Abstract
The liver is a vital organ that continuously adapts to a wide and dynamic diversity of self-antigens and xenobiotics. This involves the active contribution of immune cells, particularly by the liver-resident macrophages, the Kupffer cells (KCs), which exert a variety of central functions in liver homeostasis and disease. As such, KCs interact with their microenvironment to shape the hepatic cellular landscape, control gut-derived signal integration, and modulate metabolism. On injury, the rapid recruitment of bone marrow monocyte-derived macrophages alters this status quo and, when unrestrained, drastically compromises liver homeostasis, immune surveillance, and tissue organization. Several factors determine the functional roles of liver macrophages in these processes, such as their ontogeny, activation/polarization profile and, importantly, spatial distribution within the liver. Loss of tolerance and adaptability of the hepatic immune environment may result in persistent inflammation, hepatic fibrosis, cirrhosis, and a tumorigenic niche promoting liver cancer. In this review, we aim at providing the most recent breakthroughs in our understanding of liver macrophage biology, particularly their diversity and adaptability in the hepatic spatiotemporal context, as well as on potential therapeutic interventions that may hold the key to tackling remaining clinical challenges of varying etiologies in hepatology.
INTRODUCTION
The liver is the largest solid organ in the human body. The liver is uniquely located within the vascular system, receiving oxygenated blood supply from the hepatic artery and intestine-supplied nutrient-rich blood from the portal vein. As such, it exerts crucial functions in the metabolism and systemic blood detoxification while serving as the gateway for gut-derived signals entering the liver through the portal vein. Gut-derived signals include nutrients, microbiota-related products, food-borne antigens, and toxic xenobiotics (eg, drugs and alcohol).1 A substantial part of the blood filtering functions of the liver are carried out by highly specialized tissue-resident macrophages, also known as Kupffer cells (KCs). KCs represent the largest reservoir of tissue-resident macrophages in the human body. They are located within the fenestrated blood sinusoids and remain relatively immobile during homeostasis, sensing and catching blood particles, pathogen-associated molecular patterns, and damage-associated molecular patterns.2 Their cytoplasmic expansions reach out through the sinusoids, enabling them to establish direct membrane contacts with hepatocytes. During homeostasis, KCs are predominantly favoring the liver immune tolerance (reviewed in the work by Musrati et al 3,4). Despite the KC predominance within the parenchyma, it is increasingly recognized that the liver contains at least two additional macrophage populations during homeostasis: bile duct–associated and subcapsular macrophages with distinct ontogeny and functions.5,6 Yet, alternative cellular origins of liver macrophages remain to be demonstrated, particularly in view of potential interspecies peculiarities. Importantly and due to their potent phagocytic capacities, KCs represent the primary innate immune cell responders during liver injury.7 The liver KC pool may be replenished through local proliferation of KCs, a mechanism shown to be IL-6 dependent in mouse partial hepatectomy.8 Yet, their first-line roles also mean they are primarily affected by severe or chronic adverse events. Consequently, KC depletion has been reported in a wide range of human diseases and animal models.4 KCs themselves are not all alike. Indeed, recent reports pointed out an inherent heterogeneity through at least two KC compartments of the same embryonic origin, and more subtypes are expected to be identified in the near future. As such, KC2 was identified in mice as a minor population of CD206hiESAM+ KCs, expressing LSEC-associated genes Mrc1, Esam, and Lyve1, and a gene signature suggesting key roles in lipid metabolism and lipid-associated oxidative stress through CD36 as evidenced by siRNA delivery to liver KCs.9 Furthermore, the same CD206hiESAM+ KC2 population was evidenced to be required for an effective T-CD8+ response through IL-2–mediated antigen cross-presentation in murine livers.10 In humans, KCs are less well characterized, and no single marker can unequivocally distinguish embryonically derived from monocyte-derived liver macrophages. However, human embryonic KCs are (mostly) positive for CD49a+, T-cell/transmembrane immunoglobulin and mucin domain containing 4+, and V-set immunoglobulin-domain-containing 4+ (a macrophage complement receptor required for phagocytosis of circulating pathogens+) and less prompt to respond to lipopolysaccharides exposure by increasing inflammatory cytokine release as compared to their monocyte-derived counterpart.11
Intriguingly, when the KC self-maintenance does not suffice, monocyte-derived macrophages (MoMFs) possess the ability to replenish the liver macrophage pool and become KC-like liver macrophages as demonstrated in mouse studies and findings that remain to be confirmed in human settings.12,13,14 Yet, it has been demonstrated that those KC-like macrophages differ from KC, in particular by inducing a stronger proinflammatory response on liver rechallenge and by altering the liver free fatty acid metabolism by limiting lipid toxicity through enhanced triglyceride storage in mice.15,16 Interestingly, C-C chemokine receptor (CCR)2-blockade prevented this shift in the overall liver macrophage landscape and could prevent the establishment of liver macrophages prone to proinflammatory responses in rodent models of liver fibrosis.17,18 Furthermore, MoMFs were also demonstrated to be key contributors to the immune cell influx subsequent to portal area tissue injury, as demonstrated notably in acute and chronic bile duct injury models or chronic human liver diseases.19,20,21 The role of MoMFs, however, remains complex since they were both shown to increase proinflammatory and fibrogenic pathways and to be critical in the initiation of tissue repair mechanisms (reviewed in the work by Hassan et al22).
Peritoneal macrophages, which differentiate locally in the peritoneal cavity and express the transcription factor GATA binding protein 6 (GATA6) in mice, can represent another source of macrophages for the liver.23 Indeed, superficial and sterile thermal injury of the liver in mice induces a rapid, focal recruitment of GATA6+ peritoneal macrophages involved in inflammatory response moderation and tissue repair.24 The relevance of peritoneal macrophages in human liver diseases remains to be characterized.
As such, it is accepted that “liver macrophages” may exhibit a broad range of features in terms of cell of origin, functions, and morphological traits. In practice, macrophage-related immune responses are oftentimes found on liver biopsies and are mostly characterized using conventional staining and bright field microscopy imaging. For instance, localized aggregation of macrophages and other immune cells called a granuloma, evidences a local adaptive immune response activation and can be found in various conditions such as autoimmune or infectious diseases.25 Foamy macrophages, expressing classical macrophage markers such as F4/80 and notably surrounding injured hepatocytes in crown-like structures during steatotic liver diseases, were also evidenced to be associated with disease progression when expressing high levels of macrophage scavenger receptor 1.26,27 A variety of granulomas can be identified, including lipogranulomas associated with liver steatosis and nonspecific microgranulomas. Ceroid-laden macrophages, observed by periodic acid Schiff with diastase staining and suggestive of their phagocytic capacities, are observed in conditions marked by chronic fibrosis or steatosis in humans, rats, and mice, although their roles in disease progression remain to be clarified for their integration into the most recent definitions of functional macrophage subtypes.28,29,30 Indeed, recent technological advances further expanded our horizon on previously unexplored dimensions for liver macrophage characterization. This includes spatially resolved and timely defined cellular definitions, the analysis of multidimensional data sets of varying natures (eg, proteomic, transcriptomic, metabolic), as well as the integration of complex microenvironment parameters, including thorough characterization of the neighboring or distant cells with distinct cell or disease complexities. This literature review aims at embracing the broad diversity of “liver macrophage” in tissue homeostasis and upon varying diseases.
New dimensions for liver macrophage phenotyping
Until recently, most investigators would have considered “immune cell phenotyping” as assessing several (surface) markers, such as clusters of differentiation, defining cell subpopulations and allowing to assume cell functionalities. Nowadays, and mostly thanks to tremendous technological advances, it is possible to further dissect key functionalities in situ and predict cellular interactions at a much broader scale than ever. These advances allowed for a wide broadening of our understanding of macrophage biology, with direct implications for our toolbox to investigate liver disease driving mechanisms and consequently, to identify novel targetable cellular processes for disease management (Figure 1).
