Abstract
The vaccinia virus H3L open reading frame encodes a 324-amino-acid immunodominant membrane component of virus particles. Biochemical and microscopic studies demonstrated that the H3L protein was expressed late in infection, accumulated in the cytoplasmic viral factory regions, and associated primarily with amorphous material near immature virions and with intracellular virion membranes. Localization of the H3L protein on the surfaces of viral particles and anchorage via the hydrophobic tail were consistent with its extraction by NP-40 in the absence of reducing agents, its trypsin sensitivity, its reactivity with a membrane-impermeable biotinylation reagent, and its immunogold labeling with an antibody to a peptide comprising amino acids 247 to 259. The H3L protein, synthesized in a coupled in vitro transcription/translation system, was tightly anchored to membranes as determined by resistance to Na2CO3 (pH 11) extraction and cytoplasmically oriented as shown by sensitivity to proteinase K digestion. Further studies demonstrated that membrane insertion of the H3L protein occurred posttranslationally and that the C-terminal hydrophobic domain was necessary and sufficient for this to occur. These data indicated that the H3L protein is a member of the C-terminal anchor family and supported a model in which it is synthesized on free ribosomes and inserts into the membranes of viral particles during their maturation.
Poxviruses comprise a large family of complex DNA viruses that can replicate in the cytoplasm of vertebrate or invertebrate cells. The genome of vaccinia virus, the prototype poxvirus, contains nearly 200 open reading frames (ORFs) of which a large number are virion components (14, 21). Two distinct types of infectious virions, the intracellular mature virions (IMV) and the extracellular enveloped virions (EEV), have been isolated. EEV differ from IMV by the presence of an additional membrane derived from modified Golgi or endosomal cisternae (10, 12, 20, 29, 33). The mechanism of formation of the IMV membrane is poorly understood. In most electron micrographs, crescent membranes or their precursors that form in the presence of the drug rifampin appear to be composed of a single bilayer that is unattached to any cellular organelle (6, 9, 11). Sodeik et al. (31), however, reported that the crescent and immature virion (IV) envelopes comprise two closely apposed membranes that are derived from the cellular intermediate compartment that connects the endoplasmic reticulum (ER) to the Golgi network. Support for this model came from the colocalization of several viral membrane proteins (A17L, A14L, and A13L) with intermediate compartment markers in infected cells (15, 28).
For most eukaryotic integral membrane proteins, the translocation process occurs after a short hydrophobic sequence in the ribosome-bound nascent chain interacts with a signal recognition particle that then docks at the ER (3). Frequently, the N-terminal signal peptide is cleaved from the membrane protein during translocation although in other cases it is retained as a membrane anchor. In addition, many integral membrane proteins are N-glycosylated during transit through the ER. Thus far, of the 12 proteins that have been associated with the IMV membrane (2, 13, 32), 4 have been shown to cotranslationally insert into membranes in vitro (1, 15, 28, 30). Nevertheless, there is no direct evidence that any proteins in the IMV membrane have undergone signal peptide cleavage or N-glycosylation, modifications that would be a signature of ER trafficking. This situation leaves open the possibility that at least some integral membrane proteins are incorporated into viral membranes from the cytoplasm rather than the ER. Such posttranslational association is not unprecedented as there exists a class of eukaryotic integral membrane proteins that are translocated posttranslationally via a C-terminal hydrophobic insertion sequence (17).
We decided to examine the protein encoded by the H3L ORF because previous studies indicated that it is associated with IMV and extractable with a nonionic detergent (13, 32, 39). Here we provide evidence that the H3L protein is expressed late in infection, is exposed on the surfaces of IMV, and can posttranslationally insert into membranes via a C-terminal, hydrophobic anchor sequence.
(This work was carried out in partial fulfillment of Ph.D. requirements of the Curso de Pós Graduação em Microbiologia, Universidade Federal de Minas Gerais, Belo Horizonte, MG, Brazil.)
MATERIALS AND METHODS
Cells and viruses.
BS-C-1, HeLa, and RK13 cells were grown in Earle's modified Eagle's medium (Quality Biologicals Inc.) containing 10% fetal bovine serum. Vaccinia virus (strain WR) was propagated in HeLa cells and titered by plaque assay on BS-C-1 monolayers as described previously (8).
Antibodies.
A synthetic peptide representing amino acids 247 to 259 encoded by the H3L ORF was conjugated to keyhole limpet hemocyanin and used to repeatedly immunize a rabbit to generate polyclonal antibody 6010, which is referred to as the H3L peptide antibody. Louis Potash provided antiserum 8191 (vaccinia virus antibody) from a rabbit that was repeatedly injected with purified vaccinia virus.
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Proteins were separated on 4 to 20% polyacrylamide gradient SDS gels (Owl Separation Systems) in Tris-glycine-SDS buffer except when stated otherwise. The resolved proteins were transferred by electrophoresis to membranes (Immobilon-P; Millipore). The membranes were blocked for 1 h in TTBS (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.05% Tween 20) containing 2.5% (wt/vol) dried nonfat milk and then incubated with 1:500 dilutions of antibody in TTBS containing 0.5% dried milk. After washes with TTBS and milk, the membranes were incubated with antirabbit horseradish peroxidase conjugate (Amersham Life Science) in TTBS–0.5% milk as suggested by the manufacturer. After being washed, immune complexes were detected by a procedure involving use of the Super-Signal chemiluminescent detection kit (Pierce, Rockford, Ill.) and exposure to X-Omat film (Kodak). Alternatively, anti-rabbit immunoglobulin G (IgG) alkaline phosphatase conjugate (Promega) was used as the secondary antibody.
Immunofluorescence microscopy.
Procedures were similar to those described previously (38). HeLa cells were grown on glass coverslips and infected with 5 PFU of vaccinia virus/cell. After 8 h, the cells were washed in phosphate-buffered saline (PBS) and fixed in 3% paraformaldehyde for 20 min. The coverslips were washed in PBS, and the paraformaldehyde was quenched in 50 mM ammonium chloride. After being washed with PBS, the cells were permeabilized with PBS containing 0.05% saponin (Calbiochem). All subsequent wash steps were carried out with this solution. The cells were incubated with H3L peptide antibody in a 1:200 dilution, washed, and then incubated with rhodamine-conjugated anti-rabbit IgG (Dako Corp.). Cells were incubated with a monoclonal antibody (RL77) to protein disulfide isomerase (PDI) followed by fluorescein isothiocyanate-conjugated anti-rabbit IgG (Dako Corp.). Cells were washed and then stained with 5 μl of Hoechst 33258 (Pierce)/ml. Final washes were with PBS and then twice with distilled water. Coverslips were mounted in slides using Fluoromount G (Southern Biotechnology Associates) and observed by confocal microscopy. Images were collected on a Leica TCS NT laser scanning confocal microscope with an attached UV laser; each channel was collected separately, and then the channels were merged.
Electron microscopy.
