Abstract
Background:
Cell-specific and developmental mechanisms contribute to expression of the cystic fibrosis transmembrane conductance regulator (CFTR) gene; however, its developmental regulation is poorly understood. Here we use human induced pluripotent stem cells differentiated into pseudostratified airway epithelial cells to study these mechanisms.
Results:
Changes in gene expression and open chromatin profiles were investigated by RNA-seq and ATAC-seq, and revealed that alterations in CFTR expression are associated with differences in stage-specific open chromatin. Additionally, two novel open chromatin regions, at +19.6 kb and +22.6 kb 3′ to the CFTR translational stop signal, were observed in definitive endoderm (DE) cells, prior to an increase in CFTR expression in anterior foregut endoderm (AFE) cells. Chromatin studies in DE and AFE cells revealed enrichment of active enhancer marks and occupancy of OTX2 at these sites in DE cells. Loss of OTX2 in DE cells alters histone modifications across the CFTR locus and results in a 2.5-fold to 5-fold increase in CFTR expression. However, deletion of the +22.6 kb site alone does not affect CFTR expression in DE or AFE cells.
Conclusions:
These results suggest that a network of interacting cis-regulatory elements recruit OTX2 to the locus to impact CFTR expression during early endoderm differentiation.
Keywords: airway epithelial development, cis-regulatory elements, cystic fibrosis, gene regulation, iPSC differentiation
1 |. INTRODUCTION
Cystic fibrosis (CF) is a common, life-limiting, autosomal-recessive disorder that is characterized by mutations in the CF transmembrane conductance regulator (CFTR) gene. Loss of CFTR protein, and its associated ion and fluid transport functions, in specialized epithelial cells causes a disease phenotype in many organ systems. CF impacts the airway, pancreas, intestine, and male genital tract, among other sites, though respiratory failure due to chronic infection and inflammation is the main cause of mortality.
The precise cellular location of CFTR expression in the airway epithelium of adult human lungs is a topic of some controversy, in part arising from the use of different protocols. Earlier work using mRNA in situ hybridization and immunocytochemistry with various antibodies to CFTR showed generally low levels of expression in the airway surface epithelium, except in submucosal gland ducts.1,2 Later, immunofluorescence clearly showed CFTR protein localized to ciliated cells in the airway surface epithelium.3 More recently, single cell RNA-sequencing has enabled the definition of the rare airway cells expressing high levels of CFTR that were observed in earlier studies as pulmonary ionocytes.4,5 However, the majority of cells in the surface epithelium of the airway express much less CFTR than those in the corresponding cell layer of several other tissues, including the intestine and pancreas.6,7
Many of the cis-regulatory elements (CREs) that control cell type-selective expression of CFTR were identified previously and extensively characterized (reviewed in References 8-12). CFTR is also developmentally regulated; however, less is known about the underlying regulatory mechanisms, in part due to lack of accessibility to relevant biological samples. CFTR expression in human lung tissues is detected as early as week 10 of gestation, with maximal expression evident during the second trimester, after which it decreases to the much lower abundance characteristic of postnatal and adult tissues.6,7,13–16 Although chronic rounds of infection and inflammation ultimately lead to the irreversible lung damage seen in CF patients, the lungs of CF infants and CF fetuses may also show signs of early pathology that predates these insults to the lung epithelium.17 Hence, elucidating developmental regulatory mechanisms for CFTR is of high priority. Advances in protocols to differentiate human induced pluripotent stem cells (iPSCs) into pseudostratified airway epithelial cells,18–25 some of which express functional CFTR protein, provide a model to investigate the mechanisms governing CFTR expression in the developing airway.
In order to identify novel developmental CREs for CFTR, we differentiated human iPSCs (hiPSCs) into airway-like epithelial cells grown at air-liquid interface (ALI). This differentiation protocol results in the generation of proximal airway-like cells, which express known markers of airway epithelial cells, including basal cells (KRT5), ciliated cells (FOXJ1), goblet cells (ARG2 and MUC5AC), and secretory cells (SCGB3A2).25 Peaks of open chromatin, which are often associated with CREs, were monitored over ~650 kb flanking the CFTR locus at seven timepoints along the differentiation time course. CFTR mRNA levels evaluated in parallel. We identified two, novel developmental CREs within the topologically associated domain (TAD) encompassing the CFTR locus. These sites, which lie close to the 3′ end of the gene, appear transiently in definitive endoderm (DE) cells, while the CFTR locus is inactive, but are lost in anterior foregut endoderm (AFE) cells coincident with activation of the locus. Here we examine the function and mechanism of action of these developmental CREs, and demonstrate a potent role for orthodenticle homeobox 2 (OTX2) in the transcriptional network coordinating CFTR expression during early human lung development.
2 |. RESULTS
2.1 |. Open-chromatin profiling of CFTR locus during iPSC to ALI differentiation reveals novel 3′ CREs
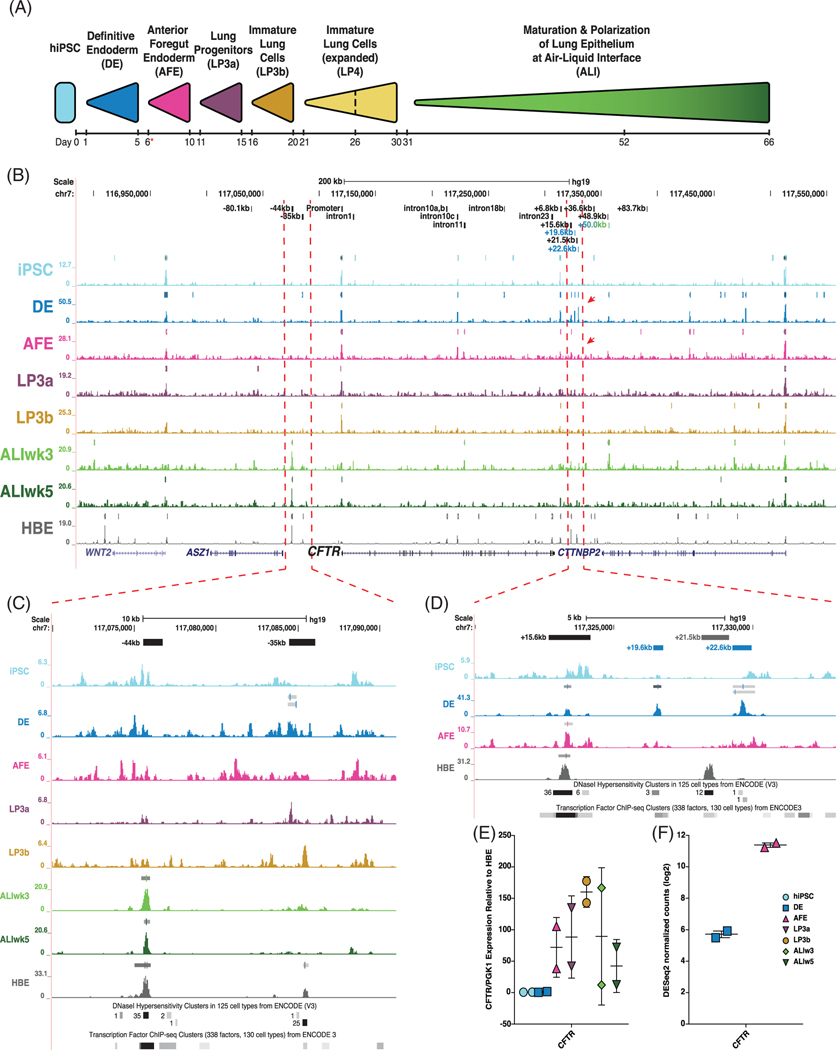
Two hiPSC lines, ND2.0 and CWRU205, were differentiated into airway epithelial cells grown at ALI, following previously published protocols.19,25 Cells were collected at seven stages throughout differentiation for ATAC-seq and RNA-seq: undifferentiated iPSC, DE, AFE, lung progenitor cells 3a, 3b (LP3a, LP3b), and at ALI after weeks 3 and 5 (ALIw3 and ALIw5) (Figure 1A). ATAC-seq profiles from irreproducible discovery rate analysis comparing both lines is shown for the ~650 kb surrounding the CFTR locus in Figure 1B. For comparison, the ATAC-seq profile for primary human bronchial epithelial cells grown at ALI (HBE-ALI) is also shown. The CFTR promoter is accessible at all stages of differentiation, as evidenced by the associated peak of open chromatin (Figure 1B), irrespective of CFTR transcript expression (Figure 1E). Notably, airway-specific CREs at −44 kb and −35 kb,26–29 which are strongly observed in HBE-ALI, appear primarily in the differentiated ALI stages (where −44 kb is more prominent, though transient expression of −35 kb may be seen at LP3b) (Figure 1C). These data suggest that the CFTR locus acquires an airway-like open chromatin profile during the course of iPSC to ALI differentiation.
FIGURE 1.
