vitro characterization of HIV-1-specific CD4+ T cells by demonstrating the in vivo persistence of selected CDR3-bearing CD4+ T-cell clones in SIV-infected macaques. These observations suggest that the development of better assay systems, such as those making use of the major histocompatibility complex tetramer technology, should make possible the characterization of the precise specificities and functions of AIDS virus-specific CD4+ T cells in infected individuals.
The magnitudes of the clonal expansions of SIV-specific CD4+ T cells observed in the present study differ from those seen in CD8+ T-cell subpopulations in SIVmac-infected macaques. Up to threefold expansions of selected Vβ family-expressing subpopulations in CD8+ PBLs can be detected in SIVmac- or simian-human immunodeficiency virus-infected macaques by PCR-based Vβ family quantitation (6). Similarly, substantial numbers of CD8+ T cells bound to a tetrameric major histocompatibility complex class I-peptide complex are apparent in PBLs of SIVmac-infected macaques and HIV-1-infected humans (19, 23). In contrast, the clonal expansion of CD4+ T cells in SIV-infected macaques was detected by CDR3 profile and sequence analyses, but not in studies of Vβ family expression (3). These different magnitudes of expansion may be attributable to the fact that CD4+ T cells are more susceptible than CD8+ T cells to SIV-induced destruction. These differences may also indicate that the mechanisms underlying the clonal expansions of CD4+ and CD8+ T cells are distinct.
The present studies suggest that SIV infections may be capable of driving a prolonged expansion rather than simply a depletion of certain Vβ-expressing CD4+ T-cell subpopulations during the progressive decline of total CD4+ PBL counts. In fact, the dominance of certain clones was maintained even after viral loads returned to baseline following inoculation of macaques with SIVmac-infected cells. It is likely that the replication rate of SIVmac and the turnover rate of T cells were transiently quite high immediately following the inoculation of the infected monkeys with the infected cells. Yet, despite the marked decline of plasma SIV RNA levels after this superinfection, the immune stimulation by viral antigen was sufficient to maintain the expansion of selected clones of CD4+ T cells. There are at least two possible explanations for the persistent dominance of the clonal SIV-specific CD4+ T-cell subpopulations. The prolonged dominance of clones of SIV-specific CD4+ T cells may result from a balance between the proliferation and destruction rates of these clones, with the proliferation rate of the SIV-specific CD4+ T-cell clones being similar to or higher than the rate of SIV-mediated destruction of CD4+ T lymphocytes in SIV-infected macaques. In fact, an increased turnover rate of total CD4+ T cells has recently been demonstrated in SIV-infected macaques (21, 27). The rapid turnover of SIV-specific CD4+ T-cell clones without clonal exhaustion could certainly result in a maintenance of the dominance of these cells within particular Vβ families. On the other hand, these SIV-specific CD4+ T-cell clones may not be susceptible to SIV-induced destruction. SIV-specific CD4+ T cells may acquire the ability to resist SIV infection if they do not express the chemokine receptors required for SIV entry (7, 15). In addition, unique cytokines produced by these CD4+ T-cell clones may preclude a productive SIV infection of these cells (25). This notion is supported by a recent study demonstrating that some cytokines can override activation-dependent HIV-1 infection of CD4+ T lymphocytes (9).
The finding that SIV-stimulated CD4+ T cells persist in virus-infected macaques implies that these cells may contribute to immune containment of SIV. No clear correlation between the detection of dominant CD4+ T-cell clones and viral clearance has been demonstrated in SIV-infected monkeys. However, the return of the elevated plasma SIV RNA level to the set point coincided in this study with an increased dominance of selected CDR3-bearing CD4+ T-cell subpopulations following superinfection with SIV-infected cells (Fig. 5C). This result is consistent with the suggestion that virus-driven CD4+ T cells contribute to antiviral immune responses. These virus-specific CD4+ T cells may play a role in maintaining or enhancing the response of virus-specific CD8+ T cells, although such T-helper function is not effective enough to fully control the virus infection. Treatment with IL-2, a major cytokine produced by CD4+ T cells, has been shown to increase the level of circulating CD4+ T lymphocytes in HIV-1-infected humans on highly active antiretroviral therapy (HAART) (8). If CD4+ T cells mediate antiviral activity, immune intervention targeting virus-specific CD4+ T cells in conjunction with HAART may lead to more-efficient control of HIV-1-induced disease.