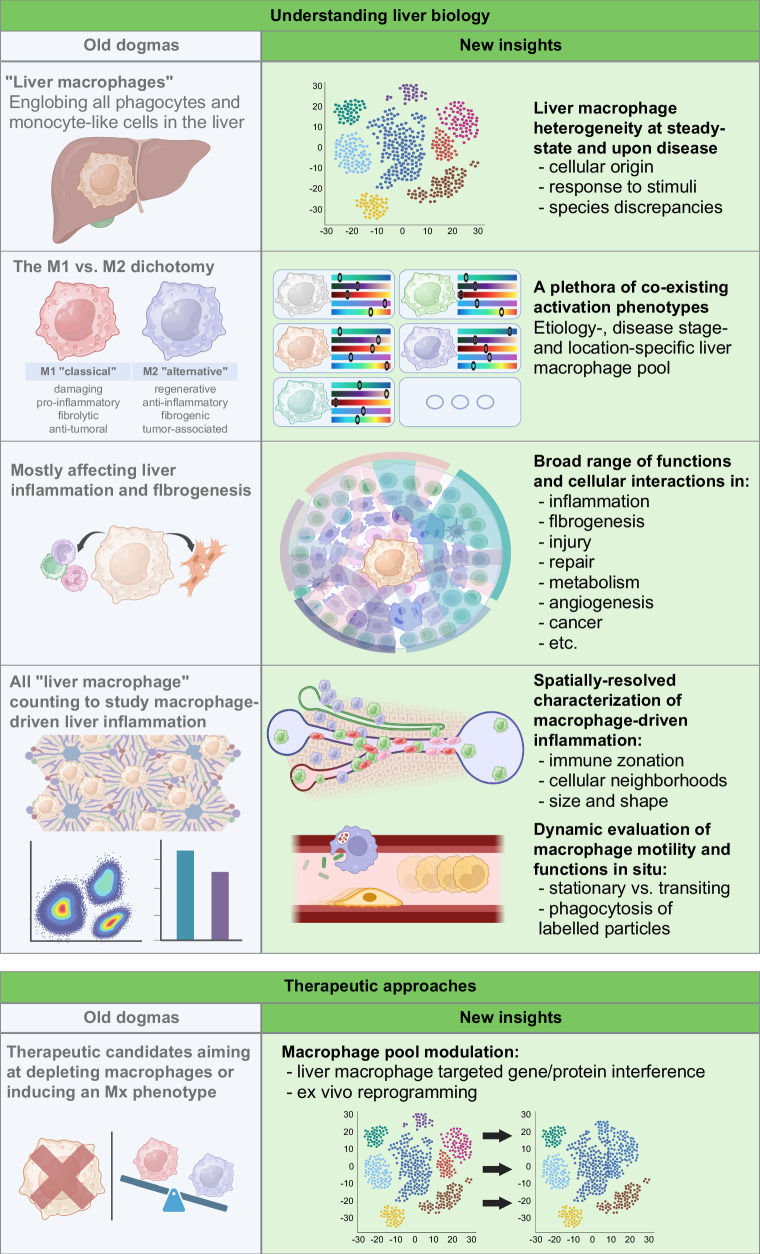
FIGURE 1.
Opposing old dogmas and new insights on liver macrophage biology and therapeutic approaches. Determining shifts in our understanding of macrophage biology led to a long-needed redefinition of the liver macrophage landscape. In particular, the field evolved from considering liver macrophages as a homogeneous pool of phagocytes to recognizing their extended diversity in origins and fate and functions in homeostasis and disease. This results in a paradigm shift from immunosuppressive to immunomodulating therapeutic approaches.
Location and timing as new phenotypic markers with relevance for disease progression
Innate immune cells are oftentimes defined as the first line of defense against pathogens or tissue injuries. This statement poses three conditions, though: that (A) the right cell (subset) would be present at (B) the appropriate location and (C) at the right time (eg, for communicating with other cells) (Figure 2). Consequently, we now see the rise of spatial biology exemplified by landmark studies deciphering not only protein or transcript expression but also cell location as a defining phenotypical marker. As such, macrophages located near the bile ducts recently received substantial attention. In conditions of cholestatic injury, these macrophages are oftentimes termed “bile duct–associated macrophages” to emphasize their localization in proximity to ductular cells, as observed in both human and mouse livers.21,31,32 In metabolic dysfunction–associated steatotic liver disease (MASLD, formerly termed NAFLD), macrophages also accumulate around ductular cells in relation to disease severity in patients.33 By a combination of spatial transcriptomics and proteomics, those macrophages were termed as lipid-associated macrophages due to their resemblance to macrophages accumulating throughout the liver during steatosis6 and due to their transcriptomic similarity to adipose tissue macrophages in obesity in both humans and mice.34 Other authors defined these phagocytes as scar-associated macrophages and notably identified them through their high expression in CD9 and triggering-receptor-expressed-on-myeloid-cells-2 (TREM2) in human disease.35 A consistent finding in all studies listed above is that bile duct–associated macrophages observed during disease likely derive from recruited, bone marrow–derived monocytes. Indeed, in mice bile duct–associated macrophages show low or no expression of CLEC4F and Timd4 and are oftentimes defined as Trem2 and secreted phosphoprotein 1 (Spp1) positive.21,33 Inline, embryonic-derived KCs progressively undergo cell death and are replaced by T-cell/transmembrane immunoglobulin and mucin domain containing 4 (Tim4)− recruited monocytes in a mouse MASLD model. Interestingly, those newly recruited monocytes adopt a transcriptome resembling that of KCs, a phenomenon attributed to diet-regulated transcription factors driving KC identity gene induction through liver-X-receptor signaling alterations.35 Similarly, we evidenced that the portal area-confined expansion of bile duct–associated macrophages was a hallmark shared between a wide variety of chronic human liver diseases ranging from MASLD to alcohol-associated hepatitis and primary sclerosing cholangitis (PSC). Intriguingly, similar findings were observed in a mouse model of PSC and acute bile duct injury.19,33 Importantly, we also evidenced drastic differences in the spatial localization of recruited monocytes between human MASLD and the diet-induced obesity-MASH mouse model. In the latter, monocyte-derived macrophages accumulate throughout the liver parenchyma and not solely in the portal area.33 More than serving as a potential means to evaluate disease stages, portal accumulation of CCR2+ monocyte-derived macrophages, in response to targeted biliary epithelial cell injury, has been shown to drive portal inflammation and fibrogenesis, as well as cholestasis in mice.19,36 These observations point toward the need for a better understanding of the roles of portal monocyte-driven inflammation in liver diseases, particularly those with a portal area component.
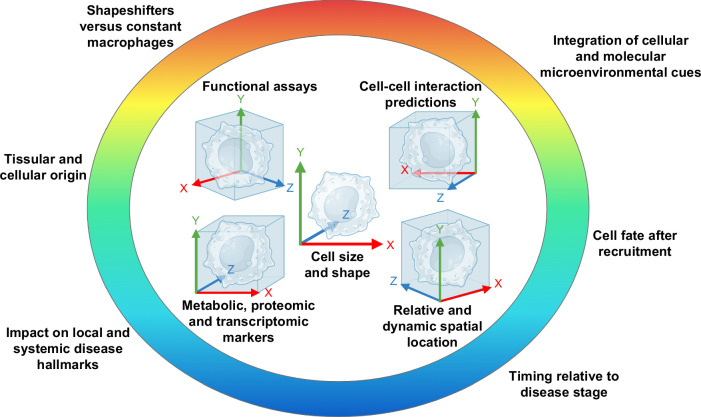
FIGURE 2.
Multidimensional dissection of liver macrophages. Recent insights into liver macrophage diversity drastically expanded the number of dimensions in which those highly versatile immune cells may be characterized. Beyond classical and alternative phenotype hallmarks, liver macrophages may now be defined in situ, in vitro, and even in silico through additional metadata such as their morphological peculiarities, functional assets, neighboring, and interactome, among other traits. Technological advances (hardware and software) growing at an unprecedented pace allow for this accelerating expansion of the liver macrophage universe.