Open-chromatin profiling of cystic fibrosis transmembrane conductance regulator (CFTR) locus during induced pluripotent stem cell (iPSC) to air-liquid interface (ALI) differentiation. A, Differentiation schematic of iPSC to lung epithelial cells at air-liquid interface. Red asterisk denotes when cells are transferred onto permeable inserts. Cells from differentiations of two donor lines (ND2.0 and CWRU205) were taken at days 0, 5, 10, 15, 20, 30, 52, and 66 for ATAC-seq and RNA-seq and reverse transcription quantitative PCR (RT-qPCR) analysis (adapted from Reference 25). B, UCSC Genome Browser view of ATAC-seq irreproducible discovery rate (IDR) tracks across the CFTR locus at different stages of iPSC to ALI differentiation, with primary human bronchial epithelial cells grown at ALI (HBE-ALI) shown in gray for comparison. IDR peaks are shown directly above open chromatin tracks for each stage. Known CFTR cis-regulatory elements (CREs) are shown at the top in black, together with novel developmental CFTR CREs in colors corresponding to the differentiation stage where the peak was observed. Red arrows denote appearance and disappearance of novel CREs. C,D, Enlarged ATAC-seq profiles surrounding: C, CFTR airway-selective CREs at −44 kb and −35 kb. D, Novel DE CREs located 3′ to the CFTR locus. Known DNaseI hypersensitivity clusters and transcription factor (TF) chromatin immunoprecipitation (ChIP)-seq cluster tracks from the UCSC Genome Browser shown below ATAC-seq profiles. E, RT-qPCR of CFTR mRNA in ND2.0 and CWRU205 shown relative to an average of three HBE cultures grown on plastic, normalized to PGK1. E, CFTR RNA-seq DEseq2 normalized counts in ND2.0 and CWRU205 in DE and AFE stages
CFTR expression during differentiation of iPSC to ALI was compared to HBE grown on plastic using reverse transcription quantitative PCR (RT-qPCR) (Figure 1E). In iPSC and DE cells, CFTR expression remains low, increases substantially in AFE to LP3b cells, before decreasing in ALI cells.
Two novel CFTR open chromatin regions at +19.6 kb and +22.6 kb from the CFTR translational stop site are observed in DE cells only (Figure 1B,D). These sites are detected in the presence of the +15.6 kb CRE, which was previously shown to be an enhancer-blocking insulator that does not bind CCCTC-binding factor (CTCF).30,31 The +19.6 kb and +22.6 kb sites flank an element of unknown function at +21.5 kb CRE, which is not evident in DE or AFE cells, but was shown to physically interact with the CFTR promoter in airway epithelial primary and cell lines.26–28 Notably, of 125 cell-types profiled for DNaseI hypersensitive clusters by the ENCODE Consortium, +19.6 kb and +22.6 kb regions are only open in four cell types (Figure 1D).32 The CRE at +19.6 kb is detected in undifferentiated hESCs, and endoderm-derived esophageal and retinal pigment epithelial cells, while the CRE at +22.6 kb is observed in ectoderm-derived epidermal melanocytes and undifferentiated hESCs (Table 1). Additionally, ENCODE ChIP-seq datasets have no detectable binding of surveyed TFs at the +22.6 kb CRE, and very low overall enrichment of few factors in limited cell types at +19.6 kb (Figure 1D).32
TABLE 1.
UCSC Genome Browser DNase clusters from 125 cell types in ENCODE (v3) detail
| +15.6 kb CREa Cell Type | +19.6 kb CREb Cell Type | +21.5 kb CREc Cell Type | +22.6 kb CREd Cell Type | |
|---|---|---|---|---|
| HRCEpiC | Jurkat | HRPEpiC | HEEpiC | H7-hESC |
| RPTEC | FibroP | H7-hESC | SAEC | Melano |
| HAEpiC | Huh-7.5 | HEEpiC | PrEC | |
| SAEC | H9ES | HRCEpiC | ||
| Caco-2 | HBMEC | pHTE | ||
| HEEpiC | HPDE6-E6E7 | HPDE6-E6E7 | ||
| H7-hESC | HRPEpiC | NHEK | ||
| HRE | Osteobl | HRE | ||
| PrEC | HCF | RPTEC | ||
| HeLa-S3 | MCF-7 | RWPE1 | ||
| HVMF | Melano | HMEC | ||
| HCPEpiC | HA-sp | LNCaP | ||
| Huh-7 | HCFaa | |||
| HCM | HIPEpiC | |||
| HPAF | HMEC | |||
| pHTE | WI-38 | |||
| A549 | H1-hESC | |||
| HeLa-S3 | HFF-Myc |
Note: Bold—Cell types with +21.5 kb open chromatin that overlap with those having open chromatin at +15.6 kb, +19.6 kb, or +22.6 kb.
Abbreviation: CRE, cis-regulatory element.
+15.6 kb open chromatin: hg19 chr7:117322806–117323495.
+19.6 kb open chromatin: hg19 chr7:117326361–117326630.
+21.5 kb open chromatin: hg19 chr7:117328266–117328570.
+22.6 kb open chromatin: hg19 chr7:117329486–117329635 and chr7:117329641–117329790.
The detection of the +19.6 kb and +22.6 kb sites in DE cells, when CFTR expression is very low, and their loss upon upregulation of CFTR transcription in AFE cells (Figure 1E,F), suggest these sites may harbor CREs which regulate CFTR expression in early stages of iPSC to ALI differentiation.
2.2 |. In silico analyses identify candidate TFs for regulating the +19.6 kb and +22.6 CREs
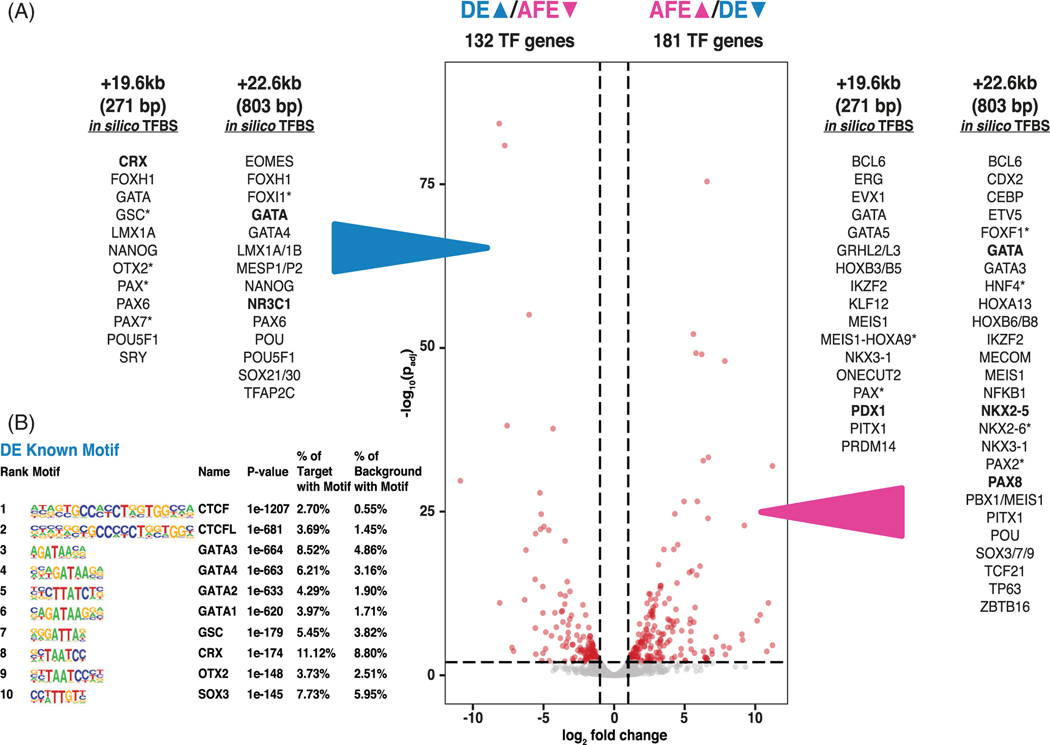
Next, we performed in silico analyses to identify potential transcription factors (TFs) that might bind to the +19.6 kb and +22.6 kb CREs. Examination of differentially expressed genes encoding TFs33 identified 132 that were significantly upregulated in DE compared to AFE cells and 181 that were significantly upregulated in AFE compared to DE cells (Figure 2A, Table S1). Next, TF binding motifs were predicted in silico for the +19.6 kb and +22.6 kb CREs using MatInspector and TRAP,34 and selected for motifs that correspond to differentially expressed TFs in DE and AFE cells (Figure 2A). Of the 85 TFs predicted to occupy the +19.6 kb CRE by MatInspector, 7 TFs are more abundant in DE and 14 in AFE. In equivalent predictions from the TRAP analysis 53 TFs were predicted to bind to the +19.6 CRE, of these, 4 were higher in DE and 5 more abundant in AFE. Of the 116 TFs predicted to bind the +22.6 kb CRE by MatInspector, 13 TFs are more highly expressed in DE and 23 in AFE. Similarly, in TRAP analysis, of the 30 TFs predicted to bind, 2 TFs were higher in DE and 6 higher in AFE. Candidate TFs predicted to occupy the DE-specific +19.6 kb and +22.6 kb CREs included CRX, OTX2, GATA factors, LMX1A/B, and others, all of which were more abundant in DE than AFE. Using HOMER motif enrichment, GATA4 (p = 1e−663), CRX (p = 1e−174), and OTX2 (p = 1e−148) motifs were identified in the top 10 overrepresented known motifs in global peaks of DE open chromatin (Figure 2B). Though present, these three factors were much less enriched in AFE peaks (GATA4: p = 1e−12; CRX: p = 1e−50; OTX2: p = 1e−8; data not shown25). Our further studies focused on GATA4 and OTX2, as although CRX and OTX2 are both members of the OTX family of biocoid-like homeodomain TFs and bind the same consensus sequence, CRX expression is highly restricted to the retina.35,36
FIGURE 2.
