Abstract
Background:
lNUAK1 is strongly associated with organ fibrosis, but its causal mechanism for modulating lipid metabolism and hepatic inflammation underlying MASH has not been fully clarified.
Method:
In our study, human liver tissues from patients with MASH and control subjects were obtained to evaluate NUAK1 expression. MASH models were established using C57BL/6 mice. Liver damage and molecular mechanisms of the NUAK1-Caspase 6 signaling were tested in vivo and in vitro.
Results:
In the clinical arm, NUAK1 expression was upregulated in liver samples from patients with MASH. Moreover, increased NUAK1 was detected in mouse MASH models. NUAK1 inhibition ameliorated steatohepatitis development in MASH mice accompanied by the downregulation of hepatic steatosis and fibrosis. Intriguingly, NUAK1 was found to facilitate Caspase 6 activation and trigger pyroptosis in MASH-stressed livers. Disruption of hepatocytes Caspase 6 decreased MASH-induced liver inflammation with upregulated TAK1 but diminished RIPK1. Moreover, we found that NUAK1/Caspase 6 axis inhibition could accelerate the interaction between TAK1 and RIPK1, which in turn led to the degradation of RIPK1.
Conclusions:
In summary, our study elucidates that NUAK1-Caspase 6 signaling controls inflammation activation in MASH through the interaction between TAK1 and RIPK1, which is crucial for controlling pyroptosis and promoting the progression of MASH.

INTRODUCTION
MASLD is the predominant type of chronic liver diseases, affecting around 25% of the general global population.1,2 MASH, which represents the progressive stage of MASLD, is primarily characterized by liver inflammation and expected to progress to fibrosis and cirrhosis, finally resulting in liver failure or HCC.3 Despite intense efforts to unravel the mechanisms underlying the progression of MASH, no approved, disease-modifying therapies are available.4,5
AMPK (5ʹadenosine monophosphate–activated protein kinase)–related kinase (NUAK1) belongs to the AMPK protein kinase family. It is also known as AMPK-related kinase 5. Importantly, it has been observed that NUAK1 plays a critical role in responding to various forms of cellular stresses.6,7 In contrast to the widely studied AMPK, the precise targets governed by NUAK1 remain to be determined. In the HCC model, inhibition of NUAK1 triggered the collapse of cellular ATP levels, which drives multiple proapoptotic responses.8 Additional studies have highlighted the potential involvement of NUAK1 in neurodevelopmental disorder regulation.9 Courchet et al10 found that haploinsufficiency of NUAK1 contributed to defects in cortical connectivity formation and a wide variety of abnormal behavioral traits. However, the biochemical processes that reveal the inflammatory action of NUAK1 are not as clear as those of the other dependencies described above.
Caspases are related aspartic-serine proteases originally identified for orchestrating the initiation or execution of apoptotic cell death under different biological processes.11 Among these caspases, the precise function of Caspase 6 has remained elusive for several decades. Despite its classification as an apoptotic executioner and connections to multiple neurodegenerative diseases,12,13 exact mechanisms of the activation and cleavage of Caspase 6 remain uncertain. Accumulating evidence has recognized a unique role for Caspase 6 in the regulation of programmed cell death and inflammasome activation. During an influenza A virus infection, Caspase 6 assisted in the activation process of NLRP3 inflammasome independently of its protease activity.14 Recent literature reported that depletion of Caspase 6 improved liver damage and fibrosis in MASH mouse model.15 Another study showed that ubiquitously expressed Caspase 6 acted as a supporting role in preventing the production of proinflammatory cytokines by cleaving RIPK1.16 However, it remains enigmatic as to whether and how the NUAK1 signaling may regulate Caspase 6 and govern pyroptosis in MASH-induced inflammatory modulation.
In this study, we unveiled a novel regulatory mechanism involving the NUAK1-Caspase 6–mediated pyroptosis in the development of MASH. Our findings demonstrate that NUAK1 plays a crucial role in mediating MASH development in Caspase 6–dependent manner. NUAK1-Caspase 6 inhibition enhances RIPK1-TAK1 interaction, which in turn alleviates pyroptosis in response to MASH-triggered liver inflammation.
METHODS
Human liver samples
Liver specimens were procured from patients with benign liver diseases who subsequently underwent hepatectomy. Postoperative pathological diagnosis was conducted by 3 experienced pathologists. Patients with MASLD were classified into 2 histological types based on the MASLD activity score. Patients who scored ≥5 on MASLD activity score were diagnosed with MASH. Normal liver specimens were obtained from 18 patients with hepatic hemangioma but without MASH. The exclusion criteria included the presence of alcohol abuse, hepatitis virus infection, or malignant tumors.
Animals
Conventional Caspase 6 deletion mice were generated using the CRISPR/Cas system. From the transcript ENSMUST00000029626, a total of 7 exons were identified. The region ranging from exon 2 to exon 7 was selected as the target site for the gene modification. A combination of the Cas9 enzyme and guide RNA were coinjected into fertilized eggs to generate the knockout (KO) mice (Cyagen Biosciences) (Supplemental Figure S5, http://links.lww.com/HC9/A947). The mice were housed in a specific pathogen-free facility under 12 hours of light and 12 hours of darkness. Then, they were euthanized under isoflurane anesthesia. Samples were excised and either immediately frozen in liquid nitrogen or fixed for further analysis.
Experimental procedures
Four-week-old male mice were fed with high-fat diet (HFD) (D12451, Research Diets, the ingredient is shown in Supplemental Table S1, http://links.lww.com/HC9/A947) or normal chow diet for 24 weeks (Supplemental Figure S1, http://links.lww.com/HC9/A947). In another MASH model, 8-week-old male mice were fed with a methionine-choline–deficient diet (Research Diets) or normal chow diet for 6 weeks. In HFD mouse model, NUAK1 inhibitor HTH-01-015 (20 mg/kg, SML1446, Sigma-Aldrich) or vehicle (10% DMSO) was administrated by i.p. injection twice daily in the last 9 weeks. HTH-01-015 is a specific NUAK1 kinase inhibitor. It could suppress NUAK1 activity through the inhibition of myosin phosphate–targeting subunit 1 (MYPT1) phosphorylation.