The portal area receives particular attention in recent studies. As such, there has been evidence for an immune zonation, particularly for liver macrophages, that is interposed to hepatocyte zonation on a central to portal area axis, and this zonation pattern is changing after birth, at least in mice.37,38 For instance, the restricted zonation of Trem2- and osteopontin (Spp1)-expressing macrophages to portal areas seems to have consequences in chronic liver diseases marked with portal fibrosis and inflammation including PSC.21,32 Portal area-located MoMFs and not KCs exert the most prominent changes in particular increased Spp1 expression in mice, when bile duct ligation was combined with dextran sodium sulfate–induced colitis, as compared to bile duct ligation alone.21 KCs located closer to portal areas were evidenced to be central players in the control of gut-derived pathogens in mice.38 This zonation was absent in germ-free mice unless orally exposed to lipopolysaccharides and totally or partially absent in myeloid differentiation primary response protein (Myd88−/−) and toll-like receptor 4 (Tlr4)−/− mice, respectively. The authors demonstrated that liver sinusoidal endothelial cells were responsible for this phenotype. These findings further support the role of chronic microbiota-derived antigen sensing by liver cells in driving KC zonation. Beyond anatomic location, direct effects of the diet on macrophage zonation were evidenced in a murine dietary MASLD model.35 In a western diet and fructose feeding model of steatohepatitis (metabolic dysfunction-associated steatohepatitis [MASH]), Clec4f+ Trem2+ macrophages present in crown-like structures are positive for the proliferation marker Ki67 and exhibit an increased expression of CD207, a pathogen binding receptor, as compared to homeostatic conditions.39 More specifically, an analysis of co-varying genes within specific experimental groups indicated a possible coupling between zone-3 detoxification processes, regeneration, and inflammation. Consistent with earlier studies discussed above, marked reductions of Cd163 and macrophage receptor with collagenous structure (Marco) KC-associated gene expressions were further evidenced in this MASH mouse model.
Besides location, timing is increasingly regarded as a key parameter in liver macrophage studies as well. Advanced transcriptomic analyses support the hypothesis of a circadian rhythm-dependent human liver macrophage activation pattern.40 In particular, macrophages were shown to be either prone to reactivity or to tolerance during daytime or nighttime, respectively.41 This brings attention to the timing of experimentation or sampling as crucial parameters when studying macrophages, a phenomenon not only described for macrophage biology but also for liver regeneration after partial hepatectomy, with potential clinical implications.42 Moreover, and importantly, sex-specific and gender-specific differences have been identified in macrophage responses, particularly in metabolism-related liver diseases. For instance, male mice fed a high-fat diet had more pronounced macrophage accumulation in adipose tissue.43 Furthermore, female bone marrow–derived murine macrophages isolated from high-fat diet-fed mice exhibited lower migratory properties than male cells, a result independent of whether or not the mice were lean or obese and independent from estrogen. Macrophages from both sexes expressed similar basal levels of CCR2, yet macrophages generated from high-fat diet-fed male mice expressed higher levels of CCR2 and leptin receptors in response to free fatty acid.43 Contrastingly, multidrug resistance 2 gene (Mdr2)-deficient female mice show higher CCR2-positive macrophage recruitment, along with more pronounced liver disease phenotype and better therapeutic response to corticosterone than male counterparts.44,45 Many investigations remain necessary to fully decipher the implications of gender on metabolic and immune-mediated liver diseases.
Cell-cell interactions
Cell-cell interactions—involving not only immune cells but also parenchymal and nonparenchymal hepatic cell populations—are essential for driving liver disease progression or proper resolution from injury.46,47 The rise of single cell–resolved transcriptomic and proteomic analyses, together with bioinformatics tools evolving at a fast pace, allowed for the elaboration of predictive algorithms that may decipher complex cellular interactions. This is particularly illustrated by CellChat and CellPhoneDB identifying receptor-ligand pairs from donor and recipient cells.48,49 These approaches have a high hypothesis-generating potential, as exemplified by recent studies.50 For instance, it has been suggested using CellChat and additional tools that blocking the macrophage-T cell interactions might prevent the establishment of a tolerogenic environment in hepatocellular carcinoma, and that the chemokine—chemokine receptor axes such as C-X-C motif chemokine ligand (CXCL) 16-CXCR6, chemokine (C-C motif) ligand (CCL)6-CCR2, and CCL5-CCR5 would be crucial molecular partners for T cell-macrophage cross talk during Schistosomiasis japonicum–induced liver fibrosis in mice.51,52 These findings, combined with evidence of spatial neighboring and temporal synchronicity, open new perspectives for advanced immune microenvironment understanding. Recently, CellChat has been upgraded to allow for spatially resolved transcriptomic-based cellular interaction analysis.53 Hence, further combination of cellular cross talk and spatial data is to be expected in the very near future in the field of liver macrophages (or their respective subsets).
Yet, experimental confirmation for such predictions of cell-cell interactions is crucial. A recent study dissected the roles of MoMFs during concanavalin-A–induced acute injury resolution in mice.54 Accordingly, it was evidenced that two distinct populations of MoMFs were involved, one controlling tissue scarring by phagocytosing cellular debris and activating fibroblasts through platelet-derived growth factor subunit B, the other one by inducing SRY-box transcription factor 9 expression in the hepatocytes lining the necrotic areas through Jagged 1/Neurogenic locus notch homolog protein 2, thus preventing tissue injury spread.54
Liver cancer and metastases represent other conditions in which cell-cell interactions are crucial, and multiple intrahepatic and extrahepatic macrophage pools are involved. The liver represents a preferred metastatic site for colorectal cancer.55 Liver metastasis growth rates were reduced in mice with preserved circulating monocytes and KC population but depleted for peritoneal macrophages, either by clodronate-loaded liposome injection or in macrophage-specific Gata6 knockout animals.56 The proposed mechanism, as demonstrated by intravital imaging, is that peritoneal macrophages directly interact with metastatic cells in the liver, which leads to their upregulation of the T-cell–suppressing molecule programmed cell death ligand-1 but not antigen presentation–related proteins and their polarization toward an alternative, protumoral phenotype, altogether preventing T-CD8+ cytotoxic lymphocyte activation. In addition, locally released IL-10 induces programmed cell death ligand-1 on hepatic myeloid cells, thereby attenuating CD8-dependent cytotoxicity against metastatic lesions in mouse models.57
Following the spread of sequencing technologies, a number of algorithms have emerged to dissect bulk and single-cell transcriptomic data. As one example of such applications, tools predicting cell-cell interactions by comparing the expression levels of ligand-receptor pairs highlighted the detrimental prognostic value of the SPP1-angiopoietin-like protein 2 duet overexpression by macrophages and cancer-associated fibroblasts, respectively, in human colorectal cancer metastasis.58 Yet, deciphering the biological roles of SPP1 (gene encoding for osteopontin) has been more challenging, as reflected by recent and earlier studies. SPP1-expressing liver macrophages were suggested to interact with CD44-expressing hepatocellular carcinoma cells and to inhibit CD8+ T cell cytotoxicity in humans.59,60 Interestingly, cholangiocarcinoma cells were also reported as a potent source of SPP1.61 Furthermore, liver lipid-associated macrophages were shown to express higher levels of SPP1 during human pancreatic cancer metastases.62 Similarly, reduced MoMF recruitment and low SPP1 expression were indicative of bariatric surgery benefits over diet management for the prevention of MASLD-related inflammation, fibrosis, and hepatocellular carcinoma in patients.63 Inline, higher levels of SPP1 were indicative of a poor outcome in PSC, and SPP1 neutralizing antibodies were protective in Mdr2–/– mice and in the bile duct ligation murine model of obstructive cholestasis.21 Similarly, SPP1-expressing macrophages are enriched in MASLD.64,65 Besides, myeloid-specific ablation of Spp1 led to an increase in liver inflammation and liver crown-like structures in mice fed a high-fat, fructose, and cholesterol diet, while knock-in mice overexpressing Spp1 in MoMF or KCs had lower NAS scores and increased oncostatin-M expression in macrophages, pointing toward a protective role of macrophage osteopontin through oncostatin-M.64 Contrastingly, Spp1 knockout in mouse MoMFs was evidenced to increase their proinflammatory response to lipopolysaccharides and to increase liver inflammation in the bile duct ligation mouse model of obstructive cholestasis, thus highlighting the importance of disease-specific and model-specific contextualization.21 These contradictory phenotypes were proposed to be the result of choosing either a MoMF-targeted gene silencing approach versus antibody-based SPP1 neutralization, preventing its effects on all target cells, including fibrogenic and biliary epithelial cells.21,64,66 Overall, the emerging insights into the key role of SPP1 in liver inflammation simultaneously show us the strengths of advanced bioinformatics in identifying relevant cell-cell interaction pathways and remind us that extensive in vivo validations remain necessary to capture the complexity of biological mechanisms taking place in healthy and diseased contexts.