In silico predicted transcription factor binding sites (TFBS) at +19.6 kb and +22.6 kb cis-regulatory elements (CREs) correspond to definitive endoderm (DE) > anterior foregut endoderm (AFE) differentially expressed TFs. A, Volcano plot of weighted fold change as a function of P-values for TF genes during DE to AFE transition. DEGs passing a fold change of 2 and adjusted P value of .01 are shown in red. In silico TFBS predictions for the +19.6 kb and +22.6 kb CREs were performed using MatInspector and transcription factor affinity prediction (TRAP). Predicted binding sites for DE differentially upregulated TFs (left) and AFE differentially upregulated TFs (right) are listed (data are from MatInspector unless marked *, which denotes TRAP; bold font denotes predictions from both analyses). B, HOMER analysis showing top 10 overrepresented binding motifs for known transcription factors under open chromatin peaks in DE cells (adapted from Reference 25)
2.3 |. OTX2 represses CFTR expression in DE cells
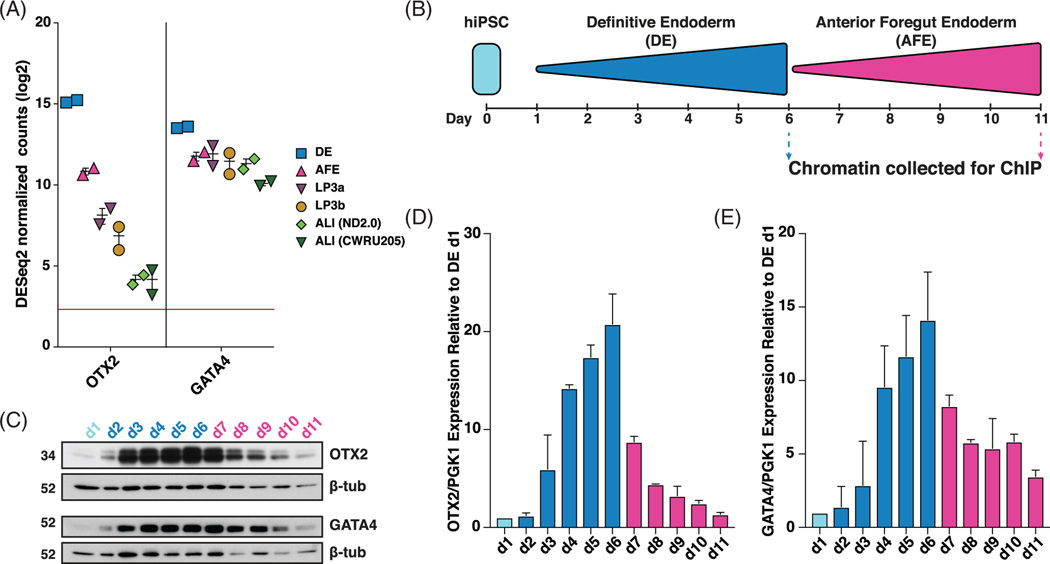
OTX2 and GATA4 gene expression is highest in DE, and decreases at each subsequent stage in iPSC to ALI differentiation (Figure 3A). Closer examination of OTX2 and GATA4 protein and gene expression over the first 11 days of the differentiation, encompassing iPSC (day 1), DE (days 2–6) and AFE (days 7–11) (Figure 3B), in ND2.0 cells showed that OTX2 and GATA4 expression peaks at day 6, and decreases as cells differentiate to AFE (Figure 3C-E).
FIGURE 3.
Orthodenticle homeobox 2 (OTX2) and GATA4 expression peaks in definitive endoderm (DE) cells. A, OTX2 and GATA4 RNA-seq DEseq2 normalized counts in ND2.0 and CWRU205 in indicated stages of induced pluripotent stem cell (iPSC) to air-liquid interface (ALI) differentiation. B, Experimental schematic of 11-day iPSC, DE and anterior foregut endoderm (AFE) differentiation. Cells were taken at each timepoint for protein and gene expression. C-E, Chromatin immunoprecipitation (ChIP) was done at day 6 for DE differentiation and day 11 for AFE differentiation (Figure 5). C, Western blots show the time course of OTX2 and GATA4 protein abundance from day 1 to day 11 of differentiation, with β-tubulin (β-tub) as control. Data shown are from the ND2.0 line (n = 2). D,E, Time course of, D, OTX2 and, E, GATA4 gene expression, normalized to PGK1 from day 1 to day 11 of differentiation in ND2.0 (n = 2). Results were consistent in the CWRU205 line
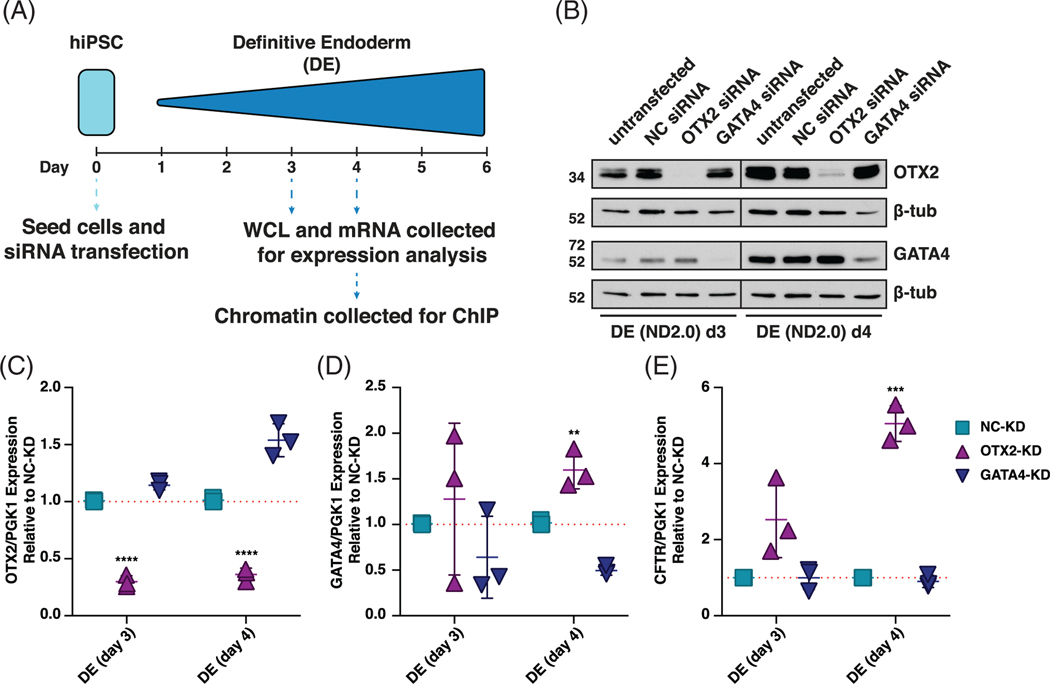
Next, OTX2 and GATA4 were depleted in DE cells by reverse transfecting iPSCs with specific siRNAs, and differentiating them into DE. Cells were collected at days 3 and 4 of differentiation, time points at which OTX2 and GATA4 expression have increased substantially from iPSC levels, and used for protein and mRNA analysis (Figure 4A). By day 6 of DE differentiation, depletion of both OTX2 and GATA4 had waned (data not shown). OTX2 and GATA4 protein and transcript levels are significantly decreased at days 3 and 4 of DE differentiation following their depletion (Figure 4B-D). Loss of GATA4 had no effect on OTX2 transcript or protein levels, while loss of OTX2 slightly increased GATA4 protein and transcript levels at day 4 (Figure 4D). Notably, loss of OTX2, results in a 2.5-fold and 5-fold increase in CFTR transcript levels at days 3 and 4 of DE differentiation, respectively (Figure 4E). Loss of GATA4 protein and transcript had no effect on CFTR transcript levels at days 3 and 4 of DE differentiation (Figure 4E). Together these data suggest that OTX2, represses CFTR expression in DE cells, through direct or indirect interaction with the CFTR locus.
FIGURE 4.
