Abstract
Transcription of the cystic fibrosis transmembrane conductance regulator (CFTR) gene is regulated by both ubiquitous and cell-type selective cis-regulatory elements (CREs). These CREs include extragenic and intronic enhancers that bind lineage-specific transcription factors, and architectural protein-marked structural elements. Deletion of the airway-selective −35 kb enhancer in 16HBE14o− lung epithelial cells was shown earlier to disrupt normal enhancer-promoter looping at the CFTR locus and nearly abolish its expression. Using a 16HBE14o− clone that lacks the endogenous −35 kb CRE, we explore the impact of relocating the functional core of this element to an ectopic site in intron 1. The −35 kb sequence establishes a de novo enhancer signature in chromatin at the insertion site, and augments CFTR expression, albeit not fully restoring WT levels. The relocated −35 kb enhancer also initiates de novo chromatin contacts with the CFTR promoter and other known CFTR CREs. These results are broadly relevant to therapeutic gene editing.
INTRODUCTION
Enhancers are defined as regulatory elements that function independently of location or orientation with respect to gene promoters, and are required for normal spatiotemporal control of gene expression. Early characterization of enhancer activity was established predominately through non-chromatinized plasmid-based reporter assays. Recent advances in genome engineering and next-generation sequencing techniques provide opportunities to manipulate enhancer sequences in vivo. Active enhancers are marked by enrichment of histone marks such as acetylation of lysine 27 on histone H3 (H3K27ac), are located in regions of open chromatin, and bind sequence specific transcription factors (TFs). These molecular features are often lost or reduced in contexts where the enhancer is inactive (reviewed in [1]).
Furthermore, chromosome conformation capture (3C) techniques enable the study of enhancer interactions in three dimensions and in real time [2]. Enhancers tend to interact with promoters and other CREs within the same topologically associating domain (TAD). TAD boundaries are generally marked by CCCTC binding factor (CTCF) occupancy, which provides an additional layer of gene regulation by isolating regions of the genome from each other [3, 4]. Some enhancers may regulate multiple genes as sub-TAD boundaries may change during development or in different cell-types, thus allowing for an enhancer to interact with a gene promoter that was previously inaccessible [5]. Alternatively, multiple studies have demonstrated a role of “enhancer adoption” or “enhancer hijacking” as a basis for some human disease, especially cancers [6, 7]. These events result in the association of genes with an ectopic enhancer, often due to mutations in TAD boundary elements or chromosomal rearrangements. However, few studies have explored the impact of relocating enhancers within a gene locus or a TAD [8, 9].
The cystic fibrosis transmembrane conductance regulator (CFTR) gene is a large, intricately regulated gene that resides within a 260 kb TAD, isolating it from its 5′ and 3′ neighbors, ASZ1 and CTTNBP2 respectively. The TAD boundaries are marked by CTCF and cohesin occupancy at CREs located −80.1 kb upstream of the promoter and +48.9 kb downstream of the last coding exon. Through the use of different cell-type and developmental CREs, CFTR expression is restricted both spatially and developmentally [10, 11]. Homozygous loss of CFTR, a small cAMP-activated chloride ion channel, results in the life limiting autosomal recessive disorder cystic fibrosis (CF), which affects 70,000 people worldwide. Development of CFTR modulator drugs has revolutionized trea™ent for nearly 90% of people with CF (pwCF) who harbor defective proteins. However, 10% of pwCF have mutations that abolish protein production and thus are not responsive to current modulator therapies (reviewed in [12]). For these individuals, there is a major effort to develop genetic therapies, particularly approaches which can correct multiple CF-causing mutations. These protocols may utilize heterologous promoters to drive expression of CFTR transgenes from safe-harbor loci [13, 14] or target the endogenous locus. The complex expression and large size of the gene make CFTR a difficult target for gene-editing therapeutics. Recent efforts have largely focused on gene editing to correct/repair the endogenous CFTR locus so that its normal regulatory machinery is not disrupted. Zinc finger nucleases (ZFNs) and CRISPR-Cas9 gene editing protocols were used successfully to introduce whole or partial WT CFTR cDNAs into the endogenous locus in mutant CFTR cells and CFTR function was restored [15–17].
In airway epithelial cells, CFTR expression is regulated in part by two airway-selective enhancers at −44 kb and −35 kb from the transcriptional start site [18–20]. We showed previously that loss of either element in airway epithelial cells nearly abolished CFTR expression, and disrupted the normal 3D looping structure [21]. Our goal here was to characterize the effect of moving the −35 kb enhancer from its normal location 5′ to the gene promoter to an intronic site within the CFTR locus. Here we show that −35kb sequence maintains enhancer activity at a different location within the CFTR locus, and initiates changes in the 3D structure of the locus that facilitate its direct interaction with the gene promoter from the new site. This element may have applications in therapeutic gene editing approaches that utilize endogenous CFTR regulatory elements to restore cell type-specific CFTR expression.
MATERIALS AND METHODS
Cell culture
Human bronchial epithelial 16HBE14o− [22] cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM), low glucose, supplemented with 10% fetal bovine serum.
CRISPR-mediated HDR
The parental cell line, 16HBE14o− Δ−35 kb, used for generating 16HBE14o− ins-35 kb cells was described previously [21]. The HDR template, with 506 bp (5′) and 593 (3′) homology arms (HAs) flanking the 497 bp −35 kb enhancer core was generated using Gibson Assembly Master Mix (New England Biolabs, E2611) and primers generated by the NEBuilder Assembly Tool v2.2.7. The HDR template was subsequently cloned into the pSCB vector and verified by Sanger sequencing. The plasmid containing the donor template was linearized with PuvI prior to transfection into cells to facilitate recombination. A single guide RNA was identified using Benchling (benchling.com) to target intron 1 at the integration site and ordered as gBlocks (Integrated DNA Technologies), cloned into pSCB, and sequenced. 16HBE14o− Δ−35 kb cells were co-transfected with pMJ920 plasmid (WT GFP-tagged Cas9 plasmid) (Addgene #42234), the cloned gRNA plasmid, and the linearized plasmid containing the HR donor with Lipofectamine3000 (ThermoFisher) following the manufacturer’s protocol. Single GFP + cells were manually diluted to 96-well plates following fluorescence activated cell sorting 48 h post-transfection. Clones were expanded and screened by PCR and sequencing to confirm integration of the template into CFTR intron 1. gBlock sequences and Gibson assembly and screening primers are listed in Table S1.