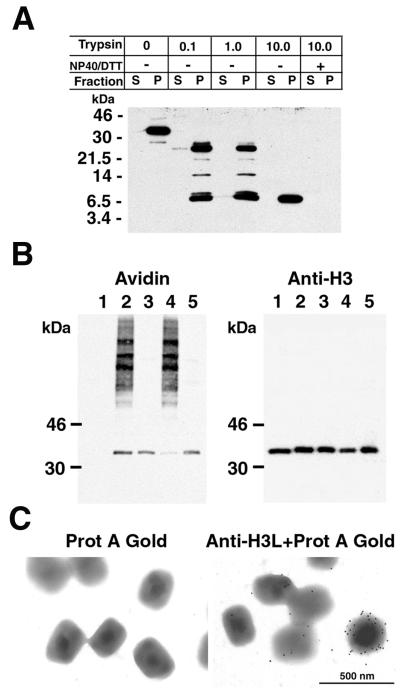
RK13 cells were grown in 60-mm-diameter dishes and infected with vaccinia virus at a multiplicity of 10. After 24 h, the cells were prepared for freezing as previously described except that the final fixation step was in 4% paraformaldehyde (38). Ultrathin sections were cut using a Leica/Reichert Ultracut FSC microtome, collected on Formvar-coated grids, and stained using standard protocols. Grids were incubated with H3L peptide antibody and then with protein A conjugated to 10-nm-diameter colloidal gold particles (Department of Cell Biology, Utrecht University School of Medicine, Utrecht, The Netherlands). Immunostained sections were viewed using a Philips CM100 transmission electron microscope.
Drops of sucrose gradient-purified virus particles were placed on grids. After 10 min the grids were incubated with 0.2% glycine in PBS, blocked with 0.1% bovine serum albumin in PBS, washed, placed on a drop of 1:500-diluted H3L peptide antibody for 30 min, rinsed, and incubated with protein A conjugated to 10-nm colloidal gold particles. The grids were then washed, and the virus particles were fixed with 2% paraformaldehyde and stained with 4% uranyl acetate for 3 min. Grids were washed, dried, and viewed in the Philips CM100 transmission electron microscope (26).
Sucrose and CsCl gradient purification.
Vaccinia virus particles were purified by centrifugation through a sucrose cushion and two successive sucrose gradient sedimentations as described previously (7). Approximately 109 virus particles were loaded on a preformed 11-ml gradient of 1.30 to 1.20 g of CsCl per ml in 10 mM Tris-HCl, pH 9.0. The tubes were centrifuged for 2 h at 25°C in a Beckman SW41 rotor at 32,000 rpm (180,000 × g). After centrifugation, 0.5-ml fractions were collected from the top of the tube. Each fraction was diluted with 1.0 ml of Tris-HCl, pH 9.0, and centrifuged in a microcentrifuge at full speed for 30 min. Pellets were resuspended in 0.1 ml of Tris buffer. The number of virus particles in each fraction was calculated by multiplication of the absorbance at 260 nm by 1.2 × 1010.
NP-40 detergent extraction of virions.
Sucrose gradient-purified vaccinia virus particles (5 × 105) were incubated in 10 mM Tris-HCl, pH 9.0, alone or with 0.5% NP-40 or 0.5% NP-40 plus 50 mM dithiothreitol (DTT). Samples were incubated for 1 h at 37°C and then centrifuged for 30 min at 4°C in a microcentrifuge at full speed. The supernatant and pellet fractions were recovered and analyzed by Western blotting.
Trypsin digestion of intact virions.
Aliquots of purified vaccinia virus WR were incubated at either 4 or 37°C with 0, 0.1, 1.0, or 10.0 mg of trypsin (Sigma-Aldrich, St. Louis, Mo.) per ml of Tris buffer (pH 6.8). Virus that had been disrupted using 1% NP-40 and 10 mM DTT was also incubated with 10.0 mg of trypsin/ml to ensure that the protein was not resistant to digestion. After 30 min, digestions were stopped by the addition of phenylmethylsulfonyl fluoride (Roche Molecular Biochemicals, Indianapolis, Ind.), and virus particles were separated from soluble material by centrifugation. Pelleted and soluble material was resuspended to equivalent volumes in Tricine sample buffer containing final concentrations of 1% SDS and 50 mM DTT. The proteins were separated by electrophoresis on a 10 to 20% Tricine gel (Novex, San Diego, Calif.) and transferred to a nitrocellulose membrane. Samples were analyzed by Western blotting using H3L peptide antibody followed by horseradish peroxidase-conjugated donkey anti-rabbit Ig (Amersham Life Sciences, Piscataway, N.J.), and the signal was developed using Supersignal West Pico chemiluminescent substrate (Pierce) and captured on Biomax Light film (Eastman Kodak).
Biotinylation of vaccinia virus surface proteins.
Sucrose gradient-purified virus particles (109), diluted in 50 μl of PBS, were incubated with 0.5 mg of sulfo–N-hydroxylsuccinimide–LC–biotin (Pierce)/ml for 30 min at room temperature as described by the manufacturer. The reaction mixtures were placed on a 0.1-ml cushion of 36% sucrose and centrifuged at full speed in a microcentrifuge for 30 min in order to remove unincorporated biotin reagent. The pellet was then directly analyzed by SDS-PAGE or first extracted with NP-40 as described above. As a control, purified particles were treated with 0.5% NP-40 and the soluble proteins were biotinylated as described above and subjected to SDS-PAGE. Blots were washed, blocked for 60 min at room temperature in TTBS containing 0.3% casein (I-Block; Tropix), and then incubated for 30 min with NeutrAvidin horseradish peroxidase conjugate (Pierce) diluted 1:20,000 in blocking solution. The membranes were washed three times for 5 min in blocking solution, and the biotin-avidin complexes were detected with the Super-Signal chemiluminescence detection kit followed by exposure to X-Omat film. The blots were then stripped by incubation twice for 30 min in 0.2 M glycine–0.1% SDS–1.0% Tween 20 and probed with the H3L peptide antibody or vaccinia virus antibody and detected as described under “SDS-PAGE and immunoblotting.”
Plasmid templates for in vitro transcription.