Hepatocellular function assay
Serum levels of alanine aminotransferase and aspartate aminotransferase were detected by Colorimetric Assay Kit (K752 and K753, Biovision).
Triglyceride and cholesterol measurement
Samples were homogenized, neutralized, and then centrifugated to collect supernatant. Triglyceride and total cholesterol levels were analyzed using Triglyceride (MAK043, Sigma-Aldrich) or Cholesterol Determination Kit (MAK266, Sigma-Aldrich) according to the manufacturer’s instructions.
ELISA assay
Murine serum and cell culture supernatants were collected specifically for cytokine analysis. The levels of TGF-β (EK0515, BOSTER), IL-1β (EK0394, BOSTER), and IL-18 (RAB0308, Sigma-Aldrich) were determined using the ELISA kits according to the manufacturer’s instructions.
Liver histological analysis
Liver samples were preserved in 10% phosphate-buffered formalin for 24 hours, subsequently being embedded in paraffin wax. Paraffin sections (5 μm) were stained with hematoxylin and eosin staining as reported.17 Frozen sections were stained with a 0.3% Oil Red O solution for 30 minutes. Subsequently, the sections were counterstained with hematoxylin for 1 minute. The degree of liver fibrosis was evaluated utilizing Masson staining (G1006, Servicebio).
Immunohistochemistry analysis
Liver sections (5 μm) were fixed with paraformaldehyde and embedded in paraffin. Then, sections were incubated at 4°C overnight with primary anti-NUAK1 antibody (CST, 47677), anti-Ly6G antibody (Santa Cruz, sc-53515), anti-α-SMA (CST, 19245), and anti-p-RIPK1 (Proteintech, 66854-1-Ig). Then slides were incubated at 37°C for 60 minutes with secondary antibody. The reaction was developed with diaminobenzidine staining. Cell nuclei were stained with hematoxylin.
Immunofluorescence analysis
Samples were incubated with the primary antibodies, including anti-HNF-4α (Abcam, ab41898), anti-CD11b (CST, 17800), anti-TAK1 (Santa Cruz, sc-7967), anti-NLRP3 (Invitrogen, 768319), or anti-ASC (CST, 67824) at 4°C overnight. Samples were washed with PBS and incubated with fluorescent secondary antibody in the dark for 60 minutes. Nuclear staining was conducted using DAPI. Images were captured with a fluorescence microscope (Olympus).
Quantitative real time-PCR
RNA, homogenized in TRIzol Reagent (Life Technologies), was extracted using RNA extraction kits (Qiagen). Subsequently, 1 µg of the purified RNA was used to facilitate reverse transcription PCR, generating complementary DNA through reverse transcription with random primers. ΔΔCt real-time PCR with SYBR green quantitative PCR kit (Thermo Fisher) was used to qualify the transcript levels of various target molecules. The 7500 FAST Real-Time PCR System was used to conduct real-time PCR analysis. The specific mRNA primers are listed in Supplemental Table S2, http://links.lww.com/HC9/A947. The expression of target genes was calculated relative to that of β-actin, which served as an internal control.
Western blots
Proteins were extracted from tissues or cells using nondenaturing lysing buffer (Thermo Scientific) with freshly added protease inhibitors (Roche). Supernatants were collected by centrifugation and then analyzed using BCA (Thermo Scientific). Proteins were separated by electrophoresis (sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels) and further transferred to the nitrocellulose membrane (Millipore). Antibodies including anti-NUAK1 (Cell Signaling Technology, 44585), anti-MYPT1 (CST, 2634), anti-p-MYPT1 (Santa Cruz, sc-377531), anti-active Caspase 6 (GeneTex, GTX59553), anti-Caspase 6 (CST, 9762), anti-TAK1 (CST, 5206), anti-RIPK1 (CST, 3493), anti-p-RIPK1 (CST, 39341), anti-GSDMD-N (CST, 10137), anti-cleaved-Caspase 1 (CST, 89332), anti-NLRP3 (Invitrogen, 768319), and anti-β-actin (CST, 4970) were used for probe. Finally, protein visualization was facilitated by the use of horseradish peroxidase–conjugated secondary antibodies (Fisher Scientific) in conjunction with Chemiluminescent Substrate (Thermo Scientific).
Caspase 6 activity assay
Cell/tissue samples were assessed by Caspase 6 Colorimetric Assay Kit (Biovision, K115).
Cell transfection
The AML12 strain of mouse hepatocytes (1 × 106) was transfected with CRISPR/Cas9 Caspase 6 KO, TAK1 KO, RIPK1 KO, GSDMD KO, CRISPR Caspase 6, TAK1, RIPK1, or GSDMD activation, and CRISPR control vector (Santa Cruz) through using Lipofectamine 3000 reagent (L3000075, Invitrogen) in antibiotic-free medium. After 6 hours, the cultured medium was replaced with a fresh medium and treated with 300 μm palmitic acid (PA) for 24 hours followed by incubation with HTH-01-015 (100 nM) for 24 hours (PBS as vehicle control).
Immunoprecipitation analysis
Hepatocytes were subjected to lysis in NP-40 buffer supplemented with protease inhibitors. The lysates were subjected to overnight incubation at 4°C on a rotator, with TAK1 antibody (CST, 5206), RIPK1 (CST, 3493), or control IgG and protein A/G beads. Following immunoprecipitation, the beads underwent 3 wash cycles with lysis buffer and were dissociated from proteins by employing a protein loading buffer containing sodium dodecyl sulfate at 100°C for 5 minutes. The proteins were subsequently analyzed through standard immunoblotting procedures.
Statistical analysis
Data are presented as the mean ± SD. Statistical variance among groups was assessed using a 1-way ANOVA coupled with the Bonferroni post hoc test. To compare the 2 groups, we used the 2-tailed Student t test. GraphPad Prism software, version 5.0 (GraphPad Software), was employed for data analysis. A p value of <0.05 was deemed statistically significant.
Study approval
All experimental procedures involving animals and human biopsies were sanctioned by the Ethics Committee of Tianjin Medical University General Hospital. Informed consent for human tissue analysis was obtained before surgery.
