Abstract
The fusion activity of chimeras of influenza virus hemagglutinin (HA) (from A/fpv/Rostock/34; subtype H7) with the transmembrane domain (TM) and/or cytoplasmic tail (CT) either from the nonviral, nonfusogenic T-cell surface protein CD4 or from the fusogenic Sendai virus F-protein was studied. Wild-type or chimeric HA was expressed in CV-1 cells by the transient T7-RNA-polymerase vaccinia virus expression system. Subsequently, the fusion activity of the expression products was monitored with red blood cells or ghosts as target cells. To assess the different steps of fusion, target cells were labeled with the fluorescent membrane label octadecyl rhodamine B-chloride (R18) (membrane fusion) and with the cytoplasmic fluorophores calcein (molecular weight [MW], 623; formation of small aqueous fusion pore) and tetramethylrhodamine-dextran (MW, 10,000; enlargement of fusion pore). All chimeric HA/F-proteins, as well as the chimera with the TM of CD4 and the CT of HA, were able to mediate the different steps of fusion very similarly to wild-type HA. Quite differently, chimeric proteins with the CT of CD4 were strongly impaired in mediating pore enlargement. However, membrane fusion and formation of small pores were similar to those of wild-type HA, indicating that the conformational change of the ectodomain and earlier fusion steps were not inhibited. Various properties of the CT which may affect pore enlargement are considered. We surmise that the hydrophobicity of the sequence adjacent to the transmembrane domain is important for pore dilation.
The fusion of influenza viruses with target membranes is mediated by the spike membrane glycoprotein, HA. In the viral membrane, HA is organized as a homotrimer, each monomer consisting of two disulfide-linked subunits, HA1 and HA2 (14, 38). The HA-mediated fusion is triggered by acidic pH, which causes the HA ectodomain to be converted to a fusogenic conformation, thereby exposing the hydrophobic N terminus of the HA2 subunit (37), the so-called fusion sequence. X-ray crystallographic studies of a fragment of the ectodomain suggest that the loop of the HA2 subunit connecting two α-helices in the neutral form (39) becomes a part of an extended trimeric coiled coil at low pH (5, 6, 30, 36). Recent reconstruction of the three-dimensional structure of the complete HA ectodomain at low pH by cryoelectron microscopy revealed that the preserved trimeric shape of the ectodomain may serve to guide the orientation of the coiled coil, thereby moving the fusion sequence to the tip of HA toward the target membrane (3). Destabilization of the target membrane by this sequence is then believed to initiate membrane fusion (11, 19).
Expression of HA in the plasma membrane of various cell lines preserves its fusion activity. In studies with these systems, it was revealed that the HA ectodomain and its low-pH-triggered conformational change is sufficient to induce membrane mixing (17, 21–24). Indeed, the HA ectodomain membrane anchored by GPI mediates the merger between the outer layers of the contacting membranes, resulting in a metastable hemifusion intermediate (17, 20, 21, 24). These studies concluded that for full fusion, i.e., merger of the inner leaflet and efficient formation as well as enlargement of pores, TM is required. Recently, it was reported that the fusion sequence of HA2 plays a role in later fusion steps as well (31).
Previous studies suggested that the specific sequence of the TM of HA-wt is not a requirement for pore formation. Parallel replacement of both the TM and the CT of the HA-wt by related domains of another enveloped virus fusion protein does not inhibit fusion activity (10, 32). However, it was left open whether both domains form a functional entity. Recently, Melikyan et al. (23) addressed this concern by constructing HA chimeras containing the TM and CT of a protein unrelated to fusion, the pIgR. They showed significant inhibition of fusion when only one domain was replaced. Simultaneous replacement did not affect fusion, suggesting that the TM and the CT are not functionally independent. On the other hand, we found that replacement of only one domain of the HA by the corresponding domain from the fusion protein of Sendai virus, the F-protein, did not affect membrane fusion or the formation of small aqueous fusion pores (34). Although this would argue against a functional entity of the two domains, we could not rule out the possibility that the substitution of those domains of HA by related domains from another viral fusion protein preserves the entity. Therefore, in the present work we studied the fusion activity of chimeric HAs containing in a combinatorial fashion the TM and CT of a totally unrelated nonviral and nonfusogenic protein, the T-cell surface glycoprotein CD4.
We expressed HA-wt (subtype H7) and the various chimeric protein constructs in monkey kidney cells (CV-1) using the transient T7-RNA-polymerase vaccinia virus expression system (2, 13, 29, 34) and measured the low-pH-induced fusion of those cells with RBCs and RBC ghosts, respectively, as targets. We further assessed the ability of chimeric HA with the TM and/or CT from Sendai virus F protein (34) to mediate pore widening. For all chimeras tested, we found membrane mixing (redistribution of the lipid analogue R18) and formation of small aqueous fusion pores (redistribution of the low-MW cytoplasmic label calcein) after fusion was triggered; however, subsequent dilation of the pore, allowing the flow of the higher-MW fluorophore TMR-D into the cytoplasm of the transfected cells, was impaired for the chimeras carrying the cytoplasmic domain of CD4.
MATERIALS AND METHODS
Abbreviations used in this paper.
aa, amino acids; ANOVA, analysis of variance; CT, cytoplasmic tail; DMEM, Dulbecco's modified Eagle's medium; DMEM+, DMEM supplemented with 5% FCS; DMEM−, DMEM without FCS; FACS, fluorescence-activated cell sorter; FITC, fluorescein isothiocyanate; FCS, heat-inactivated fetal calf serum; GPI, glycosylphosphatidylinositol; HA, influenza virus hemagglutinin; HA-wt, wild-type HA; MW, molecular weight; PBS, phosphate-buffered saline; PBS++, PBS containing 1 mM CaCl2 and 1 mM MgCl2; pIgR, polyimmunoglobulin receptor; RBC, human red blood cell; RIPA, radioimmunoprecipitation assay; RT, room temperature; R18, octadecyl rhodamine B chloride; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; TM, transmembrane domain; TMR-D, dextran-conjugated tetramethylrhodamine; WGA, wheat germ agglutinin.
Materials.