Copies of the H3L ORF were obtained by PCR with vaccinia virus DNA as a template. The oligonucleotide primers contained NcoI and BamHI restriction sites at the 5′ and 3′ ends for cloning in plasmid vectors. The primers used, with restriction endonuclease sites underlined, were GGGCCATGGCGGCGGCGAAAACTCCTGTTATTGTTGTGCC and GGGGGATCCTTAGATAAATGCGGTAACGAATGTTCCTGTAAGGAACC. The resulting DNA was digested with NcoI and BamHI (Gibco-BRL) and cloned into the pVOTE.2 plasmid (36) containing a bacteriophage T7 promoter, a modified Escherichia coli lac operator, and an encephalomyocarditis virus cap-independent leader sequence to form pFFH3L. Portions of the H3L ORF with nucleotides encoding amino acids 5 to 15 or 270 to 324 deleted were inserted into the pVOTE.2 plasmid in a similar manner. Another construct with a minigene encoding amino acids 252 to 324 was also constructed. The natural methionine codon at position 252 was used for initiation, and the codon for lysine 253 was changed to a glycine codon in order to accommodate an NcoI restriction site. The PCR primer pairs for those constructions were GCCACCATGGCGGCGGCGAGACTTCCATCAGAAACATTTCCTAATGTTCATGAG and GGG GGATCCTTAGATAAATGCGGTAACGAATGTTCCTGTAAGGAACC, GGGCCATGGCGGCGGCGAAAACTCCTGTTATTGTTGTGCC and GGGGGATCCTTATTATGGATAACGTTTAGTAGCTGCCGTTCCTATTCTAGACCAAAAATTCGG, and GGGCCATGGGACCGAATTTTTGGTCTAGAATAGGAACG and GGGGGATCCTTAGATAAATGCGGTAACGAATGTTCCTGTAAGGAACC.
In vitro transcription and translation.
The reticulocyte lysate-based TNT quick-coupled transcription/translation system (Promega) was used as directed by the manufacturer. To each 40 μl of TNT mixture 50 μCi of [35S]methionine and 1 μg of a plasmid with a T7 promoter-regulated ORF was added. After 90 min at 30°C, the reaction mixtures were centrifuged in a microcentrifuge for 20 min at full speed. The supernatants were collected and analyzed by SDS-PAGE. The gels were dried and exposed to X-Omat film.
Some transcription/translation reactions were carried out in the presence of 2.5 μl of canine pancreatic microsomal membranes (Promega). After the reaction, the mixture was centrifuged for 20 min at full speed in a microcentrifuge. The supernatant was recovered and kept on ice. In order to remove proteins loosely bound to the microsomes, the pellet was suspended in 0.2 ml of PBS and centrifuged again. (In some experiments, the pellet was then suspended in 0.1 ml of Na2CO3 solution, pH 11, kept on ice for 20 min, and then recentrifuged.) The pellets were then resuspended in 50 μl of PBS, layered on top of 75 μl of 0.5 M sucrose, and centrifuged at full speed for 20 min (15). The microsome-associated proteins were then analyzed by SDS-PAGE or first suspended in PBS and treated for 1 or 10 min with 0.5 mg of protease K (Gibco-BRL)/ml in the absence or presence of Triton X-100 (1%) detergent at room temperature. In other experiments, the sucrose cushion-purified membranes were extracted with Triton X-114 and the distribution of the H3L protein aqueous and detergent phases was determined as described previously (26).
To demonstrate posttranslational membrane insertion, transcription/translation reactions were carried out in the absence of microsomal membranes. Then cycloheximide (200 μg/ml) was added to block further translation, and 2.5 μl of microsomal membranes was added. After 30 min at 30°C, the membranes were collected, washed, and analyzed as described above.
RESULTS
Temporal synthesis of the H3L protein.
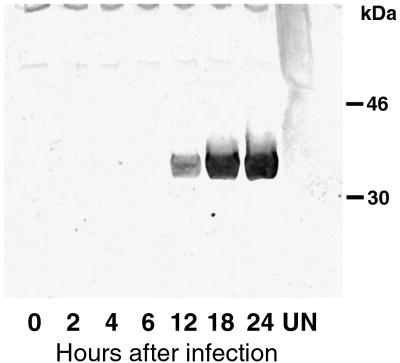
Virion proteins, including membrane components, are generally expressed late in infection at the time of particle assembly. Inspection of the DNA sequence near the start of the H3L ORF revealed a TAAATG motif typical of late promoters (27). To detect the H3L protein, we made a polyclonal antibody to a peptide proximal to the C-terminal hydrophobic tail. Late expression of the H3L protein in vaccinia virus-infected BS-C-1 cells was demonstrated by SDS-PAGE and immunoblotting (Fig. 1). A polypeptide migrating as expected for a mass of 37.5 kDa was detected at 12 h, indicating that synthesis was initiated between 6 and 12 h. Similar mobilities were found with and without use of reducing agents, suggesting the absence of disulfide-bonded oligomers.
FIG. 1.
Synthesis of the H3L protein. BS-C-1 cells in six-well plates were mock infected (UN) or infected with 10 PFU of vaccinia virus per cell for 0, 2, 4, 6, 12, 18, or 24 h. The cells were collected by centrifugation, lysed with SDS and mercaptoethanol, passed several times through a 25- by 5-mm needle, and analyzed by SDS-PAGE. The separated proteins were transferred to a membrane and probed with rabbit polyclonal H3L peptide antibody followed by an antirabbit horseradish peroxidase conjugate. The bands detected by chemiluminescence are shown. The positions and masses of markers are indicated on the right.
Intracellular localization of H3L protein.