RESULTS
Patients with MASH and mouse models of MASH have high expression of NUAK1
To investigate the involvement of NUAK1 in MASLD, we first analyzed the expression of NUAK1 in liver samples from 18 normal patients, 18 patients with MASLD, and 18 patients with MASH (Supplemental Table S3, http://links.lww.com/HC9/A947). Compared to normal livers, NUAK1 mRNA was upregulated in patients with MASLD/MASH. In particular, NUAK1 expression in patients with MASH was higher compared to patients with MASLD (Figure 1A). Hepatic NUAK1 protein in patients with MASH was observed to be notably elevated compared to those in patients without MASH (Figures 1B, C). And NUAK1 mRNA in human MASH livers showed positively correlated with the expression of liver fibrogenic gene (TGF-β) (R2 = 0.4167, p = 0.0038) (Figure 1D). Moreover, NUAK1 expression was increased at both mRNA and protein levels in the livers of mice fed with HFD or methionine-choline–deficient diet (Figures 1E, G). Using dual-immunofluorescence staining, we observed increased NUAK1 expression in hepatocytes from mouse liver specimens (Figure 1H). In addition, the expression of NUAK1 in nonparenchymal cells from mouse HFD livers was analyzed using published single-cell RNA sequencing data18 and dual-immunofluorescence staining targeting 2 main types of fibrosis-related cells (fibroblasts and HSCs). However, our results did not show the significant difference in NUAK1 expression among these cell types (Supplemental Figure S2, http://links.lww.com/HC9/A947).
FIGURE 1.
NUAK1 is upregulated in the livers of patients with MASH or mouse MASH models. (A) NUAK1 mRNA in the liver tissues of human MASLD (NAS <5) and MASH (NAS ≥5), compared with normal individuals, as determined by qRT-PCR (n = 18 per group). (B) NUAK1 protein levels in the liver tissues of human normal individuals and MASH, as determined by western blot (n = 5 per group). (C) NUAK1 immunohistochemistry staining in liver specimens of patients with MASH and normal individuals (scale bar: 50 µm). (D) Correlation between NUAK1 mRNA and liver TGF-β expression in human MASH livers. (E–G) mRNA and protein levels of NUAK1 were detected in the livers of mice fed with HFD or MCD compared with NCD. (H) Double immunofluorescence staining of NUAK1 (green) and HNF-4α (red) in liver samples of mice fed on NCD or HFD (scale bar, 50 μm). DAPI was used to visualize nuclei (blue). ANOVA, Bonferroni multiple comparison test, n = 4–6 per group. All data represent the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: HFD, high-fat diet; MCD; methionine-choline–deficient diet; NAS, nonalcoholic fatty liver disease activity score; NCD, normal chow diet; qRT-PCR, quantitative real time-PCR.
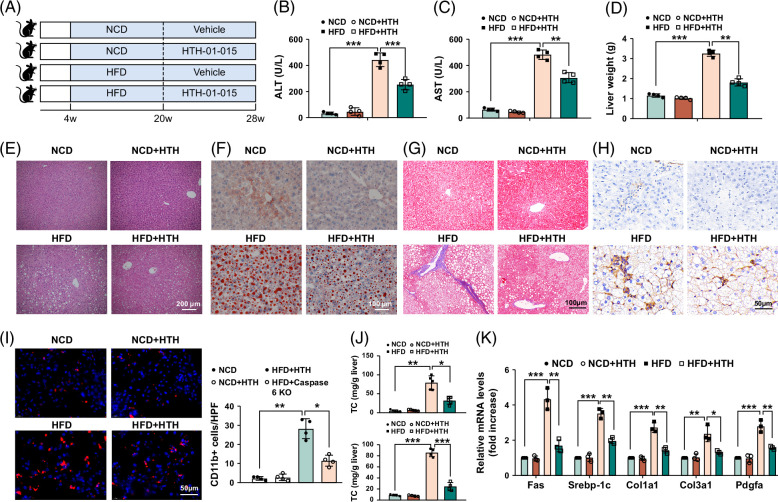
To explore the potential role of NUAK1 in MASH progression, WT mice were fed an HFD diet with or without NUAK1 inhibitor HTH-01-015 administration by i.p. injection (Figure 2A). Indeed, the phosphorylation of MYPT1 was suppressed, indicating that HTH-01-015 treatment inhibited the kinase activity of NUAK1 effectively (Supplemental Figure S3, http://links.lww.com/HC9/A947). As expected, HFD induced liver steatosis and damage as evidenced by increased levels of serum liver enzymes (alanine aminotransferase and aspartate aminotransferase) (Figures 2B, C), a significant upregulation of liver weight (Figure 2D), more stained areas by hematoxylin and eosin, Oil Red O staining (Figures 2E, F), macrophage infiltration (Figure 2I) and lower total cholesterol, triglyceride levels (Figure 2J), suggesting HFD can affect hepatic lipid metabolism. In addition, Masson staining and α-SMA staining detected more collagen fibers (Figures 2G, H) with much higher mRNA levels coding for Fas, Srebp-1c, Col1a1, Col3a1, and Pdgfa in HFD-induced livers (Figure 2K). These effects were reversed by the pharmacological inhibition of NUAK1. To corroborate these findings, we employed small-interfering RNAs to silence NUAK1 in mouse hepatocytes. NUAK1 small-interfering RNAs had similar effects to HTH-01-015, decreasing lipid accumulation and proinflammatory factor production. It also downregulated Caspase 6 activity and hepatocytes pyroptosis as well (Supplemental Figure S4, http://links.lww.com/HC9/A947). Collectively, the data presented above suggested the central role of NUAK1 in mediating MASH-induced hepatic steatosis and liver damage.
FIGURE 2.