Orthodenticle homeobox 2 (OTX2) represses cystic fibrosis transmembrane conductance regulator (CFTR) expression in definitive endoderm (DE) cells. A, Schematic of 6-day induced pluripotent stem cell (iPSC) to DE differentiation showing timing of sample collection for siRNA-mediated depletion experiments. B, Western blots showing OTX2 and GATA4 protein levels in DE d3 and DE d4 cells from ND2.0 donor. Lysates collected from untransfected cells or cells transfected with negative control siRNA (NC), OTX2 siRNA, or GATA4 siRNA. β-tubulin (β-tub) levels are shown as controls. C-E, Reverse transcription quantitative PCR (RT-qPCR) of C, OTX2, D, GATA4, E, CFTR in NC-KD, OTX2-KD, or GATA4-KD DE d3 and DE d4 cells shown relative to NC-KD, normalized to PGK1. n = 3, **** denotes P < .0001, *** denotes P < .001, and ** denotes P < .01 compared to NC-KD for the same timepoint using the Holm-Sidak multiple t test
2.4 |. OTX2 interacts with the +19.6 kb and +22.6 kb CREs in DE cells
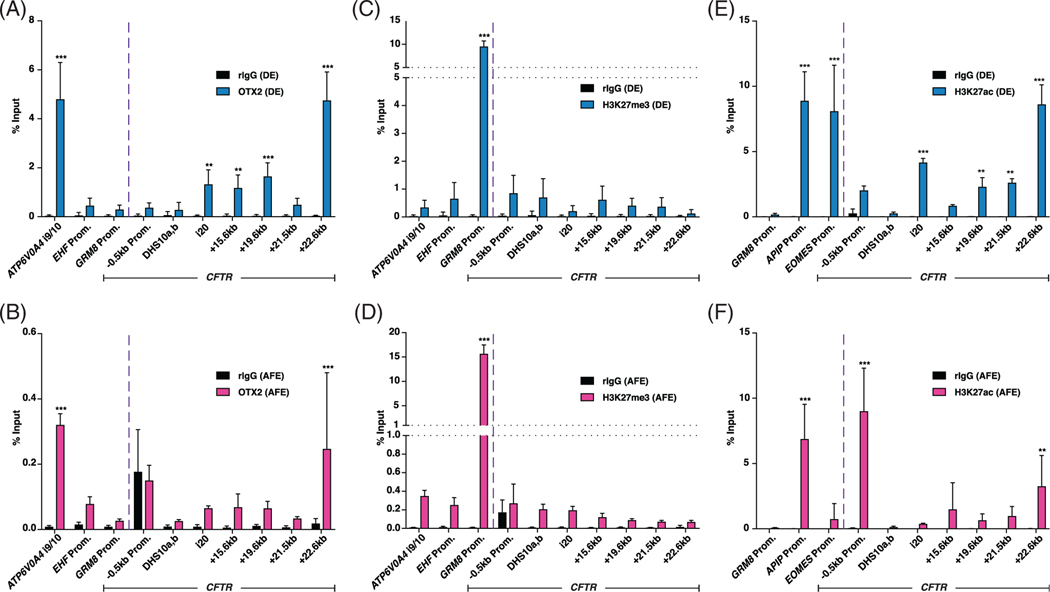
Chromatin immunoprecipitation followed by qPCR (ChIP-qPCR) was performed to identify direct interactions of OTX2 with the CFTR locus in DE and AFE cells (days 6 and 11, Figure 3B). ChIP-qPCR for OTX2 at sites across the CFTR locus identified significant OTX2 enrichment at a region in intron 20 of CFTR,37 as well as at the +15.6 kb, +19.6 kb, and +22.6 kb CREs (Figure 5A). Positive and negative control sites for the ChIP-qPCR were derived from publicly available DE OTX2 ChIP-seq data from hESCs differentiated into DE following an alternative protocol.38 Our data corroborated previous results of OTX2 enrichment (positive at ATP6V0A4 intron 9/10, negative at EHF and GRM8 promoters) (Figure 5A). The observation that OTX2 is recruited to other 3′ CREs at the CFTR locus concurrently with +19.6 kb and 22.6 kb elements is consistent with the coordinated action of multiple CREs within this large gene, as we observed previously for other cell-selective sites and TFs.39,40 While significant enrichment of OTX2 was observed at +22.6 kb and at the ATP6V0A4 i9/10 sites in AFE cells, the levels are ~10-fold lower than in DE (Figure 5B), consistent with lower abundance of OTX2 in AFE cells (Figure 3).
FIGURE 5.
Orthodenticle homeobox 2 (OTX2) and H3K27ac are enriched at cystic fibrosis transmembrane conductance regulator (CFTR) +19.6 kb and +22.6 kb cis-regulatory elements (CREs) in definitive endoderm (DE) cells. A, B, OTX2 Chromatin immunoprecipitation (ChIP) in, A, DE d6 and, B, anterior foregut endoderm (AFE) d11 ND2.0 cells. C, D, H3K27me3 ChIP in, C, DE d6 and, D, AFE d11 ND2.0 cells. E,F, H3K27ac ChIP in, E, DE d6 and, F, AFE d11 ND2.0 cells. All ChIP dataset are each normalized to percent input. n = 3. Enrichment levels: *** denotes P < .001; ** denotes P < .01 using the Holm-Sidak multiple t test
To investigate the mechanism of action the +19.6 kb and +22.6 kb CREs, we next examined repressive (H3K27me3) or active (H3K27ac) histone modifications at these sites in DE and AFE cells. While OTX2 often functions as an activating TF, it can also serve as a repressor in certain contexts.41–44 Since loss of OTX2 coincides with activation of CFTR expression in AFE cells, and transient depletion of OTX2 in DE cells causes activation of CFTR expression, we hypothesized that OTX2 might be functioning as a repressor in DE cells. However, compared to control loci, enrichment of H3K27me3 was not observed at the +19.6 kb or +22.6 kb CREs, or at any location along the CFTR locus in DE or AFE cells (Figure 5C,D), suggesting this hypothesis was not correct. Rather, the +19.6 kb and +22.6 kb CREs were enriched for H3K27ac in DE cells, though the levels were reduced upon differentiation into AFE cells (Figure 5E,F). Notably, the CFTR promoter lacked H3K27ac enrichment in DE cells where CFTR was of low abundance, and gained this active histone upon activation of expression in AFE cells (Figure 5E, F). These data suggest, that while OTX2 interacts directly with the +19.6 kb and +22.6 kb CFTR CREs in DE cells, its impact on CFTR expression in these cells may rely upon preventing the recruitment of activating factors, rather than direct repressive functions.
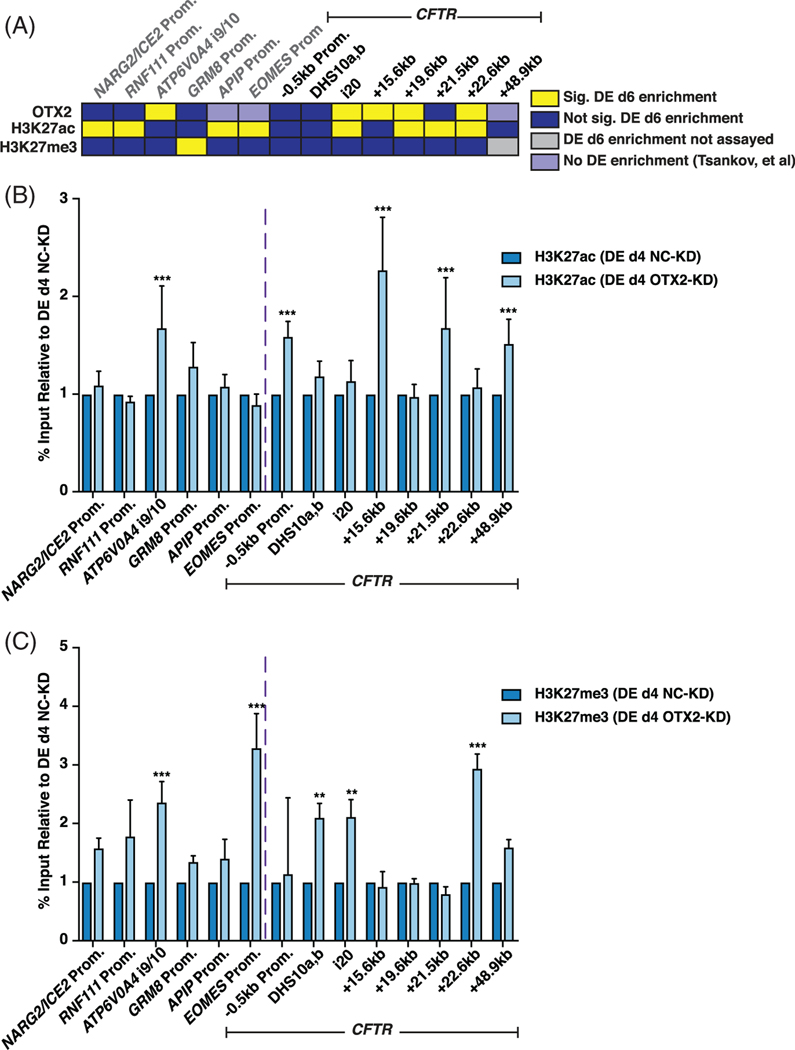
2.5 |. Loss of OTX2 protein in DE cells alters histone modifications at the CFTR locus
To further explore the mechanism by which recruitment of OTX2 to the +19.6 kb and +22.6 kb CREs might be directly regulating the chromatin dynamics of the CFTR locus, ChIP-qPCR was performed following OTX2-depletion in DE cells at day 4 of differentiation (Figure 4A). Figure 6A summarizes ChIP-qPCR data presented in Figures 5 of OTX2, H3K27ac, and H3K27me3 enrichment in DE day 6 cells. Upon reduction in OTX2, H3K27ac and H3K27me3 levels both significantly increased at the ATP6V0A4 i9/10 site, the only control site with known OTX2 enrichment in WT cells. H3K27ac and H3K27me3 levels were not statistically different at OTX2-negative control sites, irrespective of WT H3K27ac or H3K27me3 enrichment levels, with the exception of increased H3K27me3 enrichment at the EOMES promoter (Figure 6B,C). Across the CFTR locus, H3K27ac enrichment was generally unaffected by loss of OTX2, irrespective of OTX2 enrichment in WT cells, with a few notable exceptions. First, H3K27ac enrichment at the CFTR promoter increased upon loss of OTX2, consistent with the observed increase in CFTR transcription with OTX2-depletion (Figure 4E). Second, the +21.5 kb CRE, which is flanked by the +19.6 kb and +22.6 kb CREs, also showed an increase in H3K27ac enrichment upon loss of OTX2, as did the nearby +15.6 kb CRE (Figure 6B). Upon OTX2 depletion, H3K27me3 enrichment significantly increased at some OTX2-bound sites, including the +22.6 kb CRE and intron 20 (Figure 6C). H3K27me3 also increased at the OTX2 negative site in intron 10 following loss of OTX2 (Figure 6C). Based on these data, we suggest that OTX2 enrichment at the CFTR locus contributes to CFTR regulation in DE cells, in part through its role in influencing chromatin signals along the CFTR locus.