ACKNOWLEDGMENTS
This work was supported by NIH grants RR13601 (to Z.W.C.), HL64560 (to Z.W.C.), RR00165 (to Yerkes Regional Primate Research Center), and AI20729 (to N.L.L.).
REFERENCES
- 1.Berzofsky J A, Bensussan A, Cease K B, Bourge J F, Cheynier R, Lurhuma Z, Salaun J J, Gallo R C, Shearer G M, Zagury D. Antigenic peptides recognized by T lymphocytes from AIDS viral envelope-immune humans. Nature. 1988;334:706–708. doi: 10.1038/334706a0. [DOI] [PubMed] [Google Scholar]
- 2.Binley J M, Klasse P J, Cao Y, Jones I, Markowitz M, Ho D D, Moore J P. Differential regulation of the antibody responses to Gag and Env proteins of human immunodeficiency virus type 1. J Virol. 1997;71:2799–2809. doi: 10.1128/jvi.71.4.2799-2809.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z W, Kou Z, Shen L, Reimann K A, Letvin N L. Conserved T cell receptor repertoire in simian immunodeficiency virus-infected rhesus monkeys. J Immunol. 1993;151:2177–2187. [PubMed] [Google Scholar]
- 4.Chen Z W, Kou Z, Shen L, Regan J D, Lord C I, Halloran M, Lee-Parritz D, Fultz P N, Letvin N L. An acutely lethal simian immunodeficiency virus stimulates expansion of Vβ7- and Vβ14-expressing T lymphocytes. Proc Natl Acad Sci USA. 1994;91:7501–7505. doi: 10.1073/pnas.91.16.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z W, Yamamoto H, Watkins D I, Levinson G, Letvin N L. Predominant use of a T-cell receptor Vβ gene family in simian immunodeficiency virus Gag-specific cytotoxic T lymphocytes in a rhesus monkey. J Virol. 1992;66:3913–3917. doi: 10.1128/jvi.66.6.3913-3917.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z W, Kou Z, Lekutis C, Shen L, Zhou D, Halloran M, Li J, Sodroski J, Lee-Parritz D, Letvin N L. T cell receptor Vβ repertoire in an acute infection of rhesus monkeys with simian immunodeficiency viruses and a chimeric simian-human immunodeficiency virus. J Exp Med. 1995;182:21–31. doi: 10.1084/jem.182.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 8.Chun T W, Engel D, Mizell S B, Hallahan C W, Fischette M, Park S, Davey R T, Jr, Dybul M, Kovacs J A, Metcalf J A, Mican J M, Berrey M M, Corey L, Lane H C, Fauci A S. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat Med. 1999;5:651–655. doi: 10.1038/9498. [DOI] [PubMed] [Google Scholar]
- 9.Cohen O J, Kinter A, Fauci A S. Host factors in the pathogenesis of HIV disease. Immunol Rev. 1997;159:31–48. doi: 10.1111/j.1600-065x.1997.tb01005.x. [DOI] [PubMed] [Google Scholar]
- 10.Connors M, Kovacs J A, Krevat S, Gea-Banacloche J C, Sneller M C, Flanigan M, Metcalf J A, Walker R E, Falloon J, Baseler M, Stevens R, Feuerstein I, Masur H, Lane H C. HIV infection induces changes in CD4+ T cell phenotype and depletions within the CD4+ T cell repertoire that are not immediately restored by antiviral or immune-based therapies. Nat Med. 1997;3:533–540. doi: 10.1038/nm0597-533. [DOI] [PubMed] [Google Scholar]
- 11.Cottrez F, Manca F, Dalgleish A G, Arenzana-Seisdedos F, Capron A, Groux H. Priming of human CD4+ antigen-specific T cells to undergo apoptosis by HIV-infected monocytes. A two-step mechanism involving the gp120 molecule. J Clin Investig. 1997;99:257–266. doi: 10.1172/JCI119154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorochov G, Neumann A U, Kereveur A, Parizot C, Li T, Katlama C, Karmochkine M, Raguin G, Autran B, Debre P. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat Med. 1998;4:215–221. doi: 10.1038/nm0298-215. [DOI] [PubMed] [Google Scholar]
- 13.Groux H, Torpier G, Monte D, Mouton Y, Capron A, Ameisen J C. Activation-induced death by apoptosis in CD4+ T cells from human immunodeficiency virus-infected asymptomatic individuals. J Exp Med. 1992;175:331–340. doi: 10.1084/jem.175.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellerstein M K, McCune J M. T cell turnover in HIV-1 disease. Immunity. 1997;7:583–589. doi: 10.1016/s1074-7613(00)80379-9. [DOI] [PubMed] [Google Scholar]