NUAK1 inhibition alleviates liver steatosis and fibrosis on HFD-induced MASH in mice. Four-week-old mice were fed with HFD for 24 weeks, accompanied by treatment of NUAK1 inhibitor HTH-01-015 (20 mg/kg). (A) Schematic protocol of the experiment. (B, C) Liver function was evaluated by serum ALT and AST levels (U/L). (D) Liver weight was evaluated. (E) Representative histological staining (H&E) of liver tissue (scale bar, 200 μm). (F) Oil Red O staining of liver tissues (scale bar, 100 μm). (G) Masson staining of liver tissues and fibrosis score (scale bar, 100 μm). (H) Immunohistochemistry staining of α-SMA of liver tissues (scale bar, 50 μm). (I) Immunofluorescence staining of CD11b of liver tissues (scale bar, 50 μm). (J) Liver TC and TG. (K) qRT-PCR analysis of Fas, Srebp-1c, Col1a1, Col3a1, and Pdgfa in fatty livers. ANOVA, Bonferroni multiple comparison test, n = 4–6 per group. All data represent the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; H&E, hematoxylin and eosin; HFD, high-fat diet; qRT-PCR, quantitative real time-PCR; TC, total cholesterol; TG, triglyceride.
The inhibition of NUAK1 suppressed Caspase 6 activity and induced TAK1 activation and cell death in mouse MASH model
We then analyzed whether NUAK1 could influence Caspase 6/TAK1/RIPK1 pathway and pyroptosis in the HFD-induced MASH model. Indeed, compared to the normal chow diet group, HFD increased Caspase 6 activity and TAK1 expression, resulting in pyroptosis activation in fatty livers. Moreover, NUAK1 inhibitor HTH-01-015 pretreatment reduced cleaved-Caspase 6, RIPK1, p-RIPK1, GSDMD-N, cleaved-Caspase 1, and NLRP3 but augmented TAK1 expression in fatty livers (Figure 3A). Consistent with this result, reduced p-RIPK1 expression by immunohistochemistry staining and augmented Caspase 6 activity were also observed in the HTH-01-015–treated fatty livers (Figures 3B, C). In addition, as shown in Figures 3D–F, HTH-01-015 decreased the expression of IL-1β, IL-6, TNF-α, and TGF-β and the release of IL-1β and IL-18, compared to the HFD groups.
FIGURE 3.
NUAK1 inhibition suppressed Caspase 6 activity and induced TAK1 activation and cell death in mouse MASH models. Four-week-old mice were fed with HFD for 24 weeks, accompanied by the treatment of NUAK1 inhibitor HTH-01-015. (A) Western blot analysis and relative density ratio of active Caspase 6, TAK1, RIPK1, p-RIPK1, GSDMD-N, cCaspase 1, and NLRP3. (B) Immunohistochemistry staining of p-RIPK1 in fatty livers (scale bars, 50 μm). (C) Caspase 6 activity in the liver lysate. (D) qRT-PCR analysis of IL-1β, IL-6, TNF-α, and TGF-β in fatty livers. (E) ELISA analysis of serum IL-1β levels. (F) ELISA analysis of serum IL-18 levels. ANOVA, Bonferroni multiple comparison test, n = 4–6 per group. All data represent the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: HFD, high-fat diet; qRT-PCR, quantitative real time-PCR.
Disruption of Caspase 6 activates the TAK1 and inhibits RIPK1 and pyroptosis activation in fatty livers
To further confirm the causal relationship between NUAK1 and Caspase 6 in the development of MASH, we evaluated whether deletion of Caspase 6 could regulate the RIPK1/TAK1 pathway and pyroptosis in mouse MASH model. Unlike WT livers, Caspase 6 KO livers lacked Caspase 6 expression (Figure 4A). Yet, under normal chow conditions, there was no apparent liver dysfunction in Caspase 6 KO mice. Compared with the WT group, disruption of Caspase 6 significantly promoted hepatic steatosis and fibrosis as evidenced by decreased histological damage, lower liver weight, reduced hepatic triglyceride content, and mRNA expression coding for Fas, Srebp-1c, Col1a1, Col3a1, and Pdgfa (Figures 4B–F). Notably, a significant reduction in both Ly6G-positive cells and CD11b-positive cells was observed in the Caspase 6 KO group (Figure 4G). Furthermore, compared to the WT controls, the Caspase 6 KO group showed lower mRNA levels of IL-1β, IL-6, TNF-α, and TGF-β in fatty livers. Interestingly, NUAK1 activation markedly increased the expression of RIPK1 and the essential mediators for pyroptosis (GSDMD-N and cleaved-Caspase 1) while reducing the TAK1 level in the mouse MASH model. However, Caspase 6 KO diminished RIPK1, p-RIPK1, and pyroptosis protein levels and enhanced TAK1 expression (Figure 4I). In line with these results, reduced levels of p-RIPK1 expression in Caspase 6 KO fatty livers were also identified through immunofluorescence staining (Figure 4J).
FIGURE 4.
Caspase 6 deficiency activates TAK1 and inhibits RIPK1 and pyroptosis in fatty livers. Four-week-old Caspase 6 KO mice were fed with HFD for 24 weeks. (A) Caspase 6 expression was detected by western blot assay in fatty livers. (B) Photomicrographs of H&E staining, Oil Red O staining, and Masson staining in liver sections (scale bars, 200, 100, and 100 μm). (C) Liver weight was evaluated. (D) Liver function was evaluated by serum ALT and AST levels (U/L). (E) Liver TG. (F) qRT-PCR analysis of Fas, Srebp-1c, Col1a-1, Col3a-1, and Pdgfa in fatty livers. (G) Immunohistochemistry staining of Ly6G+ neutrophils and immunofluorescence staining of CD11b in liver tissues (scale bar, 50 μm). (H) qRT-PCR analysis of IL-1β, IL-6, TNF-α, and TGF-β in fatty livers. (I) Western blot analysis and relative density ratio of TAK1, RIPK1, p-RIPK1, GSDMD-N, cCaspase 1, and NLRP3 in liver tissues. (J) Immunofluorescence staining of p-RIPK1 in fatty livers (scale bars, 50 μm). ANOVA, Bonferroni multiple comparison test, n = 4–6 per group. All data represent the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; H&E, hematoxylin and eosin; HFD, high-fat diet; KO, knockout; qRT-PCR, quantitative real time-PCR; TG, triglyceride.