The fluorophores R18, calcein-AM, calcein, and TMR-D (molecular mass, 10 kDa) were purchased from Molecular Probes (Eugene, Oreg.), and Tran35S-label (>1,000 Ci/mmol) was from ICN Radiochemicals (Irvine, Calif.). Enzymes used in molecular cloning were obtained from New England Biolabs (Schwalbach/Taunus, Germany). DMEM, DMEM without l-methionine and without l-glutamine, l-glutamine, 2YT medium, and Lipofectin were purchased from Life Technologies Inc. (Karlsruhe, Germany). AmpliTaq DNA polymerase was obtained from Perkin-Elmer (Vaterstetten, Germany). Oligonucleotide primers used for sequencing and PCR were synthesized by Life Technologies. FCS and 0.05% trypsin–0.02% EDTA solution PBS were obtained from Biochrom KG (Berlin, Germany), and secondary antibody was obtained from Dakopatt (Hamburg, Germany). Protein A-Sepharose, neuraminidase (type V from Clostridium perfringens), trypsin (type XIII from bovine pancreas, tolylsulfonyl phenylalanyl chloromethyl ketone, and WGA (lectin from Triticum vulgaris) were purchased from Sigma (St. Louis, Mo.), and the DNA sequencing kit was from Stratagene (La Jolla, Calif.). Cell culture dishes were purchased from Nunc (Biochrom KG).
Construction of chimeric genes.
The various HAs expressed are marked by their composition (ectodomain/transmembrane domain/cytoplasmic tail) as H/H/H (HA-wt subtype 7) and H/H/C, H/C/H, H/C/C, H/H/F, H/F/H, and H/F/F (chimeras). H represents the respective domain of HA-wt, C represents the corresponding sequences from CD4, and F represents those of the domain of the C-tail mutant of Sendai virus glycoprotein F in which cysteine residues replace Ser-530 and Gly-534 (29). The ectodomain of all these proteins is that of HA (sequence not shown). The sequences of the TM region for HA-wt, for CD4 and for the F protein are VILWFSFGASCFLLLAIAMGLVFICV (26 aa), ALIVLGGVAGLLLFIGLGIFFCV (23 aa), and TVITIIVVMVVILVVIIVIIIVL (23 aa), respectively, and those of the CT are KNGNMRCTICI-COOH (11 aa), RCRHRRRQAERMSQIKRLLSEKKTCQCPHRFQKTCSPI-COOH (38 aa), and YRLRRCMLMCNPDERIPRDTOTLEPKIRHMOTNGGFDAMAEKR- COOH (43 aa), respectively.
All basic DNA procedures were done as described by Sambrook et al. (33). The gene of HA-wt (from influenza virus A/fpv/rostock/34, subtype H7) was present in a pTM1 vector (pTM1/HA). The gene encoding human CD4 in the vector pECEM was kindly provided by M. Veit (Freie Universität Berlin, Berlin, Germany). The vector pECEM containing CD4 was BamHI digested, and the CD4 gene was introduced into the multiple-cloning site of pTM1 (pTM1/CD4). Based on these two pTM1 plasmids, the chimeras H/C/C, H/C/H, and H/H/C were generated by standard PCR protocols and the overlap extension technique (the thermal cycler, enzyme, buffers, and deoxynucleoside triphosphates were from Perkin-Elmer). The internal sense primers were 5′-TGGCTACAAAGATGCCCTGATTGTGCTGGGGGGC-3′ (H/C/C) and 5′-TTCATATGTGTGAGGTGCCGGCACCGAAGGCGC-3′ (H/H/C); the antisense primers were 5′- GCACAATCAGGGCATCTTTGTAGCCACTACTC-3′ (H/C/C) and 5′-GTGCCGGCACCTCACACATATGAAAACAAGGC-3′ (H/H/C). The external primers were 5′-GCAGGGGATACAAAATGAACACTC-3′ (sense) and 5′-TTAGGCCTCTCGACCTGCAG-3′ (antisense). H/C/C in pTM1 and HA-wt in pTM1 were used to produce H/C/H by the same PCR technique with the internal primers 5′-TTCTTCTGTGTCAAGAACGGAAACATGCGGTGC-3′ (sense) and 5′-GTTTCCGTTCTTGACACAGAAGAAGATGCCTAGC-3′ (antisense) and the previously described external primers. The sequences of the chimeric genes in pTM1 were confirmed by double-stranded DNA sequencing. The chimeras H/F/F, H/F/H, and H/H/F were described previously (34).
Cell culture and vaccinia virus-based expression of foreign genes.
CV-1 cells were grown at 37°C and 5% CO2 in DMEM+. Transient expression of HA-wt as well as the chimeric proteins in CV-1 cells was performed as described previously (13, 34). Subconfluent monolayers of CV-1 cells (about 80% confluent, in 3.5 cm dishes) were washed twice with DMEM−, infected at a multiplicity of infection of 10 PFU per cell by vaccinia virus vTF7-3 (13), which expresses T7-RNA-polymerase, and incubated in DMEM− for 1.5 h at 37°C and 5% CO2. To adjust the surface expression levels, different amounts of plasmid DNA were used for transfection. A 2-μg (for HA-wt, H/F/F, H/F/H, and H/H/F) or 4-μg (for H/C/C, H/C/H and H/H/C) portion of plasmid vector DNA in 100 μl of DMEM− was mixed with preincubated Lipofectin (10 μg in 90 μl of DMEM−; 45-min preincubation at room temperature) and incubated for 15 min at RT. DMEM− (400 μl) was added, and the mixture was used for transfection of the vaccinia virus-infected CV-1 cells after removal of the virus inoculum.
Analysis of expression levels by immunofluorescence (FACS analysis).
CV-1 cells grown in 6-cm dishes (to about 80% confluence) were infected and transfected as described above by using three times the amounts of plasmid DNA, vaccinia virus, Lipofectin, and medium used for 3.5-cm dishes. At 30 min posttransfection, ammonium chloride was added to a final concentration of 10 mM. At 4 h posttransfection, the serum-free medium was replaced by DMEM–10% FCS–10 mM ammonium chloride, and the cells were kept overnight at 32°C and 5% CO2. They were then washed three times with ice-cold PBS. The cells were removed from the dishes by trypsin treatment (0.05% trypsin–0.02% EDTA in PBS), resuspended, and fixed with 1.5% paraformaldehyde in PBS for 1 h on ice. Immunolabeling was performed with a polyclonal antibody from rabbit (anti-H7) as the primary antibody and an FITC-conjugated porcine anti-rabbit-immunoglobulin as the secondary antibody.
The immunofluorescent cells were analyzed by flow cytometry with a FACScan device and Lysys II software (Becton Dickinson, Heidelberg, Germany) or by a flow cytometer device (Partec GmbH, Münster, Germany) with integrated software.
Metabolic labeling experiments and immunoprecipitation.