RNA preparation and reverse transcription quantitative PCR (RT-qPCR)
Total RNA was extracted using TRIzol (Thermo Fisher, 15596018) following the manufacturer’s protocol. cDNA was prepared using Taqman Reverse Transcription Reagents with random hexamers (Thermo Fisher, N8080234) and qPCR performed with Taqman Fast Advanced Master Mix (Thermo Fisher, 4444557) with primers shown in Table S1.
Western blot
Whole cell lysates for CFTR protein measurements were prepared as we described previously [23] and resolved by standard SDS-PAGE protocols. Membranes were probed with antibodies specific for CFTR (Cystic Fibrosis Foundation, CFF-596, lot # TJ20200121100285, 1:2000), β-tubulin (Sigma-Aldrich, T4026, lot # 128M4790V, 1:20,000), and anti-mouse-HRP (Agilent/Dako, P0447, lot # 20051789, 1:8000 or 1:10,000), and proteins were detected using ECL Western Blotting Substrate (Pierce).
Omni assay for transposase accessible chromatin and deep sequencing (OMNI ATAC-seq)
Omni-ATAC-seq was performed on 50,000 cells as described previously [24] with minor modifications [21]. Library size distributions were visualized by TapeStation (Agilent) and quantified using the KAPA Library Quantification Kit (Roche). Libraries were pooled and sequenced on a NextSeq 550 at high output (Illumina) using 75 bp single reads. Raw files were trimmed using Sickle [25] Available at https://github.com/najoshi/sickle) and aligned to the hg19 or modified hg19 genomes using the BWA aligner [26]. Aligned reads were marked for duplicates using Picard and coverage tracks were generated using deepTools bamCoverage [27].
4C-seq
4C-seq libraries were generated as described previously [28]. Crosslinked chromatin from 1 × 107 cells was digested using NlaIII and DpnII or Csp6I as the primary or secondary restriction enzymes, respectively. 4C experiments were performed at least twice from separate passages of each cell line. Viewpoint primer sequences and enzyme pairs are shown in Table S1. Quantification of 4C-seq reads were generated using the pipe4C pipeline v1.1 [28] with default parameters. Read density tracks were subtracted using the deepTools bigwigCompare tools [27].
Ussing chamber analysis
1× 105 cells were seeded, in triplicate, on permeable supports (Corning 3801) coated with type I collagen (1:60 dilution; Purecol 5005-B). Cells were grown in DMEM + 10% FBS in both upper and lower chambers for 7 days. Culture inserts were transferred to Ussing chambers for electrophysiological measurement. Short circuit current and transepithelial resistance measurements were taken as described previously [29]. At the indicated times amiloride (100 μM, apical), forskolin (10 μM, basolateral), and CFTR inhibitor Inh-172 (10 μM, apical) were added.
Estimation of copy number and CFTR splicing analyses
−35 kb copy number was estimated using multiplex PCR with primers listed in Table S1 and Taq polymerase (New England Biolabs, M0273). CFTR splicing analysis was performed with primers listed in Table S1 using cDNA and Taq polymerase (New England Biolabs, M0273).
Chromatin immunoprecipitation – quantitative PCR (ChIP-qPCR)
ChIP was performed using standard methods as previously described [30]. Immunoprecipitations were performed with antibodies against H3K27ac (Millipore-Sigma, 07–360, lot # 36988620) and RNA Polymerase II RPB1 (Abcam, ab76123, lot # GR3269158–2), or rabbit IgG (Abcam, 12–370, lot # 3533045). Enrichments were analyzed as percent input using PowerSYBR™ Green PCR Master Mix (Thermo Fisher Scientific) with primers listed in Table S1.
Statistics
For RT-qPCR and ChIP-qPCR, error bars in graphs represent standard deviation (S.D.), with each biological replicate data point representing the average of two technical replicates. Statistical analyses were performed in GraphPad Prism 9 and used an unpaired t-test for RT-qPCR and the Holm-Sidak multiple t test for ChIP-qPCR. Replicate numbers and P-values are noted in figure legends.