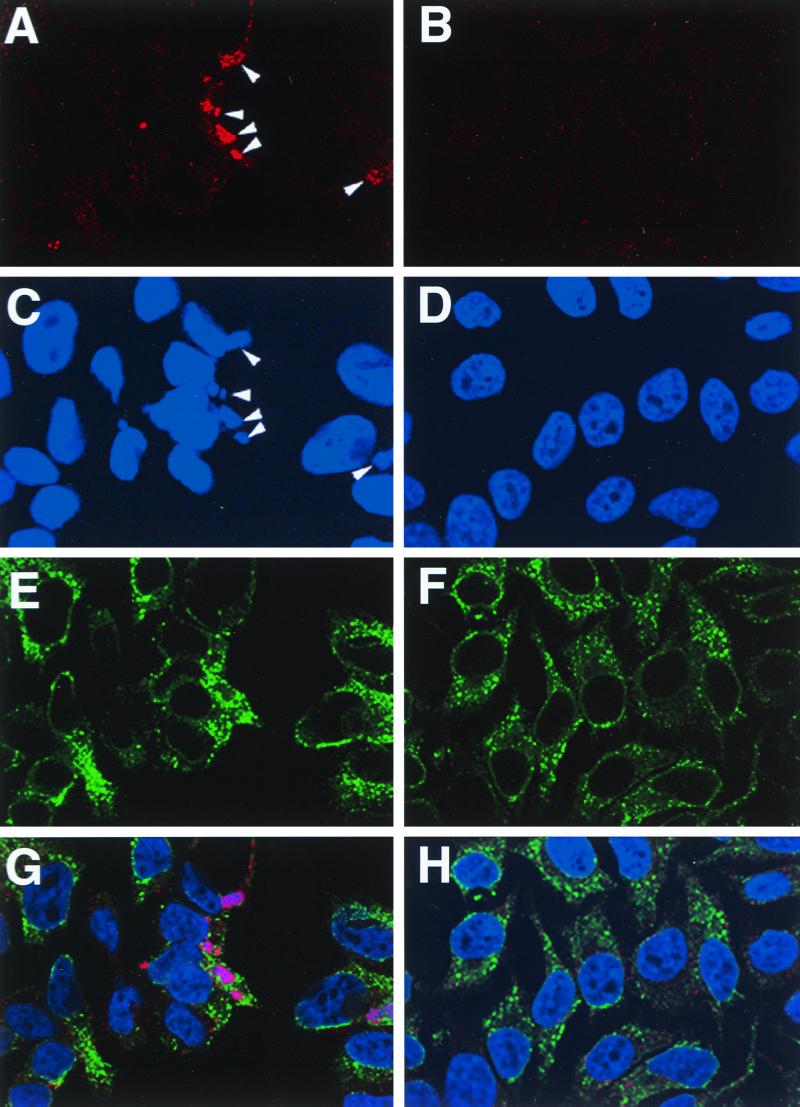
Immunofluorescence microscopy was used to determine the intracellular location of the H3L protein. At 8 h after infection, prior to the accumulation of large numbers of mature virus particles, HeLa cells were fixed, permeabilized, and incubated successively with the H3L peptide antibody and rhodamine conjugated to anti-rabbit IgG. Additionally, the cells were stained with an antibody to PDI and with Hoechst reagent in order to visualize the ER and DNA, respectively. In uninfected cells, the Hoechst dye was exclusively localized to the nucleus (Fig. 2D), whereas it was also present as discrete factory areas of the cytoplasm in cells infected for 8 h with vaccinia virus (Fig. 2C). The H3L protein, detected by rhodamine fluorescence (Fig. 2A), colocalized with the Hoechst-stained factories at this time (Fig. 2C and G). PDI staining of uninfected cells outlined the nucleus and filled the cytoplasm in a reticular pattern characteristic of the ER (Fig. 2F and H). In infected cells, a similar reticular pattern was observed except that the stain also outlined the cytoplasmic factories from which it appeared to be excluded (Fig. 2E and G). Although there was light cytoplasmic H3L staining detected outside of the factories (Fig. 2A), distinctive ER or Golgi network fluorescence was not discerned (Fig. 2E and G). At later times after infection, the factories were more dispersed and small, punctate H3L staining bodies that could represent virions were observed near the periphery of the cell (not shown).
FIG. 2.
Localization of the H3L protein in infected cells. Confocal microscopy of uninfected HeLa cells (B, D, F, and H) and cells infected with vaccinia WR virus (A, C, E, and G). After 8 h, cells were fixed, permeabilized, and triple labeled with anti-H3L peptide antibody and rhodamine-conjugated anti-rabbit IgG, anti-PDI and fluorescein isothiocyanate-conjugated anti-mouse IgG, and Hoechst dye. Fluorescence due to the H3L antibody (A and B), DNA (C and D), and PDI (E and F) and a merged image of all three (G and H) are shown. Arrowheads, viral factories.
IMV membrane association.
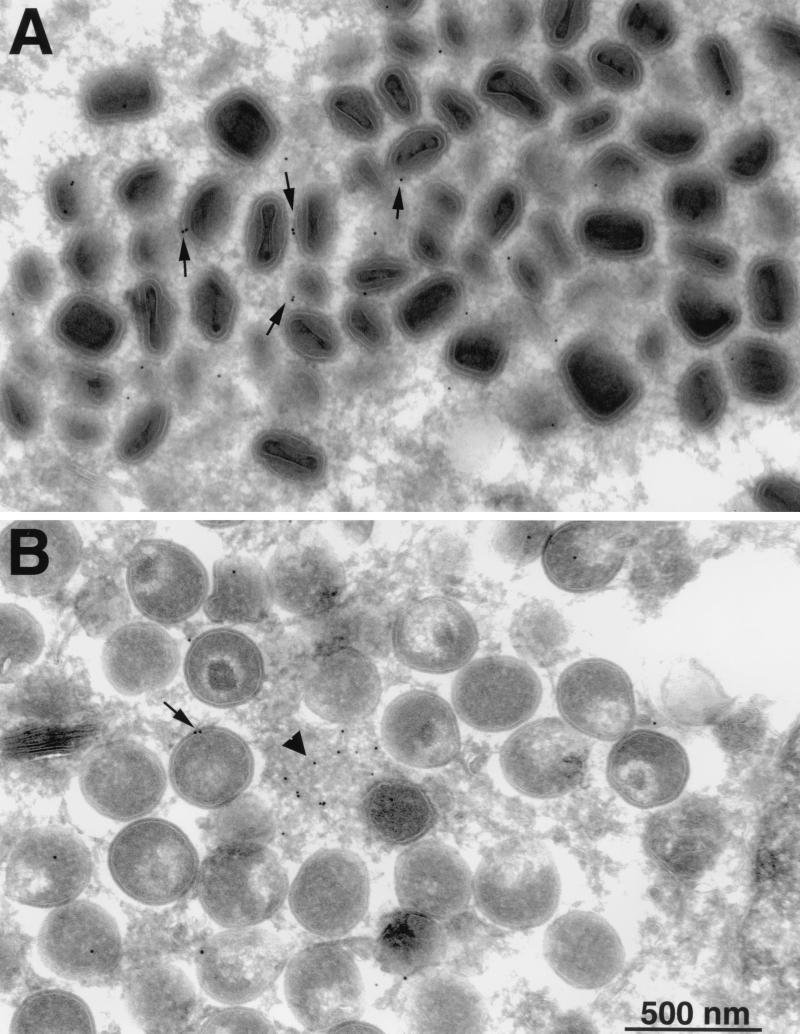
The association of the H3L protein with IMV was demonstrated by immunoelectron microscopy. Thin sections of infected cells were incubated with H3L peptide antibody followed by protein A conjugated to gold particles. Because immature and mature virus particles tend to be located in different regions of the cell, two fields are shown. In factory areas containing clusters of crescents and IV, gold grains were seen on amorphous material but comparatively few were coincident with viral membranes (Fig. 3B). In contrast, there were more gold grains overlying IMV membranes (Fig. 3A). This impression was confirmed by counting gold grains on identifiable viral structures. Thus, the numbers of grains were one to three on 6 of 92 crescents, one to three on 68 of 469 IV, and one to four on 206 of 564 mature virus particles. The background was determined to be insignificant by counting gold grains on sections of infected cells that had been stained with protein A-gold alone. The background numbers of grains were one on 2 of 571 crescents and IV and one on 5 of 688 mature particles. The background was also found to be insignificant when cells that had been infected with an H3L deletion mutant described in the accompanying paper (5) were stained with the H3L antibody and protein A-gold (data not shown). The pattern of immunogold labeling suggested that the H3L protein accumulated in depots in factory areas and was incorporated into viral membranes during the maturation of virus particles.
FIG. 3.