CV-1 cells were infected and transfected as described above but with methionine-deficient DMEM for transfection. At 30 min posttransfection, 3 μl of Tran35S-label (>1,000 Ci/mmol) was added. After radiolabeling for 3.5 h (at 37°C and 5% CO2), the dishes were transferred to ice. The cells were washed with PBS, lysed with RIPA buffer (1% Triton X-100, 1% deoxycholate, 0.1% SDS, 0.15 M sodium chloride, 20 mM Tris, 10 mM EDTA, 10 mM iodoacetamide) for 5 min, resuspended, and centrifuged in Eppendorf tubes at 1,000 × g for 5 min. The cell debris was removed, and the supernatant was incubated with anti-H7 antibody overnight at 4°C. For precipitation of the expressed HA molecules, the samples were incubated with protein A-Sepharose for 2 h at RT and centrifuged. The Sepharose pellet was washed three times with RIPA buffer. Sample buffer (nonreducing: 50 mM Tris, 2% SDS, 0.1% bromophenol blue, 10% glycerol; the reducing buffer also contained 10 mM mercaptoethanol) was added, and the samples were boiled for 3 min. After centrifugation, the immunoprecipitates were analyzed by SDS-PAGE in a 12% polyacrylamide gel and visualized by autoradiography (1 day) of the vacuum-dried gels with Kodak X-Omat AR films. Quantification of fluorograms was carried out with an Epson GT-9000 scanner and Biometra ScanPack 2.0 software.
Preparation of cells for fusion assays. (i) Preparation of HA-expressing cells.
CV-1 cells were treated as described above for expression. At 30 min after transfection, ammonium chloride (final concentration, 10 mM) was added to the medium to prevent inactivation of expressed HA molecules by acidification during intracellular transport (26). As a control, CV-1 cells infected with vaccinia virus and treated by the transfection method but without DNA were used. The cells were incubated for another 3.5 h at 37°C and 5% CO2. After replacement of the medium by DMEM with 10% FCS and 10 mM ammonium chloride, the cells were kept overnight at 32°C and 5% CO2. The expressing CV-1 cells were washed with prewarmed DMEM− (37°C), treated for 15 min at RT with prewarmed DMEM containing 1 U of neuraminidase per ml and 10 μg of trypsin per ml, and given a final wash with PBS++.
(ii) Preparation and labeling of RBC.
Human blood (erythrocyte concentrate) was washed three times with PBS (centrifugation at 2,000 × g for 10 min), and erythrocytes were resuspended in PBS (50% hematocrit). A 20-μl volume of R18-solution (1 mg/ml in ethanol) were added to 200 μl of the RBC suspension in 5 ml of PBS under gentle vortexing. After addition of 10 ml of PBS and incubation of the suspension for 30 min at RT in the dark, 25 ml of DMEM+ was added to the suspension to absorb unbound label. After a further incubation for 20 min at RT in the dark, the RBC suspension was washed in 40 ml of PBS and centrifuged. The RBC pellet was resuspended in 2 ml of PBS, and after addition of the calcein-AM solution (50 μg freshly dissolved in 50 μl of dimethyl sulfoxide), the cells were incubated for 45 min at 37°C in the dark. Subsequently, the suspension was centrifuged and washed in 20 ml of PBS. After resuspension of the pellet in 10 ml of PBS, the cells were incubated at 37°C for 20 min in the dark for cleavage of remaining calcein-AM inside the cell. The suspension of R18- and calcein-labeled RBCs was washed five times in 30 ml of PBS, resuspended in 5 ml of PBS++, and stored on ice until used for binding to the (chimeric) HA-expressing CV-1 cells.
(iii) Preparation and labeling of RBC ghosts.
Blood (2 ml) of 50% hematocrit (prepared as described above) was mixed with 30 ml of ice-cold hemolysis buffer (4 mM MgSO4, 1 mM CaCl2, 1.2 mM acetic acid, 50 mM vanadate) and pelleted. The unsealed ghosts thus obtained were resuspended in 2 ml of hemolysis buffer containing 1 mg of calcein and 5 mg of TMR-D and incubated on ice for 15 min. Tenfold-concentrated PBS and 3.3 mg of ATP in 100 μl of PBS were added before the suspension was brought to pH 7.3 by titration with sodium hydroxide. The whole procedure was performed on ice. Subsequently, the suspension was incubated for 1 h at 37°C in the dark, washed four times with 30 ml of PBS and, for fusion experiments, resuspended in 5 ml of PBS++.
Fusion assay and fluorescence microscopy.
For binding, 1 ml of labeled RBCs or RBC ghosts with a hematocrit of 0.4 and 5%, respectively, was added to the monolayer of HA-expressing cells and incubated for 10 min at RT. The samples were washed with PBS++ to remove unbound RBCs and RBC ghosts, respectively. PBS++ was replaced by sodium acetate buffer (20 mM NaC2H3O2, 150 mM NaCl, 1 mM CaCl2, 2 mM MgCl2 [pH 5.0]) to trigger fusion. Fusion was monitored at RT by redistribution of the loaded fluorescent labels from RBCs or RBC ghosts to the membrane and cytoplasm, respectively, of the (chimeric) HA-expressing CV-1 cells. Label redistribution was observed under the fluorescence microscope (Axiovert 100 with a 40× LD Achroplan objective [Zeiss]). Calcein fluorescence was monitored with a “blue” filter set (450–490 excitation filter; lp 520 emission filter). R18 and TMR-D fluorescence was visualized using a “green” filter set (510–560 excitation filter; lp 590 emission filter). Photomicrographs were obtained with an MC 80 microscope camera (Zeiss) on an Ektachrome EPH P1600× color reversal film for push processing (Kodak) at 3,200 ASA. The sodium acetate buffer was removed after 4 min and replaced by PBS++ to prevent cell damage by acidic conditions.
Binding of RBCs to CV-1 cells by lectin and fusion assay.
To exclude fusion induction by vaccinia virus proteins, control experiments were performed. Cells expressing only vaccinia virus proteins or additionally HA-wt were incubated for 5 min at RT with WGA (50 μg/ml) in PBS++, and 1 ml of labeled RBCs or labeled RBC ghosts (0.4 and 5% hematocrit, respectively) was added. After further incubation for 10 min at RT in the dark under occasional gentle agitation, unbound cells were removed by five washes with PBS++. Fusion was monitored by fluorescence microscopy as described above.
RESULTS
Expression of HA-wt and the chimeras H/C/C, H/C/H, and H/H/C.