RESULTS
The −35 kb CFTR enhancer is functional when relocated to an intronic site in the gene
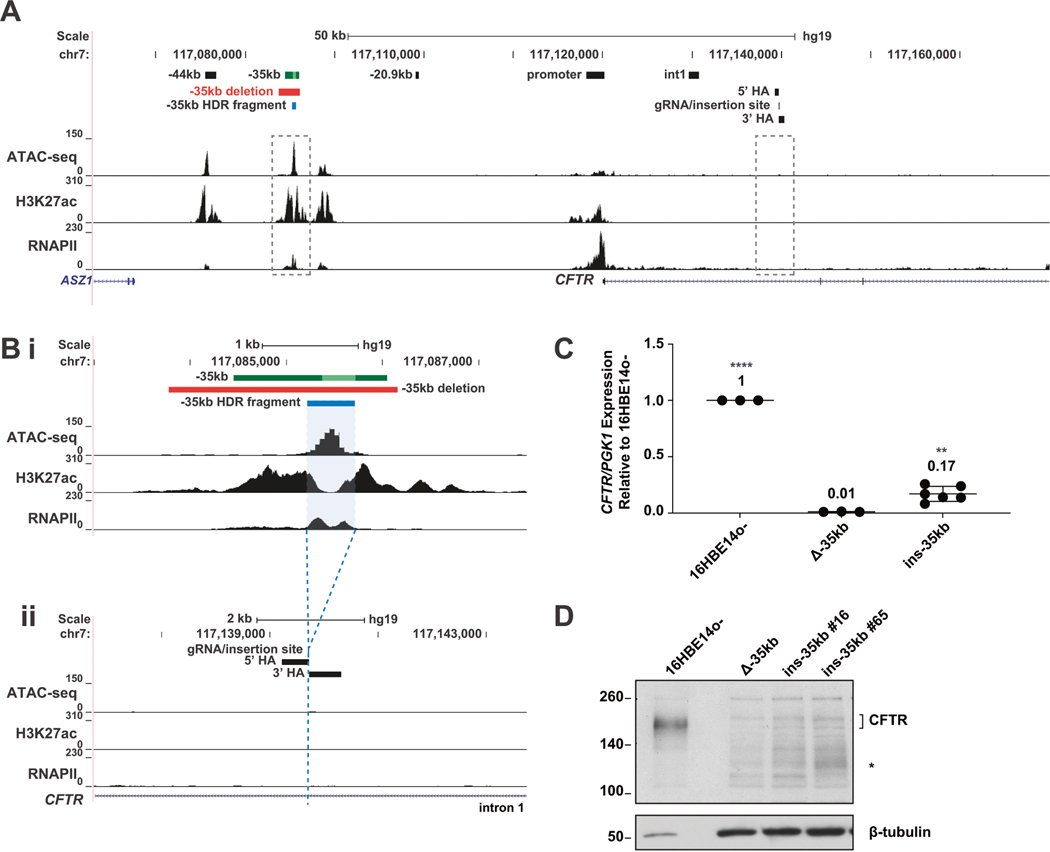
Deletion of the −35 kb airway-selective enhancer in 16HBE14o− airway epithelial cells (subsequently referred to as 16HBE14o− Δ−35 kb cells) resulted in loss of CFTR expression, reorganization of the CFTR locus, and a reduction of RNA polymerase II (RNAPII) recrui™ent [21]. Using a CRISPR-mediated homology-directed repair (HDR) strategy, we inserted 450 bp of the −35 kb sequence (encompassing the 350 bp enhancer core [19]), into intron 1 of CFTR in 16HBE14o− Δ−35 kb cells (hg19 chr7:117139728–117139729). The location in intron 1 was chosen for the following reasons: (1) it was devoid of H3K27ac and RNAPII enrichment [21], (2) no open chromatin is observed at this site in 16HBE14o− cells [21], and (3) it is 10 kb downstream of a known intestine-selective enhancer sequence [31–34] (Fig. 1A, B). Three clones containing the −35 kb insertion on both CFTR alleles, named 16HBE14o− ins-35kb were obtained (Fig. S1). Two clones (clones #6 and #16) each contained 2 copies of the insertion, while one clone (clone #26) contained multiple tandem copies and so was excluded from the further analyses, unless explicitly stated (Fig. S2A). The insertion of the −35 kb non-coding sequence into intron 1 did not disrupt normal splicing of the CFTR transcript (Fig. S2B, C). CFTR expression in the clones was 17% that of 16HBE14o−, and a significant increase over 16HBE14o− Δ−35kb cells (14%) (Fig. 1C).
Fig. 1. Relocation of the −35 kb enhancer partially restores CFTR expression in 16HBE14o− cells.
A The genomic coordinates of the 5′ end of the CFTR locus are shown on hg19 at the top and below are open chromatin mapping (ATAC-seq), H3K27ac and RNAPII enrichment (ChIP-seq) in 16HBE14o− cells (ChIP-seq data from [21]). Magnified regions shown in panel (B) are boxed with dotted line. B Enlarged region of panel (A) detailing the −35 kb region (subpanel i), the −35 kb enhancer (dark green bar with 350 bp core enhancer [19] shown in lime green), the −35 kb enhancer deletion (red bar), and −35 kb fragment (blue bar) subsequently relocated to intron 1, with homology arms (HA) noted (subpanel ii). C CFTR mRNA expression normalized to PGK1, shown relative to 16HBE14o− WT cells (n = 3). Data for 2 Δ−35 kb ins i1 clones are pooled. **** denotes P < .0001 and ** denotes P < 0.01 compared to 16HBE14o− Δ−35kb cells using an unpaired t-test. D CFTR protein expression by western blot analysis, normalized to β-tubulin.
Furthermore, partial restoration of CFTR protein expression was seen by western blot in the three clones containing the −35 kb enhancer sequence inserted in intron 1, in comparison to 16HBE14o− Δ−35 kb cells. However, the protein levels were much lower than in WT 16HBE14o− cells (Figs. 1D and S1D). In addition to some restoration of the mature full length CFTR protein in the insertion clones, we also noted 2 smaller forms of CFTR (marked by * in Figs. 1D and S1D), which likely correspond to alternative forms described previously [35, 36].
To determine whether the CFTR protein produced in the insertion clones restored CFTR channel activity we performed Ussing chamber measurements on epithelial monolayers. Consistent with the protein expression data, partial restoration of CFTR channel activity was seen in the insertion clones, as measured by their response to forskolin and CFTR(inh)-172, (Fig. S1E) in comparison to 16HBE14o− WT and Δ−35 kb cells. These data indicate that relocation of the −35 kb enhancer to an ectopic site in the CFTR locus is able to partially restore CFTR expression and function when compared to WT cells.