FIGURE 6.
Loss of orthodenticle homeobox 2 (OTX2) in definitive endoderm (DE) cells affects histone modifications at cystic fibrosis transmembrane conductance regulator (CFTR) cis-regulatory elements (CREs). A, OTX2, H3K27ac, and H3K27me3 enrichment in DE day 6 cells summarized from Figure 5A,C,E. B, C: B, H3K27ac and, C, H3K27me3 enrichment in DE d4 ND2.0 cells following siRNA-mediated depletion of OTX2-KD or negative control (NC-KD) (see Figure 3B for experimental timeline). Data normalized to percent input and shown relative to DE d4 NC-KD values for same primer set. B, n = 3; C, n = 2; *** denotes P < .001; ** denotes P < .01 comparing normalized OTX2-KD enrichment levels to normalized NC-KD enrichment levels at each site using the Holm-Sidak multiple t test
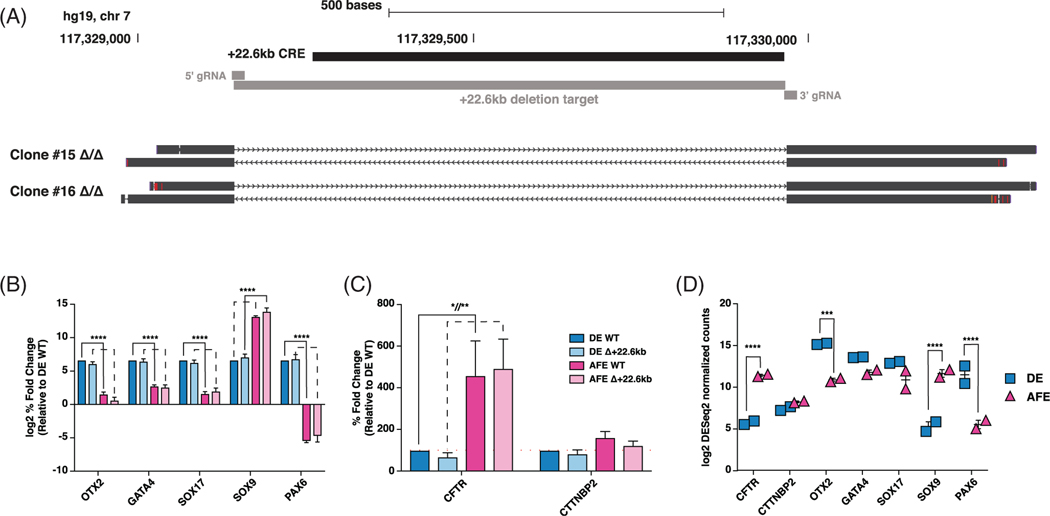
2.6 |. Deletion of the +22.6 kb CRE does not affect CFTR expression in DE or AFE cells
To investigate whether the +22.6 kb CRE had a functional impact on regulating CFTR expression in DE or AFE cells, this site was removed from iPSCs by CRISPR/Cas9-mediated nonhomologous end joining (NHEJ), and clonal lines with homozygous deletions were established (Figure 7A). The CRISPR/Cas9-derived modification, and subsequent clonal selection, had no effect on the expression OTX2, GATA4, or important markers of differentiation in DE or AFE cells (SOX17, SOX9, PAX6) (Figure 7B). Marker gene expression recapitulated the expression trends measured by RNA-seq in nontargeted, WT DE and AFE cells (Figure 7C,D). However, while CFTR expression was upregulated in the transition from DE to AFE cells in targeted and deletion clones, the loss of the +22.6 kb CRE, did not significantly impact CFTR mRNA abundance at either stage (Figure 7C). Importantly, expression of cortactin binding protein 2 (CTTNBP2), the gene adjacent to the +22.6 kb CRE on the 3′ side, was also unaltered in the deletion clones (Figure 7C). Together, these data suggest that the +22.6 kb CRE alone does not have the capacity to regulate CFTR expression during the transition from DE to AFE cells.
FIGURE 7.
Deletion of +22.6 kb cis-regulatory element (CRE) does not alter cystic fibrosis transmembrane conductance regulator (CFTR) expression in definitive endoderm (DE) or anterior foregut endoderm (AFE) cells. A, UCSC Genome Browser view of +22.6 kb CRE CRISPR/Cas9-mediated deletion schematic and sequencing alignments of two homozygous deletion clones. B, C, Reverse transcription quantitative PCR (RT-qPCR) of B., orthodenticle homeobox 2 (OTX2), GATA4, SOX17, SOX9, and PAX6; C, CFTR and cortactin binding protein 2 (CTTNBP2) in WT and +22.6 kb deleted DE and AFE cells. WT data combine CWRU205 WT, and two nontargeted clones; +22.6 kb deletion data combine two homozygous deletion clones. Expression is shown relative to DE WT, normalized to PGK1, n = 3. D, RNA-seq DEseq2 normalized counts in ND2.0 and CWRU205 in DE and AFE stages. **** denotes P < .0001, ** denotes P < .01, and * denotes P < .05 using a two-way analysis of variance (ANOVA) and Sidak’s multiple comparisons test. Not significant is not shown
3 |. DISCUSSION
Here, we profiled CFTR expression in an iPSC to ALI model of airway epithelial cells differentiation, with the aim of uncovering the mechanisms and CREs that regulate this process. We demonstrated that CFTR expression in this model system recapitulates the developing human and sheep lung, whereby expression increases during development, and then decreases following the second trimester, with continued low postnatal expression.6,7,13–15,45 However, it is not clear whether this profile is the result of a change in CFTR expression in individual cell types or an alteration in the relative abundance of CFTR-expressing cells. The task of accurately monitoring the identity of CFTR-expressing cells in the developing lung will require single cell RNA-seq and/or single molecule fluorescence in situ hybridization. We observed that CFTR expression becomes significantly upregulated in the AFE stage, and then decreases through to culture of airway epithelial cells at ALI. Notably, we observed two novel open chromatin regions at the 3′ end of the CFTR locus that were only detected in DE cells, thus prior to CFTR upregulation in AFE.
The two new DE-specific CREs, +19.6 kb and +22.6 kb downstream of the CFTR translational stop site, are located within the CFTR TAD boundary (−80.1 kb to +48.9 kb) in a 3′ region that contains both cell-type selective CREs and others that are seen in multiple cell types.11 This TAD separates CFTR from its neighboring genes, ankyrin repeat, SAM and basic leucine zipper domain containing 1 (ASZ1) and CTTNBP2, which are both transcribed from the antisense strand. Upstream of the DE CREs is the +15.6 kb CRE, which is observed in DE, AFE, donor derived HBE-ALI (Figure 1D), and multiple other epithelial cell types (Table 1), irrespective of CFTR expression.26 We showed previously that the +15.6 kb CRE houses a CTCF-independent enhancer blocking insulator, which is often marked by H3K4me1 and p300 enrichment.30,31 Notably, the presence of this element does not prevent interactions of CREs located further 3′ within the TAD with the CFTR promoter.
Interestingly, the +19.6 kb and +22.6 kb CREs immediately flank the CRE at +21.5 kb, which is predominantly observed in cells of epithelial origin, including primary airway cells (Figure 1, Table 1).27 Notably, of the 12 cell types in which the +21.5 kb site is open according to ENCODE data, nine cell types also have open chromatin at the +15.6 kb CRE, though only one (HEEpiC, esophageal epithelial cells) additionally shows open chromatin at +19.6 kb and +22.6 kb (Table 1).32 However, the open chromatin signal for +21.5 kb is dominant in this region in the HEEpiC cells. Together, these data suggest that the +19.6 kb and +22.6 kb CREs are generally not coincident with the +21.5 kb CRE. As the +19.6 kb and +22.6 kb CREs are mainly detected in pre-differentiated cells, while +21.5 kb is seen in differentiated epithelial cells, the +19.6 kb and +22.6 kb sites may play a role in establishing local chromatin structure for later recruitment into regulatory mechanisms.
OTX2 has mainly been studied in relation to its important role in brain development and its role in type 3 and 4 medulloblastomas.46–49 It predominantly functions as an activating factor through direct interactions with DNA, and exerts its repressive function mainly in an indirect manner.41–44 OTX2 has also been reported to have pioneer TF activity, whereby its overexpression can open closed chromatin and enhance the recruitment and deposition of H3K27ac at known OTX2-bound active enhancers.44 Additionally, while many OTX2-bound sites are active enhancers with H3K27ac enrichment, many intergenic sites occupied by OTX2 lack H3K27ac, and are thought to help keep chromatin in a bivalent state.43 Notably, OTX2 ChIP-seq in medulloblastoma cell lines, hES-derived DE cells, and MSCs with OTX2-overexpression, all show OTX2 enrichment at +19.6 kb CFTR CRE, even though CFTR and CTTNBP2 are not expressed or very lowly expressed in many of these cell types.38,44 Additionally, enrichment for histone marks across the CFTR promoter and the intergenic region between CFTR and CTTNBP2, including H3K27ac, H327me3, and H3K4me1 are lacking in these cell types.44,50 Interestingly, of the sites assayed for OTX2 enrichment across the CFTR locus, only the element at +15.6 kb is bound by OTX2 in the absence of H3K27ac enrichment. Additionally, upon loss of OTX2, the +15.6 kb CRE significantly gained H3K27ac enrichment, implicating a mechanism that prevents its activation.