Immunogold electron microscopy. BS-C-1 cells that had been infected for 24 h were fixed in paraformaldehyde, cryosectioned, and incubated with H3L peptide antibody and then with 10-nm gold particles conjugated to protein A. (A) Field containing large numbers of IMV. (B) Field containing large numbers of IV. Arrows, gold particles associated with IMV or IV; arrowhead, gold particles in amorphous material in the factory area.
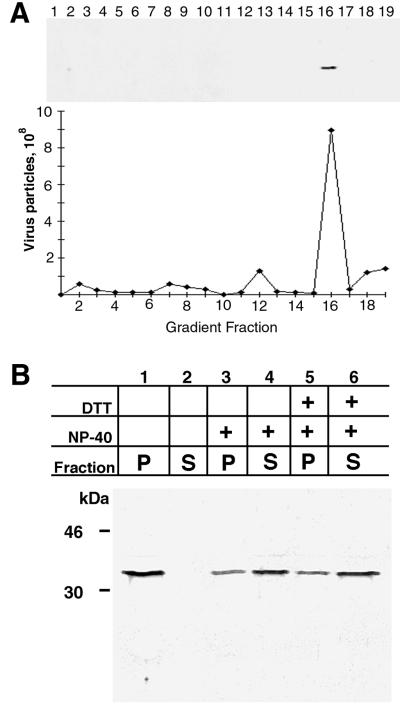
The association of the H3L protein with IMV was also demonstrated by SDS-PAGE and immunoblotting of virions purified from infected-cell lysates by successive rate zonal sucrose and CsCl density gradient centrifugations. The H3L protein was coincident with the fraction containing IMV (Fig. 4A). Vaccinia virions, purified through two successive sucrose gradient centrifugations, were treated with solutions containing NP-40 detergent with or without DTT. The released and unreleased proteins were separated by centrifugation and analyzed by SDS-PAGE and immunoblotting. Most of the H3L protein was extracted with NP-40 (Fig. 4B), indicating a membrane location. Unlike what was found for many viral membrane proteins, a reducing agent did not enhance release, suggesting a location near the surface. Nevertheless, some H3L protein was resistant to extraction with or without reducing agent.
FIG. 4.
Association of the H3L protein with purified vaccinia virus particles and extraction with nonionic detergent. (A) Western blot of purified virus particles. Sucrose gradient-purified intracellular virions were centrifuged in a performed CsCl gradient. Fractions (0.5 ml) were collected from the top of the tube, diluted, and centrifuged to pellet virus particles. The pellets were suspended, and part of the suspension was used to calculate the number of virus particles from the absorbance at 260 nm; the remainder was analyzed by SDS-PAGE and immunoblotted with H3L peptide antibody followed by anti-rabbit IgG horseradish peroxidase conjugate. CsCl fraction numbers are indicated. (B) Extraction of H3L protein with NP-40 detergent. Purified vaccinia virions were incubated in Tris buffer containing 0.5% NP-40 or 0.5% NP-40 and 50 mM DTT. After centrifugation, the supernatant (S) and pellet (P) fractions were analyzed by SDS-PAGE and immunoblotting with the H3L peptide antibody. The masses and positions of markers are indicated at the left.
Surface location and topology.
Three independent methods were used to investigate the surface location and topology of the H3L protein. Sucrose gradient-purified virus particles were treated with trypsin and then collected by centrifugation. The supernatant and pellet fractions were analyzed by SDS-PAGE and Western blotting with H3L peptide antibody. At trypsin concentrations of 0.1 to 1.0 mg/ml, two major digestion products of about 26 and 8 kDa were resolved (Fig. 5A). With 10 mg of trypsin/ml, only the smaller species remained. Both species were present exclusively in the pellet fraction, suggesting that they retained the membrane anchor sequence. The limit digestion was not due to intrinsic trypsin resistance of the H3L protein since more-complete digestion occurred in the presence of NP-40 (Fig. 5A). The data are compatible with a trypsin-sensitive site in the vicinity of lysine 96 and arginine 97 to give a partial digestion product of 26 kDa and another site near arginine 248 to give a complete digestion product of 8.8 kDa, both of which could retain the antibody epitope between amino acids 247 and 259 and the C-terminal hydrophobic domain from amino acids 270 to 300, which presumably serves as the membrane anchor.
FIG. 5.
Topology of the H3L protein. (A) Trypsin sensitivity of the virion-associated H3L protein. Sucrose gradient-purified virus particles were treated with trypsin and then collected by centrifugation. Equivalent portions of the supernatant and pellet fractions were analyzed by SDS-PAGE and Western blotting with the H3L peptide antibody. Masses of protein markers are indicated on the left. (B) Biotinylation of membrane-bound H3L protein. Sucrose gradient-purified virus particles were incubated with sulfo-NHS-LC-biotin for 30 min. At the end of the reaction, the mixture was layered over a sucrose cushion and centrifuged to separate virus particles from residual unlinked biotin. A portion of the biotinylated virus particles was analyzed by SDS-PAGE (lane 2); another portion was extracted with NP-40 and separated into soluble (lane 3) and insoluble (lane 4) fractions. Lane 1, nonbiotinylated virions; lane 5, soluble proteins that were biotinylated after NP-40 extraction and centrifugation. The electrophoretically separated proteins were transferred to a membrane and probed with NeutrAvidin (avidin). The membrane was then stripped and immunoblotted with the anti-H3L antibody followed by anti-rabbit IgG horseradish peroxidase conjugate (anti-H3). (C) Decoration of vaccinia virus particles with the antibody to the H3L protein. Sucrose gradient-purified vaccinia virions were adsorbed to grids and incubated with the H3L peptide antibody and protein A-gold (right) or with protein A-gold alone (left). After negative staining, the grids were examined by electron microscopy.
If the H3L protein is anchored to the viral membrane by the C-terminal hydrophobic domain, then 21 of the 23 lysines should be accessible to sulfo-NHS-LC-biotin, a water-soluble ester that is unable to penetrate lipid bilayers and that can couple to exposed primary amines. Purified IMV were incubated with the coupling reagent and then centrifuged through a sucrose cushion to remove unbound biotin. One portion of the particles was treated directly with SDS, whereas another was first extracted with NP-40 detergent without reducing agent to obtain the superficial membrane-associated components. The proteins were resolved by SDS-PAGE, transferred to a nylon membrane, and then treated with an avidin-horseradish peroxidase conjugate. A prominent biotinylated band of the expected size for the H3L protein was detected in the total virion extract and was the major product released by NP-40 (Fig. 5B). This biotinylated band was coincident with H3L protein, as determined by stripping the blot and reprobing it with a specific antibody (Fig. 5B). The small amount of H3L protein that was not extracted with NP-40 was faintly labeled with biotin (Fig. 5B). Whether this fraction of H3L protein has a more internal location or is poorly exposed due to technical reasons is not known. The nature of the high-molecular-weight bands that were not extracted with NP-40 and that did not react with the H3L antibody is unknown. However, the finding raises the possibility that the labeling of some internal proteins occurred.