Changes to chromatin structure and 3D organization of the CFTR locus upon relocation of the −35 kb enhancer
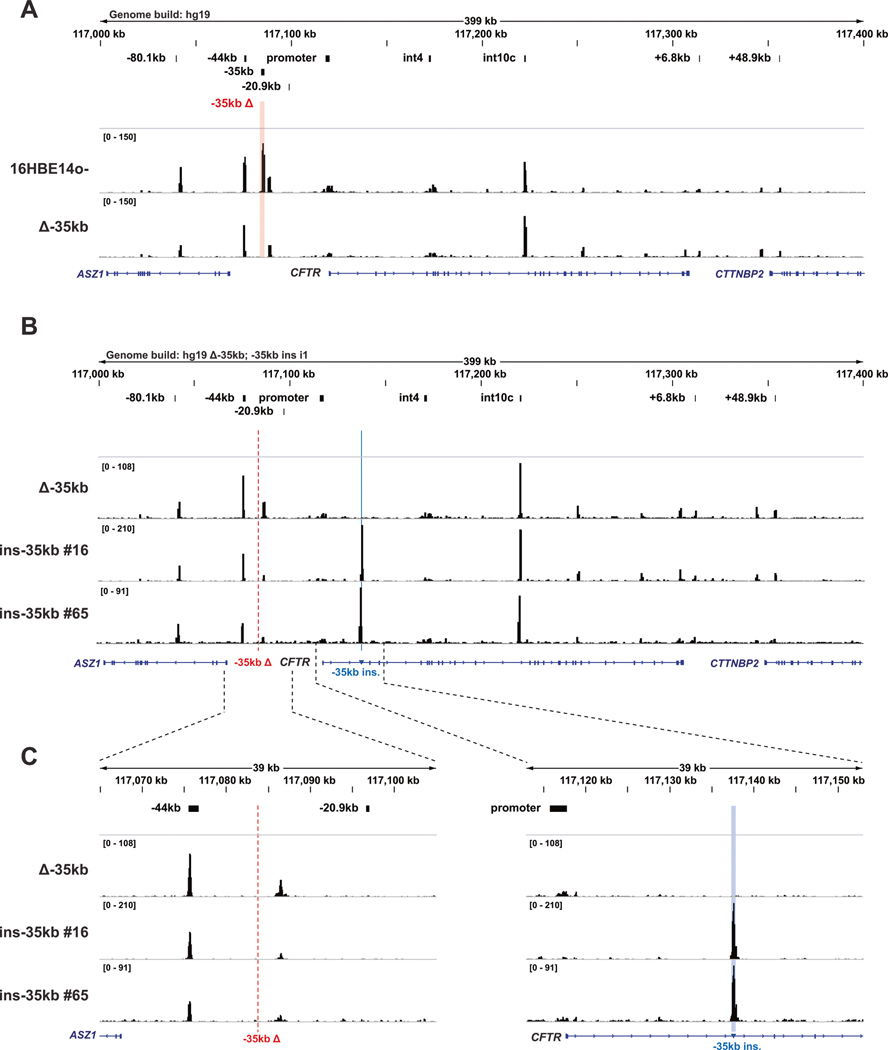
To evaluate whether chromatin surrounding the ectopic −35 kb sequence remains open outside of its normal environment, assay for transposase accessible chromatin with high throughput sequencing (ATAC-seq) was performed on modified and parental cells. In 16HBE14o− Δ−35 kb cells, no gross changes in open chromatin across the CFTR locus are observed compared to WT parental cells, except for the loss of open chromatin flanking the deleted −35 kb region (Fig. 2A). However, upon relocation of the −35 kb sequence, a striking gain in open chromatin is seen at the insertion site in 16HBE14o− ins-35 kb cells, which shows no open chromatin in unmodified 16HBE14o− and 16HBE14o− Δ−35kb cells (Fig. 2B, C). With the exception of this gain of de novo open chromatin at the insertion site, little overall change to the open chromatin profile of 16HBE14o− cells is observed across the rest of the CFTR locus in 16HBE14o− ins-35kb cells (Fig. 2B, C).
Fig. 2. Relocation of the CFTR −35kb enhancer element creates open chromatin at the insertion site in intron 1.
A Open chromatin mapping of 16HBE14o− WT and 16HBE14o− Δ−35kb cells across the CFTR locus, mapped to the hg19 genome. B Open chromatin mapping of 16HBE14o− Δ−35 kb and ins-35 kb clones, mapped to a modified hg19 genome lacking the sequence removed for the −35 kb deletion (red dotted line) and incorporating the sequence inserted into intron 1 (blue line). C Enlarged regions of panel (B) showing the genomic region surrounding the −35 kb deletion site (left) and −35 kb insertion site (right).
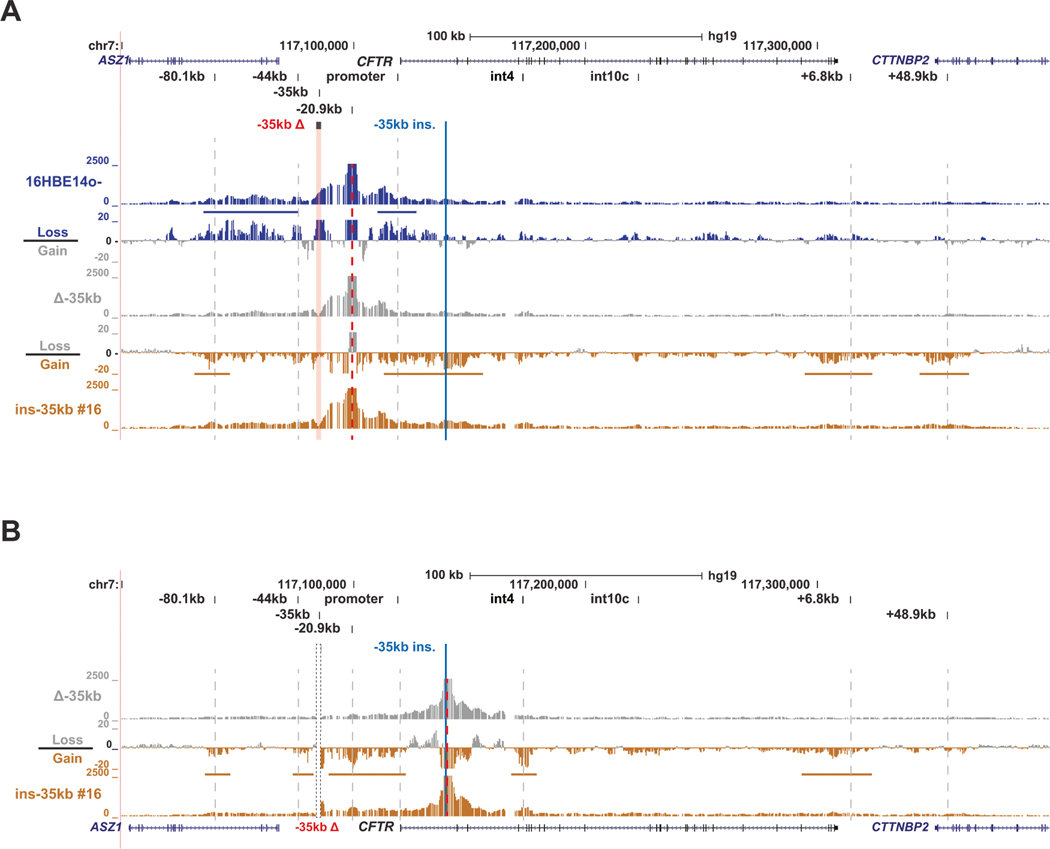
We next investigated whether the relocated −35 kb core sequence is able to physically interact with the CFTR promoter and CREs that normally interact with it at its endogenous location in WT 16HBE14o− cells. To achieve this, we performed 4C-seq on WT 16HBE14o−, 16HBE14o− Δ−35 kb, and 16HBE14o− ins-35 kb cells. We previously described a set of 4C-seq viewpoints that enable comprehensive analysis of structural features of the CFTR locus [37]. First, a viewpoint at −20.9 kb was used, a site that corresponds to a CTCF-bound, enhancer blocking insulator element [38] that plays an important role in the structural organization of the CFTR locus. By subtracting the interaction profile of the 16HBE14o− Δ−35 kb cells from 16HBE14o− WT cells, a loss of interactions between the −20.9 kb and the CFTR promoter is observed, as well as a general loss of interactions of −20.9 kb with upstream CREs including the region surrounding the −35 kb deletion, the −44 kb enhancer, and the 5′ TAD boundary at −80.1 kb (Fig. 3A).