Together these studies support the hypothesis that OTX2 recruitment to the +15.6 kb, +19.6 kb, and +22.6 kb CFTR CREs does not establish active enhancers at these sites, but rather is important for maintaining appropriate differentiation stage-specific histone modifications in this region.
Importantly, ChIP-seq of OTX2 in hES-derived DE cells shows OTX2 enrichment at multiple sites in the CFTR locus, including introns 10, 20, and 21 (legacy nomenclature), as well as at the +19.6 kb and +22.6 kb regions. The intron 10 OTX2 site is at a known region of open chromatin in the CFTR locus (DHS10c, legacy); however, the function of this site is currently unclear, though it does not have enhancer activity in many cell types.37,39,51 While we cannot exclude the possibility that the +19.6 kb and +22.6 kb sites are important for CTTNBP2 regulation rather than CFTR, multiple lines of evidence would suggest their function is related to CFTR expression. First, CFTR is a DEG in the DE to AFE transition, whereas CTTNBP2 is neither a DEG (Figure 7E) nor does its expression change substantially following OTX2-KD (data not shown). Second, examination of 5C-seq data on ENCODE32,52, which measures long-range chromatin interactions in hESCs and HeLa-S3 cells, shows that the +19.6 kb to +22.6 kb region interacts with upstream chromatin (including a site close to −25 kb, with respect to the CFTR promoter), and not regions downstream in CTTNBP2 locus. Third, using a viewpoint at the +22.6 kb CRE to examine the interactions of this element by 4C-seq in DE and AFE cells, an association with regions between the −35 kb and −20.9 kb CFTR CREs was seen, but no interactions were evident with the CTTNBP2 locus (Figure S1).
Though we present evidence here for a direct effect of OTX2 on CFTR expression, this TF may also exert an indirect effect on the locus in vivo through the control of other TFs that regulate CFTR. In this way, the OTX2 transcriptional network could have a profound effect on CFTR expression during airway epithelial differentiation.
4 |. EXPERIMENTAL PROCEDURES
4.1 |. CFTR nomenclature
CFTR introns and exons are numbered using legacy nomenclature,53 for consistency with our previous work.11
4.2 |. Cell culture
iPSC lines CR0000008 (Cell line ID: ND2.0) https://www.nimhgenetics.org/stem_cells/crm_lines.php was purchased from RUCDR Infinite Biologics (www.rucdr.org). CWRU205 was obtained from Dr Paul J. Tesar (Dept. of Genetics and Genome Sciences, Case Western Reserve University). Both lines were from male donors. Differentiation was performed as previously described.21,25
Donor-derived HBE cells were obtained from the Marsico Lung Institute CF Center Tissue Procurement and Cell Culture Core (Chapel Hill, NC) cultured according to the published protocol.54,55
4.3 |. ATAC-seq and RNA-seq data
ATAC-seq and RNA-seq data were generated previously25 and are accessible from the GEO database series GSE136859.
4.4 |. RNA preparation and RT-qPCR
Total RNA was extracted using TRIzol (Invitrogen) following the manufacturer’s protocol. RT-qPCR was done using standard protocols56 with primers shown in Table 2.
TABLE 2.
Primers and gBlocks
| SYBRGreen assay | ||
|---|---|---|
|
| ||
| Gene | Forward sequence (5′ > 3′) | Reverse sequence (5′ > 3′) |
| OTX2 | TTCGGGTATGGACTTGCTGC | GATGTCTGGGTACCGGGTCT |
| GATA4 | TCTACATGAAGCTCCACGGG | TGAAGGAGCTGCTGGTGTC |
| SOX17 | AAGGGCGAGTCCCGTATC | TTGTAGTTGGGGTGGTCCTG |
| SOX9 | GAGGAAGTCGGTGAAGAACG | ATCGAAGGTCTCGATGTTGG |
| PAX6 | TCTTTGCTTGGGAAATCCG | CTGCCCGTTCAACATCCTTAG |
| CTTNBP2 | CCCCGTGGGACTTTATGAGG | GCCTTCTTGAGGACCTGACA |
| PGK1 | TAACAAGCTGACGCTGGAC | GCAGCCTTAATCCTCTGGTT |
| TaqMan assay | ||
|
| ||
| Gene | Sequence (5′ > 3′) | |
| CFTR | AGCTGTCAAGCCGTGTTCTAGATA ATGAGGAGTGCCACTTGCAAA /56-FAM/CACACGAAA/ZEN/TGTGCCAATGCAAGTCCTT/3IABkFQ/ |
|
| PGK1 | TAACAAGCTGACGCTGGAC GCAGCCTTAATCCTCTGGTT /56-JOEN/CGACTCTCA/ZEN/TAACGACCCGCTTCC/3IABkFQ/ |
|
| ChIP-qPCR | ||
|
| ||
| Amplicon site | Forward sequence (5′ > 3′) | Reverse sequence (5′ > 3′) |
| 18s | CGGCTACCACATCCAAGGAA | GCTGGAATTACCGCGGCT |
| NARG2/ICE2 promoter | GGAAAGGGGACGCAAAGAGA | TCCTACCCCTGCTTTGCTTG |
| RNF111 promoter | CACGTACAGTTTTGGTGGCG | AGTAATGCGTCGAGAGCCAG |
| ATP6V0A4 i9/10 | AAGGTGCCAAAAATGGAGTG | AGGAGAGTTCAAGGCCAGGT |
| GRM8 promoter | CCAATCAAGCTCCAATGCCC | TGTGAGAGAAATAGCGGGCG |
| APIP promoter | TTATCTTGTGGGCTTCCCCG | AATTAGCTCCGCGGCTTACA |
| EOMES Promoter | TTTCGCCGGAATTTAGGGGG | ATGACTAATGCGCCAAGGGT |
| EHF promoter | TCCCGGTTGGTGGCTTATTG | GGCACCACGGGTGTTATCAA |
| CFTR −0.5 kb promoter | GTTCTCCCGCCGGTGG | CAGTCGCGGCCTCTCTTTAG |
| CFTR DHS10a,b | TGCTTTATTGAATGGCATTACCTCTA | AGATGCTTGTGGTAAGGGAGGAG |
| CFTR i20 | AGGATGCCAGACAGGGAGAT | ACCTTTTGCCCCTCAGCTTT |
| CFTR +15.6 kb | ATCCATTTTCTTCAAGTCTCTCTCCAT | GGAATGAGGATTGTTTATGATTTGG |
| CFTR +19.6 kb | CTGCAAGTATTTCTTGGATGCTGT | GTAACCCATTTGGCTTGAGGG |
| CFTR +21.5 kb | GGCTAGTTCCTTGACCTGAACAA | TGGAAGTTCCTCCATTTTGAATG |
| CFTR +22.6 kb | GCCAGGGAATGAGTCTTGTG | GCCTCCTGTTCTTCCCAAAT |
| CFTR +48.9 kb | GGCATCAGCCAGTCAAGGTTT | AGCAGAGGGCAAGTGGTACTT |
| +22.6 kb deletion | ||
|
| ||
| Name | Sequence (5′ > 3′) | |
| +22.6 kb 5′ gBlock | TGTACAAAAAAGCAGGCTTTAAAGGAACCAATTCAGTCGACTGGATCCGGTACCAAGGTCGGGCA GGAAGAGGGCCTATTTCCCATGATTCCTTCATATTTGCATATACGATACAAGGCTGTTAGAGAGATAAT TAGAATTAATTTGACTGTAAACACAAAGATATTAGTACAAAATACGTGACGTAGAAAGTAATAATTTCTT GGGTAGTTTGCAGTTTTAAAATTATGTTTTAAAATGGACTATCATATGCTTACCGTAACTTGAAAGTATT TCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCGTTCAAGACTACTTGCACGG GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGA GTCGGTGCTTTTTTTCTAGACCCAGCTTTCTTGTACAAAGTTGGCATTA |
|
| +22.6 kb 3′ gBlock | TTGTACAAAAAAGCAGGCTTTAAAGGAACCAATTCAGTCGACTGGATCCGGTACCAAGGTCGGGCAGGA AGAGGGCCTATTTCCCATGATTCCTTCATATTTGCATATACGATACAAGGCTGTTAGAGAGATAATTAG AATTAATTTGACTGTAAACACAAAGATATTAGTACAAAATACGTGACGTAGAAAGTAATAATTTCTTGG GTAGTTTGCAGTTTTAAAATTATGTTTTAAAATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTC GATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCGTACTCACACCGTCGCCAGA GTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGA GTCGGTGCTTTTTTTCTAGACCCAGCTTTCTTGTACAAAGTTGGCATTA |
|
| +22.6 kb EF | TGGCACTCTCCTTCTGTGTG | |
| +22.6 kb ER | CAGGGCATCTTCTGGCTTAC | |
| +22.6 kb IF | GCCAGGGAATGAGTCTTGTG | |
| +22.6 kb IR | ACTCATTCCCTGGCTTCCTT | |
| 4C-seq primers | ||
|
| ||
| Name | Reading primer (5′ > 3′) | Nonreading primer (5′ > 3′) |
| CFTR +22.6 kb viewpoint | TACACGACGCTCTTCCGATCTCAAA CATCCTGGTAAACCATG |
ACTGGAGTTCAGACGTGTGCTCTTCCGATCTGC ACTTATCACTAGCTAATGT |
Abbreviations: CFTR, cystic fibrosis transmembrane conductance regulator; ChIP-qPCR, Chromatin immunoprecipitation followed by qPCR; CTTNBP2, cortactin binding protein 2; OTX2, orthodenticle homeobox 2.