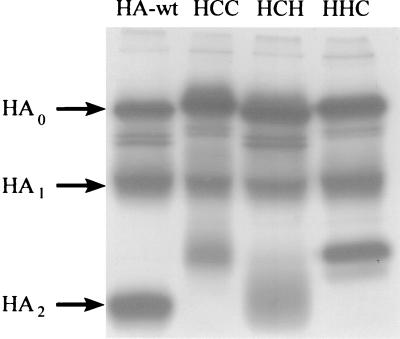
HA-wt and the constructs H/C/C, H/C/H, and H/H/C were expressed in CV-1 cells by the transient T7-RNA-polymerase vaccinia virus system, metabolically labeled with [35S]cysteine-[35S]methionine, and immunoprecipitated as described above. The autoradiograph (Fig. 1) of an SDS-polyacrylamide gel obtained under reducing conditions (see Materials and Methods) revealed the biosynthesis of uncleaved precursors of HA0 and of HA/CD4 chimeras as well as the two cleavage-generated subunits of those precursors. Similar results were obtained for the respective F chimera (not shown), as reported previously (34). Differences in the mobility between HA-wt, H/C/H, H/H/C, and H/C/C indicate different molecular weights based on the various lengths of the CT originating either from HA-wt or from CD4. As expected, this became most evident by comparison between the subunit HA2 and the corresponding cleavage product of chimeras. Densitometric analysis of the autoradiographs (data not shown) revealed that the efficiency of expression and the cleavage efficiency of HA/CD4 chimeras were similar to those obtained for HA-wt.
FIG. 1.
Expression of HA and HA-derived chimeras of CD4 in CV-1 cells. The genes of HA-wt, H/C/C, H/C/H, and H/H/C were cloned into the plasmid vector pTM1 and expressed with the transient T7-RNA-polymerase vaccinia virus expression system (see Materials and Methods). The gene products were metabolically labeled with [35S]cysteine-[35S]methionine. Cell extracts were immunoprecipitated with polyclonal antibody directed against influenza virus HA (H7) and subjected to SDS-PAGE under reducing conditions on a 12% polyacrylamide gel. The upper bands represent the precursor molecules (HA0 and the analogues of the chimeras H/C/C, H/C/H, and H/H/C); the two lower bands represent the subunits HA1 and HA2 (or analogues of the respective chimera) (marked on the left). The exposure time for the autoradiography was 1 day.
To confirm regular processing of HA-wt and the HA/CD4 chimeras after synthesis in the cell, we performed experiments including endo-β-N-acetylglucosaminidase H and peptide N-glycosidase F digestion. We found that HA-wt and the chimeras acquired endo-β-N-acetylglucosaminidase H resistance, indicating a proper processing through the intracellular pathway (data not shown). The known palmitoylation of the two cysteine residues of the CT of CD4 (8) is preserved in the chimeras H/H/C, H/C/H, and H/C/C (E. Ponimaskin, C. E. Weber, and M. F. G. Schmidt, unpublished observation).
To compare the surface expression of HA-wt with that of the chimeras, immunolabeled CV-1 cells (see Materials and Methods) were subjected to FACS analysis. The results (shown in Table 1) indicate a somewhat lower transfection efficiency in CV-1 cells for the various HA/CD4 chimeras than for HA-wt between 82% [H/C/C] and 95% [H/C/H] with respect to HA-wt). The surface expression of those chimeras was about 70% of that of HA-wt. One-way ANOVA (α = 0.05) revealed no significant difference between HA/CD4 chimeras for transfection efficiency or for surface expression. In agreement with our previous study (34), the transfection efficiency of HA/F chimeras was on the order of that of HA/CD4 chimeras while the surface expression was almost the same as that of HA-wt (data not shown).
TABLE 1.
FACS analysis of CV-1 cellsa
| Protein | Relative transfection efficiency (%)b | Relative mean fluorescence index (%)c |
|---|---|---|
| HA-wt | 100 | 100 |
| H/H/C | 90.8 ± 7.9 | 65.8 ± 3.7 |
| H/C/H | 95.6 ± 9.6 | 74 ± 4.5 |
| H/C/C | 82.1 ± 8.3 | 66.2 ± 8.0 |
CV-1 cells expressing HA-wt (H/H/H) or the chimeras H/C/C, H/C/H, and H/H/C were doubly immunolabeled and analyzed by FACS. The primary antibody was a polyclonal antibody against HA (H7). The secondary antibody was an FITC-conjugated anti-rabbit IgG. About 15,000 single cells were measured in each experiment. The measurement was calibrated by using vaccinia virus-infected CV-1 cells.
Transfection efficiency was determined by calculating all cells which show fluorescence activity specifically above that of the vaccinia virus-infected control with respect to the overall number of measured cells and is given as a percentage of the H/H/H (HA-wt) transfection efficiency value, set at 100%. Each value represents the results of four independent experiments (mean ± standard error). No significant differences between data for the chimeras were found (ANOVA, α = 0.05).
The mean fluorescence index is a measure of the amount of protein per expressing cell localized in its membrane and thus is a useful indicator of the relative surface density of the expressed protein. The relative mean fluorescence index was calculated with respect to H/H/H (HA-wt), set at 100%. Each value represents the results of four independent experiments (mean ± standard error). No significant differences between data for the chimeras were found (ANOVA, α = 0.05).
Fusion assays. (i) Redistribution of the membrane label R18.
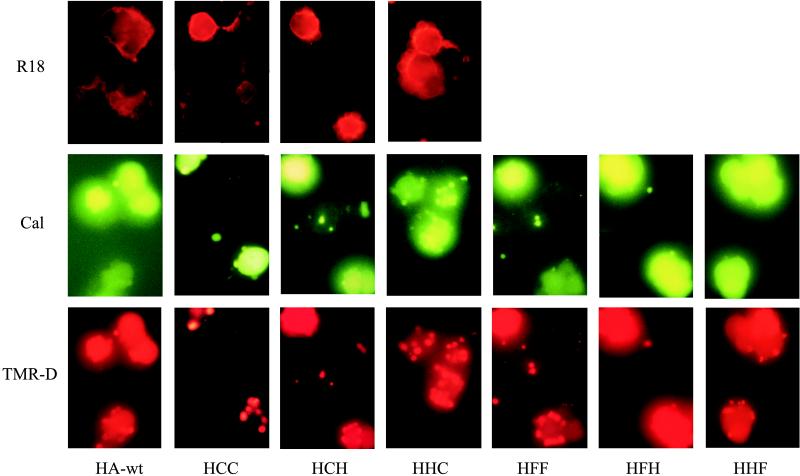
We studied membrane fusion between CV-1 cells expressing HA-wt, H/C/C, H/C/H, or H/H/C and R18-labeled RBCs at pH 5.0 (Fig. 2). R18 redistribution for HA-wt as well as for all the chimeras was first observed between 1.5 and 2 min after the pH trigger was set. Between 3 and 4.5 min after lowering of the pH, almost all HA-wt- or chimera-expressing cells with bound RBCs showed R18 labeling, indicating efficient membrane mixing. As revealed by seven independent experiments, no significant difference of the membrane fusion activity between HA-wt and the chimeras was found. This also applies to the chimeras H/F/F, H/F/H, and H/H/F (data not shown), which is in line with our previous study (34).