Fig. 3. The −35kb enhancer influences 3D structure of the CFTR locus independent of its location.
A 4C-seq quantification data shown for 16HBE14o– WT (blue), 16HBE14o– Δ−35 kb (grey) and 16HBE14o– ins-35 kb clone #16 (gold) cells with the −20.9 kb CRE as the viewpoint (red dotted line). At the top the hg19 genomic coordinates are shown. Key CFTR CREs are shown above the read quantification tracks of the 4C-seq interaction profiles. Between these tracks for each cell type are the corresponding subtraction tracks for each pair, in log2 scale, with colors indicating the cells from which losses or gains in interactions were measured. Horizontal colored bars denote key gains or losses discussed in the results. B 4C-seq data shown for 16HBE14o− Δ−35 kb (grey) and 16HBE14o− ins-35 kb clone #16 (gold) cells with a viewpoint adjacent to the intron 1 insertion site (red dotted line). Read quantification and subtraction tracks shown as in panel (A).
Restoration of the −35 kb sequence to intron 1 (16HBE14o− ins-35 kb), results in novel interactions between −20.9 kb and the area surrounding the −35 kb insertion in intron 1 (Fig. 3A). As the 4C-seq sequence files were aligned to the WT hg19 genome, the gain in interactions between the −20.9 kb viewpoint and −35 kb in the double modified cells (16HBE14o− ins-35 kb) is indicative of −20.9 kb interacting with −35 kb physically located in intron 1. Additionally, increased interactions were observed between −20.9 kb and the 5′ TAD boundary and increased interactions with downstream elements such as the +6.8 kb insulator and the +48.9 kb 3′ TAD boundary in 16HBE14o− ins-35 kb cells as compared to 16HBE14o− Δ−35kb cells (Fig. 3A).
To investigate whether the relocated −35 kb sequence is able to establish de novo interactions with other important CFTR CREs, we generated a novel 4C-seq viewpoint located immediately adjacent to the −35 kb insertion site in intron 1. This viewpoint shows a gain in interactions in the 16HBE14o− ins-35 kb cells compared to 16HBE14o− Δ−35 kb cells between intron 1 and the promoter region, and also key CFTR CREs, such as the 5′ and 3′ TAD boundaries, the −44 kb enhancer, downstream CREs (Fig. 3B). A gain in interactions was also observed between intron1 anda site in intron 4 (Fig. 3B), which is marked by H3K4me1 during development [39] and sometimes interacts with intronic CREs, though its function is unknown [34, 37]. Together, these data indicate that the −35 kb enhancer initiates open chromatin in a non-endogenous location within the CFTR locus and can establish de novo chromatin interactions with the gene promoter and other CFTR CREs in this genomic context.
Gain of an active enhancer signature upon relocation of the −35kb element to intron 1 of CFTR
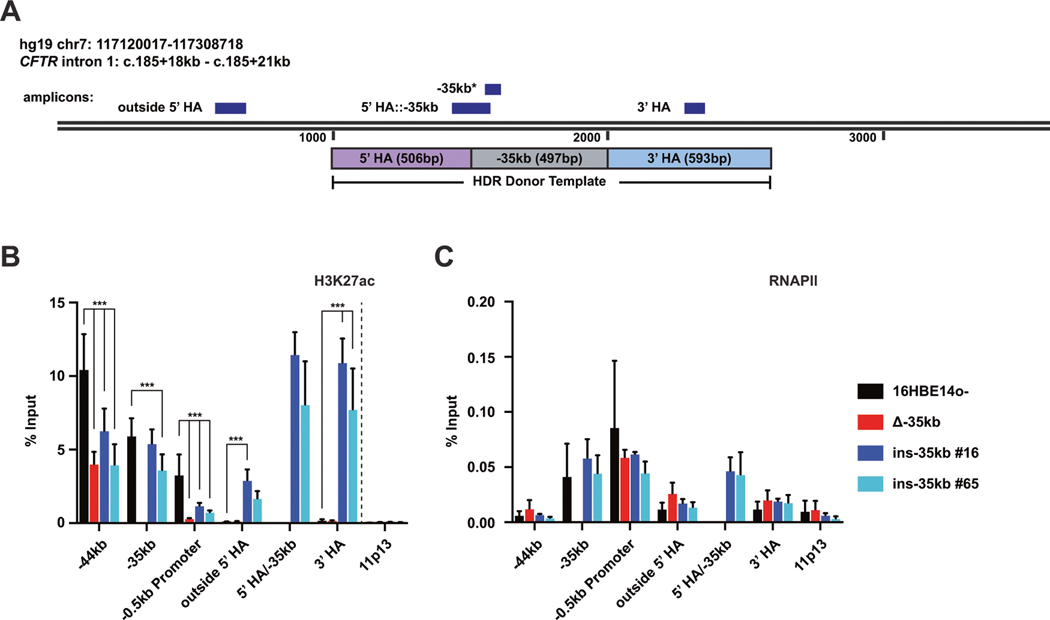
To further assess changes to local CFTR chromatin following relocation of the −35 kb enhancer to intron 1 of CFTR, we performed chromatin immunoprecipitation (ChIP) -qPCR for the active enhancer mark H3K27ac and also RNA polymerase II (RNAPII) in 16HBE14o− ins-35kb cells. In WT 16HBE14o− cells, the −35 kb element shows strong enrichment of H3K27ac, as well as modest occupancy of RNAPII, while the insertion site in intron 1 lacks this chromatin signature (Fig. 1 [21]). Here we used a panel of PCR primer sets at the −44 kb and −35 kb enhancers, the CFTR promoter, and at multiple sites in intron 1 to investigate changes in active chromatin marks upon modification of the CFTR locus (Fig. 4A). Of note, the intron 1::−35 kb primer set uses a forward primer in intron 1 and a reverse primer in −35 kb, and therefore only amplifies a product in 16HBE14o− ins-35 kb cells. Upon relocation of −35 kb to intron 1, a significant gain in H3K27ac enrichment is seen with primers surrounding the insertion site (Fig. 4B). Similar levels of H3K27ac enrichment is seen at the intron1::−35 kb junction as well at the site of the 3′ HA. No changes of H3K27ac are seen at the chr11p13 negative control site. Levels of H3K27ac enrichment at the −44 kb enhancer and the CFTR promoter (Fig. 3B) are directly correlated with CFTR expression in these cells (Fig. 1C). RNAPII occupancy across intron 1 does not significantly change upon the −35 kb relocation at the sites assayed, however RNAPII occupies the −35 kb sequence to the same extent independent of its location in the CFTR locus (Fig. 3C). Together these data suggest that −35 kb can establish an enhancer chromatin signature irrespective of its location within the CFTR locus.