To further investigate the topology of the H3L protein, purified IMV were incubated with the H3L peptide antibody followed by protein A conjugated to gold particles or, as a control, with the protein A-gold alone. Electron-microscopic images showed gold particles decorating the outside of virions that had been incubated with antibody, whereas few particles were associated with virions that had been treated only with the protein A-gold (Fig. 5C). The variable number of grains per virion likely reflected the relatively poor accessibility of the antibody to the epitope. This might also explain our inability to effectively neutralize IMV with the H3L peptide antibody (data not shown).
Taken together, our data suggest that the H3L protein is largely on the exterior surfaces of IMV and is anchored to the membrane by its C-terminal hydrophobic domain.
Association of in vitro-synthesized H3L protein with microsomes.
Coupled transcription/translation in rabbit reticulocyte lysates supplemented with canine pancreatic microsomes was used to study the mechanism of insertion of the H3L protein into membranes. The template consisted of a plasmid containing the H3L ORF under control of the bacteriophage T7 promoter. In the absence of microsomes, a radioactively labeled band corresponding in size to the H3L protein was present exclusively in the supernatant fraction after centrifugation (Fig. 6A). In contrast, approximately one-half of the protein was in the pellet fraction when microsomes were present (Fig. 6A; data not shown). The association with microsomes seemed specific since under the same conditions a control protein, luciferase, remained entirely in the supernatant fraction (data not shown).
FIG. 6.
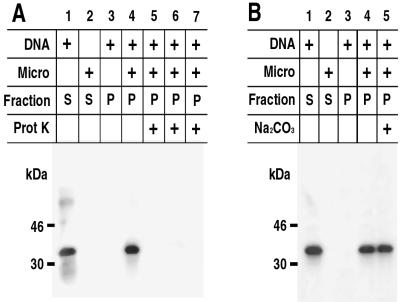
In vitro synthesis of the H3L protein and association with microsomal membranes: sensitivity to Na2CO3 and proteinase K. (A) Proteinase sensitivity of the membrane-associated H3L protein. The H3L ORF, regulated by a bacteriophage T7 promoter, was transcribed and translated in a reticulocyte lysate in the presence of [35S]methionine. Reactions were carried out in the absence (lane 2) or presence (lanes 1 and 3 to 7) of a DNA template or in the absence (lanes 1 and 3) or presence (lanes 2 and 4 to 7) of canine microsomal membranes (micro). The mixtures were layered over a sucrose cushion and centrifuged and supernatant (S) and pellet (P) fractions were obtained. Some of the pellet fractions were treated with proteinase K alone for 1 min (lane 5) or 2 min (lane 6) or with Triton X-100 plus proteinase K (lane 7). The samples were analyzed by SDS-PAGE and autoradiography. (B) Resistance of membrane-bound H3L protein to Na2CO3 extraction. Reactions were carried out and analyzed as for panel A except that in lane 5 the microsomes were incubated in Na2CO3 and then centrifuged through a sucrose cushion before SDS-PAGE. The masses and positions of markers are indicated on the left.
We previously used proteinase digestion to determine the topology of the microsomal-membrane-associated A17L protein (1). Under these conditions, only protein on the cytoplasmic face of the membrane was susceptible to proteinase K. The membrane-associated H3L protein was digested by proteinase K into fragments too small to be retained in the gel (Fig. 6A), suggesting that most of it faced the cytoplasm.
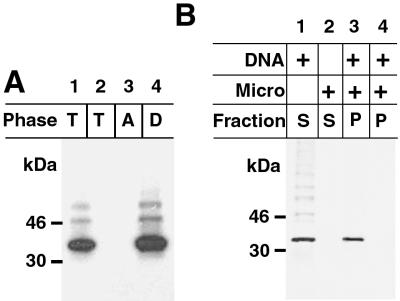
Further experiments were performed to determine whether the H3L protein was integrally associated with the microsomal membrane. Nonanchored proteins can be dissociated from membranes with Na2CO3 (pH 11). However, the H3L protein was still recovered in the membrane fraction after such treatment (Fig. 6B). The ability of a protein to insert into a membrane bilayer has been correlated with its partitioning in Triton X-114 (4). The H3L protein was synthesized in the absence of microsomes and then extracted with Triton X-114. After temperature-induced phase separation, the protein was found exclusively in the detergent phase (Fig. 7A). These data support the integral association of the H3L protein with membranes.
FIG. 7.
Hydrophobicity and posttranslational insertion of the H3L protein into membranes. (A) Partition of the H3L protein in the Triton X-114 detergent phase. The H3L protein was synthesized in vitro in the absence of microsomal membranes as described in the legend to Fig. 6. Triton X-114 detergent was added to the reaction mixture, and the mixture was subjected to temperature-induced phase separation. The [35S]methionine-labeled proteins in the total extract (T) and in the aqueous (A) and detergent (D) phases were analyzed by SDS-PAGE and autoradiography. Lane 2, control in which no template was added. (B) Posttranslational insertion of the H3L protein into membranes. Transcription/translation in the absence or presence of DNA or microsomal membranes was carried out as described in the legend to Fig. 6. In lane 3, 200 μg of cycloheximide/ml was added to stop translation before the addition of microsomal membranes to demonstrate posttranslational membrane insertion of H3L. In lane 4, cycloheximide was added at the start of the reaction to demonstrate complete inhibition of translation. The masses and positions of markers are indicated at the left.
Insertion of the H3L protein into microsomes occurs posttranslationally.
The majority of integral membrane proteins are cotranslationally inserted into microsomal membranes. However, there is a class of proteins that insert posttranslationally (17). To determine whether the H3L protein belongs to the pre- or posttranslational-insertion class, transcription/translation was performed in the absence of microsomes. Cycloheximide was added to prevent further translation, and then the incubation was continued in the presence of microsomes. After centrifugation, the H3L protein was found in the microsomal-pellet fraction (Fig. 7B), indicating posttranslational insertion. Membrane insertion still occurred when the translation reaction mixture was passed through a Sephadex G-50 column prior to addition of microsomes (data not shown), suggesting that an ATP energy source was not required.
Localization of the membrane anchor domain.