FIG. 2.
Redistribution of the fluorescent label during fusion. HA-wt-, H/C/C-, H/C/H-, H/H/C-, H/F/F-, H/F/H-, and H/H/F-expressing cells were bound to either R18-labeled RBC (top row) or calcein (Cal)- and TMR-D-labeled RBC ghosts (middle and bottom rows, respectively). Fusion was triggered at RT by replacing neutral buffer (PBS++ [pH 7.4]) with acidic buffer (sodium acetate buffer [pH 5.0]). Fluorographs obtained taken 5 min (HA-wt) or 15 min (chimeras) after the pH trigger. For details, see Results.
(ii) Redistribution of the aqueous labels calcein and TMR-D.
R18 redistribution between transfected cells and RBCs is indicative of hemifusion of membranes. To address whether our chimeras trigger complete membrane fusion, i.e., fusion of the exoplasmic as well as the cytoplasmic leaflets, and subsequent formation of an aqueous fusion pore, the redistribution of aqueous cytoplasmic fluorescent labels was employed. To this end, we analyzed the fusion of HA-wt- and chimera-expressing CV-1 cells with RBC ghosts doubly labeled with the aqueous labels calcein (MW 623) and TMR-D (MW 10,000). The use of labels with different sizes allowed us to monitor both the formation and the enlargement of the aqueous fusion pore. Based on our observation that binding of two or more ghosts to control CV-1 (i.e., vaccinia virus-infected but not transfected cells) was very rare, we consider that CV-1 cells with two or more bound ghosts are cells expressing HA-wt or chimeric proteins.
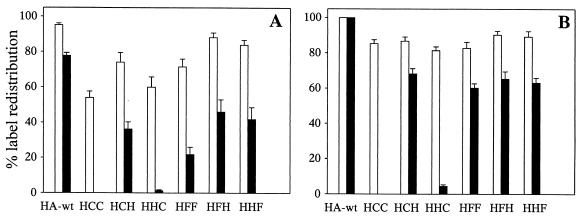
The redistribution of cytoplasmic labels was monitored by fluorescence microscopy (Fig. 3) 5 and 20 min after the fusion trigger. To quantify the redistribution of the label at each time point, five sectors (of 0.2 mm2) of fusing RBC–CV-1 cell complexes were selected and the following parameters were determined. NTMR-D represents the number of CV-1 cells labeled with the fluorophore TMR-D. Notably, we never observed leakage of TMR-D from CV-1 cells over the time course of the experiment. NC reflects the number of CV-1 cells with two or more bound RBC ghosts but without any redistribution of TMR-D from bound ghosts to CV-1 cells. The total number of HA-wt- or chimera-expressing cells per sector (the 100% value) is then given by the sum of NC and NTMR-D. NCAL corresponds to the number of CV-1 cells which have undergone calcein redistribution. However, a specific problem was encountered with the determination of NCAL. We often observed leakage of calcein from CV-1 cells upon fusion. As a consequence, NCAL would have been underestimated by measuring the number of calcein-labeled CV-1 cells. To circumvent this, NCAL was determined from the difference between the total number of expressing CV-1 cells (NC + NTMR-D) and the number of CV-1 cells with two or more bound calcein-labeled ghosts but without any redistribution of calcein to CV-1 cells.
FIG. 3.
Formation of an aqueous fusion pore, assessed by using fluorescent labels of different MW. Calcein- and TMR-D-labeled RBC ghosts were bound to CV-1 cells expressing HA-wt and the chimeras H/C/C, H/C/H, H/H/C, H/F/F, H/F/H, and H/H/F. Fusion was triggered by replacing neutral buffer (pH 7.4) with acidic buffer (pH 5.0) at RT. The diagrams show the percentage of HA-wt- or chimera-expressing CV-1 cells with calcein redistribution (white columns) and TMR-D redistribution (black columns) (for details, see Results) at 5 min (A) and 20 min (B) after the pH trigger. Each column represents the results of seven independent experiments (mean and standard error of estimate).
Typical fluorographs of calcein and TMR-D redistribution upon fusion between ghosts and HA-wt- or chimeric-protein-expressing CV-1 cells are shown in Fig. 2. The results of seven independent experiments are summarized in Fig. 3. Here, the percentage of expressing CV-1 cells which have undergone formation of an aqueous fusion pore as detected by calcein or TMR-D redistribution is shown. HA-wt and all chimeric constructs containing the TM and/or CT of CD4 (H/H/C, H/C/H, and H/C/C) mediated the flow of calcein from the lumen of labeled RBC ghosts into the cytoplasm of CV-1 cells, albeit with quantitative differences, especially in the early phase (5 min) after triggering of fusion. For HA-wt, more than 90% of expressing CV-1 cells showed redistribution of calcein. For the chimeric proteins, about 60% of H/C/C- and H/H/C-expressing and about 75% of H/C/H-expressing CV-1 cells were labeled with calcein. Nevertheless, after prolonged incubation (20 min), more than 80% of CV-1 cells expressing chimeric constructs had fused with RBC ghosts. Similar results were obtained when intact RBCs labeled with calcein (see Materials and Methods) were used as a target instead of ghosts.
When measuring TMR-D redistribution, we found that replacement of the HA-wt TM with the corresponding CD4 domain did not affect the enlargement of the fusion pore, although it was delayed with respect to HA-wt. After 20 min about 70% of H/C/H-expressing CV-1 cells were labeled with TMR-D. After the same period, almost all HA-wt-expressing cells showed TMR-D redistribution. In contrast, replacement of the CT of HA-wt by the respective domain of CD4 abolished the formation of large aqueous fusion pores. Almost no redistribution of TMR-D was found for H/C/C and H/H/C-expressing cells, even 20 min after triggering of fusion.
While we have reported previously (34) that replacement of the TM and/or CT by the respective domains of the F protein from Sendai virus (H/H/F, H/F/H, and H/F/F) did not affect the redistribution of calcein between CV-1 cells and RBCs (Fig. 2 and 3), we can now add that the dilation of the aqueous fusion pore is not inhibited. As is clear from Fig. 2 and 3, all mutants containing F domains triggered the labeling of CV-1 cells by TMR-D. A delay of calcein and TMR-D redistribution was observed for HA/F-chimeras compared to HA-wt.
(iii) Vaccinia virus-infected cells.