4.5 |. RNA-seq analysis
RNA-seq data was processed as previously described to generate differential gene expression lists.25 Differentially expressed gene results were filtered with a fold change of 2 and Benjamini-Hochberg adjusted P-value of .01 for human TF genes.33
4.6 |. In silico TF binding site predictions
MatInspector version 8.4.1 (Genomatix, Munich, Germany) was used to predict TF binding sites (Matrix Library 11.0) within the +19.6 kb and +22.6 kb CFTR CREs using default matrix search parameters (core similarity, 0.75; matrix similarity, optimized). The same regions were analyzed with TF Affinity Prediction (TRAP) Web Tool34 using the default search parameters (TRANSFAC 2010.1 vertebrates matrix, human-promoters background model, and Benjamini-Hochberg correction, P-value <.05). Results from both analyses were filtered for differentially expressed TF genes.
4.7 |. Western blot
Whole cell lysates were collected using RIPA buffer (1% [vol/vol] IGEPAL CA-630 [Sigma-Aldrich]; 0.5% [wt/vol] sodium deoxycholate; 0.1% [wt/vol] SDS; 1X cOmplete protease inhibitor Cocktail [Roche] in PBS). Protein concentration was determined by DC Protein Assay (BioRad) and 8 to 10 μg lysates were resolved by SDS-PAGE using standard methods and probed with the following anti-bodies: anti-OTX2 (Proteintech, 13497–1-AP, lot # 0039785, 1:1000), anti-GATA4 (R&D Systems, MAB2607, lot #CCQC0116081, 1:1000), anti-β-tubulin (Sigma-Aldrich, T4026), anti-rabbit-HRP (Sigma-Aldrich, A0545, 1:10 000), and anti-mouse-HRP (Agilent/Dako, P0447, lot # 20051789, 1:10 000). Proteins were detected using ECL Western Blotting Substrate (Pierce).
4.8 |. Chromatin immunoprecipitation-qPCR
ChIP was performed using standard methods as previously described.26 Immunoprecipitations were performed with antibodies against OTX2 (Proteintech, 13497–1-AP, lot # 0039785), H3K27ac (Millipore-Sigma, 07–360, lot # 3071583), H3K27me3 (Millipore-Sigma 07–449, lot # 3146226), and normal rabbit IgG (Abcam, 12–370, lot # 3059594). Enrichments were analyzed percent input using PowerSYBR Green PCR Master Mix (Thermo Fisher Scientific) with primers listed in Table 2.
4.9 |. siRNA-mediated transient transfections
iPSCs (ND2.0) were reverse transfected with Lipofectamine RNAiMAX (ThermoFisher) following the manufacturer’s protocol with Silencer Select OTX2 siRNA (Invitrogen, 439240 [s9931], lot# AS02D09A) or Silencer Select Negative-Control #2 siRNA (Invitrogen, 4390846 lot# AS022CDS). For lysate and expression analysis, 10 pmol siRNA was transfected into 200 000 iPSC (ND2.0) seeded into hESC-qualified Matrigel (Corning) coated 24-well plates. For chromatin preparation, 200 pmol siRNA was transfected into ~4.2 × 106 to 5.5 × 106 cells per hESC-qualified Matrigel (Corning) coated 10 cm plate. Cells were plated and transfected in mTeSR1 complete media (StemCell Technologies) supplemented with 10 μM Y-27632 (Tocris/Biotechne). Media was changed daily with STEMdiff DE media (StemCell Technologies) and cells collected for RNA and lysate at 72 and 96 hours posttransfection (days 3 and 4) and at 96 hours for chromatin preparation.
4.10 |. CRISPR-mediated deletions
A pair of gRNAs flanking the CFTR +22.6 kb CRE were identified using Benchling (benchling.com) and ordered as gBlocks from Integrated DNA Technologies, cloned into pSCB, and sequenced. Then, 7.5 × 104 cells were seeded onto hESC-qualified Matrigel coated six-well plates 24 hours prior to transfection in mTeSR1 complete media supplemented with 10 μM Y-27632. Cells were transfected with Lipofectamine3000 (ThermoFisher) at a 1:1:1 ratio with pMJ920 plasmid (WT GFP-tagged Cas9 plasmid) (Addgene #42234) and the cloned gRNA plasmids following the manufacturer’s protocol in fresh mTeSR1 complete media. Media was changed daily and cells were prepared for fluorescence-activated cell sorting of GFP-positive cells 48 hours after transfection. Single cells were plated onto hESC-qualified Matrigel coated 96-well plates and recovered into mTeSR1 complete media supplemented with CloneR (StemCell Technologies) following the manufacturer’s protocol. Clones were screened for deletion as previously described.57 gBlock sequences and primers are listed in Table 2.
4.11 |. Circular chromosome conformation capture with deep sequencing (4C-seq) analysis
4C-seq libraries were prepared and analyzed from DE and AFE cells as previously described.58 NlaIII and Csp6I were used as the primary and secondary restriction enzymes, respectively. Primer sequences used to generate 4C-seq libraries the +22.6 kb viewpoint are shown Table 2. The sequencing data were processed using the R package pipe4C.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr Paul Tesar for the CWRU205 line, the NIMH Repository and the Genomics Resource (RGR) at Rutgers University for ND2.0 (CR0000008) iPSC line; Dr Pieter Faber and colleagues at the University of Chicago Genomics Core for sequencing; also Dr Scott Randell and colleagues at the Marsico Lung Institute Tissue Procurement and Cell Culture Core, for donor-derived HBE cultures (supported in part by CFF and NIH grant DK065988). This work was supported by the Cystic Fibrosis Foundation (Harris15/17XX0, Harris16G0, and Harris18P0) (A. H.) and NIH R01HL094585 (A. H.).
Funding information
Cystic Fibrosis Foundation, Grant/Award Number: Harris15/17XX0, Harris16G0, and Harris18P0; National Heart, Lung, and Blood Institute, Grant/Award Number: R01HL094585
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
CONFLICT OF INTEREST
The authors declare no competing interests. The funders had no role in the design of the study, in the collection, analyses, or interpretation of the data, in the writing of the manuscript, or in the decision to publish the results.
REFERENCES
- 1.Crawford I, Maloney PC, Zeitlin PL, et al. Immunocytochemical localization of the cystic fibrosis gene product CFTR. Proc Natl Acad Sci U S A. 1991;88(20):9262–9266. 10.1073/pnas.88.20.9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelhardt JF, Yankaskas JR, Ernst SA, et al. Submucosal glands are the predominant site of CFTR expression in the human bronchus. Nat Genet. 1992;2(3):240–248. 10.1038/ng1192-240. [DOI] [PubMed] [Google Scholar]
- 3.Kreda SM, Mall M, Mengos A, et al. Characterization of wild-type and deltaF508 cystic fibrosis transmembrane regulator in human respiratory epithelia. Mol Biol Cell. 2005;16(5):2154–2167. 10.1091/mbc.e04-11-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montoro DT, Haber AL, Biton M, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560(7718):319–324. 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plasschaert LW, Zilionis R, Choo-Wing R, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560(7718):377–381. 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris A, Chalkley G, Goodman S, Coleman L. Expression of the cystic fibrosis gene in human development. Development. 1991;113(1):305–310. [DOI] [PubMed] [Google Scholar]
- 7.Trezise AE, Chambers JA, Wardle CJ, Gould S, Harris A. Expression of the cystic fibrosis gene in human foetal tissues. Hum Mol Genet. 1993;2(3):213–218. [DOI] [PubMed] [Google Scholar]
- 8.McCarthy VA, Harris A. The CFTR gene and regulation of its expression. Pediatr Pulmonol. 2005;40(1):1–8. 10.1002/ppul.20199. [DOI] [PubMed] [Google Scholar]
- 9.Ott CJ, Blackledge NP, Leir SH, Harris A. Novel regulatory mechanisms for the CFTR gene. Biochem Soc Trans. 2009;37(Pt 4):843–848. 10.1042/BST0370843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillen AE, Harris A. Transcriptional regulation of CFTR gene expression. Front Biosci (Elite Ed). 2012;4:587–592. 10.2741/401. [DOI] [PubMed] [Google Scholar]
- 11.Gosalia N, Harris A. Chromatin Dynamics in the Regulation of CFTR Expression. Genes (Basel). 2015;6(3):543–558. 10.3390/genes6030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swahn H, Harris A. Cell-selective regulation of CFTR gene expression: relevance to gene editing therapeutics. Genes (Basel). 2019;10(3). 10.3390/genes10030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCray PB Jr, Wohlford-Lenane CL, Snyder JM. Localization of cystic fibrosis transmembrane conductance regulator mRNA in human fetal lung tissue by in situ hybridization. J Clin Invest. 1992;90(2):619–625. 10.1172/JCI115901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tizzano EF, Chitayat D, Buchwald M. Cell-specific localization of CFTR mRNA shows developmentally regulated expression in human fetal tissues. Hum Mol Genet. 1993;2(3):219–224. 10.1093/hmg/2.3.219. [DOI] [PubMed] [Google Scholar]
- 15.Gaillard D, Ruocco S, Lallemand A, Dalemans W, Hinnrasky J, Puchelle E. Immunohistochemical localization of cystic fibrosis transmembrane conductance regulator in human fetal airway and digestive mucosa. Pediatr Res. 1994;36(2):137–143. 10.1203/00006450-199408000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Marcorelles P, Montier T, Gillet D, Lagarde N, Ferec C. Evolution of CFTR protein distribution in lung tissue from normal and CF human fetuses. Pediatr Pulmonol. 2007;42(11):1032–1040. 10.1002/ppul.20690. [DOI] [PubMed] [Google Scholar]