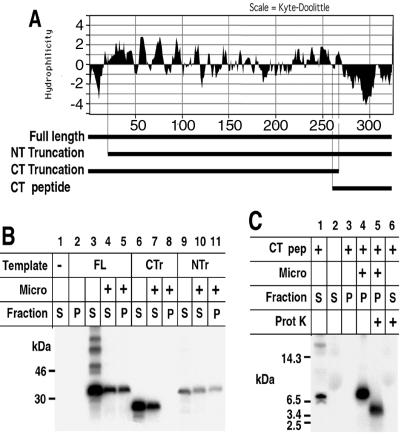
The H3L protein contains short N-terminal and long C-terminal hydrophobic domains (Fig. 8A). To determine which domain was responsible for membrane targeting, we constructed ORFs encoding the N- and C-terminally truncated proteins depicted in Fig. 8A and carried out transcription/translation in the presence of microsomal membranes. The full-length membrane protein and the species with the N-terminal deletion were found in both the membrane pellet and supernatant fractions, whereas the species with the C-terminal truncation was entirely in the supernatant (Fig. 8B). These results indicated that the hydrophobic C terminus was essential for membrane association. To extend this observation, a minigene encoding a product largely composed of the C-terminal hydrophobic domain was constructed (Fig. 8A) and expressed in the presence or absence of microsomes. When the reaction was carried out in the presence of microsomal membranes, the C-terminal polypeptide was associated with the pellet fraction (Fig. 8C). We presume that the membrane association occurred posttranslationally, although this was not specifically examined. The membrane-bound material was then treated with proteinase K and analyzed on a gel capable of resolving small peptides. The peptide was reduced in size, indicating that part of it was susceptible to digestion (Fig. 8C). The protected segment was likely buried in the membrane because the free polypeptide synthesized in the absence of microsomes was totally digested by proteinase K (Fig. 8C). Thus, the H3L protein can associate with membranes via its C-terminal hydrophobic domain.
FIG. 8.
Membrane insertion of truncated H3L proteins. (A) Hydrophilicity plot of H3L protein. Bars beneath the plot, full-length H3L protein, an N-terminal (NT) truncation, a C-terminal (CT) truncation, and a C-terminal peptide. (B) Insertion of truncated H3L proteins into membranes. Transcription/translation in the presence or absence of microsomal membranes and analysis of supernatant and pellet fractions by SDS-PAGE and autoradiography were carried out as described in the legend to Fig. 6. Lane 1, no DNA was added to the reaction mixture; lanes 2 to 5, the full-length (FL) protein was synthesized; lanes 6 to 8, the C-terminal truncated species (CTr) was synthesized; lanes 9 to 11, the N-terminal truncated species (NTr) was synthesized. Soluble (S) and pellet (P) fractions were analyzed by SDS-PAGE and autoradiography. (C) Insertion of a C-terminal peptide (CT pep) into membranes. Transcription/translation reactions in the absence or presence of microsomal membranes were carried out using a template encoding the C-terminal peptide. Where indicated (+), proteinase K treatment was carried out after the separation of supernatant and pellet fractions. Masses and positions of markers are indicated at the left.
DISCUSSION
Biochemical studies indicated that the H3L protein was expressed late in infection and was incorporated into the IMV. The protein accumulated in the cytoplasmic factory regions, where it was visualized as amorphous deposits in the vicinity of viral membranes and as components of immature and mature virus particles. The density of gold grains was approximately 2.5 times higher in IMV than in IV, suggesting that the H3L protein becomes membrane associated during virus maturation rather than at the initial stages of membrane formation. Several lines of evidence indicated that the H3L protein is attached to the surfaces of IMV. These included efficient extraction with NP-40 in the absence of reducing agents, trypsin sensitivity, biotinylation by a membrane-impermeable reagent, and immunogold labeling of intact particles. Independent evidence for surface localization was recently reported by Lin et al. (18). The trypsin resistance of the C-terminal peptide and the exposure of an epitope within amino acids 247 to 259 suggested that the H3L protein is anchored through its C-terminal hydrophobic domain. The presence of the H3L protein in virus factory areas and the absence of characteristic patterns of ER or Golgi network labeling with the H3L-specific antibody raised the possibility of an unconventional means of viral membrane insertion directly from the cytoplasm.
In the second part of this study, we used an in vitro transcription/translation system to analyze the mechanism of membrane association. The H3L ORF was transcribed by bacteriophage T7 RNA polymerase in a reticulocyte lysate supplemented with canine pancreatic microsomes. The H3L protein was intimately associated with the microsomes because it resisted extraction with Na2CO3 (pH 11) but was released with nonionic detergent. The protein was accessible to proteinase K, indicating that most of it was external to the membrane. Inspection of the H3L amino acid sequence suggested the absence of a functional N-terminal signal peptide that would allow the association of the ribosome-bound nascent chain with microsomes. In accordance with this, we found that membrane association could occur posttranslationally, after cycloheximide treatment and gel filtration of the translation reaction mixture. Analysis of truncated forms of the H3L protein indicated that the C-terminal hydrophobic domain was necessary and sufficient for membrane insertion. The proteinase resistance of the membrane-bound C-terminal fragment was consistent with its penetration into the lipid bilayer.
Posttranslational insertion is not unprecedented, as this occurs with a class of eukaryotic proteins with C-terminal membrane anchors (17). Membrane insertion occurs independently of the classical SRP/Sec61p pathway and follows release of the protein from the ribosome (23). The prototype of this class is cytochrome b5, and other members include the SNARE protein synaptobrevin, polyoma virus middle T antigen, and Epstein-Barr virus BHRF-1 protein (16). These proteins, like H3L, are cytoplasmically oriented and anchored by a C-terminal hydrophobic segment. Also as for H3L, the C-terminal hydrophobic segment is sufficient for membrane insertion (37). ATP was found to be required for translocation of synaptobrevin (16), whereas H3L insertion was independent of this energy source. The ATP requirements of other members of this family are unknown.
Other vaccinia virus proteins might also be inserted into viral membranes by a mechanism similar to that of H3L. One candidate is the product of the vaccinia virus L1R ORF. The L1R protein has a hydrophobic C terminus, associates with viral factory areas rather than ER or Golgi membranes, and is located on the surfaces of IMV (24, 38). Another candidate is the D8L protein, which has a hydrophobic C terminus and which is exposed on the surfaces of IMV (19, 22, 30). The product of the A27L ORF is also located on the surfaces of IMV and has an unconventional targeting mechanism that does not involve the ER (25, 30). However, the structure of the A27L protein is quite different from that of H3L, and it is believed to associate with viral membranes indirectly through protein-protein interactions (34, 35).
Taken together, our in vivo and in vitro data support a model in which the H3L protein is synthesized on free ribosomes and accumulates in viral factory areas, where it becomes associated with the membranes of maturing viral particles. Since the H3L protein can associate with microsomal membranes in vitro, it seems likely that an additional viral component or receptor provides specificity for viral membranes. In the accompanying paper (5), we show that expression of the H3L protein is important for efficient assembly of mature virus particles.
ACKNOWLEDGMENTS
We thank Christine White and Joanna Shisler for discussions and advice, Brian Ward and Owen Schwartz for help with confocal microscopy, and Norman Cooper for providing cells. Special thanks to Erna G. Kroon for encouragement and making possible the interaction between the Laboratory of Viral Diseases and the Laboratorio de Virus, ICB, UFMG, Brazil.