To exclude the possibility that vaccinia virus-specific products induce fusion, we studied in parallel whether label redistribution occurred also between labeled RBCs or RBC ghosts and vaccinia virus-infected CV-1 cells (no transfection with vector DNA [see Materials and Methods]). In this experiment, we observed no redistribution of fluorescent labels to CV-1 cells, neither for R18 nor for calcein plus TMR-D, even if RBCs or ghosts were bound to CV-1 cells by WGA before the pH trigger was set. The CV-1 cells and RBCs or RBC ghosts, as well as the labeling of the latter, remained stable and unaffected over a period of 3 h. Since additional control experiments exclude a possible inhibition of fusion by WGA in the medium (see also reference 34), redistribution of the various fluorescent labels between the transfected CV-1 cells and RBCs or RBC ghosts, described above, must be attributed to the fusion activity of HA-wt or chimeric proteins.
DISCUSSION
Impaired fusion pore enlargement of HA chimeras.
In the present study we assessed the significance of the TM and CT of influenza virus HA (subtype H7 from A/fpv/rostock/34) for membrane fusion. While our previous work concentrated on investigating the role of palmitoylation (12, 28) and of the CT and TM of HA by replacement of both domains by those from another fusion protein (Sendai virus F protein) (34), here we have studied the fusion activity of HA chimeras containing the corresponding domains from a totally unrelated, nonviral and nonfusogenic protein, the cellular glycoprotein CD4. We found that the CT and TM of CD4 were tolerated by HA in terms of lipid mixing (redistribution of R18) and the formation of small aqueous fusion pores (transfer of calcein). However, a different picture emerged for these constructs when the enlargement of the aqueous fusion pore was monitored with the fluorescent high-MW dextran TMR-D (MW 10,000). HA-wt readily promoted the formation of a large pore. Extending our previous study, we also found that the HA/F chimeras promoted this late fusion step, although it was delayed with respect to that in HA-wt. We take this as a strong indication that the TM and/or the CT sequences of Sendai virus F protein form a functional entity with the domains of influenza virus HA (H7) during the fusion process, leading to pore formation and growth. The same conclusion can be drawn for the HA/CD4 chimera H/C/H. However, quite differently, the chimeric constructs H/C/C and H/H/C were not able to mediate enlargement of the aqueous pore, although membrane fusion and formation of small pores were not affected (Fig. 2). This suggests that the CT, but not the TM domain of CD4 was responsible for the failure of those two chimeras to mediate pore growth. We noted that the extent of pore formation and growth of H/C/H and HA/F chimeras was lower in comparison to that of HA-wt. This may be caused by their somewhat lower expression in the plasma membrane of CV-1 cells (Table 1). Importantly, the transfection efficiency and surface expression of H/C/C and H/H/C cannot account for their inability to promote pore enlargement, since neither parameter was significantly different from that of H/C/H (Table 1).
Relevance of the TM domain of HA to fusion.
The observation that all chimeric proteins, in particular H/C/C and H/H/C, mediate membrane fusion as well as the formation of small aqueous fusion pores indicates that the CT does not affect the conformational change of the HA ectodomain required for initiating fusion as well as steps preceding the enlargement of an aqueous fusion pore. The data demonstrate that the CT affects late steps of the fusion process rather than early intermediates. The observation that the TM of a nonviral, nonfusion integral membrane protein, CD4, can functionally substitute for the corresponding HA domain confirms previous reports by others (23). Similar results have been obtained for other viral fusion proteins. Replacement of the TM of vesicular stomatitis virus G protein with that of CD4, of the viral nonfusion protein gD from herpes simplex virus, or of glycoprotein E3-11.6K from nonenveloped adenovirus preserved the ability of these constructs to mediate syncytium formation (25).
Although these observations reveal that specific sequences of the TM do not constitute general precondition for fusion, previous studies have shown that modification of this domain may have a strong impact on fusion activity. Melikyan et al. (23) reported that replacement of the highly conserved glycine residue in the TM for HA (subtype H2) at position 520 by leucine inhibited membrane fusion at the stage of lipid mixing. A similar observation has been made for vesicular stomatitis virus G protein (7). This suggests that functioning of the TM in fusion may require not yet identified sequence motifs. Taking into account the fact that glycine residues may be important for the association of transmembrane helices (4), one attractive hypothesis is that interaction of the TM within and perhaps between trimers could be essential for (complete) fusion. Indeed, previous studies suggested that HA trimers could be stabilized by the interaction of their transmembrane helices (9, 16). Although the precise sequence motifs mediating association of transmembrane helices of integral membrane proteins remain to be elucidated, the sequence LIxxGxxxGxxxT, with the most crucial part GxxxG, has been strongly implicated as a potential candidate (4). Remarkably, the TM of CD4 also contains a sequence very similar to this motif: LIVLGGVAG. Thus, further mutation analysis of the chimera H/C/H could provide a valuable tool to elucidate the role of transmembrane association in HA fusion.
Modification of the CT of HA affects the fusion process.
Amino acid residues of the cytoplasmic domain of influenza virus HA are highly conserved among the several HA subtypes, implicating its importance for influenza virus activities (15). So far it is not clear whether the CT plays any functional role in the fusion process. Influenza virus A/Udorn/72 with HA lacking its CT assembled and replicated almost as efficiently as did virions containing HA-wt (15). Deletion of the CT of HA (subtype H3) did not influence syncytium formation (35), and redistribution of a high-MW label (MW 40,000) between transfected CV-1 cells and RBCs was similar to that in HA-wt (22). In contrast, elongation of the CT of HA (subtype H7) by 6 aa caused a fivefold reduction in the extent of syncytium formation between HA-expressing CV-1 cells (27).
Recently, Melikyan et al. (23) found that replacement of the CT of HA from subtype H2 by that of the pIgR almost completely abolished the formation of an aqueous pore while replacement of the TM caused only a partial inhibition. However, formation of the aqueous fusion pore was not affected when these domains were simultaneously replaced with those of pIgR. This led to the conclusion that the TM and CT may not act independently of each other. In contrast, our data demonstrate that the inhibitory influence of the CT of CD4 on pore growth cannot be reversed by the presence of the TM of CD4. Similar results have been reported for vesicular stomatitis virus G protein (25). In this case, replacement of TM by the respective domain from CD4 did not abolish syncytium formation of G-protein-expressing COS-1 cells whereas additional replacement of the CT caused a drastic reduction in syncytium formation.
Taken together, these data suggest that the CT may not be relevant for HA-mediated fusion. However, alterations or replacement of its sequence seem to have an impact on the fusion process. Thus, whatever the function of the CT may be in viral infection and/or replication, the highly conserved sequence of the CT has to have specific structural features in order to not impair fusion.
Properties of the CT of CD4 that may affect the fusion activity of HA.