Fig. 4. The −35kb enhancer establishes an enhancer chromatin signature independent of genomic location.
A Schematic showing the location of qPCR amplicons across the intron 1 insertion regions for ChIP-qPCR experiments. Note that the −35 kb primer amplicon will detect −35 kb sequence independent of location. H3K27ac (B) and RNAPII (C) enrichment in 16HBE14o−, 16HBE14o− Δ−35 kb, and 16HBE14o− ins-35 kb clones #16 and #65 cells. Data normalized to percent input, with the chr11p13 region shown as negative control. (B), n = 3; (C), n = 2; *** denotes P < 0.001 compared to 16HBE14o− enrichment levels at each site using the Holm-Sidak multiple t test (not significant not shown).
DISCUSSION
To date, few studies have addressed the characteristics of enhancer elements that are moved to an ectopic location within the same locus [8, 9]. Our goal was to determine whether a key airway cell enhancer for the CFTR gene was functional when moved to an ectopic site in the endogenous gene, and hence could have utility in therapeutic gene editing protocols. The −35 kb airway-selective enhancer was inserted into intron 1 of CFTR in a 16HBE14o− cell line lacking this element in its endogenous location, (as described previously [21]). While −35 kb is able to establish de novo open chromatin in intron 1 (Fig. 2), generate chromatin contacts with the gene promoter and other known −35kb interacting CREs (Fig. 3, and model in Fig. 5) and leads to H3K27ac enrichment at the integration site (Fig. 4), it is not able to fully restore CFTR mRNA expression, CFTR protein or channel activity in these cells (Figs. 1 and S1). This suggests that while the −35 kb CRE is able to maintain enhancer activity independent of location, other sequence dependent factors also likely play a role in regulating CFTR expression in these cells. Similar results were obtained in experiments on the endogenous mouse Nanog locus in vivo [8]. The −5 kb Nanog enhancer was relocated to a site downstream of the Nanog 3′ end and only partial recapitulation of the −5 kb Nanog enhancer activity was thought to result from the limited length of the CRE sequence used. The fragment of the −35 kb CRE that was relocated into intron 1 in our study contains the mapped core of enhancer activity as measured by luciferase assays [19]. Of note, larger fragments containing the complete −35 kb CRE had lower enhancer activities than the core element, in those experiments. The core element we have used here fully encompasses the peak of open chromatin at this site, though it does not fully cover the flanking peaks of H3K27ac enrichment at the endogenous site (Fig. 1B). In this case it is possible that the partial rather than complete restoration of enhancer activity at the new site may also result from inability of the small relocated CRE sequence to completely reconfigure surrounding chromatin that was previously inactive. Also, it may be relevant that only in 16HBE14o− cells, an additional small peak of open chromatin is seen at −33 kb, (adjacent to the −35 kb site), which is also modestly enriched for H3K27ac and RNAPII (Fig. 1A). The function of this sequence is unknown, and though the chromatin remains open at this site in 16HBE14o− Δ−35 kb and ins-35 kb cells (Fig. 2), it may be functionally associated with full activity of the −35 kb CRE and dependent on their close proximity in WT cells. In addition, the presence of H3K27ac enrichment at the −44 kb enhancer may be of mechanistic relevance. Deletion of the −35 kb enhancer from the 16HBE14o- cell line is also accompanied by a reduction in H3K27ac occupancy at the −44 kb site, and the levels remain low upon relocation of the −35 kb enhancer to intron 1 (Fig. 4B). As noted earlier, deletion of the −44 kb enhancer from 16HBE14o-cells also resulted in reduced CFTR expression and alteration of the 3D organization of the locus [21]. Hence, the fact that the −35 kb enhancer does not fully restore CFTR expression in the 16HBE14o-ins-35kb cells may result from the disruption of the functional relationship of these 2 enhancers when they are in close proximity.