Flavio G. da Fonseca was supported by “Fundação Coordenação de Aperfeiçoamento de Pessoal de Nível Superior,” Brazil, and by an intramural training award from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Betakova T, Wolffe E J, Moss B. Membrane topology of the vaccinia virus A17L envelope protein. Virology. 1999;261:347–356. doi: 10.1006/viro.1999.9870. [DOI] [PubMed] [Google Scholar]
- 2.Betakova T, Wolffe E J, Moss B. Vaccinia virus A14.5L gene encodes a hydrophobic 53-amino-acid virion membrane protein that enhances virulence in mice and is conserved among vertebrate poxviruses. J Virol. 2000;74:4085–4092. doi: 10.1128/jvi.74.9.4085-4092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blobel G, Dobberstein B. Transfer of proteins across membranes. II. Reconstruction of functional rough microsomes from heterologous components. J Cell Biol. 1975;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 5.da Fonseca F G, Weisberg A, Wolffe E J, Moss B. Effects of deletion or stringent repression of the H3L envelope gene on vaccinia virus replication. J Virol. 2000;74:7518–7528. doi: 10.1128/jvi.74.16.7518-7528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dales S, Mosbach E H. Vaccinia as a model for membrane biogenesis. Virology. 1968;35:564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- 7.Earl P L, Cooper N, Wyatt S, Moss B, Carroll M W. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1998. pp. 16.16.1–16.16.3. [Google Scholar]
- 8.Earl P L, Moss B. Generation of recombinant vaccinia viruses. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1991. pp. 16.17.1–16.17.16. [Google Scholar]
- 9.Grimley P M, Rosenblum E N, Mims S J, Moss B. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J Virol. 1970;6:519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiller G, Weber K. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol. 1985;55:651–659. doi: 10.1128/jvi.55.3.651-659.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollinshead M, Vanderplasschen A, Smith G L, Vaux D J. Vaccinia virus intracellular mature virions contain only one lipid membrane. J Virol. 1999;73:1503–1517. doi: 10.1128/jvi.73.2.1503-1517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichihashi Y, Matsumoto S, Dales S. Biogenesis of poxviruses: role of A-type inclusions and host cell membranes in virus dissemination. Virology. 1971;46:507–532. doi: 10.1016/0042-6822(71)90056-0. [DOI] [PubMed] [Google Scholar]
- 13.Jensen O N, Houthaeve T, Shevchenko A, Cudmore S, Ashford T, Mann M, Griffiths G, Locker J K. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J Virol. 1996;70:7485–7497. doi: 10.1128/jvi.70.11.7485-7497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson G P, Goebel S J, Paoletti E. An update on the vaccinia virus genome. Virology. 1993;196:381–401. doi: 10.1006/viro.1993.1494. [DOI] [PubMed] [Google Scholar]
- 15.Krijnse-Locker J, Schleich S, Rodriguez D, Goud B, Snijder E J, Griffiths G. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J Biol Chem. 1996;271:14950–14958. doi: 10.1074/jbc.271.25.14950. [DOI] [PubMed] [Google Scholar]
- 16.Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport T A. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 1995;14:217–223. doi: 10.1002/j.1460-2075.1995.tb06994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutay U, Hartmann E, Rapoport T A. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 18.Lin C L, Chung C S, Heine H G, Chang W. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J Virol. 2000;74:3353–3365. doi: 10.1128/jvi.74.7.3353-3365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maa J-S, Rodriguez J F, Esteban M. Structural and functional characterization of a cell surface binding protein of vaccinia virus. J Biol Chem. 1990;265:1569–1577. [PubMed] [Google Scholar]
- 20.Morgan C. Vaccinia virus reexamined: development and release. Virology. 1976;73:43–58. doi: 10.1016/0042-6822(76)90059-3. [DOI] [PubMed] [Google Scholar]
- 21.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2637–2671. [Google Scholar]
- 22.Niles E G, Seto J. Vaccinia virus gene D8 encodes a virion transmembrane protein. J Virol. 1988;62:3772–3778. doi: 10.1128/jvi.62.10.3772-3778.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rapoport T A. Transport of proteins across the endoplasmic reticulum membrane. Science. 1992;258:931–936. doi: 10.1126/science.1332192. [DOI] [PubMed] [Google Scholar]
- 24.Ravanello M P, Hruby D E. Characterization of the vaccinia virus L1R myristylprotein as a component of the intracellular virion envelope. J Gen Virol. 1994;75:1479–1483. doi: 10.1099/0022-1317-75-6-1479. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez J F, Paez E, Esteban M. A 14,000-Mr envelope protein of vaccinia virus is involved in cell fusion and forms covalently linked trimers. J Virol. 1987;61:395–404. doi: 10.1128/jvi.61.2.395-404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roper R L, Payne L G, Moss B. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene J. Virol. 1996;70:3753–3762. doi: 10.1128/jvi.70.6.3753-3762.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosel J L, Earl P L, Weir J P, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986;60:436–439. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salmons T, Kuhn A, Wylie F, Schleich S, Rodriguez J R, Rodriguez D, Esteban M, Griffiths G, Locker J K. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J Virol. 1997;71:7404–7420. doi: 10.1128/jvi.71.10.7404-7420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sodeik B, Cudmore S, Ericsson M, Esteban M, Niles E G, Griffiths G. Assembly of vaccinia virus: incorporation of p14 and p32 into the membrane of the intracellular mature virus. J Virol. 1995;69:3560–3574. doi: 10.1128/jvi.69.6.3560-3574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi T, Oie M, Ichihashi Y. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology. 1994;202:844–852. doi: 10.1006/viro.1994.1406. [DOI] [PubMed] [Google Scholar]
- 33.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- 34.Vazquez M I, Esteban M. Identification of functional domains in the 14-kilodalton envelope protein (A27L) of vaccinia virus. J Virol. 1999;73:9098–9109. doi: 10.1128/jvi.73.11.9098-9109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vázquez M-I, Rivas G, Cregut D, Serrano L, Esteban M. The vaccinia virus 14-kilodalton (A27L) fusion protein forms a triple coiled-coil structure and interacts with the 21-kilodalton (A17L) virus membrane protein through a C-terminal α-helix. J Virol. 1998;72:10126–10137. doi: 10.1128/jvi.72.12.10126-10137.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward G A, Stover C K, Moss B, Fuerst T R. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc Natl Acad Sci USA. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitley P, Grahn E, Kutay U, Rapoport T A, von Heijne G. A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J Biol Chem. 1996;271:7583–7586. doi: 10.1074/jbc.271.13.7583. [DOI] [PubMed] [Google Scholar]
- 38.Wolffe E J, Vijaya S, Moss B. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology. 1995;211:53–63. doi: 10.1006/viro.1995.1378. [DOI] [PubMed] [Google Scholar]
- 39.Zinoviev V V, Tchikaev N A, Chertov O, Malygin E G. Identification of the gene encoding vaccinia virus immunodominant protein p35. Gene. 1994;147:209–214. doi: 10.1016/0378-1119(94)90067-1. [DOI] [PubMed] [Google Scholar]