The broad range of macrophage interactions with other cells keeps on expanding. The combination of nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and necrosome activation trigger hepatic infiltration monocyte-derived macrophages with fibrogenic and carcinogenic phenotypes in mice, a process suggested to originate from CCL20 secretion by hepatocyte with sublethal necroptosis.67 Contrastingly, in vivo genome-wide CRISPR-Cas9 knockout screening led to the identification of the erythroid membrane-associated protein-galectin-9-dectin-2 axis as a driver of KC-mediated cancer cell phagocytosis, thereby limiting liver metastasis in mice.68 This study further showed that human patients with low erythroid membrane-associated protein expression on tumors had increased numbers of liver metastases. Another recent example is that “anti-inflammatory” MoMFs are prone to express higher levels of the potent fibrogenic cytokine, TGF-β1. These MoMFs also represent a major source of the angiogenic VEGF in murine livers.69 As such, “anti-inflammatory” MoMFs may be seen as key players in tissue repair mechanisms necessary for organ function recovery after injury. Yet, uncontrolled release of those factors may lead to fibrosis and to the arising of a tumorigenic environment (reviewed in the work by Medrano-Bosch et al70). Liver macrophage zonation, participating in the establishment of the sinusoidal KC niche, is dependent on hepatic stellate cell-derived signals lost during inflammation (eg, in MASH) and concomitant activation of stellate cells.39 Indeed and as opposed to the healthy condition, in murine MASH, distinct populations of MoMFs and KCs share similitudes in their transcriptional signatures and seem to converge toward a Cd207hi or a Trem2hi KC-like cell phenotype, although further evidence is required to establish a clear contribution from cells of both liver-resident and circulating macrophage origins.39
Neutrophils are, alongside MoMFs, first responders of the innate immune system involved in pathogen clearance. Whether neutrophil activation is detrimental or beneficial in liver diseases is as debatable as for MoMFs. Indeed, neutrophil-released reactive oxygen species polarize MoMFs toward an anti-inflammatory phenotype, and this is key in promoting mouse liver repair in acetaminophen-induced acute liver injury.71 Neutrophil can further polarize macrophages toward a repair phenotype by releasing micro-RNA 223, promoting injury resolution in mouse models.72
On a similar note, macrophages are key in sensing bacteria-derived metabolites, which in turn shape the macrophage responses. In particular, microbiome, mycobiome, and virome analyses highlighted that striking differences between wildlings and laboratory mice are highly relevant for the study of potent therapeutic candidate responses, as wildlings better reflected human immune responses and, noteworthy, better fitted with actual clinical trials than conventional laboratory animals.73
In vivo imaging visualizes macrophage functionality
In light of the intricate cellular players present within the liver microenvironment and the systemic circulation, and despite major progress in in vitro modeling, there are physiologically relevant reasons demanding in situ investigations, ideally in a living organism (ie, animal model). As such, recent advances in intravital time-lapse microscopy allow for the study of dynamic changes in liver sinusoidal branching together with the analysis of the liver macrophage pool in rodents.74 Meticulous studies using intravital imaging in mice identified the ability of recruited MoMFs to fuse and form syncytia or “giant” macrophage cells to restore the blood filtering functions lost during KC depletion.2 Subsequent to fibrosis induction by carbon tetrachloride injections in mice, the authors evidenced CD44-dependent recruitment of MoMFs, which were shown to form KC-like syncytia to restore the liver macrophage-dependent blood filtering capacities.2 The formation of such syncytia was demonstrated to be dependent on myeloid-specific CD36 expression by using conditional knockout animals. These data may, in part, explain the remarkable impairment of phagocytic function of macrophages in human liver cirrhosis and may be seen as a detrimental switch in the liver immunological milieu abilities to combat infections.75 Time-lapse intravital imaging further evidenced morphological changes in liver macrophages, such as a lower number of membrane protrusions after high-fat diet feeding in mice.76 Importantly, these changes in macrophages were associated with increased mortality after a second hit injury in steatotic mice, explained by a reduced ability of liver macrophages to capture blood-borne E. coli. The authors concluded on the importance of further characterizing the immunological milieu in steatotic livers at different stages of MASLD. Indeed, while TLR4-mediated macrophage monitoring of circulating pathogen-associated molecular patterns is crucial for tolerogenesis, macrophage over-activation leads to chronic inflammation and disease progression.76
Novel sophisticated ex vivo models to investigate liver macrophages
Ground-breaking technological advances have been emerging at a very fast pace in the recent decade. As noted in the sections above, these resulted in paradigm shifts and drastically expanded our understanding of macrophage biology. Consequently, they further highlighted our need for finely tuneable, high-end, physiologically relevant, and ethically robust models for tailored investigations.
Immune-competent in vitro assays for the study of liver macrophages
In vitro approaches present several advantages for molecular investigations and drug candidate testing. Yet, until recently, in vitro models of liver diseases were mostly hepatocyte-like cell-based and devoid of a competent immune cell compartment. This may be explained by the fact that primary liver immune cell cultivation presents a few challenges. Indeed, high donor-dependent heterogeneity of primary liver macrophages is further complicated by a biased enrichment in particular liver macrophage subpopulations by varying isolation or culture methods. As such, it was shown that primary liver macrophage plating (eg, for functional assays) leads to a depletion of low-adherent proinflammatory subpopulations.77 Hence, using the whole fraction of primary liver macrophages for phenotyping may be more representative of the in vivo situation.
As an alternative to primary cell isolation and culture, Groeger et al78 generated induced pluripotent stem cell–derived human hepatocytes and isogenic macrophages. This system allowed them to explore the functional cross talk between the two cell types involved in type 2 diabetes and MASLD. This approach notably identified macrophage-derived TNF-α and IL1β as the main drivers of insulin resistance in hepatocytes. In a more elaborate approach, the group of Takebe developed an induced pluripotent stem cell–based steatohepatitis model that includes hepatocytes, biliary cells, fibroblasts, and macrophages and is referred to as human liver organoid.79 Oleic acid exposure resulted in elevated inflammation and further induced KC-like macrophages to become more proinflammatory while increasing THP-1 monocytic cell migration to the human liver organoid. Yet, according to the authors’ comments, technical issues such as batch-to-batch variability remain to be tackled particularly during response to stimuli. An alternative approach that does not rely on the supply of extracellular matrix components and growth factors also generated a population of macrophages in an exclusively three-dimensional culture protocol.80 Key hepatocyte metabolic functions were evidenced, although the authors concede their model mostly resembles a developing instead of a mature organ. As another option, advanced perfusable “liver-on-a-chip” approaches may be advantageous for modeling immune-related mechanisms, as static hepatic cell populations can be exposed to interactions with circulating blood leukocytes as well as circulating blood-borne compounds and shear stress.81,82,83 The extent to which these models can predict in vivo responses, for instance, for early testing of therapeutic drugs for liver diseases, remains to be evaluated.