- 15.Hill C M, Deng H, Unutmaz D, KewalRamani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinter A L, Bende S M, Hardy E C, Jackson R, Fauci A S. Interleukin 2 induces CD8+ T cell-mediated suppression of human immunodeficiency virus replication in CD4+ T cells and this effect overrides its ability to stimulate virus expression. Proc Natl Acad Sci USA. 1995;92:10985–10989. doi: 10.1073/pnas.92.24.10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kou Z, Hallouan M, Shen L, Lee-Parritz D, Simon M, Sehgal P, Shen Y, Chen Z W. In vivo effects of a bacterial superantigen in higher nonhuman primates: profound dynamics of TCR repertoire and a dichotomy of responses in different anatomic compartments. J Immunol. 1998;160:5170–5180. [PubMed] [Google Scholar]
- 18.Krowka J F, Stites D P, Jain S, Steimer K S, George-Nascimento C, Gyenes A, Barr P J, Hollander H, Moss A R, Homsy J M, et al. Lymphocyte proliferative responses to human immunodeficiency virus antigens in vitro. J Clin Investig. 1998;83:1198–1203. doi: 10.1172/JCI114001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroda M J, Schmitz J E, Barouch D H, Craiu A, Allen T M, Sette A, Watkins D I, Forman M A, Letvin N L. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J Exp Med. 1998;187:1373–1381. doi: 10.1084/jem.187.9.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macphee R D, Duncan S R, Sattler F R, Theofilopoulos A N. T cell receptor (TCR) BV gene repertoires and clonal expansions of CD4 cells in patients with HIV infections. Clin Exp Immunol. 1997;107:21–30. doi: 10.1046/j.1365-2249.1997.d01-886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohri H, Bonhoeffer S, Monard S, Perelson A S, Ho D D. Rapid turnover of T lymphocytes in SIV-infected rhesus macaques. Science. 1998;279:1223–1227. doi: 10.1126/science.279.5354.1223. [DOI] [PubMed] [Google Scholar]
- 22.Novembre F J, De Rosayro J, O'Neil S P, Anderson D C, Klumpp S A, McClure H M. Isolation and characterization of a neuropathogenic simian immunodeficiency virus derived from a sooty mangabey. J Virol. 1998;72:8841–8851. doi: 10.1128/jvi.72.11.8841-8851.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 24.Pitcher C J, Quittner C, Peterson D M, Connors M, Koup R A, Maino V C, Picker L J. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- 25.Poli G, Kinter A L, Justement J S, Bressler P, Kehrl J H, Fauci A S. Retinoic acid mimics transforming growth factor beta in the regulation of human immunodeficiency virus expression in monocytic cells. Proc Natl Acad Sci USA. 1992;89:2689–2693. doi: 10.1073/pnas.89.7.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg E S, Billingsley J M, Caliendo M A, Broswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 27.Rosenzweig M, DeMaria M A, Harper D M, Friedrich S, Jain R K, Johnson R P. Increased rates of CD4+ and CD8+ T lymphocyte turnover in simian immunodeficiency virus-infected macaques. Proc Natl Acad Sci USA. 1998;95:6388–6393. doi: 10.1073/pnas.95.11.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz D, Sharma U, Busch M, Weinhold K, Matthews T, Lieberman J, Birx D, Farzedagen H, Margolick J, Quinn T, et al. Absence of recoverable infectious virus and unique immune responses in an asymptomatic HIV+ long-term survivor. AIDS Res Hum Retrovir. 1994;10:1703–1711. doi: 10.1089/aid.1994.10.1703. [DOI] [PubMed] [Google Scholar]
- 29.Wahren B, Morfeldt-Månsson L, Biberfeld G, Moberg L, Sönnerborg A, Ljungman P, Werner A, Kurth R, Gallo R, Bolognesi D. Characteristics of the specific cell-mediated immune response in human immunodeficiency virus infection. J Virol. 1987;61:2017–2023. doi: 10.1128/jvi.61.6.2017-2023.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou D, Chalifoux R, Lee-Parritz D, Simon M, Schen Y, Chen Z W. Mycobacterium bovis BCG enhances pathogenicity of simian immunodeficiency virus infection and accelerates progression to AIDS in macaques. J Immunol. 1999;162:2204–2216. [PubMed] [Google Scholar]