The most obvious difference between the CTs of HA and CD4 is in their lengths, with the CD4 CT being more than three times as long as the HA CT. In a previous study it was suggested that elongation of the CT of HA-wt may cause a reduction of membrane fusion activity and an impaired fusion pore formation (27). However, if elongation of the CT were the sole cause of the inhibition of pore growth, we would expect a similar inhibition for chimeras containing the CT of the F protein, which is four times as long as that of influenza virus HA-wt. However, this was not the case (Fig. 2 and 3). Furthermore, Melikyan et al. (23) observed the transfer of a large soluble aqueous dye (MW, 40,000) similar to that in HA-wt for a chimera whose TM and CT were replaced by the respective domains of the Env protein of Rous sarcoma virus. The CT of Env is 28 aa, about 2.5 times as long as that of HA-wt. On the other hand, shortening the CT of the pIgR used in that study to 11 aa did not render the respective chimeric HA competent to mediate content mixing (i.e., pore enlargement). Thus, the length of the CT of H/C/C and H/H/C is most probably not the cause of the observed impairment of pore enlargement.
A highly conserved feature of the CT of influenza virus HA is its pronounced hydrophobicity. As we have suggested previously, the hydrophobicity of the CT may be important for pore enlargement (12). The hydrophobicity is determined (i) by the highly conserved palmitoylation of the three cysteine residues which are localized in the TM and CT of HA-wt and (ii) by the amino acids of the CT, as visualized in a hydrophobicity plot (Fig. 4). Recently, we have shown that acylation-deficient mutants of HA interfered with the late fusion steps involved in syncytium formation (12) but did not affect early events such as membrane fusion and the subsequent formation of small pores (12, 28). However, the known palmitoylation of the two cysteine residues located in the CT close to the TM are preserved in the HA/CD4 chimeras used (Ponimaskin et al., unpublished). This precludes a deficiency in palmitoylation of CT as a cause of the impairment of pore growth.
FIG. 4.
Hydrophobicity plots of the cytoplasmic domains of influenza virus HA, Sendai virus F protein, vesicular stomatitis virus (VSV) G protein, human immunodeficiency virus type 1 (HIV-1) gp41, herpes simplex virus type 1 (HSV) gD, the deleted CT of pIgR, and human CD4. Hydrophobicity was calculated using the hydrophobicity scale of Abraham and Leo (1) by moving a window of 5 residues along the protein sequence in the direction from the N terminus to the C terminus (18).
Hydrophobicity analysis (1) reveals that the CT of CD4 close to the TM is quite hydrophilic in comparison to that of HA-wt, which has a hydrophobic stretch close to the membrane border (Fig. 4). While comparison of only these two sequences may not substantiate the importance of the hydrophobic property of the CT for complete fusion, other indications support such a hypothesis. The hydrophobic profiles of the respective sequences of the CTs from vesicular stomatitis virus G protein and Sendai virus F protein are very similar to that of HA-wt (Fig. 4), and, as shown here, the chimeras H/F/F and H/H/F support complete fusion. Also, in the hydrophobicity profile of gp41, the fusion-involved transmembrane protein of human immunodeficiency virus type 1, a TM-proximal stretch of hydrophobic amino acid residues becomes visible, although it is shifted slightly in the distal direction. On the other hand, the hydrophobicity plots of the CT of pIgR and of gD from herpes simplex virus show a more hydrophilic profile, similar to that of CD4 (Fig. 4). Chimeras of HA containing the CT of pIgR and of vesicular stomatitis virus G protein containing the CT and TM of either gD or CD4, respectively, were reported to be unable to form fusion pores (23) and to mediate syncytium formation (25), respectively (see above). Taken together, the results point to a critical role of the hydrophobicity of the TM-proximal region of viral fusion-mediating proteins for pore enlargement during the fusion process.
How does the hydrophobicity of the cytoplasmic tail promote pore dilation?
Based on the observation that GPI-anchored HA causes the formation of a metastable hemifusion diaphragm after triggering the conformational change of the ectodomain (17, 21, 24) the “elastic-coupling” model has been proposed to explain the complete membrane fusion and the subsequent formation of an aqueous fusion pore (21, 23). This model predicts that the stiff linkage between the ectodomain and the TM of HA causes destabilization and eventually rupture of the hemifusion diaphragm, leading to full membrane fusion and pore formation. As we discussed previously (12), an intriguing feature of the model could be an orientational shift of the TM within the viral (plasma) membrane from a vertical to a tilted position. The latter requires at least partial immersion of the CT into the lipid phase. Reduction of the hydrophobicity of the CT either by deacylation (12) or by replacement of the amino acid sequence could hinder its immersion and the tilting of the TM. This would interfere with the fusion process. This hypothesis is also consistent with the observation that fusion is not impaired in the case of HA lacking its CT (15). It is also feasible that modification of the CT may give rise to a strong interaction between HA trimers, forming a small fusion pore, which would inhibit pore enlargement. Future experiments are warranted to test these hypotheses and to determine how the hydrophobicity of the CT influences only the late step of fusion pore growth. Experiments involving recombinant modifications focused on the CT segment close to the TM of fusogenic proteins should contribute to our understanding of pore enlargement during membrane fusion.
ACKNOWLEDGMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft to M.F.G.S. (SFB 366) and A.H. (SFB 312).
We are grateful to Wolf-Dietrich Döcke and Florian Kern (Institute for Clinical Immunology, Charité, Berlin) and to Karin Müller (Institut für Fortpflanzung landwirtschaftlicher Nutztiere e.V., Schönow, Germany) for advice and use of their FACS facilities.