In silico models and advanced bioinformatics toolboxes for the study of liver macrophage biology
An exemplary study that aimed at generating and exploiting large data sets identified transcriptomic patterns across a multitude of tissue-specific myeloid cells, including dendritic cells and KCs.84 This mononuclear phagocyte network analysis particularly evidenced varying and conserved roles of myeloid cells in metabolism regulation across the organism. As such, macrophages located in nervous tissues were shown to be relatively metabolically quiescent, whereas microglia exert similarities with adipose tissue macrophages when it comes to lipid-associated gene expressions. Yet, a similar comparative analysis on MASLD livers remains to be performed and could enlighten some peculiarities and metabolic repolarization between KCs and MoMFs in a steatotic environment. An ever-increasing dimensionality of newly acquired data requires novel means of analysis to guide interpretation. Beyond becoming the new norm for large dataset analysis, in silico approaches have proved to be able to generate novel knowledge, as exemplified by the prediction of bile duct morphogenesis mechanisms.85,86 As another example, multiplex immunohistochemistry has been deployed to define liver macrophage zonation, and an in silico model was established to evaluate the importance of KC positioning for portal vein–derived bacteria clearance.38 Importantly, these findings have been substantiated by intravital microscopy data. This process is driven by chronic exposure to pathogen-associated molecular patterns and mediated by Myd88 signaling. This combined approach of in vivo and in silico models notably enabled the investigators to elaborate on the strategic positioning of KCs to effectively catch pathogens before their spreading.
More elaborate models allow for the prediction of liver macrophage changes over time in defined settings. As such, Mdm2 inducible deficiency was used as a model of acute senescence-driven liver injury in mice, and to collect data on the associated microenvironment changes.87 These data were then integrated to build an in silico mechanistic model. This approach led the authors to the conclusion that there is a threshold for the initial senescence that, once passed, leads to irreversible injury due to unresolved macrophage-driven inflammation. Noteworthy, the model was also able to predict senescence-induced proinflammatory signals originating from endothelial cells, myofibroblast activation, and extracellular matrix deposition in a dose-dependent manner.87
Machine-learning has been used to generate a predictive algorithm for macrophage reactivity and tolerance that relies on a 338-gene signature correlating with disease stage and outcome in a variety of organs, including the liver.40 Mouse interstrain immune cell transcriptional variations are long known. Recently, a study took advantage of this “confounder” to identify specific epigenetic alterations that could predispose to enhanced liver macrophage response to injury. By deploying advanced algorithms, the authors first established that the mouse interstrain variability was similar to the interindividual genetic variations in humans.88 One of these relevant differences was the potent leptin signaling in KCs from BALB/cJ mice, known to exert a relatively more immunotolerant phenotype. Importantly, the leptin receptor is one of the genes that is not expressed by newly homed monocytes in the liver, which could be indicative of cellular origin and partially drive their inflammation-prone response to secondary hits. Additionally, the authors evidenced that KCs in C57BL/6J mice had increased trans-acting chromatin activity at elements predicted to bind NF-κB, which could explain their tendency to react strongly in inflammatory models.88
Recent liver macrophage-centered implications for translational research
As our knowledge of liver macrophages expands, it has become clear that an immunomodulatory rather than a depleting approach holds promise to tackle the remaining challenges in inflammatory liver disease management. Hence, two angles are being explored, which consist of either orienting macrophage phenotypes prior to their (re-)injection into patients or directly in situ.
Ex vivo macrophage engineering
The safety of ex vivo matured autologous MoMFs reinjection in patients with compensated cirrhosis has been demonstrated.89 Inline with this, an ongoing clinical trial studying macrophage therapy for liver cirrhosis (MATCH) aims at evaluating the benefits of such an approach for patients with cirrhosis of diverse etiologies, with as a primary endpoint of reducing the MELD score.90 The results are still awaited and will provide crucial insights into the potential benefits. Similar to chimeric antigen receptor (CAR) T cells, induced pluripotent stem cell–derived CAR macrophages with a strong proinflammatory phenotype are being developed as a potential antitumor approach. As such, CAR macrophages with toll-like receptor 4 intracellular toll/IL-1R (TIR) domain-containing CARs displayed a potent antitumor effect by being able to phagocytose cancer cells.91 Those cells remain to be tested in the liver tumor microenvironment, yet CAR macrophages represent appealing candidates in immunosuppressive primary liver tumors. Along this thought, ex vivo expansion protocols have been developed to retain the specific epigenetic profile of alveolar macrophages in order to use them as cellular therapeutics in pulmonary disease without adverse reprogramming after transfer.92 Potentially, all those approaches might benefit from a combination with ex vivo macrophage labeling. It now appears possible to track nanodroplet-loaded macrophages following injection through acoustic ultrasound imaging.93 This labeling does not alter in vivo macrophage phagocytic capacities and holds the potential of evaluating the successful migration of macrophages to the sites of tumor or injury in clinical settings.
In situ macrophage reprogramming
The gold standard of macrophage phenotype modulation would be an in situ targeted repolarization without the need for extensive ex vivo protocols. Lentiviral vector-mediated engineering of liver macrophages was used to induce the local expression of interferon-alpha. Combined with cytotoxic T-lymphocyte associated protein 4 immune checkpoint blockade, this approach proved to impair colorectal and pancreatic ductal adenocarcinoma liver metastases in mice, notably by increasing antigen-presenting cell functionalities and by promoting MHC-II–restricted cytotoxic T cell response.94
Ring finger protein 41 (RNF41) is known to inhibit proinflammatory factor expressions, and its expression is reduced in liver macrophages from patients with cirrhosis.95 Using dendrimer-graphite nanoparticles–conjugated plasmids, investigators upregulated Rnf41 expression in liver macrophages in the carbon tetrachloride–induced and thioacetamide-induced mouse models of liver fibrosis.95 Such an Rnf41 overexpression was shown to ameliorate liver fibrosis and increase hepatocyte proliferation, along with increased expression of anti-inflammatory markers. Accordingly, macrophage Rnf41 depletion aggravated the mouse phenotypes and reduced their survival.
Expected future developments—the rise of learning machines
Recent and future breakthroughs invariably generate a plethora of novel research and clinical avenues. With the current and unprecedented pace of technological advances in all analytical methods and broader access to large data sets and bioinformatics resources, we can look forward to a series of mind-shifting discoveries on liver macrophages and their associated biological processes in the coming years. While the deployment of artificial intelligence-based algorithms and large language models is subject to debate, one must acknowledge that our communities will need to adapt and, hopefully for the best, take advantage of such technologies. To that end, there is both growing interest and raising concerns on relying on deep-learning-based and deep generative models to explore new research directions.96
CONCLUSIONS
Already about 5 years ago, the emergence of single-cell RNA-sequencing technology and the functional insights from sophisticated experimental models changed the concept of hepatic macrophages, replacing old dogmas on polarized subtypes (eg, M1/M2) with new insights on their heterogeneity and adaptability.4 Although these findings mostly derive from animal models, the rise of novel technologies applied to patient-derived tissues or cells will surely consolidate or refine our current insights. Within as little as five years, tremendous further progress has been made in our understanding of liver macrophage biology. This unprecedented pace of discoveries led to a significant broadening of our perspectives on clinical opportunities for virtually all liver conditions. Noteworthy, these advances are mirrored by similar breakthroughs in other hepatic immune and nonimmune cell populations’ biology or the discovery of novel biomarkers for instance through hypothesis-free omics approaches.97 Our research community task is now to standardize and integrate this ever-growing mass of knowledge and generate concrete translations to tackle remaining clinical challenges.
Acknowledgments
FUNDING INFORMATION
This work was funded by the German Research Foundation (DFG Ta434/8-1, SFB/TRR 296 and SFB1382, Project-ID 403224013).
CONFLICTS OF INTEREST
Frank Tacke consults, advises, is on the speakers’ bureau and received grants from Gilead. He consults, advises, and is on the speakers’ bureau for AbbVie. He consults, advises, and received grants from AstraZeneca and MSD. He consults and advises Allergan, GSK, Alnylam, Bristol-Myers Squibb, Intercept, Inventiva, Pfizer, Novartis, Novo Nordisk, and Sanofi. He is on the speakers’ bureau for Falk and Orphalan. The remaining author has no conflicts to report.