- 17.Bedrossian CW, Greenberg SD, Singer DB, Hansen JJ, Rosenberg HS. The lung in cystic fibrosis. A quantitative study including prevalence of pathologic findings among different age groups. Hum Pathol. 1976;7(2):195–204. 10.1016/s0046-8177(76)80023-8. [DOI] [PubMed] [Google Scholar]
- 18.Somers A, Jean JC, Sommer CA, et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28(10):1728–1740. 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong AP, Bear CE, Chin S, et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat Biotechnol. 2012;30 (9):876–882. 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Firth AL, Dargitz CT, Qualls SJ, et al. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2014;111(17):E1723-E1730. 10.1073/pnas.1403470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong AP, Chin S, Xia S, Garner J, Bear CE, Rossant J. Efficient generation of functional CFTR-expressing airway epithelial cells from human pluripotent stem cells. Nat Protoc. 2015;10 (3):363–381. 10.1038/nprot.2015.021. [DOI] [PubMed] [Google Scholar]
- 22.Huang SX, Green MD, de Carvalho AT, et al. The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat Protoc. 2015;10(3):413–425. 10.1038/nprot.2015.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rankin SA, Han L, McCracken KW, et al. A retinoic acid-hedgehog cascade coordinates mesoderm-inducing signals and endoderm competence during lung specification. Cell Rep. 2016;16(1):66–78. 10.1016/j.celrep.2016.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins F, Kramer P, Jacob A, et al. Prospective isolation of NKX2–1-expressing human lung progenitors derived from pluripotent stem cells. J Clin Invest. 2017;127(6):2277–2294. 10.1172/JCI89950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerschner JL, Paranjapye A, Yin S, et al. A functional genomics approach to investigate the differentiation of iPSCs into lung epithelium at air-liquid interface. J Cell Mol Med. 2020;24: 9853–9870. 10.1111/jcmm.15568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ott CJ, Blackledge NP, Kerschner JL, et al. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci U S A. 2009;106(47):19934–19939. 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z, Ott CJ, Lewandowska MA, Leir SH, Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J Cell Mol Med. 2012;16(6):1321–1330. 10.1111/j.1582-4934.2011.01439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Leir SH, Harris A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol Cell Biol. 2013;33(15):2843–2853. 10.1128/MCB.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Leir SH, Harris A. Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am J Respir Cell Mol Biol. 2015;52(3):387–396. 10.1165/rcmb.2014-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackledge NP, Carter EJ, Evans JR, Lawson V, Rowntree RK, Harris A. CTCF mediates insulator function at the CFTR locus. Biochem J. 2007;408(2):267–275. 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackledge NP, Ott CJ, Gillen AE, Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009;37(4): 1086–1094. 10.1093/nar/gkn1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haeussler M, Zweig AS, Tyner C, et al. The UCSC genome browser database: 2019 update. Nucleic Acids Res. 2019;47(D1): D853–D858. 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert SA, Jolma A, Campitelli LF, et al. The human transcription factors. Cell. 2018;172(4):650–665. 10.1016/j.cell.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 34.Thomas-Chollier M, Hufton A, Heinig M, et al. Transcription factor binding predictions using TRAP for the analysis of ChIP-seq data and regulatory SNPs. Nat Protoc. 2011;6(12):1860–1869. 10.1038/nprot.2011.409. [DOI] [PubMed] [Google Scholar]
- 35.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91(4):531–541. 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Miki T, Iwanaga T, et al. Identification, tissue expression, and functional characterization of Otx3, a novel member of the Otx family. J Biol Chem. 2002;277(31):28065–28069. 10.1074/jbc.C100767200. [DOI] [PubMed] [Google Scholar]
- 37.Phylactides M, Rowntree R, Nuthall H, Ussery D, Wheeler A, Harris A. Evaluation of potential regulatory elements identified as DNase I hypersensitive sites in the CFTR gene. Eur J Biochem. 2002;269(2):553–559. [DOI] [PubMed] [Google Scholar]
- 38.Tsankov AM, Gu H, Akopian V, et al. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518(7539):344–349. 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mouchel N, Henstra SA, McCarthy VA, Williams SH, Phylactides M, Harris A. HNF1alpha is involved in tissue-specific regulation of CFTR gene expression. Biochem J. 2004; 378(Pt 3):909–918. 10.1042/BJ20031157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerschner JL, Gosalia N, Leir SH, Harris A. Chromatin remodeling mediated by the FOXA1/A2 transcription factors activates CFTR expression in intestinal epithelial cells. Epigenetics. 2014;9(4):557–565. 10.4161/epi.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai RY, Staedtke V, Lidov HG, Eberhart CG, Riggins GJ. OTX2 represses myogenic and neuronal differentiation in medulloblastoma cells. Cancer Res. 2012;72(22):5988–6001. 10.1158/0008-5472.CAN-12-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bunt J, Hasselt NE, Zwijnenburg DA, et al. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer. 2012;131(2):E21–E32. 10.1002/ijc.26474. [DOI] [PubMed] [Google Scholar]
- 43.Bunt J, Hasselt NA, Zwijnenburg DA, Koster J, Versteeg R, Kool M. OTX2 sustains a bivalent-like state of OTX2-bound promoters in medulloblastoma by maintaining their H3K27me3 levels. Acta Neuropathol. 2013;125(3):385–394. 10.1007/s00401-012-1069-2. [DOI] [PubMed] [Google Scholar]
- 44.Boulay G, Awad ME, Riggi N, et al. OTX2 activity at distal regulatory elements shapes the chromatin landscape of group 3 medulloblastoma. Cancer Discov. 2017;7(3):288–301. 10.1158/2159-8290.CD-16-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broackes-Carter FC, Mouchel N, Gill D, Hyde S, Bassett J, Harris A. Temporal regulation of CFTR expression during ovine lung development: implications for CF gene therapy. Hum Mol Genet. 2002;11(2):125–131. [DOI] [PubMed] [Google Scholar]
- 46.Adamson DC, Shi Q, Wortham M, et al. OTX2 is critical for the maintenance and progression of Shh-independent medulloblastomas. Cancer Res. 2010;70(1):181–191. 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boon K, Eberhart CG, Riggins GJ. Genomic amplification of orthodenticle homologue 2 in medulloblastomas. Cancer Res. 2005;65(3):703–707. [PubMed] [Google Scholar]
- 48.Suda Y, Matsuo I, Aizawa S. Cooperation between Otx1 and Otx2 genes in developmental patterning of rostral brain. Mech Dev. 1997;69(1–2):125–141. 10.1016/s0925-4773(97)00161-5. [DOI] [PubMed] [Google Scholar]
- 49.Acampora D, Boyl PP, Martinez-Barbera JP, Annino A, Signore M, Simeone A. Otx genes in evolution: are they involved in instructing the vertebrate brain morphology? J Anat. 2001;199(Pt 1–2):53–62. 10.1046/j.1469-7580.2001.19910053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loh KM, Ang LT, Zhang J, et al. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell. 2014;14(2): 237–252. 10.1016/j.stem.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith DJ, Nuthall HN, Majetti ME, Harris A. Multiple potential intragenic regulatory elements in the CFTR gene. Genomics. 2000;64(1):90–96. 10.1006/geno.1999.6086. [DOI] [PubMed] [Google Scholar]
- 52.Dostie J, Richmond TA, Arnaout RA, et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16(10):1299–1309. 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsui LC, Dorfman R. The cystic fibrosis gene: a molecular genetic perspective. Cold Spring Harb Perspect Med. 2013;3(2): a009472. 10.1101/cshperspect.a009472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH. Well-differentiated human airway epithelial cell cultures. Methods Mol Med. 2005;107:183–206. 10.1385/1-59259-861-7:183. [DOI] [PubMed] [Google Scholar]
- 55.Randell SH, Fulcher ML, O’Neal W, Olsen JC. Primary epithelial cell models for cystic fibrosis research. Methods Mol Biol. 2011; 742:285–310. 10.1007/978-1-61779-120-8_18. [DOI] [PubMed] [Google Scholar]
- 56.Yang R, Browne JA, Eggener SE, Leir SH, Harris A. A novel transcriptional network for the androgen receptor in human epididymis epithelial cells. Mol Hum Reprod. 2018;24(9):433–443. 10.1093/molehr/gay029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.NandyMazumdar M, Yin S, Paranjapye A, et al. Looping of upstream cis-regulatory elements is required for CFTR expression in human airway epithelial cells. Nucleic Acids Res. 2020; 48(7):3513–3524. 10.1093/nar/gkaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krijger PHL, Geeven G, Bianchi V, Hilvering CRE, de Laat W. 4C-seq from beginning to end: a detailed protocol for sample preparation and data analysis. Methods. 2020;170:17–32. 10.1016/j.ymeth.2019.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.