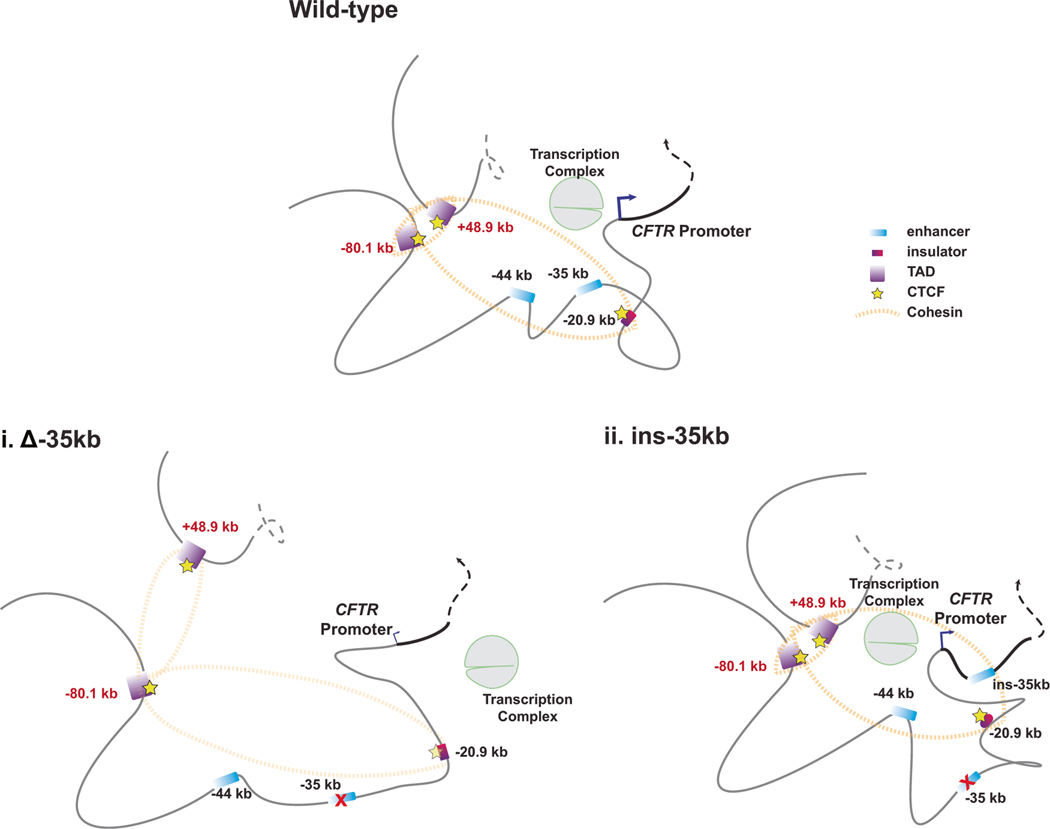
Fig. 5. Model to summarize the impact on CFTR locus 3D structure of relocating the −35kb enhancer to a site in intron 1.
In airway epithelial cells removal of the −35kb enhancer (i) is associated with a substantial loss of CFTR expression and disruption of interactions between TAD boundaries, another enhancer at −44 kb, an insulator element at −20.9 kb, and the CFTR promoter. Relocation of the −35 kb enhancer to intron 1 of CFTR (ii), partially restores CFTR expression, and is associated with de novo interactions with the relocated −35 kb enhancer, the CFTR promoter, and other CREs.
We obtained one cell clone (16HBE14o− ins-35kb #26) that contained more than one copy of −35 kb inserted into intron 1 of one CFTR allele (Fig. S2B) and had higher CFTR expression than WT 16HBE14o− cells (Fig. S1C). This clone also contained more CFTR protein and showed slightly elevated CFTR channel activity compared to the two single copy insertion clones (Fig. S1D, E). This suggests that amplified copy numbers of the −35 kb CRE could coordinate higher expression levels, consistent with the amplification of super-enhancer sequences causing the over-expression of cancer-related genes [40]. Notably, 16HBE14o− ins-35kb #26 cells also showed much more open chromatin at the insertion site and also increased H3K27ac enrichment at the CFTR promoter compared to the other clones generated in this study (Fig. S3).
It is of interest that both clones 16HBE14o− ins-35kb #26 and #65 showed faster migrating forms of CFTR on western blots (marked by *s in Fig. 1D and S1D), in addition to low levels of mature CFTR. Based upon the predicted molecular weights of these forms they likely correspond to alternative functional forms of CFTR that initiate translation at ATG residues in exon 4 of CFTR [35, 36].
It is promising that the −35 kb CRE sequence is able to establish de novo chromatin contacts with the CFTR promoter and other CREs at the locus, notably the TAD boundaries, and the −44 kb enhancer, when moved to an intronic location in the gene. Of note, we also observed a strong interaction between the relocated −35 kb CRE sequence and the region surrounding introns 4–6 of the CFTR locus. In WT 16HBE14o− cells, a site in intron 4 (hg19 chr7: 117172267–117173466) is weakly open as measured by ATAC-seq (Fig. 2A), however, it is not enriched for the active histone mark H3K27ac [21], and therefore not predicted to have enhancer activity. A site in CFTR intron 6 was shown to be the location of a mono-allelic integration of SV40, which was used for the initial immortalization (SV40ori-) of the 16HBE14o− cell line [22, 41]. This intron 6 site also displays a low level of open chromatin in these cells and robust RNAPII occupancy, but lacks H3K27ac enrichment [21]. It has not been possible to generate a good 4C-seq viewpoint for the −35 kb CRE due to local sequence limitations, so we cannot exclude any interactions between this site and introns 4–6 in WT cells. However, as the −20.9 kb insulator is known to interact with the intron 4 region in multiple cell types [34, 37], this region likely plays a role in organizing the 3D looping of the CFTR locus. The work described here has important implications for the future development of CF gene editing therapeutics, especially those designed to correct multiple CFTR mutations through genomic integration of a partial CFTR cDNA into the endogenous locus.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Shih-Hsing Leir and Tom Kelley for helpful discussion and the Epithelial Cell Core at the CWRU CF Center (Cystic Fibrosis Foundation RDP R447-CR11) for Ussing chamber measurements. Also, Dr. Vian Peshdary for reagent generation, and the CWRU School of Medicine Genomics Core for sequencing.
FUNDING INFORMATION
This work was supported by the Cystic Fibrosis Foundation (Davis19XX0) and NIH HL094585 and HD068901 (both to AH).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
ADDITIONAL INFORMATION
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41434–022-00378–7.
DATA AVAILABILITY
Data are deposited at NCBI GEO accession GSE203560.