Footnotes
Abbreviations: CAR, chimeric antigen receptor; Cas9, CRISPR-associated protein 9; CCL, chemokine (C-C motif) ligand; CCR, C-C chemokine receptor; KC, Kupffer cell; MARCO, macrophage receptor with collagenous structure; MASH, metabolic dysfunction-associated steatohepatitis; MASLD, metabolic dysfunction–associated steatotic liver disease; Mdr2, multidrug resistance 2 gene; MoMF, monocyte-derived macrophage; Myd88, myeloid differentiation primary response protein; PSC, primary sclerosing cholangitis; RNF41, ring finger protein 41; SPP1, secreted phosphoprotein 1; TLR, toll-like receptor; TREM2, triggering-receptor-expressed-on-myeloid-cells-2.
Contributor Information
Adrien Guillot, Email: adrien.guillot@charite.de.
Frank Tacke, Email: frank.tacke@charite.de.
REFERENCES
- 1. Tilg H, Adolph TE, Trauner M. Gut-liver axis: Pathophysiological concepts and clinical implications. Cell Metab. 2022;34:1700–18. [DOI] [PubMed] [Google Scholar]
- 2. Peiseler M, Araujo David B, Zindel J, Surewaard BGJ, Lee WY, Heymann F, et al. Kupffer cell-like syncytia replenish resident macrophage function in the fibrotic liver. Science. 2023;381:eabq5202. [DOI] [PubMed] [Google Scholar]
- 3. Musrati MA, De Baetselier P, Movahedi K, Van Ginderachter JA. Ontogeny, functions and reprogramming of Kupffer cells upon infectious disease. Front Immunol. 2023;14:1238452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guillot A, Tacke F. Liver macrophages: Old dogmas and new insights. Hepatol Commun. 2019;3:730–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sierro F, Evrard M, Rizzetto S, Melino M, Mitchell AJ, Florido M, et al. A liver capsular network of monocyte-derived macrophages restricts hepatic dissemination of intraperitoneal bacteria by neutrophil recruitment. Immunity. 2017;47:374–88 e376. [DOI] [PubMed] [Google Scholar]
- 6. Guilliams M, Bonnardel J, Haest B, Vanderborght B, Wagner C, Remmerie A, et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–96 e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guilliams M, Scott CL. Liver macrophages in health and disease. Immunity. 2022;55:1515–29. [DOI] [PubMed] [Google Scholar]
- 8. Ait Ahmed Y, Fu Y, Rodrigues RM, He Y, Guan Y, Guillot A, et al. Kupffer cell restoration after partial hepatectomy is mainly driven by local cell proliferation in IL-6-dependent autocrine and paracrine manners. Cell Mol Immunol. 2021;18:2165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bleriot C, Barreby E, Dunsmore G, Ballaire R, Chakarov S, Ficht X, et al. A subset of Kupffer cells regulates metabolism through the expression of CD36. Immunity. 2021;54:2101–16 e2106. [DOI] [PubMed] [Google Scholar]
- 10. De Simone G, Andreata F, Bleriot C, Fumagalli V, Laura C, Garcia-Manteiga JM, et al. Identification of a Kupffer cell subset capable of reverting the T cell dysfunction induced by hepatocellular priming. Immunity. 2021;54:2089–100 e2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martrus G, Goebels H, Langeneckert AE, Kah J, Flomm F, Ziegler AE, et al. CD49a Expression identifies a subset of intrahepatic macrophages in humans. Front Immunol. 2019;10:1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bonnardel J, T’Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, et al. Stellate cells, hepatocytes, and endothelial cells imprint the Kupffer cell identity on monocytes colonizing the liver macrophage niche. Immunity. 2019;51:638–54 e639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scott CL, Zheng F, De Baetselier P, Martens L, Saeys Y, De Prijck S, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7:10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bleriot C, Dupuis T, Jouvion G, Eberl G, Disson O, Lecuit M. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity. 2015;42:145–58. [DOI] [PubMed] [Google Scholar]
- 15. Tran S, Baba I, Poupel L, Dussaud S, Moreau M, Gelineau A, et al. Impaired Kupffer cell self-renewal alters the liver response to lipid overload during non-alcoholic steatohepatitis. Immunity. 2020;53:627–40 e625. [DOI] [PubMed] [Google Scholar]
- 16. Kinoshita M, Uchida T, Sato A, Nakashima M, Nakashima H, Shono S, et al. Characterization of two F4/80-positive Kupffer cell subsets by their function and phenotype in mice. J Hepatol. 2010;53:903–10. [DOI] [PubMed] [Google Scholar]
- 17. Guo Y, Zhao C, Dai W, Wang B, Lai E, Xiao Y, et al. C-C motif chemokine receptor 2 inhibition reduces liver fibrosis by restoring the immune cell landscape. Int J Biol Sci. 2023;19:2572–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lefere S, Devisscher L, Tacke F. Targeting CCR2/5 in the treatment of nonalcoholic steatohepatitis (NASH) and fibrosis: opportunities and challenges. Expert Opin Investig Drugs. 2020;29:89–92. [DOI] [PubMed] [Google Scholar]
- 19. Guillot A, Guerri L, Feng D, Kim SJ, Ahmed YA, Paloczi J, et al. Bile acid-activated macrophages promote biliary epithelial cell proliferation through integrin alphavbeta6 upregulation following liver injury. J Clin Invest. 2021;131:e132305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature. 2019;575:512–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Muynck K, Heyerick L, De Ponti FF, Vanderborght B, Meese T, Van Campenhout S, et al. Osteopontin characterizes bile duct-associated macrophages and correlates with liver fibrosis severity in primary sclerosing cholangitis. Hepatology. 2024;79:269–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hassan GS, Flores Molina M, Shoukry NH. The multifaceted role of macrophages during acute liver injury. Front Immunol. 2023;14:1237042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buechler MB, Kim KW, Onufer EJ, Williams JW, Little CC, Dominguez CX, et al. A Stromal niche defined by expression of the transcription factor WT1 mediates programming and homeostasis of cavity-resident macrophages. Immunity. 2019;51:119–30 e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J, Kubes P. A reservoir of mature cavity macrophages that can rapidly invade visceral organs to affect tissue repair. Cell. 2016;165:668–78. [DOI] [PubMed] [Google Scholar]
- 25. Mironova M, Gopalakrishna H, Rodriguez Franco G, Holland SM, Koh C, Kleiner DE, et al. Granulomatous liver diseases. Hepatol Commun. 2024;8:e0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Govaere O, Petersen SK, Martinez-Lopez N, Wouters J, Van Haele M, Mancina RM, et al. Macrophage scavenger receptor 1 mediates lipid-induced inflammation in non-alcoholic fatty liver disease. J Hepatol. 2022;76:1001–12. [DOI] [PubMed] [Google Scholar]
- 27. Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117:175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seehafer SS, Pearce DA. You say lipofuscin, we say ceroid: Defining autofluorescent storage material. Neurobiol Aging. 2006;27:576–88. [DOI] [PubMed] [Google Scholar]
- 29. Rantakari P, Patten DA, Valtonen J, Karikoski M, Gerke H, Dawes H, et al. Stabilin-1 expression defines a subset of macrophages that mediate tissue homeostasis and prevent fibrosis in chronic liver injury. Proc Natl Acad Sci U S A. 2016;113:9298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fallowfield JA, Mizuno M, Kendall TJ, Constandinou CM, Benyon RC, Duffield JS, et al. Scar-associated macrophages are a major source of hepatic matrix metalloproteinase-13 and facilitate the resolution of murine hepatic fibrosis. J Immunol. 2007;178:5288–95. [DOI] [PubMed] [Google Scholar]
- 31. Trussoni CE, LaRusso NF. Macrophages make a difference in cholestatic liver diseases - but how? J Hepatol. 2023;79:1349–51. [DOI] [PubMed] [Google Scholar]