The NUAK1/Caspase 6 signaling is required for the crosstalk between TAK1 and RIPK1 in MASH-induced hepatocytes
To explore the functional role of Caspase 6 in regulating RIPK1-TAK1 interaction, hepatocytes were transfected with CRISPR-mediated Caspase 6 activation plasmids (p-CRISPR-CAS6 Act) and then incubated with HTH-01-015. We found that p-CRISPR-CAS6 Act increased lipid accumulation (Figure 5A) and lactate dehydrogenase (LDH) release (Figure 5B). Moreover, increased mRNA coding for IL-1β, IL-6, TNF-α, and TGF-β (Figure 5C) and IL-1β secretion (Figure 5D) were observed compared with control vector–treated cells. Caspase 6 activation inhibited TAK1 but enhanced RIPK1, p-RIPK1, GSDMD-N, cleaved-Caspase 1, and NLRP3. To determine whether TAK1-RIPK1 colocalization is regulated by the NUAK1/Caspase 6 axis, double immunofluorescence staining was conducted and showed the reduced colocalization of RIPK1-TAK1 in Caspase 6–activated hepatocytes (Figure 5G). Coimmunoprecipitation assays revealed that Caspase 6 activation could restrain the binding of TAK1 to RIPK1 (Figure 5H).
FIGURE 5.
The NUAK1/Caspase 6 signaling is required for the crosstalk between TAK1 and RIPK1 in MASH-induced hepatocytes. Mouse hepatocytes were treated with HTH-01-015 and CRISPR-Caspase 6 activation vectors, followed by PA stimulation. (A) Oil Red O staining of PA-stimulated hepatocytes (scale bar, 50 and 20 μm). (B) LDH releasing from hepatocytes. (C) qRT-PCR analysis of IL-1β, IL-6, TNF-α, and TGF-β in hepatocytes. (D) ELISA analysis of IL-1β levels in culture medium from PA-stimulated hepatocytes. (E) Caspase 6 activity in hepatocytes. (F) Western blot analysis and relative density ratio of aCaspase 6, TAK1, RIPK1, p-RIPK1, GSDMD-N, cCaspase 1, and NLRP3 in PA-stimulated hepatocytes. (G) Immunofluorescence staining of TAK1 (red) and RIPK1 (green) in hepatocytes after treatment with HTH-01-015. DAPI was used to visualize nuclei (blue) (scale bars, 20 and 10 μm). (H) Immunoprecipitation analysis of TAK1 and RIPK1 in hepatocytes induced by PA. ANOVA, Bonferroni multiple comparison test, n=4–6 per group. All data represent the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: LDH, lactate dehydrogenase; PA, palmitic acid; qRT-PCR, quantitative real time-PCR.
TAK1 acts as a key mediator to degrade RIPK1 and attenuate pyroptosis and inflammatory response in hepatocytes
To dissect the mechanistic role of TAK1 in the modulation of RIPK1 degradation in MASH-mediated inflammatory response, CRISPR plasmids were applied to regulate the expression of TAK1 in hepatocytes. Indeed, compared with the control group, TAK1 deletion caused induction of lipid accumulation (Figure 6A) and LDH release (Figure 6B) in PA-induced hepatocytes, accompanied by the enhanced secretion of IL-1β (Figure 6C). Moreover, RIPK1, p-RIPK1, GSDMD-N, cleaved-Caspase 1, and NLRP3 protein levels were increased in hepatocytes pretreated with CRISPR-TAK1 KO vectors (Figure 6D). Further confirmation of this was obtained through immunofluorescence staining, which demonstrated p-RIPK1 increase (Figure 6E) in hepatocytes after transfected with the CRISPR-TAK1 KO plasmids. Consistent with the effects of TAK1 deficiency in the hepatocytes, PA-stimulated steatosis was controlled in TAK1 activation hepatocytes, as indicated by lipid accumulation suppression (Figure 6F) and less secretion of LDH (Figure 6G) and IL-1β (Figure 6H). The expression of pyroptosis-related proteins was downregulated at the same time (Figure 6I). Immunofluorescence staining showed that the induction of TAK1 inhibited p-RIPK1 (Figure 6J) after transfection with CRISPR-TAK1 activation vector. Taken together, our data indicate that TAK1 is a key mediator necessary for the degradation of RIPK1 and controlling pyroptosis and inflammatory response against MASH.
FIGURE 6.
TAK1 acts as a key mediator to degrade RIPK1 and attenuate pyroptosis and inflammatory response in hepatocytes. Mouse hepatocytes were transfected with TAK1 KO or activation vector, then treated with HTH-01-015, followed by PA stimulation. (A) Oil Red O staining of PA-stimulated hepatocytes (scale bars, 10 and 5 μm. (B) LDH releasing from hepatocytes. (C) ELISA analysis of IL-1β levels in culture medium from PA-stimulated hepatocytes. (D) Western blot analysis and relative density ratio of TAK1, RIPK1, p-RIPK1, GSDMD-N, cCaspase 1, and NLRP3 in PA-stimulated hepatocytes. (E) Immunofluorescence staining of p-RIPK1 (green) in hepatocytes (scale bars, 20 and 10 μm). (F) Oil Red O staining of PA-stimulated hepatocytes (scale bars, 10 and 5 μm). (G) LDH releasing from hepatocytes. (H) ELISA analysis of IL-1β levels in culture medium from PA-stimulated hepatocytes. (I) Western blot analysis and relative density ratio of TAK1, RIPK1, p-RIPK1, GSDMD-N, cCaspase 1, and NLRP3 in PA-stimulated hepatocytes. (J) Immunofluorescence staining of p-RIPK1 (red) in hepatocytes (scale bars, 20 and 10 μm). ANOVA, Bonferroni multiple comparison test, n=4–6 per group. All data represent the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: KO, knockout, LDH, lactate dehydrogenase; PA, palmitic acid.