REFERENCES
- 1.Abraham D J, Leo A J. Extension of the fragment method to calculate amino acid zwitterion and side chain partition coefficients. Proteins Struct Funct Genet. 1987;2:130–152. doi: 10.1002/prot.340020207. [DOI] [PubMed] [Google Scholar]
- 2.Bagai S, Lamb R A. Truncation of the COOH-terminal region of the paramyxovirus SV5 fusion protein leads to hemifusion but not complete fusion. J Cell Biol. 1996;135:73–84. doi: 10.1083/jcb.135.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böttcher C, Ludwig K, Herrmann A, van Heel M, Stark H. Structure of influenza haemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett. 1999;463:255–259. doi: 10.1016/s0014-5793(99)01475-1. [DOI] [PubMed] [Google Scholar]
- 4.Brosig B, Langosch D. The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. 1998;7:1052–1056. doi: 10.1002/pro.5560070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–42. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Skehel J J, Wiley D C. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA2 to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci USA. 1999;96:8967–8972. doi: 10.1073/pnas.96.16.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleverley D Z, Lenard J. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc Natl Acad Sci USA. 1998;95:3425–3430. doi: 10.1073/pnas.95.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crise B, Rose J K. Identification of palmitoylation sites on CD4, the human immunodeficiency virus receptor. J Biol Chem. 1992;267:13593–13597. [PubMed] [Google Scholar]
- 9.Doms R, Helenius A. Properties of a viral fusion protein. In: Ohki S, Doyle D, Flanagan T D, Hui S W, Helenius A, editors. Molecular mechanisms of membrane fusion. New York, N.Y: Plenum Press; 1987. pp. 385–398. [Google Scholar]
- 10.Dong J, Roth M G, Hunter E. A chimeric avian retrovirus containing the influenza virus gene has an expanded host range. J Virol. 1992;66:7374–7382. doi: 10.1128/jvi.66.12.7374-7382.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durrer P, Galli C, Hoenke S, Corti C, Glück R, Vorherr T, Brunner J. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 12.Fischer C, Schroth-Diez B, Herrmann A, Garten W, Klenk H-D. Acylation of the influenza hemagglutinin modulates fusion activity. Virology. 1998;248:284–294. doi: 10.1006/viro.1998.9286. [DOI] [PubMed] [Google Scholar]
- 13.Fuerst T R, Niles E G, Studier F W, Moss B. Eucaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gething M-J, Henneberry J, Sambrook J. Fusion activity of the hemagglutinin of influenza virus. Curr Top Membr Transp. 1988;32:337–364. [Google Scholar]
- 15.Jin H, Leser G P, Lamb R A. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 1994;13:5504–5515. doi: 10.1002/j.1460-2075.1994.tb06885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemble G W, Henis Y A, White J M. GPI- and transmembrane-anchored influenza hemagglutinin differ in structure and receptor binding activity. J Cell Biol. 1993;122:1253–1265. doi: 10.1083/jcb.122.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemble G W, Danieli T, White J M. Lipid-anchored influenza hemagglutinin promotes hemifusion, but not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 18.Korte T, Ludwig K, Herrmann A. pH-dependent hydrophobicity profile of hemagglutinin of influenza virus and its possible relevance in virus fusion. Biosci Rep. 1992;12:397–406. doi: 10.1007/BF01121503. [DOI] [PubMed] [Google Scholar]
- 19.Lüneberg J, Martin I, Nüssler F, Ruysschaert J-M, Herrmann A. Structure and topology of the influenza virus fusion peptide in lipid bilayers. J Biol Chem. 1995;270:27606–27614. doi: 10.1074/jbc.270.46.27606. [DOI] [PubMed] [Google Scholar]
- 20.Markosyan R M, Cohen F C, Melikyan G B. The lipid-anchored ectodomain of influenza virus hemagglutinin (GPI-HA) is capable of inducing nonenlarging fusion pores. Mol Biol Cell. 2000;11:1143–1152. doi: 10.1091/mbc.11.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melikyan G B, White J M, Cohen F S. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J Cell Biol. 1995;131:679–691. doi: 10.1083/jcb.131.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melikyan G B, Jin H, Lamb R A, Cohen F S. The role of the cytoplasmic tail region of influenza virus hemagglutinin in formation and growth of fusion pores. Virology. 1997;235:118–128. doi: 10.1006/viro.1997.8686. [DOI] [PubMed] [Google Scholar]
- 23.Melikyan G B, Lin S, Roth M G, Cohen F S. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol Biol Cell. 1999;10:1821–1836. doi: 10.1091/mbc.10.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nüssler F, Clague M J, Herrmann A. Meta-stability of the hemifusion intermediate induced by glycosylphosphatidylinositol-anchored influenza hemagglutinin. Biophys J. 1997;73:2280–2291. doi: 10.1016/S0006-3495(97)78260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odell D, Wanas E, Yan J, Ghosh H P. Influence of membrane anchoring and cytoplasmic domains on the fusogenic activity of vesicular stomatitis virus glycoprotein G. J Virol. 1997;71:7996–8000. doi: 10.1128/jvi.71.10.7996-8000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohuchi M, Cramer A, Vey M, Ohbuchi R, Garten W, Klenk H-D. Rescue of vector-expressed fowl plague virus hemagglutinin in biologically active form by acidotropic agents and coexpressed M2 protein. J Virol. 1994;68:920–926. doi: 10.1128/jvi.68.2.920-926.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohuchi M, Fischer C, Ohuchi R, Herwig A, Klenk H-D. Elongation of the cytoplasmic tail interferes with the fusion activity of influenza virus hemagglutinin. J Virol. 1998;72:3554–3559. doi: 10.1128/jvi.72.5.3554-3559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philipp H C, Schroth B, Veit M, Krumbiegel M, Herrmann A, Schmidt M F G. Assessment of fusogenic properties of influenza virus hemagglutinin deacylated by site-directed mutagenesis and hydroxylamine treatment. Virology. 1995;210:20–28. doi: 10.1006/viro.1995.1313. [DOI] [PubMed] [Google Scholar]
- 29.Ponimaskin E, Schmidt M F G. Domain-structure of cytoplasmic border region is main determinant for palmitoylation of influenza virus hemagglutinin (H7) Virology. 1998;249:325–335. doi: 10.1006/viro.1998.9303. [DOI] [PubMed] [Google Scholar]
- 30.Qiao H, Pelletier S L, Hoffman L, Hacker J, Armstrong R T, White J M. Specific single or double proline substitutions in the “spring-loaded” coiled-coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J Cell Biol. 1998;141:1335–1347. doi: 10.1083/jcb.141.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao H, Armstrong R T, Melikyan G B, Cohen F S, White J M. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol Biol Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth M G, Doyle C, Sambrook J, Gething M-J. Heterologous transmembrane and cytoplasmic domains direct functional chimeric influenza virus hemagglutinins into the endocytic pathway. J Cell Biol. 1986;102:1271–1283. doi: 10.1083/jcb.102.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Schroth-Diez B, Ponimaskin E, Reverey H, Schmidt M F G, Herrmann A. Fusion activity of transmembrane and cytoplasmic domain chimeras of the influenza virus glycoprotein hemagglutinin. J Virol. 1998;72:133–141. doi: 10.1128/jvi.72.1.133-141.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simpson D A, Lamb R A. Alterations to influenza virus hemagglutinin cytoplasmic tail modulate virus infectivity. J Virol. 1992;66:790–803. doi: 10.1128/jvi.66.2.790-803.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skehel J J, Wiley D C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 37.White J M, Wilson I A. Anti-peptide antibodies detect steps in a protein conformational change: low-pH activation of the influenza virus hemagglutinin. J Cell Biol. 1987;105:2887–2896. doi: 10.1083/jcb.105.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiley D C, Skehel J J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–394. doi: 10.1146/annurev.bi.56.070187.002053. [DOI] [PubMed] [Google Scholar]
- 39.Wilson I A, Skehel J J, Wiley D C. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–377. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]