- 32. Peiseler M, Tacke F. Bile duct-associated macrophages enter the spotlight in inflammatory cholestatic liver disease. Hepatology. 2024;79:257–60. [DOI] [PubMed] [Google Scholar]
- 33. Guillot A, Winkler M, Silva Afonso M, Aggarwal A, Lopez D, Berger H, et al. Mapping the hepatic immune landscape identifies monocytic macrophages as key drivers of steatohepatitis and cholangiopathy progression. Hepatology. 2023;78:150–66. [DOI] [PubMed] [Google Scholar]
- 34. Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell. 2019;178:686–98 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seidman JS, Troutman TD, Sakai M, Gola A, Spann NJ, Bennett H, et al. Niche-specific reprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity. 2020;52:1057–74 e1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guicciardi ME, Trussoni CE, Krishnan A, Bronk SF, Lorenzo Pisarello MJ, O’Hara SP, et al. Macrophages contribute to the pathogenesis of sclerosing cholangitis in mice. J Hepatol. 2018;69:676–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hildebrandt F, Andersson A, Saarenpaa S, Larsson L, Van Hul N, Kanatani S, et al. Spatial Transcriptomics to define transcriptional patterns of zonation and structural components in the mouse liver. Nat Commun. 2021;12:7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gola A, Dorrington MG, Speranza E, Sala C, Shih RM, Radtke AJ, et al. Commensal-driven immune zonation of the liver promotes host defence. Nature. 2021;589:131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bendixen SM, Jakobsgaard PR, Hansen D, Hejn KH, Terkelsen MK, Bjerre FA, et al. Single cell-resolved study of advanced murine MASH reveals a homeostatic pericyte signaling module. J Hepatol. 2024;80:467–81. [DOI] [PubMed] [Google Scholar]
- 40. Ghosh P, Sinha S, Katkar GD, Vo D, Taheri S, Dang D, et al. Machine learning identifies signatures of macrophage reactivity and tolerance that predict disease outcomes. EBioMedicine. 2023;94:104719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Timmons GA, O’Siorain JR, Kennedy OD, Curtis AM, Early JO. Innate rhythms: Clocks at the center of monocyte and macrophage function. Front Immunol. 2020;11:1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wei S, Zheng Q, Pan Y, Xu Y, Tang J, Cai X. Interplay between liver circadian rhythm and regeneration after PHx. Genomics. 2022;114:1–8. [DOI] [PubMed] [Google Scholar]
- 43. Chen KE, Lainez NM, Coss D. Sex differences in macrophage responses to obesity-mediated changes determine migratory and inflammatory traits. J Immunol. 2021;206:141–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petrescu A, Grant S, Frampton G, Kain J, Hadidi K, Williams E, et al. Glucocorticoids cause gender-dependent reversal of hepatic fibrosis in the MDR2-knockout mouse model. Int J Mol Sci. 2017;18:2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krishnan A, Katsumi T, Guicciardi ME, Azad AI, Ozturk NB, Trussoni CE, et al. Tumor necrosis factor-related apoptosis-inducing ligand receptor deficiency promotes the ductular reaction, macrophage accumulation, and hepatic fibrosis in the Abcb4(-/-) mouse. Am J Pathol. 2020;190:1284–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wallace SJ, Tacke F, Schwabe RF, Henderson NC. Understanding the cellular interactome of non-alcoholic fatty liver disease. JHEP Rep. 2022;4:100524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peiseler M, Schwabe R, Hampe J, Kubes P, Heikenwalder M, Tacke F. Immune mechanisms linking metabolic injury to inflammation and fibrosis in fatty liver disease-novel insights into cellular communication circuits. J Hepatol. 2022;77:1136–60. [DOI] [PubMed] [Google Scholar]
- 48. Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–506. [DOI] [PubMed] [Google Scholar]
- 50. Andrews TS, Nakib D, Perciani CT, Ma XZ, Liu L, Winter E, et al. Single-cell and spatial transcriptomics characterisation of the immunological landscape in the healthy and PSC human liver. J Hepatol. 2024;80:730–43. [DOI] [PubMed] [Google Scholar]
- 51. Mo Z, Liu D, Chen Y, Luo J, Li W, Liu J, et al. Single-cell transcriptomics reveals the role of macrophage-naive CD4 + T cell interaction in the immunosuppressive microenvironment of primary liver carcinoma. J Transl Med. 2022;20:466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y, Li J, Li H, Jiang J, Guo C, Zhou C, et al. Single-cell RNA sequencing to dissect the immunological network of liver fibrosis in Schistosoma japonicum-infected mice. Front Immunol. 2022;13:980872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jin S, Plikus MV, Nie Q. CellChat for systematic analysis of cell-cell communication from single-cell and spatially resolved transcriptomics. bioRxiv. 2023. doi: 10.1101/2023.11.05.565674 [DOI] [Google Scholar]
- 54. Feng D, Xiang X, Guan Y, Guillot A, Lu H, Chang C, et al. Monocyte-derived macrophages orchestrate multiple cell-type interactions to repair necrotic liver lesions in disease models. J Clin Invest. 2023;133:e166954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. He K, Wang Z, Luo M, Li B, Ding N, Li L, et al. Metastasis organotropism in colorectal cancer: Advancing toward innovative therapies. J Transl Med. 2023;21:612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hossain M, Shim R, Lee WY, Sharpe AH, Kubes P. Gata6(+) resident peritoneal macrophages promote the growth of liver metastasis. Nat Commun. 2022;13:4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shiri AM, Zhang T, Bedke T, Zazara DE, Zhao L, Lücke J, et al. IL-10 dampens antitumor immunity and promotes liver metastasis via PD-L1 induction. J Hepatol. 2024;80:634–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu X, Qin J, Nie J, Gao R, Hu S, Sun H, et al. ANGPTL2+cancer-associated fibroblasts and SPP1+macrophages are metastasis accelerators of colorectal cancer. Front Immunol. 2023;14:1185208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Liu Y, Zhang L, Ju X, Wang S, Qie J. Single-cell transcriptomic analysis reveals macrophage-tumor crosstalk in hepatocellular carcinoma. Front Immunol. 2022;13:955390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang S, Qian L, Li Z, Li Y, Bai J, Zheng B, et al. Integrated multi-omics landscape of liver metastases. Gastroenterology. 2023;164:407–23 e417. [DOI] [PubMed] [Google Scholar]
- 61. Song G, Shi Y, Meng L, Ma J, Huang S, Zhang J, et al. Single-cell transcriptomic analysis suggests two molecularly subtypes of intrahepatic cholangiocarcinoma. Nat Commun. 2022;13:1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang S, Fang W, Zhou S, Zhu D, Chen R, Gao X, et al. Single cell transcriptomic analyses implicate an immunosuppressive tumor microenvironment in pancreatic cancer liver metastasis. Nat Commun. 2023;14:5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen S, Tang L, Guillot A, Liu H. Bariatric surgery associates with nonalcoholic steatohepatitis/hepatocellular carcinoma amelioration via SPP1 suppression. Metabolites. 2022;13:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Han H, Ge X, Komakula SSB, Desert R, Das S, Song Z, et al. Macrophage-derived osteopontin (SPP1) protects from nonalcoholic steatohepatitis. Gastroenterology. 2023;165:201–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Syn WK, Choi SS, Liaskou E, Karaca GF, Agboola KM, Oo YH, et al. Osteopontin is induced by hedgehog pathway activation and promotes fibrosis progression in nonalcoholic steatohepatitis. Hepatology. 2011;53:106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Coombes JD, Swiderska-Syn M, Dolle L, Reid D, Eksteen B, Claridge L, et al. Osteopontin neutralisation abrogates the liver progenitor cell response and fibrogenesis in mice. Gut. 2015;64:1120–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vucur M, Ghallab A, Schneider AT, Adili A, Cheng M, Castoldi M, et al. Sublethal necroptosis signaling promotes inflammation and liver cancer. Immunity. 2023;56:1578–95 e1578. [DOI] [PubMed] [Google Scholar]