RIPK1 is essential for inhibiting pyroptosis activation and inflammation in response to MASH
As the NUAK1-Caspase 6 signaling can regulate both TAK1/RIPK1 crosstalk in the modulation of pyroptosis, we then asked whether RIPK1 is essential for inhibiting pyroptosis activation and inflammation in response to MASH. Consistent with the in vivo results, hepatocytes treated with PA and HTH-01-015 exhibited lower lipid accumulation and pyroptosis activation. RIPK1-activation vector administration did aggravate lipid accumulation and LDH release in hepatocytes (Figures 7A, B). The mRNA levels (IL-1β, IL-6, TNF-α, and TGF-β) and IL-1β secretion were increased (Figures 7C, D). Pyroptosis and NLRP3 inflammasome accumulation in hepatocytes were accelerated at the same time (Figures 7E, F). Hepatocyte GSDMD activation enhanced mRNA levels of inflammatory cytokines (IL-1β, IL-6, and TNF-α) (Figure 7G). In contrast, CRISPR-RIPK1 KO reduced lipid accumulation (Figure 7H), LDH release (Figure 7I), proinflammatory factor production (Figure 7J), pyroptosis activation, and NLRP3-ASC colocalization (Figures 7K, L) compared to the control vector–treated groups. In addition, GSDMD inhibition reduced the production of inflammatory cytokines (IL-1β, IL-6T NF-α, and TGF-β) (Figure 7M). These results indicate that RIPK1 is essential for suppressing pyroptosis activation and subsequent inflammation in response to MASH.
FIGURE 7.
RIPK1 is essential for regulating pyroptosis activation and inflammation in response to MASH. Mouse hepatocytes were transfected with RIPK1 KO, GSDMD KO, RIPK1 Activation, and GSDMD Activation vectors, then treated with HTH-01-015, followed by PA stimulation. (A) Oil Red O staining of PA-stimulated hepatocytes (scale bars, 50 and 20 μm). (B) LDH releasing from hepatocytes. (C) qRT-PCR analysis of IL-1β, IL-6, TNF-α, and TGF-β in hepatocytes. (D) ELISA analysis of IL-1β levels in culture medium from PA-stimulated hepatocytes. (E) Western blot analysis and relative density ratio of RIPK1, GSDMD-N, cCaspase 1, and NLRP3 in PA-stimulated hepatocytes. (F) Immunofluorescence staining of NLRP3 (red) and ASC (green) in hepatocytes (scale bars, 20 and 10 μm). (G) qRT-PCR analysis of IL-1β, IL-6, and TNF-α in hepatocytes. (H) Oil Red O staining of PA-stimulated hepatocytes (scale bars, 20 and 10 μm). (I) LDH releasing from hepatocytes. (J) qRT-PCR analysis of IL-1β, IL-6, TNF-α, and TGF-β in hepatocytes. (K) Western blot analysis and relative density ratio of RIPK1, GSDMD-N, cCaspase 1, and NLRP3 in PA-stimulated hepatocytes. (L) Immunofluorescence staining of NLRP3 (red) and ASC (green) in hepatocytes (scale bars, 20 and 10 μm). (M) qRT-PCR analysis of IL-1β, IL-6, TNF-α, and TGF-β in hepatocytes. ANOVA, Bonferroni multiple comparison test, n = 4–6 per group. All data represent the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations: KO, knockout, LDH, lactate dehydrogenase; PA, palmitic acid; qRT-PCR, quantitative real time-PCR.
DISCUSSION
In this study, we elucidate the mechanistic role of NUAK1 in the modulation of Caspase 6–driven pyroptosis in MASH-induced inflammatory response. We demonstrate that (1) NUAK1 activates pyroptosis in MASH-triggered liver inflammation; (2) NUAK1-Caspase 6 inhibition enhances TAK1 binding to RIPK1; (3) TAK1 is crucial in the regulation of RIPK1 degradation, which in turn alleviated pyroptosis; and (4) RIPK1 is essential for inducing pyroptosis activation and inflammation in response to MASH. Our results highlight the importance of the NUAK1-Caspase 6 signaling as a key regulator in controlling hepatic pyroptosis during MASH development.
NUAK1 was originally identified as a molecular scaffold in controlling cellular metabolism and responding to several stress modalities, such as oxidative and inflammatory stress.18,19 However, current knowledge on the regulatory role of NUAK1 underlying the development of MASH remains unclear. Using human, mouse, and cell systems, we proved that NUAK1 is a pivotal driver in MASH-induced liver steatosis and fibrosis. Indeed, NUAK1 expression was upregulated in patients with MASH. Moreover, pharmacological inhibition of NUAK1 suppressed pyroptosis and inflammation in mice fed HFD or methionine-choline–deficient diet, as evidenced by decreased levels of LDH, liver enzymes, and histopathological damage. The production of proinflammatory factors and pyroptosis-related protein levels were also downregulated at the same time. These results implicate that NUAK1 may present as a potential therapeutic target against MASH progression.
Caspase 6 was discovered decades ago and originally served as an executioner caspase.20 However, many studies have suggested that Caspase 6 is not essential in initiating apoptosis.21,22 Through the modification of cell cycle initiation, Caspase 6 controlled the terminal differentiation of B cells into plasma cells.23 Furthermore, Caspase 6 also undertakes vital functions in macrophage activation driven by polymorphonuclear neutrophils.24 Recent evidence has proved that the AMPK-Caspase 6 axis functioned in MASH by regulating both inflammation and energy metabolism.25 To date, how the Caspase 6 activation regulates pyroptosis remains unclear. In the murine model of influenza A virus infection, Caspase 6 interfaced with RIPK3, augmenting the interaction between ZBP1 and RIPK3, which augmented the activation of pyroptosis.14 Our results showed that Caspase 6 deficiency mitigated MASH-induced liver injury. Disruption of Caspase 6 suppressed GSDMD-N and cCaspase 1, the essential mediators for pyroptosis activation. In the progression of fibrogenesis after organ injury, NUAK1 promoted TGF-β signaling, which consequently resulted in organ scaring.18 In addition, TGF-β mediated caspase activation is essential during craniofacial development or allergic inflammation.26,27 In our mouse MASH model, NUAK1 inhibition suppressed the activity of Caspase 6 accompanied by lower TGF-β levels. It is reasonable to speculate that TGF-β is likely to serve as a bridge between NUAK1 signaling and Caspase 6 activation.