REFERENCES
- 1.Field A, Adelman K. Evaluating enhancer function and transcription. Annu Rev Biochem. 2020;89:213–34. [DOI] [PubMed] [Google Scholar]
- 2.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation center. Nature. 2012;485:381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang P, Keller CA, Giardine B, Grevet JD, Davies JOJ, Hughes JR, et al. Comparative analysis of three-dimensional chromosomal architecture identifies a novel fetal hemoglobin regulatory element. Genes Dev. 2017;31:1704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lettice LA, Daniels S, Sweeney E, Venkataraman S, Devenney PS, Gautier P, et al. Enhancer-adoption as a mechanism of human developmental disease. Hum Mutat. 2011;32:1492–9. [DOI] [PubMed] [Google Scholar]
- 7.Northcott PA, Lee C, Zichner T, Stütz AM, Erkek S, Kawauchi D, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agrawal P, Blinka S, Pulakanti K, Reimer MH, Stelloh C, Meyer AE, et al. Genome editing demonstrates that the −5 kb Nanog enhancer regulates Nanog expression by modulating RNAPII initiation and/or recrui™ent. J Biol Chem. 2021;296:100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolt CC, Lopez-Delisle L, Hintermann A, Mascrez B, Rauseo A, Andrey G, et al. Context-dependent enhancer function revealed by targeted inter-TAD relocation. bioRxiv. 2022;13:3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerschner JL, Paranjapye A, NandyMazumdar M, Yin S, Leir SH, Harris A. OTX2 regulates CFTR expression during endoderm differentiation and occupies 3’ cis-regulatory elements. Dev Dyn. 2021;250:684–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosalia N, Harris A. Chromatin dynamics in the regulation of CFTR expression. Genes (Basel). 2015;6:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ensinck M, Mottais A, Detry C, Leal T, Carlon MS. On the corner of models and cure: gene editing in cystic fibrosis. Front Pharmacol. 2021;12:677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia E, Duan R, Shi F, Seigel KE, Grasemann H, Hu J. Overcoming the undesirable CRISPR-Cas9 expression in gene correction. Mol Ther Nucleic Acids. 2018;13:699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou ZP, Yang LL, Cao H, Chen ZR, Zhang Y, Wen XY, et al. In vitro validation of a CRISPR-mediated CFTR correction strategy for preclinical translation in pigs. Hum Gene Ther. 2019;30:1101–16. [DOI] [PubMed] [Google Scholar]
- 15.Vaidyanathan S, Baik R, Chen L, Bravo DT, Suarez CJ, Abazari SM, et al. Targeted replacement of full-length CFTR in human airway stem cells by CRISPR-Cas9 for pan-mutation correction in the endogenous locus. Mol Ther. 2022;30:223–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki S, Crane AM, Anirudhan V, Barillà C, Matthias N, Randell SH, et al. Highly efficient gene editing of cystic fibrosis patient-derived airway basal cells results in functional CFTR correction. Mol Ther. 2020;28:1684–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bednarski C, Tomczak K, vom Hövel B, Weber WM, Cathomen T. Targeted integration of a super-exon into the CFTR locus leads to functional correction of a cystic fibrosis cell line model. PLoS One. 2016;11:e0161072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Leir SH, Harris A. Oxidative stress regulates CFTR gene expression in human airway epithelial cells through a distal antioxidant response element. Am J Respir Cell Mol Biol. 2015;52:387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Leir SH, Harris A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol Cell Biol. 2013;33:2843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Ott CJ, Lewandowska MA, Leir SH, Harris A. Molecular mechanisms controlling CFTR gene expression in the airway. J Cell Mol Med. 2012;16:1321–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.NandyMazumdar M, Yin S, Paranjapye A, Kerschner JL, Swahn H, Ge A, et al. Looping of upstream cis-regulatory elements is required for CFTR expression in human airway epithelial cells. Nucleic Acids Res. 2020;48:3513–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cozens AL, Yezzi MJ, Kunzelmann K, Ohrui T, Chin L, Eng K, et al. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am J Respir Cell Mol Biol. 1994;10:28–47. [DOI] [PubMed] [Google Scholar]
- 23.Cai Z, Palmai-Pallag T, Khuituan P, Mutolo MJ, Boinot C, Liu B, et al. Impact of the F508del mutation on ovine CFTR, a Cl− channel with enhanced conductance and ATP-dependent gating. J Physiol. 2015;593:2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods. 2017;14:959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi N, Fass JN. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33). 2011. https://github.com/najoshi/sickle. [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, et al. deep-Tools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krijger PHL, Geeven G, Bianchi V, Hilvering CRE, de Laat W. 4C-seq from beginning to end: a detailed protocol for sample preparation and data analysis. Methods. 2020;170:17–32. [DOI] [PubMed] [Google Scholar]
- 29.Fan Z, Perisse IV, Cotton CU, Regouski M, Meng Q, Domb C, et al. A sheep model of cystic fibrosis generated by CRISPR/Cas9 disruption of the CFTR gene. JCI Insight. 2018;3:e123529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, et al. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci USA. 2009;106:19934–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith AN, Barth ML, McDowell TL, Moulin DS, Nuthall HN, Hollingsworth MA, et al. A regulatory element in intron 1 of the cystic fibrosis transmembrane conductance regulator gene. J Biol Chem. 1996;271:9947–54. [DOI] [PubMed] [Google Scholar]
- 32.Rowntree RK, Vassaux G, McDowell TL, Howe S, McGuigan A, Phylactides M, et al. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum Mol Genet. 2001;10:1455–64. [DOI] [PubMed] [Google Scholar]
- 33.Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med. 2009;13:680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin S, NandyMazumdar M, Paranjapye A, Harris A. Cross-talk between enhancers, structural elements and activating transcription factors maintains the 3D architecture and expression of the CFTR gene. Genomics. 2022;114:110350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewandowska MA, Costa FF, Bischof JM, Williams SH, Soares MB, Harris A. Multiple mechanisms influence regulation of the cystic fibrosis transmembrane conductance regulator gene promoter. Am J Respir Cell Mol Biol. 2010;43:334–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramalho AS, Lewandowska MA, Farinha CM, Mendes F, Gonçalves J, Barreto C, et al. Deletion of CFTR translation start site reveals functional isoforms of the protein in CF patients. Cell Physiol Biochem. 2009;24:335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang R, Kerschner JL, Gosalia N, Neems D, Gorsic LK, Safi A, et al. Differential contribution of cis-regulatory elements to higher order chromatin structure and expression of the CFTR locus. Nucleic Acids Res. 2016;44:3082–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blackledge NP, Carter EJ, Evans JR, Lawson V, Rowntree RK, Harris A. CTCF mediates insulator function at the CFTR locus. Biochem J. 2007;408:267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang A, Yue F, Li Y, Xie R, Harper T, Patel NA, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell. 2015;16:386–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Choi PS, Francis JM, Imielinski M, Watanabe H, Cherniack AD, et al. Identification of focally amplified lineage-specific super-enhancers in human epithelial cancers. Nat Genetics. 2016;48:176–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valley HC, Bukis KM, Bell A, Cheng Y, Wong E, Jordan NJ, et al. Isogenic cell models of cystic fibrosis-causing variants in natively expressing pulmonary epithelial cells. J Cystic Fibrosis. 2019;18:476–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are deposited at NCBI GEO accession GSE203560.