- 68. Li J, Liu XG, Ge RL, Yin YP, Liu YD, Lu WP, et al. The ligation between ERMAP, galectin-9 and dectin-2 promotes Kupffer cell phagocytosis and antitumor immunity. Nat Immunol. 2023;24:1813–24. [DOI] [PubMed] [Google Scholar]
- 69. Bohn T, Rapp S, Luther N, Klein M, Bruehl TJ, Kojima N, et al. Tumor immunoevasion via acidosis-dependent induction of regulatory tumor-associated macrophages. Nat Immunol. 2018;19:1319–29. [DOI] [PubMed] [Google Scholar]
- 70. Medrano-Bosch M, Simon-Codina B, Jimenez W, Edelman ER, Melgar-Lesmes P. Monocyte-endothelial cell interactions in vascular and tissue remodeling. Front Immunol. 2023;14:1196033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang W, Tao Y, Wu Y, Zhao X, Ye W, Zhao D, et al. Neutrophils promote the development of reparative macrophages mediated by ROS to orchestrate liver repair. Nat Commun. 2019;10:1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Calvente CJ, Tameda M, Johnson CD, Del Pilar H, Lin YC, Adronikou N, et al. Neutrophils contribute to spontaneous resolution of liver inflammation and fibrosis via microRNA-223. J Clin Invest. 2019;129:4091–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 2019;365:eaaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hublitz KW, Stamatiades EG. Elucidating immune monitoring of tissue-resident macrophages by intravital microscopy. Methods Mol Biol. 2024;2713:337–46. [DOI] [PubMed] [Google Scholar]
- 75. Pose E, Coll M, Martinez-Sanchez C, Zeng Z, Surewaard BGJ, Catala C, et al. Programmed death ligand 1 is overexpressed in liver macrophages in chronic liver diseases, and its blockade improves the antibacterial activity against infections. Hepatology. 2021;74:296–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Diniz AB, Antunes MM, Lacerda VAS, Nakagaki BN, Freitas Lopes MA, Castro-Oliveira HM, et al. Imaging and immunometabolic phenotyping uncover changes in the hepatic immune response in the early phases of NAFLD. JHEP Rep. 2020;2:100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zimmermann A, Hänsel R, Gemünden K, Kegel-Hübner V, Babel J, Bläker H, et al. In vivo and in vitro characterization of primary human liver macrophages and their inflammatory state. Biomedicines. 2021;9:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Groeger M, Matsuo K, Heidary Arash E, Pereira A, Le Guillou D, Pino C, et al. Modeling and therapeutic targeting of inflammation-induced hepatic insulin resistance using human iPSC-derived hepatocytes and macrophages. Nat Commun. 2023;14:3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ouchi R, Togo S, Kimura M, Shinozawa T, Koido M, Koike H, et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 2019;30:374–384 e376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Harrison SP, Siller R, Tanaka Y, Chollet ME, de la Morena-Barrio ME, Xiang Y, et al. Scalable production of tissue-like vascularized liver organoids from human PSCs. Exp Mol Med. 2023;55:2005–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rezvani M, Vallier L, Guillot A. Modeling nonalcoholic fatty liver disease in the dish using human-specific platforms: Strategies and limitations. Cell Mol Gastroenterol Hepatol. 2023;15:1135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Otumala AE, Hellen DJ, Luna CA, Delgado P, Dissanayaka A, Ugwumadu C, et al. Opportunities and considerations for studying liver disease with microphysiological systems on a chip. Lab Chip. 2023;23:2877–98. [DOI] [PubMed] [Google Scholar]
- 83. Liu H, Yin G, Kohlhepp MS, Schumacher F, Hundertmark J, Abdelwahab Hassan M, et al. Dissecting acute drug-induced hepatotoxicity and therapeutic responses of steatotic liver disease using primary mouse liver and blood cells in a liver-on-a-chip model. Advanced Science. 2024:e2403516. doi: 10.1002/advs.202403516. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gainullina A, Mogilenko DA, Huang LH, Todorov H, Narang V, Kim KW, et al. Network analysis of large-scale ImmGen and Tabula Muris datasets highlights metabolic diversity of tissue mononuclear phagocytes. Cell Rep. 2023;42:112046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Van Liedekerke P, Gannoun L, Loriot A, Johann T, Lemaigre FP, Drasdo D. Quantitative modeling identifies critical cell mechanics driving bile duct lumen formation. PLoS Comput Biol. 2022;18:e1009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Passman AM, Haughey MJ, Carlotti E, Williams MJ, Cereser B, Lin ML, et al. Hepatocytes undergo punctuated expansion dynamics from a periportal stem cell niche in normal human liver. J Hepatol. 2023;79:417–32. [DOI] [PubMed] [Google Scholar]
- 87. Ashmore-Harris C, Antonopoulou E, Aird RE, Man TY, Finney SM, Speel AM, et al. Utilising an in silico model to predict outcomes in senescence-driven acute liver injury. bioRxiv. 2023. doi: 10.1101/2023.10.11.561528 [DOI] [Google Scholar]
- 88. Bennett H, Troutman TD, Zhou E, Spann NJ, Link VM, Seidman JS, et al. Discrimination of cell-intrinsic and environment-dependent effects of natural genetic variation on Kupffer cell epigenomes and transcriptomes. Nat Immunol. 2023;24:1825–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Moroni F, Dwyer BJ, Graham C, Pass C, Bailey L, Ritchie L, et al. Safety profile of autologous macrophage therapy for liver cirrhosis. Nat Med. 2019;25:1560–5. [DOI] [PubMed] [Google Scholar]
- 90. Brennan PN, MacMillan M, Manship T, Moroni F, Glover A, Graham C, et al. Study protocol: A multicentre, open-label, parallel-group, phase 2, randomised controlled trial of autologous macrophage therapy for liver cirrhosis (MATCH). BMJ Open. 2021;11:e053190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lei A, Yu H, Lu S, Lu H, Ding X, Tan T, et al. A second-generation M1-polarized CAR macrophage with antitumor efficacy. Nat Immunol. 2024;25:102–16. [DOI] [PubMed] [Google Scholar]
- 92. Subramanian S, Busch CJ, Molawi K, Geirsdottir L, Maurizio J, Vargas Aguilar S, et al. Long-term culture-expanded alveolar macrophages restore their full epigenetic identity after transfer in vivo. Nat Immunol. 2022;23:458–68. [DOI] [PubMed] [Google Scholar]
- 93. Chudal L, Santelli J, Lux J, Woodward A, Hafeez N, Endsley C, et al. In vivo ultrasound imaging of macrophages using acoustic vaporization of internalized superheated nanodroplets. ACS Appl Mater Interfaces. 2023;15:42413–23. [DOI] [PubMed] [Google Scholar]
- 94. Kerzel T, Giacca G, Beretta S, Bresesti C, Notaro M, Scotti GM, et al. In vivo macrophage engineering reshapes the tumor microenvironment leading to eradication of liver metastases. Cancer Cell. 2023;41:1892–910 e1810. [DOI] [PubMed] [Google Scholar]
- 95. Moreno-Lanceta A, Medrano-Bosch M, Fundora Y, Perramon M, Aspas J, Parra-Robert M, et al. RNF41 orchestrates macrophage-driven fibrosis resolution and hepatic regeneration. Sci Transl Med. 2023;15:eabq6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nam D, Chapiro J, Paradis V, Seraphin TP, Kather JN. Artificial intelligence in liver diseases: Improving diagnostics, prognostics and response prediction. JHEP Rep. 2022;4:100443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Thiele M, Villesen IF, Niu L, Johansen S, Sulek K, Nishijima S, et al. Opportunities and barriers in omics-based biomarker discovery for steatotic liver diseases. J Hepatol. 2024. doi: 10.1016/j.jhep.2024.03.035 [DOI] [PubMed] [Google Scholar]