As our current studies suggest that NUAK1-Caspase 6 signaling is involved in the regulation of pyroptosis in MASH-induced inflammation, the question arises as to what molecular mechanisms may endow the NUAK1-Caspase 6 axis with its capability to regulate pyroptosis in MASH-mediated immune regulation. In this study, we found that the expression of TAK1 in hepatocytes was markedly decreased, accompanied by RIPK1 upregulation in MASH model, while NUAK1 deficiency promoted TAK1 levels but suppressed RIPK1 expression. Additional evidence was provided by our in vitro study, revealing that TAK1 interacted with RIPK1 through direct binding in hepatocytes in response to HTH-01-015 treatment. Furthermore, Caspase 6 activation hindered hepatocytes TAK1-RIPK1 interaction even with NUAK1 inhibitor before treatment. Our current results indicated that NUAK1-mediated Caspase 6 activation impeded TAK1-RIPK1 interaction in MASH-induced inflammation regulation.
Another important finding of our study is that TAK1 is key for controlling RIPK1 degradation in the inflammatory action of MASH model. TAK1 serves as a significant modulator of inflammation-related pathways.28,29 TAK1 deficiency in cholangiocytes underwent spontaneous necroptosis induced primarily by TNFR1 and led to biliary damage in mice.30 Moreover, pathogen blockade of TAK1 triggered caspase-8–dependent cleavage of gasdermin D and cell death.28 RIPK1 plays critical roles in facilitating diverse forms of cell death, including RIPK1-dependent apoptosis, pyroptosis, and necroptosis.31,32,33 TAK1 deficiency drived RIPK1-dependent inflammation in hepatocytes, subsequently fostering the progression of liver fibrosis.34 Consistent with previous studies, disruption of hepatocytes TAK1 augmented RIPK1, GSDMD-N, and cCaspase 1 expression, whereas TAK1 activation suppressed RIPK1 and pyroptosis-related proteins. Taken together, these findings uncover an unprecedented role for TAK1-RIPK1 interaction in controlling pyroptosis in MASH-mediated inflammatory regulation.
Our study delineates the putative function by which NUAK1-mediated Caspase 6 activation may lead to reduced hepatocytes TAK1 but increased RIPK1. Strikingly, hepatocytes TAK1 deficiency abolished its interaction with RIPK1, which in turn reduced the degradation of RIPK1. Finally, it enhanced hepatocyte pyroptosis and MASH-driven liver inflammation.
In summary, we identify the functional role of NUAK1-Caspase 6 signaling in modulating TAK1-RIPK1 interaction during the development of MASH. Our results illustrate that NUAK1-Caspase 6 signaling controls inflammatory regulation through a direct interaction between TAK1 and RIPK1, which impedes the degradation of RIPK1 in MASH. These insights offer a prospective therapeutic target for fatty liver inflammatory injury.
Supplementary Material
DATA AVAILABILITY STATEMENT
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
AUTHOR CONTRIBUTIONS
All authors contributed to the study conception and design of the study. Material preparation and data collection were conducted by Mingwei Sheng, Shuhan Huo, and Yiqi Weng. Data analysis was performed by Mingwei Sheng and Lili Jia. The first draft of the manuscript was written by Yuanbang Lin and Mingwei Sheng. The manuscript was revised by Wenli Yu and Weihua Liu. All authors commented on the previous versions of the manuscript. All authors have read and approved the final manuscript.
FUNDING INFORMATION
This work was supported by the National Natural Science Foundation of China, grant/award numbers: 82370670, 81700569, and 81700659; the Natural Science Foundation of Tianjin, grant/award number: 21JCQNJC00080; the Science and Technology Foundation of Tianjin Health Bureau, grant/award number: TJWJ2021QN014; and the Tianjin Key Medical Discipline (Specialty) Construction Project, grant/award number: TJYXZDXK-045A.
CONFLICTS OF INTEREST
The authors have no conflicts to report.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Academic Committee of Tianjin Medical University General Hospital (ZYY-DWFL-IRB- 002(F)-01).
Footnotes
Abbreviations: AMPK, 5ʹadenosine monophosphate–activated protein kinase; HFD, high-fat diet; KO, knockout; LDH, lactate dehydrogenase; MYPT1, myosin phosphate–targeting subunit 1; PA, palmitic acid.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.hepcommjournal.com.
Contributor Information
Mingwei Sheng, Email: shengmingwei@tmu.edu.cn.
Shuhan Huo, Email: hsh03272022@163.com.
Lili Jia, Email: jialili19880801@163.com.
Yiqi Weng, Email: 465592351@qq.com.
Weihua Liu, Email: liuwh80@163.com.
Yuanbang Lin, Email: linyuanbang@tmu.edu.cn.
Wenli Yu, Email: ywl-tfch@nankai.edu.cn.
REFERENCES
- 1. The Lancet G;Hepatology . Headway and hurdles in non-alcoholic fatty liver disease. Lancet Gastroenterol Hepatol. 2020;5:93. [DOI] [PubMed] [Google Scholar]
- 2. Stefan N, Häring H-U, Cusi K. Non-alcoholic fatty liver disease: Causes, diagnosis, cardiometabolic consequences, and treatment strategies. Lancet Diabetes Endocrinol. 2019;7:313–324. [DOI] [PubMed] [Google Scholar]
- 3. Sheka AC, Adeyi O, Thompson J, Hameed B, Crawford PA, Ikramuddin S. Nonalcoholic steatohepatitis: A review. JAMA. 2020;323:1175–1183. [DOI] [PubMed] [Google Scholar]
- 4. Diehl AM, Day C. Cause, pathogenesis, and treatment of nonalcoholic steatohepatitis. N Engl J Med. 2017;377:2063–2072. [DOI] [PubMed] [Google Scholar]
- 5. Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of MASLD development and therapeutic strategies. Nat Med. 2018;24:908–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Port J, Muthalagu N, Raja M, Ceteci F, Monteverde T, Kruspig B, et al. Colorectal tumors require NUAK1 for protection from oxidative stress. Cancer Discov. 2018;8:632–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oh S, Lee J, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Nuak1 phosphorylates histone H3 threonine 11 in low glucose stress by the cooperation of AMPK and CK2 signaling. Elife. 2020;9:e64588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu L, Ulbrich J, Muller J, Wustefeld T, Aeberhard L, Kress TR, et al. Deregulated MYC expression induces dependence upon AMPK-related kinase 5. Nature. 2012;483:608–612. [DOI] [PubMed] [Google Scholar]
- 9. Lasagna-Reeves CA, de Haro M, Hao S, Park J, Rousseaux MW, Al-Ramahi I, et al. Reduction of Nuak1 decreases Tau and reverses phenotypes in a tauopathy mouse model. Neuron. 2016;92:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Courchet V, Roberts AJ, Meyer-Dilhet G, Del Carmine P, Lewis TL, Jr, Polleux F, et al. Haploinsufficiency of autism spectrum disorder candidate gene NUAK1 impairs cortical development and behavior in mice. Nat Commun. 2018;9:4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van Opdenbosch N, Lamkanfi M. Caspases in cell death, inflammation, and disease. Immunity. 2019;50:1352–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang XJ, Cao Q, Zhang Y, Su XD. Activation and regulation of caspase-6 and its role in neurodegenerative diseases. Annu Rev Pharmacol Toxicol. 2015;55:553–572. [DOI] [PubMed] [Google Scholar]
- 13. Skotte NH, Pouladi MA, Ehrnhoefer DE, Huynh K, Qiu X, Nielsen SMB, et al. Compromised IGF signaling causes caspase-6 activation in Huntington disease. Exp Neurol. 2020;332:113396. [DOI] [PubMed] [Google Scholar]
- 14. Zheng M, Karki R, Vogel P, Kanneganti TD. Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense. Cell. 2020;181:674–687 e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J, Chen C, Zhang W, Bi J, Yang G, Li E. Salsalate reverses metabolic disorders in a mouse model of non-alcoholic fatty liver disease through AMPK activation and caspase-6 activity inhibition. Basic Clin Pharmacol Toxicol. 2021;128:394–409. [DOI] [PubMed] [Google Scholar]
- 16. van Raam BJ, Ehrnhoefer DE, Hayden MR, Salvesen GS. Intrinsic cleavage of receptor-interacting protein kinase-1 by caspase-6. Cell Death Differ. 2013;20:86–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li C, Sheng M, Lin Y, Xu D, Tian Y, Zhan Y, et al. Functional crosstalk between myeloid Foxo1-beta-catenin axis and Hedgehog/Gli1 signaling in oxidative stress response. Cell Death Differ. 2021;28:1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang T, He X, Caldwell L, Goru SK, Ulloa Severino L, Tolosa MF, et al. NUAK1 promotes organ fibrosis via YAP and TGF-beta/SMAD signaling. Sci Transl Med. 2022;14:eaaz4028. [DOI] [PubMed] [Google Scholar]
- 19. Huang CY, Kuo WW, Yeh YL, Ho TJ, Lin JY, Lin DY, et al. ANG II promotes IGF-IIR expression and cardiomyocyte apoptosis by inhibiting HSF1 via JNK activation and SIRT1 degradation. Cell Death Differ. 2014;21:1262–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gray DC, Mahrus S, Wells JA. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell. 2010;142:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Berta T, Park CK, Xu ZZ, Xie RG, Liu T, Lu N, et al. Extracellular caspase-6 drives murine inflammatory pain via microglial TNF-alpha secretion. J Clin Invest. 2014;124:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ehrnhoefer DE, Skotte NH, Reinshagen J, Qiu X, Windshugel B, Jaishankar P, et al. Activation of caspase-6 is promoted by a mutant Huntingtin fragment and blocked by an allosteric inhibitor compound. Cell Chem Biol. 2019;26:1295–1305.e1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Watanabe C, Shu GL, Zheng TS, Flavell RA, Clark EA. Caspase 6 regulates B cell activation and differentiation into plasma cells. J Immunol. 2008;181:6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kobayashi H, Nolan A, Naveed B, Hoshino Y, Segal LN, Fujita Y, et al. Neutrophils activate alveolar macrophages by producing caspase-6-mediated cleavage of IL-1 receptor-associated kinase-M. J Immunol. 2011;186:403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao P, Sun X, Chaggan C, Liao Z, In Wong K, He F, et al. An AMPK-caspase-6 axis controls liver damage in nonalcoholic steatohepatitis. Science. 2020;367:652–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang X, Yokota T, Iwata J, Chai Y. Tgf-beta-mediated FasL-Fas-Caspase pathway is crucial during palatogenesis. J Dent Res. 2011;90:981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tirpude NV, Sharma A, Joshi R, Kumari M, Acharya V. Vitex negundo Linn. extract alleviates inflammatory aggravation and lung injury by modulating AMPK/PI3K/Akt/p38-NF-kappaB and TGF-beta/Smad/Bcl2/caspase/LC3 cascade and macrophages activation in murine model of OVA-LPS induced allergic asthma. J Ethnopharmacol. 2021;271:113894. [DOI] [PubMed] [Google Scholar]
- 28. Orning P, Weng D, Starheim K, Ratner D, Best Z, Lee B, et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science. 2018;362:1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Malireddi RKS, Gurung P, Kesavardhana S, Samir P, Burton A, Mummareddy H, et al. Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity-independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J Exp Med. 2020;217:jem.20191644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krishna-Subramanian S, Singer S, Armaka M, Banales JM, Holzer K, Schirmacher P, et al. RIPK1 and death receptor signaling drive biliary damage and early liver tumorigenesis in mice with chronic hepatobiliary injury. Cell Death Differ. 2019;26:2710–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lalaoui N, Boyden SE, Oda H, Wood GM, Stone DL, Chau D, et al. Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature. 2020;577:103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Newton K, Wickliffe KE, Dugger DL, Maltzman A, Roose-Girma M, Dohse M, et al. Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature. 2019;574:428–431. [DOI] [PubMed] [Google Scholar]
- 33. Zheng Z, Deng W, Bai Y, Miao R, Mei S, Zhang Z, et al. The lysosomal Rag-Ragulator complex licenses RIPK1– and caspase-8–mediated pyroptosis by Yersinia. Science. 2021;372:eabg0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tan S, Zhao J, Sun Z, Cao S, Niu K, Zhong Y, et al. Hepatocyte-specific TAK1 deficiency drives RIPK1 kinase-dependent inflammation to promote liver fibrosis and hepatocellular carcinoma. Proc Natl Acad Sci USA. 2020;117:14231–14242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.